Abstract
Nucleosome-remodelling factors containing the ATPase ISWI, such as ACF, render DNA in chromatin accessible by promoting the sliding of histone octamers. Although the ATP-dependent repositioning of mononucleosomes is readily observable in vitro, it is unclear to which extent nucleosomes can be moved in physiological chromatin, where neighbouring nucleosomes, linker histones and the folding of the nucleosomal array restrict mobility. We assembled arrays consisting of 12 nucleosomes or 12 chromatosomes (nucleosomes plus linker histone) from defined components and subjected them to remodelling by ACF or the ATPase CHD1. Both factors increased the access to DNA in nucleosome arrays. ACF, but not CHD1, catalysed profound movements of nucleosomes throughout the array, suggesting different remodelling mechanisms. Linker histones inhibited remodelling by CHD1. Surprisingly, ACF catalysed significant repositioning of entire chromatosomes in chromatin containing saturating levels of linker histone H1. H1 inhibited the ATP-dependent generation of DNA accessibility by only about 50%. This first demonstration of catalysed chromatosome movements suggests that the bulk of interphase euchromatin may be rendered dynamic by dedicated nucleosome-remodelling factors.
Keywords: chromatin, Drosophila, linker histone, nucleosome array, remodelling
Introduction
The discovery of ATP-dependent nucleosome-remodelling factors provided a solution to the old problem of how eukaryotic genomes can be utilized despite of their tight packing with histones in the form of chromatin (Becker and Horz, 2002). Typically, nucleosome-remodelling factors are multiprotein complexes that contain dedicated ATPases of the SNF2 family (Flaus and Owen-Hughes, 2004). These ATPases resemble specialized DNA translocases. Enzymes of this kind experience series of conformational changes triggered by cycles of nucleotide binding, hydrolysis and dissociation that control the binding and release of DNA, thereby effectively moving a segment of DNA relative to the enzyme. However, unlike ‘classical' translocases, nucleosome-remodelling factors not only contact DNA but also the histone moiety of nucleosomes. Translocating DNA then leads to disruption of histone–DNA contacts and hence to ‘remodelling' of canonical nucleosome structure. As a result, nucleosomes can be disassembled and reassembled (Lusser and Kadonaga, 2003; Varga-Weisz and Becker, 2006; Li et al, 2007) or moved to a neighbouring segment of DNA through a process termed ‘nucleosome sliding' (Becker, 2002). Nucleosome remodelling thus enables dynamic transitions of chromatin structure, and by this means endows chromatin with the flexibility required to access the information stored in DNA.
Much about the function of nucleosome-remodelling enzymes has been learned from studies using mono- or dinucleosome substrates. However, these simple model substrates bear little resemblance to physiological chromatin. The long oligonucleosomal fibres characterizing native chromatin are no mere linear arrays of nucleosomes, but fold into compact 30-nm fibres. The exact geometry of these fibres is still controversial (Robinson and Rhodes, 2006; Tremethick, 2007), but it seems clear that the association of the linker histone H1 stabilizes the folding of the nucleosomal array considerably (Carruthers et al, 1998; Carruthers and Hansen, 2000). In Drosophila embryonic development, the appearance of linker histones correlates with chromatin compaction (Ner and Travers, 1994). In agreement, their depletion results in a less compact chromatin structure in a number of species (Shen et al, 1995; Fan et al, 2005; Maresca et al, 2005). Despite their severe impact on chromatin structure and their high abundance in the nucleus—approximately one H1 per nucleosome in typical eukaryotic nuclei (Woodcock et al, 2006)—their effect on chromatin remodelling has only been addressed in a handful of studies and remains controversial.
With their globular domain, linker histones bind to DNA at the nucleosomal dyad and where it enters the nucleosome, forming ‘chromatosomes' (Brown et al, 2006; Robinson and Rhodes, 2006; Sheng et al, 2006). The long and highly basic C-terminus of linker histones presumably interacts with linker DNA and contributes to the stability of binding (Hendzel et al, 2004). Electron microscopy shows that H1 modulates the trajectory of the two DNA segments that emerge from the nucleosome ‘core' by organizing it into a ‘stem' structure (Bednar et al, 1998). Many structural studies of linker histone interactions with nucleosomes have made use of the variant H5, a specialized linker histone found in avian erythrocytes (Sun et al, 1990; Zhou et al, 1998). The interaction of H5 with chromatin appears to be more stable than that of H1 (Thomas and Rees, 1983). Chromatin in the presence of H5 is packed differently and is more repressive as compared with H1-containing chromatin (Sun et al, 1990), which may be due to the presence of a third DNA-binding surface in H5 (Ramakrishnan et al, 1993; Fan et al, 2005).
Since linker histones constrain the path of the linker DNA, they are expected to inhibit the dynamic detachment of DNA segments from the nucleosome (Li et al, 2005). Consistent with this idea, linker histones inhibit the spontaneous sliding of histone octamers on DNA at elevated temperatures (Pennings et al, 1994; Ura et al, 1995), a reaction that requires transient unpeeling of DNA from the histone surface (Yager and van Holde, 1984). Similarly, one would expect ATP-dependent nucleosome repositioning to be difficult in the presence of linker histones, as this is likely to involve pulling linker DNA into the nucleosome (Flaus and Owen-Hughes, 2004; Saha et al, 2006). Moreover, remodelling factors of the ISWI type and H1 may compete for the same binding site (Langst and Becker, 2001; Zofall et al, 2004; Strohner et al, 2005). Consistent with these theoretical considerations, the presence of the linker histone inhibited the ATP-dependent ability of the SWI/SNF complex to generate access to mononucleosomal DNA (Hill and Imbalzano, 2000). Likewise, on dinucleosomes, ACF-dependent remodelling was impaired by incorporation of H1 (Saeki et al, 2005). Others found that addition of H1 to nucleosomes did not inhibit SWI/SNF-dependent repositioning (Ramachandran et al, 2003).
Mononucleosomes provide a convenient assay for nucleosome remodelling, but remodelling enzymes are also able to act on nucleosomes in extended arrays (Corona et al, 1999; Boyer et al, 2000; Hassan et al, 2001). Considering the strong effect of linker histones on chromatin folding, their impact on chromatin remodelling can only be reliably investigated in nucleosome arrays. Yet, only few studies attempted to measure ATP-dependent remodelling on oligonucleosomal substrates. Horn et al (2002) found, monitoring the accessibility of DNA in nucleosome arrays, that several remodelling enzymes (human and yeast SWI/SNF, Mi-2 and Xenopus ACF) were largely inhibited in presence of linker histone H5. In contrast, Varga-Weisz et al (1995) observed striking mobility of nucleosomes in complex chromatin fully loaded with histone H1, assembled in preblastoderm Drosophila embryo extracts. This early finding suggested that H1-containing chromatin could be rendered dynamic, but due to the crude nature of the system did not pinpoint the responsible enzymes and cofactors.
Clarification of the extent to which ATP-dependent remodelling can work on H1-containing chromatin and defining the limitations for nucleosome mobility is of fundamental importance. After all, most of the eukaryotic genome is supposedly organized in H1-containing 30-nm fibres (Horowitz et al, 1994). Is the bulk of euchromatin characterized by nucleosome mobility, or only a small fraction from which H1 has been stripped and the 30-nm fibre destabilized? We therefore sought to re-investigate the effect of linker histones on ATP-dependent nucleosome mobility in a fully defined system of H1-containing chromatin. For this purpose, we reconstituted chromatosome arrays from purified components (Huynh et al, 2005) and subjected these arrays to ATP-dependent remodelling by Drosophila ACF. ACF consists of the ATPase ISWI and the associated subunit Acf1 (Ito et al, 1999; Strohner et al, 2005). In addition to assisting the assembly of chromatin with regular nucleosome spacing (Ito et al, 1997; Lusser et al, 2005), ACF can slide mononucleosomes on short DNA fragments (Eberharter et al, 2001). To our surprise we found that ACF, but not CHD1, was able to catalyse considerable ACF-dependent movements of entire chromatosomes within fully loaded arrays.
Results
Reconstitution of chromatin with stoichiometric amounts of linker histones
To investigate the effect of linker histones on chromatin remodelling, it was crucial to work with chromatin arrays containing one linker histone per histone octamer. Only then we could be certain that any remodelling on these arrays occurred on chromatosomes (nucleosomes+linker histone) and not on a fraction of nucleosomes devoid of linker histones. Following a protocol for the reconstitution of homogeneous, linker histone-containing chromatin arrays (Huynh et al, 2005), we assembled 12-mer nucleosome and chromatosome arrays on 12-mer repeats of the 601 nucleosome positioning sequence (Lowary and Widom, 1998), using histone octamers and H1 from Drosophila embryos or H5 from chicken erythrocytes. DNA fragments lacking positioning sequences—between 692 and 1113 bp, derived from the pUC18 vector—were present during all reconstitutions. They served as competitor DNA (crDNA) to bind excess histones and were removed after the assembly (Figure 1C). In pilot studies (not shown) we determined the amounts of histone octamers required to assemble homogeneous 12-mer nucleosome arrays. Titrations of linker histones are shown in Figure 1B. Addition of increasing amounts of H1 resulted in a slower migration of the resulting array on native agarose gels (best resolved on 0.7% agarose), whereas incorporation of H5 led to faster migrating arrays (best resolved on 1.4% agarose), in agreement with earlier studies (Huynh et al, 2005). The different mobility of H5- versus H1-containing chromatin demonstrates the difference in packing brought about by the H5 variant (Sun et al, 1990). Increased addition of linker histones did not lead to a further change in the mobility of the arrays; instead, excess linker histones bound to the crDNA, as seen by gel mobility shifts (Figure 1B, lanes 6 and 7, and 14 and 15). To evaluate the degree of reconstitution, all arrays were purified from excess proteins, free and nucleosomal crDNA by MgCl2 precipitation (Figure 1C; Schwarz et al, 1996). We directly determined the histone stoichiometry by gel electrophoretic analysis of the purified reconstituted chromatin (Figure 1D). The amount of each histone was calculated from the Coomassie blue stain in each band as determined by densitometry on a Li-Cor Odyssey machine. Stoichiometric, saturating levels of linker histones were reached on adding linker histones at a nominal molar ratio of 2.5 molecules of input linker histones per 601 sequence. Excess histones bound to the crDNA were removed during the purification step.
Figure 1.
Reconstitution of chromatin arrays with stoichiometric amounts of linker histone H1 or H5. (A) Overview on chromatin reconstitution and quality controls. 12-mer nucleosome arrays and chromatosome arrays containing linker histone H1 or H5 were assembled on tandem repeats of the 601 nucleosome positioning sequence according to Huynh et al (2005). To bind excess histones, crDNA with no positioning sequence was added to the assembly. (B) Arrays (6 pmol) assembled with increasing molar ratios of H1 or H5 (H1/nuc or H5/nuc) were applied on 0.7 or 1.4% native agarose gels, respectively. (C) Arrays were purified from unbound histones and competitor DNA by MgCl2 precipitation. Corresponding amounts of input, pellet and supernatant were analysed on 1.4% native agarose gel. (D) The protein content of 60 pmol nucleosomal arrays after MgCl2 precipitation was separated on 15% polyacrylamide gel and visualized by Coomassie blue staining to control the relative amounts of core and linker histones. (E) Arrays (6 pmol) were digested with AvaI at 26°C for 1 h, and the resulting fragments were resolved on native agarose gel to observe positioning sequences unbound by histone octamers (200 bp), nucleosomes (Nuc) and chromatosomes (Chrom).
As a further control for the occupancy of 601 sequences by histone octamers, arrays were digested by AvaI that cuts between those sequences (Figure 1A). Digestion of the nucleosome arrays yielded mononucleosomes, but no free 200-bp DNA fragments, which can be distinguished by native agarose gel electrophoresis (Figure 1E, lane 1). Evidently, the vast majority of 601 sequences were occupied by histone octamers. A minor fraction of subnucleosomal particles gave rise to a band migrating slightly faster than mononucleosomes. The digestion showed furthermore that nucleosomes did not occupy alternative positions to the ones dictated by the 601 sequence, since they did not occlude the AvaI site. AvaI digestion of H1-containing chromatin yielded mostly chromatosomes and only a minor fraction of nucleosomes (Figure 1E, lanes 2–4). The fraction of nucleosomes did not decrease significantly upon adding more H1, showing that saturation had been reached. Chromatosome arrays assembled with H5 were more resistant to digestion by AvaI. H5-containing chromatosomes migrated only slightly slower than nucleosomes (Figure 1E, lanes 5–7) supporting the idea that H5-containing chromatin is different from the one containing the canonical linker histone.
We conclude that the reconstituted chromatin consist of regular chromatosome arrays with stoichiometric levels of linker histones. All arrays used for remodelling reactions were quality-controlled by the methods described.
Linker histone-containing chromatin can be rendered accessible by ACF
As a quantitative measure for chromatin remodelling we monitored changes in the accessibility of nucleosomal DNA. For this purpose we reconstituted end-labelled nucleosome or chromatosome arrays, and incubated them with an excess of the restriction endonuclease AluI, which cuts within the nucleosome positioning sequence, 45 bp into the nucleosome. Without remodelling activity, about 70% of arrays were resistant to cleavage (Figure 2B), demonstrating that seven out of 10 arrays did not contain a single accessible positioning sequence. On these uninterrupted arrays the development of restriction site accessibility was now monitored in the presence of ATP and Drosophila ACF expressed from baculovirus vectors in insect cells. Arrays were incubated with AluI and with or without ACF and ATP at 26°C. After 1 h, the reactions were quenched with excess unlabelled DNA, total DNA was purified and analysed on agarose gels. Comparing the percentages of uncut DNA confirmed that ACF increased the accessibility of nucleosomal DNA in an ATP-dependent manner (Figure 2A, lane 3). A total of 80% of the resistant arrays were cleaved in the presence of ACF and ATP, but not if one of these was omitted. In presence of H1, 48% of otherwise resistant arrays could be cleaved when adding ACF and ATP, so the ATP-dependent increase in accessibility was significant, although the absolute extent of opening was reduced (Figure 2A, lane 6). When probing arrays assembled with a higher molar ratio of H1 per 601 sequence (Figure 2A, lanes 7–9), the degree of inhibition remained the same, showing that the ability to remodel was not due to sub-saturating H1 levels. Remarkably, ACF was able to promote the access of the endonuclease even to H5-containing chromatin. A total of 35–45% of resistant H5 chromatosome arrays were cut in the presence of both ACF and ATP (Figure 2A, lanes 12 and 15). These results are consistent with earlier observations from the Peterson laboratory (Horn et al, 2002) who found that xACF was able to decrease the fraction of nuclease resistant H5-containing chromatosome arrays from about 85% to 60% in an ATP-dependent reaction.
Figure 2.
Linker histone containing chromatin can be rendered accessible by ACF. (A) Nucleosome and chromatosome arrays were assembled on end-labelled DNA using different molar ratios of linker histone H1 or H5 per 601 positioning sequence (H1/nuc or H5/nuc). Arrays (0.6 pmol) were used as substrate in remodelling assays for ACF or CHD1. To monitor the accessibility of nucleosomal DNA, the enzyme AluI, was added together with 2.4 pmol ACF or an amount of CHD1 with equivalent ATPase activity. After 1 h, proteins were removed by Proteinase K and the DNA analysed on agarose gel. (B) Quantification of the results of panel A.
We were surprised by the efficiency of chromatosome remodelling by ACF, and wondered whether ACF was specifically suited for dealing with linker histones. ACF is able to assist the formation of H1-containing chromatin, whereas the nucleosome-remodelling ATPase CHD1 can only promote nucleosome assembly, but is unable to incorporate also the linker histone (Lusser et al, 2005). To explore whether the remodelling activities correlated with the assembly properties of the enzymes, we compared the activities of ACF and CHD1 in our system. To assure that the parallel reactions contained equivalent nucleosome-remodelling activity, we first standardized the enzyme inputs according to their nucleosome-stimulated ATPase activity. Similar to ACF, CHD1 enhanced the accessibility towards AluI in an ATP-dependent manner (Figure 2A, lanes 16 and 17; Figure 2B). However, in the presence of linker histones H1 or H5, no remodelling activity could be observed (Figure 2A, lanes 18–25; Figure 2B). We also directly compared enzyme amounts that are equally active on nucleosome arrays for their activity on chromatosome arrays. Similar results were obtained (Supplementary Figure 1). This indicates that the remodelling of chromatosome arrays by ACF is not the result of incomplete reconstitution. It also shows that ACF is better suited than CHD1 to remodel linker histone-containing chromatin. To investigate whether this quality of ACF is intrinsic to its ATPase subunit or relies on its associated subunit Acf1, we repeated the assay with ISWI alone. The remodelling activity of ISWI in the presence of linker histones H1 and H5 was similar to that of ACF, whereas CHD1 was inactive (Supplementary Figure 1). We, therefore, speculate that the ability to deal with chromatosome arrays may be a characteristic of all ISWI-containing remodelling complexes. In support of this notion, recent observations by Tamkun and co-workers suggest that an ISWI-containing complex, possibly the remodelling factor NURF, may affect the H1 association with chromosomes (Corona et al, 2007).
ACF repositions nucleosomes in the presence of linker histones
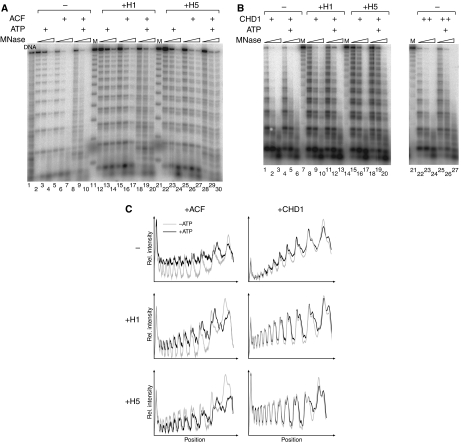
Accessibility of nucleosomal DNA provides a quantitative measure of chromatin remodelling. It does not, however, provide information about the nature of remodelling. Access to DNA may result from nucleosome repositioning as well as disruption of histone–DNA contacts (Fan et al, 2003). ACF can ‘slide' mononucleosomes along DNA (Eberharter et al, 2001), but it is not clear whether the presence of linker histones modulates the outcome of nucleosome remodelling within the array. To visualize potential nucleosome movements within the chromatosome arrays, we subjected reconstituted, end-labelled chromatin to remodelling by ACF or CHD1 and then probed nucleosome positions by partial digestion with micrococcal nuclease (MNase). MNase digestion in the absence of remodelling yielded a highly regular ladder of DNA fragments (Figure 3A, lanes 2–7), but this pattern was dramatically altered when arrays had been incubated with both ACF and ATP (Figure 3A, lanes 8–10). The cleavage profile resembled the one obtained from digesting free DNA (Figure 3A, lane 1), suggesting that nucleosome positions had been randomized throughout the entire array by ATP-dependent remodelling. These ATP-dependent changes were clearly visualized by comparing densitometry profiles of corresponding lanes (Figure 3C).
Figure 3.
ACF repositions nucleosomes in presence of linker histones. End-labelled nucleosome arrays (1.8 pmol) or chromatosome arrays (containing either H1 or H5) were incubated with ATP and 7.2 pmol ACF (A) or CHD1 (B). For panel B, lanes 1–20, an amount of CHD1 was chosen that exhibited an equal remodelling activity to that of ACF on nucleosome arrays. The fivefold amount of CHD1 was used in panel B lanes 21–27. After 1 h, DNA was partially digested by MNase (three time points), purified and separated by agarose gel electrophoresis. Lane 1 shows the MNase digestion pattern obtained from free 12-mer 601 repeats. As a marker (M), nucleosome arrays were digested by AluI. (C) Densitometry profiles of selected lanes (A, lanes 5, 8, 16, 19, 26 and 29; B, lanes 1, 4, 8, 11, 16 and 19). MNase digests of remodelling reactions performed with (black graphs) or without ATP (grey graphs) were compared; ACF, CHD1, H1 and H5 were present during the reaction as indicated.
Digestion of chromatosome arrays with MNase also yielded a regular cleavage pattern; however, additional bands appeared below the primary band already seen in nucleosome arrays (Figure 3A). These bands are particularly strong on H5-containing chromatin and are presumably due to structural changes in linker DNA upon interaction of linker histones. The pattern derived from H1-containing chromatin clearly changed upon prior remodelling by ACF, again approaching the one characteristic of free DNA (Figure 3A, lanes 18–20). The effects are particularly evident in the lower part of the gel, which raises the possibility that remodelling is enhanced at the ends. The H5-containing arrays were more resistant to nucleosome repositioning, albeit some additional bands, particularly in the more digested fraction, point to a low level of nucleosome repositioning (Figure 3A, lanes 28–30). CHD1 affected the MNase digestion pattern neither in the absence nor in the presence of linker histones (Figure 3B, lanes 1–20). Densitometry profiles of corresponding lanes from reactions conducted with or without ATP were largely overlapping (Figure 3C) showing that CHD1 slides nucleosomes much less efficiently than ACF. This conclusion was confirmed by the observation that even the fivefold amount of CHD1 did not induce any significant ATP-dependent changes in the MNase digestion pattern (Figure 3B, lanes 21–27). The result, taken together with the one in Figure 2, shows that access to oligonucleosomal DNA can be generated by two distinct strategies: nucleosome sliding (by ACF) and nucleosome remodelling without overt changes in histone octamer positions (by CHD1).
To elucidate whether the presence of histone modifications in our arrays influenced the outcome of these experiments, arrays were also assembled using recombinant Drosophila histones expressed in Escherichia coli (Supplementary Figure 2), which in contrast to those purified from Drosophila embryos did not carry any modifications. The reconstitution of linker histone-containing chromatin and the ability of ACF, ISWI and CHD1 to remodel the resulting chromatin was essentially the same as with native histones (Supplementary Figures 3 and 4).
ACF catalyses the movement of chromatosomes
Since ACF can assist the assembly of H1-containing chromatin (Lusser et al, 2005), it might also be able to catalyse the opposite reaction, that is, the eviction of H1. Therefore, it is possible that nucleosome repositioning by ACF relies on prior stripping of the linker histone. In an attempt to detect H1 presumably released from chromatin upon remodelling, we included radiolabelled mononucleosomes into the reaction, which may function as acceptors for H1. The decreased mobility of these labelled nucleosomes on native gels upon association of H1 should provide a sensitive assay for linker histone release. However, we were unable to detect any H1 transfer with this assay (not shown).
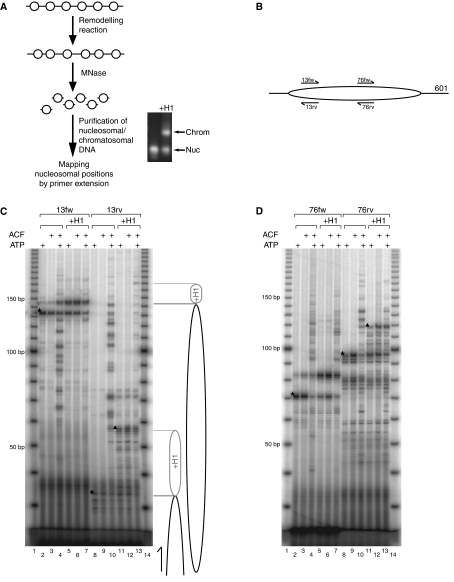
Next, we attempted to detect the movement of H1 along with nucleosomes (Figure 4A). To this end we remodelled nucleosome or chromatosome arrays with ACF as before (controls did not contain ATP or enzyme) and then digested the chromatin with MNase. The endonuclease and exonucleolytic ‘trimming' activity of MNase defines the borders of the nucleosome or chromatosome, because histones protect the DNA they organize from digestion. The resulting nucleosomes and chromatosomes were separated on a native agarose gel (Figure 4A). In agreement with the literature (Nightingale et al, 1996), MNase treatment led to displacement of a fraction of H1 from the chromatosomes, so that a mixture of nucleosomes and chromatosomes was obtained from the digestion of chromatosome arrays independent of whether they had been remodelled. To map the positions of nucleosomes upon remodelling, we excised the bands corresponding to nucleosomes and chromatosomes before and after remodelling, purified the associated DNA and determined nucleosome/chromatosome positions by primer extension. Two different positions, 13 and 76 bp into the positioning sequence, were selected and radiolabelled oligonucleotides were annealed. Forward and reverse primers named 13fw, 13rv, 76fw and 76rv (Figure 4B) were extended in separate reactions until the fragments' ends were reached. The positions of particles can be deduced from the length of the obtained DNA fragments and the primer-annealing site.
Figure 4.
ACF catalyses the movement of chromatosomes. (A) Scheme of experimental steps to map nucleosome positions after remodelling reactions. Nucleosome arrays or H1 chromatosome arrays were incubated with or without ACF and ATP. After 1 h, the arrays were digested to nucleosomes or chromatosomes by MNase. Nucleosomes and chromatosomes obtained after MNase digestion were resolved by preparative agarose gel electrophoresis. Nucleosomes (from nucleosome arrays) and chromatosomes (from chromatosome arrays) were excised from 1.1% native agarose gel and the DNA was extracted. To map the positions of nucleosomes/chromatosomes, primer extension was performed with the primers depicted in panel B. (B) Annealing positions of the primers used for primer extension reactions (13fw, 13rv, 76fw, 76rv). Arrows indicate the primers, the black line the DNA and the oval the position of the nucleosome before the remodelling reaction. (C) Primer extension reactions with primers 13fw and 13rv performed on isolated nucleosomes/chromatosomes were analysed on 7% polyacrylamide 20% urea gels. For reactions conducted with 13rv, nucleosome positions corresponding to the indicated bands are represented by drawings at the right side of the gel. (D) Same as panel C, but with primers 76fw and 76rv.
Probing nucleosomal DNA from arrays in the absence of remodelling, all four primers yielded a ladder of bands, which are most likely due to single-stranded nicks generated by MNase (Cockell et al, 1983). However, the longest, most prominent band in each reaction confirmed the nucleosome position defined by the 601 sequence (asterisks in Figure 4C and D). For example, annealing and extension of the 13rv primer resulted in the expected 33-bp band (Figure 4C, lanes 8 and 9). The DNA fragments derived from chromatosomes were 20 bp longer than nucleosomal ones at the 5′ end (detected by the reverse primers 13rv and 76rv; triangles in Figure 4C and D). This is in line with the known fact that H1 protects 20 bp of linker DNA from nuclease digestion (Simpson, 1978), and confirms the asymmetric interaction of the linker histone (Zhou et al, 1998).
Upon remodelling by ACF, additional bands were obtained from nucleosomal DNA with all primers, indicating repositioning of nucleosomes. The most dramatic effects could be observed in the case of the 13rv primer. Without remodelling, the 33-bp fragment indicative of 601-directed positioning was most prominent (Figure 4C, lanes 8 and 9). When both ACF and ATP had been added before, bands of different sizes up to about 150 bp were obtained (Figure 4C, lane 10), demonstrating repositioning of nucleosomes along the entire length of the DNA repeat. Prominent bands considerably longer than 150 bp are not expected, because the nucleosome protects only 147 bp from nuclease digestion. Remarkably, extensive nucleosome repositioning was also seen in DNA purified from chromatosomes, again pointing to movements of nucleosomes throughout the length of the 601 repeat (Figure 4C, lanes 7 and 13; Figure 4D, lanes 7 and 13). Not surprisingly, the largest bands observed were slightly longer (up to about 180 bp) due to H1 binding. Since the DNA had been derived from gel-purified chromatosomes after a remodelling reaction, we conclude that ACF can move entire chromatosomes on DNA in an ATP-dependent manner.
Discussion
Due to the abundance of linker histones in interphase chromatin, H1-containing nucleosome arrays are probably the most common and physiological substrate for ATP-dependent chromatin remodelling factors. It is therefore important to understand whether and how these complexes can deal with the linker histone. So far, the literature mostly suggested that linker histones hinder chromatin remodelling (Horn et al, 2002). Residual remodelling activity has largely been attributed to incomplete loading of the substrate with linker histones. We tried to rule out this experimental shortcoming by tightly controlling the stoichiometric incorporation of linker histones into chromatin arrays. Yet, ACF was able to induce the movement of entire chromatosome units throughout extended arrays. Importantly, the inability of CHD1 to remodel H1-containing chromatin confirms the inhibitory nature of the chromatosome array. Our data are in accordance with previous findings in a crude, undefined system that nucleosome movements can occur within H1-containing chromatin (Varga-Weisz et al, 1995), but they present, to our knowledge, the first direct demonstration of ATP-dependent chromatosome mobility in a defined chromatin array.
Our results are surprising in light of the documented impediments of linker histones on nucleosome remodelling. First, H1 binding limits the amount of free linker DNA, which is known to determine the efficiency of ACF-dependent remodelling (Yang et al, 2006; Gangaraju and Bartholomew, 2007). Second, H1 is likely to compete with ISWI-type remodellers for nucleosomal binding sites (Kagalwala et al, 2004; Yang et al, 2006; Gangaraju and Bartholomew, 2007). In addition, H1 is believed to constrain the path of DNA entering and exiting the nucleosome (Sheng et al, 2006) and may therefore hinder DNA translocation. Finally, the increased compaction promoted by linker histones might restrict the access of remodelling factors towards the chromatin fibre. According to both currently favoured models for the structure of the 30-nm fibre, the linker DNA and hence all points of access for remodelling enzymes are located inside the chromatin fibre (Robinson and Rhodes, 2006). The cation concentrations in our experiments (50 mM KCl, 1.5 mM MgCl2) will promote the compaction of the nucleosomal array (Carruthers et al, 1998; Robinson et al, 2006).
In spite of these possible constraints, we observed a considerable ACF- and ATP-dependent repositioning of chromatosomes. We considered that H1 purified from Drosophila embryos might carry modifications, decreasing its affinity for chromatin. For example, the extensive phosphorylation of linker histone C-termini interferes with DNA binding and relieves its inhibitory impact on SWI/SNF-dependent chromatin remodelling (Hill et al, 1991; Horn et al, 2002). However, mass spectrometrical analysis of histone H1 purified from Drosophila embryos did not reveal extensive phosphorylation (A Villar and A Imhof, personal communication). We therefore consider it unlikely that phosphorylation impacted the outcome of our experiments.
The inhibitory effect of histone H1 on nucleosome remodelling was apparent when CHD1 was used as a remodelling enzyme. Notably, CHD1's activity on nucleosome arrays was equal to that of ACF, ruling out a defective activity of CHD1. Rather, ACF appears particularly suited for coping with linker histones. This is supported by the observation that ACF can assist the assembly of H1-containing chromatin arrays, whereas CHD1 can only promote assembly of H1-free chromatin (Lusser et al, 2005). Recently, the ISWI-containing remodelling factor NURF has been suggested to be involved in modulating the association of H1 with chromosomes in vivo (Corona et al, 2007). The ability to slide chromatosomes may thus be a more widespread property of remodelling enzymes.
How might ACF achieve chromatosome repositioning? ACF may directly catalyse the eviction of H1 before nucleosome sliding, and a number of reports indicate that nucleosome-remodelling factors can, in principle, disrupt the DNA interactions of other proteins than core histones (Kikyo et al, 2000; Nagaich et al, 2004; Sprouse et al, 2006). Although we were unable to detect free linker histones during remodelling, our analysis does not exclude that a fraction of H1 is transiently dislocated to secondary sites on the nucleosome array or an acceptor site on ACF. In vivo, linker histone displacement may be facilitated by cooperating histone chaperones. ACF and the histone chaperone NAP1 can act in concert towards the assembly of H1-containing chromatin (Lusser et al, 2005), and it is thus conceivable that in cells ACF may cooperate with chaperones to catalyse the reverse reaction, which is the eviction of linker histones. However, since we did not include a chaperone in our experiment, alternative mechanisms have to be considered.
Chromatosome movements might already be facilitated if only the linker histone's globular domain was transiently detached from the nucleosome, while the C-terminal tail remained associated with the linker DNA. Such a scenario is reminiscent of documented changes on H1 interaction due to transcription, where selective crosslinking in Drosophila showed that the globular domain but not the C-terminal tail of linker histones was reversibly displaced from chromatin (Nacheva et al, 1989). In line with these considerations, the C-terminal tail contributes to H1 binding to DNA and determines its residence time on chromatin in living cells (Bharath et al, 2002; Hendzel et al, 2004; Catez et al, 2006).
The analysis of chromatosome positions by primer extension revealed that in our arrays H1 protects DNA from nuclease digestion only on one side of the nucleosome, suggesting an asymmetrical binding of H1 in agreement with earlier observations (Zhou et al, 1998; Brown et al, 2006). This asymmetrical interaction, combined with the repetitive nature of the 601 array, endows the entire array with directionality. Although the precise topography of the ACF–nucleosome complex is not known at present, Bartholomew and co-workers suggested on the basis of site-directed DNA affinity labelling that the related ISW2 complex interacts with linker DNA only on one side of the nucleosome (Kagalwala et al, 2004; Dang et al, 2006). We thus speculate that ACF may interact with nucleosomal linker on the side that is not contacted by the globular domain of H1, in order to initiate the remodelling reaction. Propagation of a ‘looped segment' of DNA around the histone octamer would then lead to movement of the histone octamer and concomitant displacement of the globular domain. The domain would then have to relocate and bind to the new nucleosome dyad and DNA entry point. A testable prediction of this hypothesis is that nucleosome sliding in presence of H1 would be unidirectional.
We do not know at this point whether ACF distributively targets individual nucleosomes within a nucleosome array or rather remodels neighbouring nucleosomes processively. In the latter case the fibre ends may provide points of entry. However, our restriction enzyme accessibility assays did not reveal a gradient of increased accessibility towards the ends of the array, as might be expected from such a scenario. On the other hand ACF is known to remain bound to its initial substrate during chromatin assembly (Fyodorov and Kadonaga, 2002), and we observed earlier that nucleosomes within extended arrays were repositioned by Drosophila embryonic extract in apparent synchrony (Varga-Weisz et al, 1995). Further experiments are required to clarify this issue.
Our study provides the first evidence that ATP-dependent nucleosome-remodelling factors can mobilize entire chromatosomes, even if they reside in extensive arrays. Hence, the majority of euchromatin might be characterized by mobile nucleosomes and chromatosomes.
Materials and methods
Reconstitution of nucleosome and chromatosome arrays
Nucleosome and chromatosome arrays were assembled by continuous salt dialysis as described (Huynh et al, 2005). Arrays were purified by precipitation with 5 mM MgCl2 (Schwarz et al, 1996). Native Drosophila histone octamers were purified from 0–12 h ael embryos (Simon and Felsenfeld, 1979). Recombinant Drosophila histones were expressed in E. coli and purified according to Morales et al (2004). Histone H1 was obtained from 0–12 h ael Drosophila embryos (Croston et al, 1991). Chicken histone H5 and the construct containing 12 200-bp repeats of the 601 nucleosome positioning sequence were provided by the Rhodes lab. To obtain linear 601 arrays, the insert was cut out with EcoRI and HindIII. Plasmid DNA was digested by DraI to 19, 692, 811 and 1113-bp fragments serving as competitor DNA. For radioactively labelled arrays, 20% of the 12-mer 601 repeats were labelled. For this purpose, repeats were purified from low melting agarose (Biozym) by phenol extraction. The HindIII end was labelled with α[32P]dCTP and Klenow polymerase (NEB), and excess nucleotides were removed with Roche Quick Spin Columns Sephadex G-50 according to the manufacturers' instructions.
Gel mobility-shift assays
Native agarose gels contained Seakem GTG Agarose (Biozym) gels in 0.2 × TB (18 mM Tris, 18 mM boric acid). H1 binding to chromatin arrays was analysed on 0.7% agarose gels, H5 binding to chromatin arrays on 1.4% agarose gels. Arrays were visualized by EtBr staining. To monitor occupancy of positioning sequences, 6-pmol arrays were digested by 15 U AvaI in RB50 (10 mM HEPES–KOH, pH 7.6, 50 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA) for 1 h at 26°C, run on 1.1% agarose gels and stained by SYBR® gold (Invitrogen).
Determination of histone stoichiometry
Arrays (60 pmol) were analysed on 15% polyacrylamide gels. Proteins were stained with Coomassie blue, intensities of bands were measured using the Odyssey® Infra Red Imaging System (Li-Cor).
Purification of ACF and CHD1 from Sf9 cells
Flag-ACF and flag-CHD1 were expressed in Sf9 cells using baculovirus constructs and purified as described previously (Eberharter et al, 2004). For flag-ACF, flag-Acf1 and ISWI were coexpressed, the baculoviruses containing both constructs were a gift from Dr J Kadonaga. The flag-CHD1 baculovirus was kindly provided by Dr A Lusser.
Chromatin remodelling reactions
All remodelling reactions were carried out for 1 h at 26°C. For probing nucleosomal DNA accessibility, 0.6 pmol 12-mer nucleosome or chromatosome arrays were incubated in 10 μl RB50 with or without 20 μM ATP. Reactions were started by adding 5 U AluI (NEB) along with 2.4 pmol ACF (0.3 ACF per nucleosome/chromatosome) or corresponding amounts of CHD1 (judged by ATPase activity) and stopped by adding 200 ng free DNA. Proteins were removed by Proteinase K (1 h, 37°C), DNA was precipitated with ethanol and applied on 1.3% agarose in 50 mM Tris, 384 mM glycine. Gels were dried and exposed to phosphoimager screens. The percentage of uncut DNA was determined with AIDA image analyser software. For MNase read-out reactions, 1.8 pmol arrays were incubated in 30 μl RB50, 20 μM ATP. Reactions were started by the addition of 7.2 pmol ACF or CHD1 (equal remodelling activity), stopped by 600 ng free DNA and digested by adding 4 × 10−3 U of MNase (Sigma) and CaCl2 to 3 mM for 1, 3 and 5 min (if reactions contained ACF) or for 0.5, 1 and 3 min (if reactions contained CHD1 or no remodeller). Digests were terminated by adding EDTA to 10 mM. As a marker, 0.6 pmol nucleosome arrays were digested for 1 h at 26°C with 5 U AluI. Free 12-mer 601 repeats (0.6 pmol labelled repeats+200 ng unlabelled DNA) were digested for 1 min at 26°C with 10−4 U of MNase. DNA was processed and visualized as above. 1D-evaluation of selected lanes was performed using AIDA image analyser software. If remodelling reactions were followed by primer extension, 3 pmol arrays were incubated in 15 μl RB50, 20 μM ATP with 12 pmol ACF. After 1 h, arrays were digested with 10−3 U MNase at 26°C for 20 min in the presence of 1 mM CaCl2. Nucleosomes and chromatosomes were resolved on 1.1% native agarose gels, stained with SYBR gold, excised and extracted using the Qiagen gel extraction kit. A total of 10% of the recovered DNA served as template for primer extension reactions (5 μM primers 13fw 5′-ATCTGACACGTGCCTGGA-3′, 13rv 5′-TCCAGGCACGTGTCAGAT-3′; 76fw 5′-CGTACGTGCGTTTAAGC-3′ or 76rv 5′-GCTTAAACGCACGTACG-3′ labelled radioactively by polynucleotide kinase (NEB), 3 U Taq polymerase (NEB), 60°C annealing temperature, 12 cycles), which were analysed on 7% polyacrylamide 20% urea gels in TBE.
ATPase assays
ATPase assays were performed as described (Eberharter et al, 2004) with 200 ng free or oligonucleosomal DNA and 15–25 pmol remodeller.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Anton Eberharter for fruitful discussion and critical reading of the manuscript. We thank Catherine Regnard, Andrew Routh and Andreas Hochheimer for providing recombinant histone octamers, chicken H5 and starting material for Drosophila H1 purification, respectively. We also thank Ragnhild Eskeland and Natascha Kunert for the assistance in the purification of H1 and histone octamers from Drosophila embryos. This work was supported by Deutsche Forschungsgemeinschaft through SFB 594 (TP A6) and by the European Union via the Network of Excellence ‘The Epigenome' (FP6-503433). VM is a fellow of the Munich International Max Planck Research School for Molecular and Cellular Life Sciences.
Please note that the print publication of this article was delayed due to a systems error. The Publishers sincerely apologise for any inconvenience this may have caused.
References
- Becker PB (2002) Nucleosome sliding: facts and fiction. EMBO J 21: 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB, Horz W (2002) ATP-dependent nucleosome remodeling. Annu Rev Biochem 71: 247–273 [DOI] [PubMed] [Google Scholar]
- Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA 95: 14173–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharath MM, Ramesh S, Chandra NR, Rao MR (2002) Identification of a 34 amino acid stretch within the C-terminus of histone H1 as the DNA-condensing domain by site-directed mutagenesis. Biochemistry 41: 7617–7627 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL (2000) Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J Biol Chem 275: 18864–18870 [DOI] [PubMed] [Google Scholar]
- Brown DT, Izard T, Misteli T (2006) Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol 13: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers LM, Bednar J, Woodcock CL, Hansen JC (1998) Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37: 14776–14787 [DOI] [PubMed] [Google Scholar]
- Carruthers LM, Hansen JC (2000) The core histone N termini function independently of linker histones during chromatin condensation. J Biol Chem 275: 37285–37290 [DOI] [PubMed] [Google Scholar]
- Catez F, Ueda T, Bustin M (2006) Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol 13: 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M, Rhodes D, Klug A (1983) Location of the primary sites of micrococcal nuclease cleavage on the nucleosome core. J Mol Biol 170: 423–446 [DOI] [PubMed] [Google Scholar]
- Corona DF, Langst G, Clapier CR, Bonte EJ, Ferrari S, Tamkun JW, Becker PB (1999) ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell 3: 239–245 [DOI] [PubMed] [Google Scholar]
- Corona DF, Siriaco G, Armstrong JA, Snarskaya N, McClymont SA, Scott MP, Tamkun JW (2007) ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol 5: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston GE, Lira LM, Kadonaga JT (1991) A general method for purification of H1 histones that are active for repression of basal RNA polymerase II transcription. Protein Expr Purif 2: 162–169 [DOI] [PubMed] [Google Scholar]
- Dang W, Kagalwala MN, Bartholomew B (2006) Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol 26: 7388–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Vetter I, Ferreira R, Becker PB (2004) ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J 23: 4029–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, He X, Kingston RE, Narlikar GJ (2003) Distinct strategies to make nucleosomal DNA accessible. Mol Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI (2005) Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T (2004) Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr Opin Genet Dev 14: 165–173 [DOI] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT (2002) Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418: 897–900 [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B (2007) Dependency of ISW1a chromatin remodeling on extranucleosomal DNA. Mol Cell Biol 27: 3217–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Neely KE, Workman JL (2001) Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104: 817–827 [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th'ng JP (2004) The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem 279: 20028–20034 [DOI] [PubMed] [Google Scholar]
- Hill CS, Rimmer JM, Green BN, Finch JT, Thomas JO (1991) Histone-DNA interactions and their modulation by phosphorylation of -Ser–Pro–X–Lys/Arg- motifs. EMBO J 10: 1939–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Imbalzano AN (2000) Human SWI/SNF nucleosome remodeling activity is partially inhibited by linker histone H1. Biochemistry 39: 11649–11656 [DOI] [PubMed] [Google Scholar]
- Horn PJ, Carruthers LM, Logie C, Hill DA, Solomon MJ, Wade PA, Imbalzano AN, Hansen JC, Peterson CL (2002) Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol 9: 263–267 [DOI] [PubMed] [Google Scholar]
- Horowitz RA, Agard DA, Sedat JW, Woodcock CL (1994) The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol 125: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, Rhodes D (2005) A method for the in vitro reconstitution of a defined ‘30 nm' chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol 345: 957–968 [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B (2004) Topography of the ISW2–nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J 23: 2092–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikyo N, Wade PA, Guschin D, Ge H, Wolffe AP (2000) Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 289: 2360–2362 [DOI] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001) ISWI induces nucleosome sliding on nicked DNA. Mol Cell 8: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J (2005) Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol 12: 46–53 [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Lusser A, Kadonaga JT (2003) Chromatin remodeling by ATP-dependent molecular machines. Bioessays 25: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT (2005) Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166 [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Freedman BS, Heald R (2005) Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol 169: 859–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales V, Straub T, Neumann MF, Mengus G, Akhtar A, Becker PB (2004) Functional integration of the histone acetyltransferase MOF into the dosage compensation complex. EMBO J 23: 2258–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacheva GA, Guschin DY, Preobrazhenskaya OV, Karpov VL, Ebralidse KK, Mirzabekov AD (1989) Change in the pattern of histone binding to DNA upon transcriptional activation. Cell 58: 27–36 [DOI] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford R, Hager GL (2004) Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14: 163–174 [DOI] [PubMed] [Google Scholar]
- Ner SS, Travers AA (1994) HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J 13: 1817–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale K, Dimitrov S, Reeves R, Wolffe AP (1996) Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J 15: 548–561 [PMC free article] [PubMed] [Google Scholar]
- Pennings S, Meersseman G, Bradbury EM (1994) Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci USA 91: 10275–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran A, Omar M, Cheslock P, Schnitzler GR (2003) Linker histone H1 modulates nucleosome remodeling by human SWI/SNF. J Biol Chem 278: 48590–48601 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V, Finch JT, Graziano V, Lee PL, Sweet RM (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 362: 219–223 [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Fairall L, Huynh VA, Rhodes D (2006) EM measurements define the dimensions of the ‘30-nm' chromatin fiber: evidence for a compact, interdigitated structure. Proc Natl Acad Sci USA 103: 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Rhodes D (2006) Structure of the ‘30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol 16: 336–343 [DOI] [PubMed] [Google Scholar]
- Saeki H, Ohsumi K, Aihara H, Ito T, Hirose S, Ura K, Kaneda Y (2005) Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci USA 102: 5697–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Schwarz PM, Felthauser A, Fletcher TM, Hansen JC (1996) Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry 35: 4009–4015 [DOI] [PubMed] [Google Scholar]
- Shen X, Yu L, Weir JW, Gorovsky MA (1995) Linker histones are not essential and affect chromatin condensation in vivo. Cell 82: 47–56 [DOI] [PubMed] [Google Scholar]
- Sheng S, Czajkowsky DM, Shao Z (2006) Localization of linker histone in chromatosomes by cryo-atomic force microscopy. Biophys J 91: L35–L37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon RH, Felsenfeld G (1979) A new procedure for purifying histone pairs H2A+H2B and H3+H4 from chromatin using hydroxylapatite. Nucleic Acids Res 6: 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RT (1978) Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry 17: 5524–5531 [DOI] [PubMed] [Google Scholar]
- Sprouse RO, Brenowitz M, Auble DT (2006) Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. EMBO J 25: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Langst G (2005) A ‘loop recapture' mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol 12: 683–690 [DOI] [PubMed] [Google Scholar]
- Sun JM, Ali Z, Lurz R, Ruiz-Carrillo A (1990) Replacement of histone H1 by H5 in vivo does not change the nucleosome repeat length of chromatin but increases its stability. EMBO J 9: 1651–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JO, Rees C (1983) Exchange of histones H1 and H5 between chromatin fragments. A preference of H5 for higher-order structures. Eur J Biochem 134: 109–115 [DOI] [PubMed] [Google Scholar]
- Tremethick DJ (2007) Higher-order structures of chromatin: the elusive 30 nm fiber. Cell 128: 651–654 [DOI] [PubMed] [Google Scholar]
- Ura K, Hayes JJ, Wolffe AP (1995) A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J 14: 3752–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB (2006) Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev 16: 151–156 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Blank TA, Becker PB (1995) Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J 14: 2209–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock CL, Skoultchi AI, Fan Y (2006) Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14: 17–25 [DOI] [PubMed] [Google Scholar]
- Yager TD, van Holde KE (1984) Dynamics and equilibria of nucleosomes at elevated ionic strength. J Biol Chem 259: 4212–4222 [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ (2006) The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol 13: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S (1998) Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature 395: 402–405 [DOI] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Bartholomew B (2004) Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol 24: 10047–10057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information