Abstract
In Huntington's disease (HD), mutant Huntingtin, which contains expanded polyglutamine stretches, forms nuclear aggregates in neurons. The interactions of several transcriptional factors with mutant Huntingtin, as well as altered expression of many genes in HD models, imply the involvement of transcriptional dysregulation in the HD pathological process. The precise mechanism remains obscure, however. Here, we show that mutant Huntingtin aggregates interact with the components of the NF-Y transcriptional factor in vitro and in HD model mouse brain. An electrophoretic mobility shift assay using HD model mouse brain lysates showed reduction in NF-Y binding to the promoter region of HSP70, one of the NF-Y targets. RT–PCR analysis revealed reduced HSP70 expression in these brains. We further clarified the importance of NF-Y for HSP70 transcription in cultured neurons. These data indicate that mutant Huntingtin sequesters NF-Y, leading to the reduction of HSP70 gene expression in HD model mice brain. Because suppressive roles of HSP70 on the HD pathological process have been shown in several HD models, NF-Y could be an important target of mutant Huntingtin.
Keywords: heat shock protein, Huntington's disease, NF-Y, transcription
Introduction
Huntington's disease (HD) is an autosomal-dominant, adult-onset neurodegenerative disorder characterized by chorea, psychiatric disturbances and dementia (Landles and Bates, 2004). HD is caused by a CAG repeat expansion in exon 1 of the Huntingtin (Htt) gene (The Huntington's Disease Collaborative Research Group, 1993). The product, mutant Htt containing expanded polyglutamine (polyQ), forms nuclear aggregates in neurons in specific regions including the striatum and cortex, where severe neurodegeneration is observed. Analyses of HD postmortem tissues or other HD model systems have identified many genes whose expression is altered by mutant Htt (Chan et al, 2002; Luthi-Carter et al, 2002; Kotliarova et al, 2005). Mutant Htt's effects on gene expression in mice recapitulate changes observed in human HD brains (Kuhn et al, 2007). Recently, we found that expression of sodium channel subunit β4 is severely reduced in the brains of early-phase HD model mice, which might be involved in dendritic abnormalities of neurons observed in HD (Oyama et al, 2006). Thus, alteration of gene expression is one of the key pathological changes in HD (Cha, 2000; Sugars and Rubinsztein, 2003), yet the mechanisms of this alteration have not been fully elucidated.
Recent studies have identified interactions of mutant Htt with several transcriptional factors such as CREB-binding protein (CBP), TBP, SP1, TAFII130 and p53 (Harjes and Wanker, 2003; Li and Li, 2004). CBP, containing a polyQ stretch, is incorporated into mutant Htt aggregates, which leads to sequestration of this protein and suppression of CREB-mediated CRE transcription (Steffan et al, 2000; Nucifora et al, 2001). TBP, also containing a polyQ stretch, is incorporated into mutant Htt aggregates (Suhr et al, 2001; van Roon-Mom et al, 2002; Matsumoto et al, 2006), and suppression of its DNA binding by mutant Htt was shown in an in vitro system (Schaffar et al, 2004). SP1, which contains a glutamine-rich region, and TAFII130 interact with a soluble form of mutant Htt, which leads to suppression of SP1 transcriptional activity, finally resulting in downregulation of dopamine D2 or nerve growth factor receptor (Dunah et al, 2002; Li et al, 2002). p53 is incorporated into mutant Htt aggregates, leading to repression of p53-mediated transcription (Steffan et al, 2000). Although these observations indicate that mutant Htt aggregates sequester these transcriptional factors and suppress their function, thus leading to neurodegeneration in HD, direct evidence for the involvement of altered expression of the target genes in the HD pathological process has yet to be obtained. In addition, several observations seem to be in conflict with the above sequestration model. For example, it has been shown that CBP and TBP are not depleted in HD model mouse brains (Yu et al, 2002), and CREB-mediated CRE transcription is actually increased in these brains (Obrietan and Hoyt, 2004). Increased expression of p53 or SP1 has been reported for HD model mouse brains, and the knockout of each gene alleviates the phenotype in the model mice (Bae et al, 2005; Qiu et al, 2006). Thus, the relationship between the sequestration of the above transcriptional factors and the HD pathological process is not yet clear.
Heat shock proteins (HSPs), including HSP70 and HSP40, have important roles in the folding and assembly of newly synthesized proteins and the refolding of misfolded and aggregated proteins (Mayer and Bukau, 2005). HSPs are reported to be incorporated into Htt aggregates (Jana et al, 2000). In vitro studies have suggested the suppressive role of these HSPs on aggregation of mutant Htt (Muchowski et al, 2000; Wacker et al, 2004). Furthermore, increased expression of these HSPs suppressed neurodegenerative phenotypes in mouse and Drosophila HD model systems (Chan et al, 2000; Kazemi-Esfarjani and Benzer, 2000; Fujimoto et al, 2005). These observations suggest the suppressive roles of these HSPs on the HD pathological process (Sakahira et al, 2002; Barral et al, 2004). Importantly, expression of HSPs, including HSP70, has been reported to progressively decrease in mouse models of HD (Merienne et al, 2003; Hay et al, 2004), suggesting that the suppressive roles of these HSPs are disturbed in the HD brain, although the precise mechanism of altered HSP expression has not been elucidated.
We previously established a method for direct analysis of mutant Htt aggregate-associated proteins by mass spectrometry (Doi et al, 2004). By this method, we identified components of the NF-Y transcriptional factor as candidate interaction proteins for mutant Htt aggregates. We confirmed this interaction in vitro, in cultured cells and in HD model mouse brains. Because NF-Y has been reported to regulate HSP70 transcription (Li et al, 1998; Imbriano et al, 2001), we focused on NF-Y activity and HSP70 expression in HD model mouse brain. We provided evidence supporting the idea that mutant Huntingtin aggregates sequester NF-Y components to reduce functional NF-Y, leading to the reduction of HSP70 gene expression. Considering the suppressive roles of HSP70 on the HD pathological process, NF-Y might be an important target of mutant Htt to induce or promote HD pathology.
Results
Components of the NF-Y transcriptional factor were incorporated into mutant Htt aggregates in Htt-transfected neuro2a cells
We have previously identified several ubiquitin-interacting proteins by mass spectrometry analysis of nuclear inclusions purified from neuro2a cells expressing N-terminal Huntingtin (Nhtt) containing a pathological stretch of glutamine (150Q) fused with EGFP and SV40 NLS (Nhtt150Q–EGFP–NLS) (Doi et al, 2004). Further analysis of mass spectrometry data suggested incorporation of components of NF-Y transcriptional factor in addition to a known Htt aggregate-interacting protein, CA150 (Holbert et al, 2001). However, other transcriptional factors, such as CBP, TBP or SP1, were not found in our data.
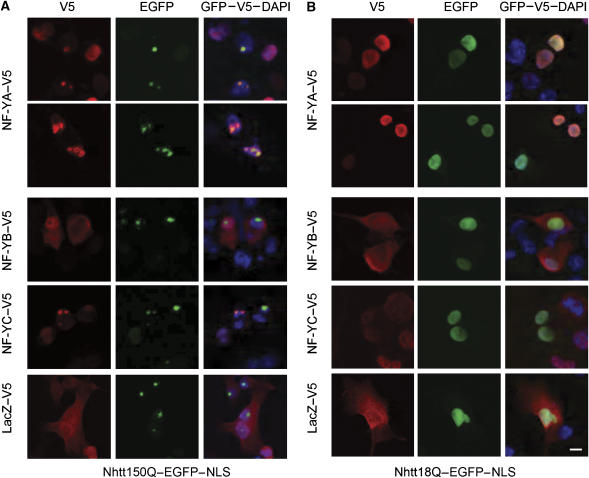
NF-Y, which is composed of three subunits, NF-YA, NF-YB, and NF-YC, binds to CCAAT sequence to regulate gene transcription (Supplementary Figure S1A; Maity and de Crombrugghe, 1998; Mantovani, 1999). To confirm the interactions between mutant Htt and NF-Y components, NF-YA, NF-YB or NF-YC tagged with V5 was overexpressed in neuro2a cells expressing Nhtt150Q–EGFP–NLS. As shown in Figure 1A, NF-YA–V5 or NF-YC–V5, but not NF-YB–V5, colocalized with Nhtt150Q–EGFP–NLS in nuclear inclusions. This colocalization was not observed when LacZ–V5 is used as a negative control (Figure 1A). In addition, these concentrated stainings of NF-YA–V5 or NF-YC–V5 were not observed in the cells expressing Nhtt protein containing nonpathological lengths of glutamine repeats (18Q) (Figure 1B). These results indicate that among NF-Y components, NF-YA and NF-YC preferentially localizes to nuclear inclusions with mutant Htt in neuro2a cells.
Figure 1.
Localization of NF-Y components in Nhtt150Q or Nhtt18Q–EGFP–NLS expressing Neuro2a cells. Neuro2a cells inducibly expressing Nhtt150Q–EGFP–NLS (A) or Nhtt18Q–EGFP–NLS (B) were transfected with expression vectors for NF-YA, NF-YB, NF-YC or LacZ tagged with V5. After 3 days, cells were fixed and stained with monoclonal anti-V5 antibody and Alexa 546-conjugated anti-mouse IgG. Nhtt proteins were detected as GFP fluorescence. Nuclei were detected with DAPI. Scale bars: 10 μm.
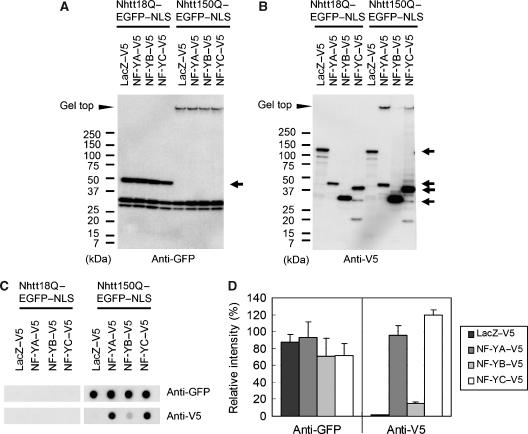
Next, we examined whether NF-YA and NF-YC form SDS-insoluble aggregates with mutant Htt in neuro2a cells by western blot analysis. Anti-GFP staining showed gel top bands in Nhtt150Q–EGFP–NLS-expressing cells, but not in Nhtt18Q–EGFP–NLS-expressing cells (Figure 2A), indicating the formation of SDS-insoluble aggregates by Nhtt150Q–EGFP–NLS. Anti-V5 staining showed the gel top bands for NF-YA or NF-YC in the cells expressing Nhtt150Q–EGFP–NLS, but not in the cells expressing Nhtt18Q–EGFP–NLS (Figure 2B). Filter trap assay confirmed the preferential incorporation of NF-YA or NF-YC into SDS-insoluble aggregates with Nhtt150Q–EGFP–NLS in neuro2a cells, whereas overexpression of NF-YA or NF-YC itself did not significantly change the amount of Nhtt150Q–EGFP–NLS aggregates (Figure 2C and D). These data indicate that NF-YA or NF-YC forms SDS-resistant protein complexes with mutant Htt without affecting mutant Htt aggregation in neuro2a cells. Co-precipitation of NF-YA or NF-YC with soluble Ntt proteins was not observed by immunoprecipitation assay using nonionic detergent (TritonX-100)-soluble fractions of transfected neuro2a cells (data not shown), suggesting that NF-YA and NF-YC preferentially interacts with aggregated form of mutant Htt in neuro2a cells. There is a glutamine-rich region in NF-YA and NF-YC but not in NF-YB (Supplementary Figure S1A; Maity and de Crombrugghe, 1998; Mantovani, 1999), suggesting this region is involved in interaction with mutant Htt aggregates. In this paper, we referred to biochemically insoluble protein aggregates as ‘aggregates'. On the other hand, histologically visible ‘aggregates' were called as inclusions.
Figure 2.
Part of NF-YA or NF-YC was incorporated into SDS-insoluble aggregates with Nhtt150Q–EGFP in Neuro2a cells. (A, B) Neuro2a cells were transfected with expression vector for NF-YA, NF-YB, NF-YC or LacZ tagged with V5 together with expression vector for Nhtt18Q–EGFP–NLS or Nhtt150Q–EGFP–NLS. After 24 h, cells were subjected to SDS–PAGE and western blot analysis using anti-GFP (A) or anti-V5 (B) antibody. Bands for expressed proteins are indicated by arrows and positions of the gel top are indicated by arrowheads. Bands for Nhtt150Q–EGFP–NLS were not observed possibly due to its insolubilization. (C, D) Same samples were subjected to filter trap assay and stained with the same antibodies (C), and trapped proteins in Nhtt150Q–EGFP–NLS expressing cells were quantified (D). Values are means±s.d. of three independent experiments.
NF-YA interacts with mutant Htt aggregates in vitro
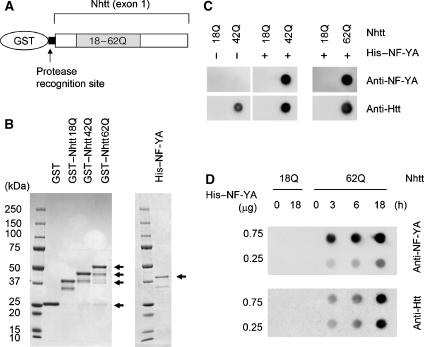
To examine whether the interaction between mutant Htt and NF-Y components is direct, we performed a binding assay using purified GST-fused Htt exon1 (Nhtt) containing 18Q, 42Q or 62Q (Figure 3A) and His-tagged NF-YA. As purification of His-tagged NF-YC was difficult because of the instability of the protein during the purification process, we used only NF-YA for the following experiments. CBB staining showed high purity of these proteins (Figure 3B).
Figure 3.
Interaction between NF-YA and aggregated Htt in vitro. (A) Scheme for the constructs of GST–Nhtt18Q, GST–Nhtt42Q or GST–Nhtt62Q. Recognition site for HRV-3C protease is indicated. (B) CBB staining of each purified protein. Bands for expressed proteins are indicated by arrows. (C) Interaction between Nhtt aggregates and His–NF-YA. GST–Nhtt–18Q, GST–Nhtt42Q or GST–Nhtt62Q treated with HRV-3C protease were incubated at 37°C for 18 h. During these processes, Nhtt42Q or Nhtt62Q, but not Nhtt18Q, formed aggregates. Each protein was co-incubated with His–NF-YA protein for 2 h at 4°C, and then subjected to filter trap assay. The aggregated proteins were detected with anti-NF-YA or anti-Htt antibody. (D) Co-aggregation of NF-YA with Nhtt62Q. After treatment with HRV-3C protease, 10 μg of Nhtt18Q or Nhtt62Q protein was co-incubated with 0.25 or 0.75 μg of His–NF-YA for the indicated time. The trapped aggregated proteins were detected as in panel C.
First, we examined the interaction between Htt aggregates and NF-YA. To generate Htt aggregates in vitro, we cleaved Nhtt proteins from GST using HRV-3C protease and incubated at 37°C for 18 h. By filter trap assay, aggregated proteins were detected by anti-Htt when we used GST–Nhtt42Q or GST–Nhtt62Q, but not GST–Nhtt18Q, as described previously (Figure 3C) (Scherzinger et al, 1997; Busch et al, 2003). When His–NF-YA was co-incubated with these Nhtt proteins, His–NF-YA was detected together with aggregated Nhtt42Q or Nhtt62Q, but not Nhtt18Q, by filter trap assay (Figure 3C), indicating that His–NF-YA directly interacts with these aggregates. Next, we checked the possibility of co-aggregation of NF-YA with mutant Htt in vitro. His–NF-YA was co-incubated with protease-treated Nhtt proteins for the indicated time, and then samples were subjected to filter trap assay. Nhtt62Q started to form aggregates after 3 h incubation (Figure 3D). His–NF-YA was also detected after 3 h incubation in a dose-dependent manner (Figure 3D). The data indicate that His–NF-YA co-aggregated with mutant Htt in vitro.
NF-YA or NF-YC antibodies stained nuclear inclusions in cortex and striatum of HD model mouse brain sections
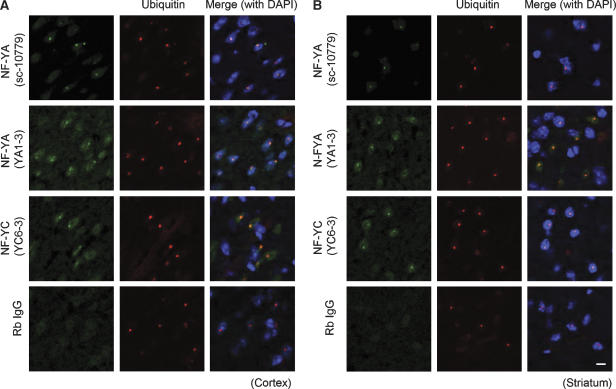
A transgenic mouse line (R6/2) expressing an Htt exon 1 that contains expanded polyglutamine residues is used extensively as an HD model (Mangiarini et al, 1996; Davies et al, 1997). To examine whether NF-YA or NF-YC interacts with mutant Htt aggregates in vivo, we stained brain sections from 12-week-old R6/2 mice with anti-NF-YA or -NF-YC antibodies, together with anti-ubiquitin, a marker for nuclear inclusions containing mutant Htt (Davies et al, 1997). Antibodies against NF-YA or NF-YC, but not IgG, showed dot-like staining colocalized with ubiquitin staining in cell nuclei in both cortex (Figure 4A) and striatum (Figure 4B). These clear dot-like stainings were not observed when we used brain sections from age-matched control mouse (data not shown). On the contrary, dot-like stainings were not observed by anti-NF-YB antibody in cortex and striatum of brain sections from 12-week-old R6/2 mice (Supplementary Figure S2). These results suggest that NF-YA and NF-YC, but not NF-YB, localize in nuclear inclusions containing mutant Htt in cortex and striatum of HD model mouse.
Figure 4.
Antibodies against NF-YA or NF-YC showed dot-like stainings colocalized with ubiquitin-positive inclusions in R6/2 mouse brain. Coronal sections (10 μm) prepared from frozen brain of 12w R6/2 mouse were fixed and stained with anti-NF-YA (sc-10779 or YA1-3), anti-NF-YC (YC 6-3) or rabbit IgG together with anti-ubiquitin (MAB1510), a marker for nuclear inclusions. Nuclear inclusions positive for ubiquitin were stained by NF-YA or NF-YC antibodies, but not rabbit IgG, in cortex (A) and striatum (B). Scale bars: 10 μm.
NF-Y binding to the HSP70 promoter was reduced in cortex and striatum of HD model mice
The proximal region of the human HSP70 promoter contains two NF-Y-binding sites (CCAAT boxes) in addition to sites for other transcriptional factors, such as SP1, HSF1 and TBP (see Figure 10A) (Wu et al, 1986; Greene et al, 1987; Morgan et al, 1987; Morgan, 1989). These transcriptional factor-binding sites were conserved in mice (Bevilacqua et al, 1997). Positive regulation of HSP70 transcription by NF-Y has been shown in Xenopus oocyte and mouse lymphoma cell lines by reporter gene assays (Li et al, 1998; Imbriano et al, 2001).
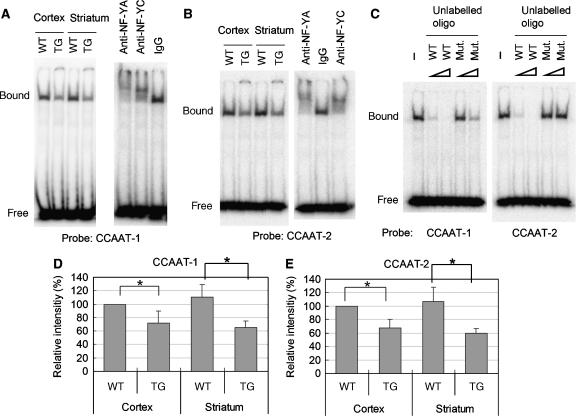
The incorporation of NF-Y components into mutant Htt aggregates in R6/2 mouse brain raises the possibility that NF-Y function was altered. To examine this, we performed electrophoretic mobility shift assay (EMSA) using transcriptional binding sites of the human HSP70 promoter as probes (Supplementary Table II). Interestingly, when we performed EMSA using the proximal CCAAT (CCAAT-1) of HSP70 promoter as a probe, we observed a reduction in DNA–protein complexes in cortex and striatum lysates of 12-week-old R6/2 mice (Figure 5A). Supershifts were observed by pre-incubation of the lysates with anti-NF-YA or -NF-YC, but not IgG (Figure 5A). Furthermore, binding was suppressed by addition of unlabelled wild-type annealed oligo much effectively than its mutant containing two mutations in the CCAAT sequence (Figure 5C; Supplementary Table II). These data indicate that proteins binding to the probe are NF-Y components. Quantified data showed that the binding of NF-Y was reduced to 30–40% of control levels in these brain regions (Figure 5D). Similar data were obtained when we used another CCAAT region (CCAAT-2) as a probe (Figure 5B, C and E). These data indicate that NF-Y binding to HSP70 promoter was reduced in the cortex and striatum of 12-week-old R6/2 mice.
Figure 5.
NF-Y binding to two CCAAT regions in HSP70 promoter was reduced in cortex and striatum of 12w R6/2 mouse brain. Cortex or striatum lysates prepared from 12-week-old R6/2 (TG) or control mouse (WT) brain was subjected to EMSA. (A) EMSA using proximal CCAAT region (CCAAT-1) as a probe (left panel). Supershift assay using anti-NF-YA, anti-NF-YC or IgG (right panel). (B) EMSA using distal CCAAT region (CCAAT-2) as a probe (left panel). Supershift assay (right panel). (C) Competition analysis using 10- or 100-fold excess unlabelled annealed oligo (WT) or its mutant (Mut.). (D, E) Quantification of the amount of DNA–protein complex. Values are means±s.d. of three independent experiments (*P<0.05).
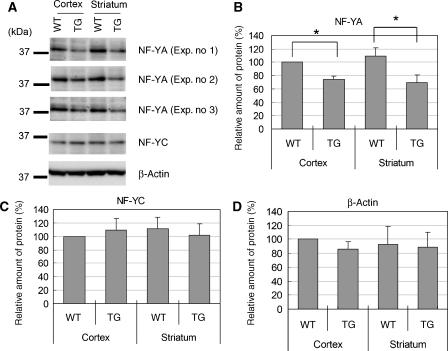
By analysing the lysates used for EMSA, we found that the amount of NF-YA was decreased in both cortex and striatum lysates of R6/2 mice (Figure 6A). Quantified data revealed a 30–40% reduction in NF-YA proteins (Figure 6B), which is highly compatible with the reduction of functional NF-Y complex estimated from the EMSA study (Figure 5). On the contrary, the amounts of NF-YC and β-actin were not altered in these lysates (Figure 6A, C and D). These results suggest that reduction of NF-Y binding to DNA was caused by a reduction of NF-YA protein itself. On the contrary, RT–PCR analysis revealed that mRNA of NF-YA or NF-YC was actually increased in the cerebrum of 8- and 12-week-old R6/2 mice, whereas mRNA levels of NF-YB in R6/2 mice was not significantly different from that in control mice (Supplementary Figure S1B–D). These data suggest that reduction of NF-YA in the lysates was not caused by reduction of its expression at the transcriptional level. These data also imply the existence of some compensatory system for functional impairment of NF-Y in vivo.
Figure 6.
The amount of NF-YA protein decreased in the lysates of R6/2 mice. (A) The lysates used for EMSA from R6/2 (TG) or control mice (WT) were subjected to western blot analysis using antibodies against NF-YA (YA1-3), NF-YC (YC5-3) and β-actin. In the case of NF-YA, data from three experiments are shown. (B–D) Quantification of the amount of NF-YA, NF-YC or β-actin. Values are means±s.d. of three independent experiments (*P<0.05).
We next examined the DNA binding of SP1, HSF1 and TBP. In contrast to NF-Y, DNA binding of SP-1 was not reduced in R6/2 cortex or striatum (Supplementary Figure S3A–C). In the case of HSF1, these brain tissue lysates did not show clear shifted bands, but such bands were seen when we used lysates of neuro2a cells treated with heat shock (Supplementary Figure S3D). This suggests that the amount of active HSF1 is very low in these lysates. EMSA for TBP did not work well (data not shown), possibly because of technical difficulties. Western blot analysis revealed that the amount of SP-1, HSF1 or TBP was not significantly changed among the lysates used for EMSA (Supplementary Figure S4A–D). Taken together, our data suggest that among the transcriptional factors reported to interact with the proximal region of the HSP70 promoter, only NF-Y is affected in the cortex and striatum of 12-week-old R6/2 mice.
Reduction in NF-Y, but not SP1, binding to DNA was also observed in cortex of 8-week-old R6/2 mouse brain (Supplementary Figure S5A–D) in which reduction in HSP70 protein was observed by our experiments (Figure 7), as well as that of others (Merienne et al, 2003; Hay et al, 2004). In addition, we could obtain similar data when we used cortex lysates prepared from 36- and 38-week-old R6/1 mice, another mutant Htt transgenic mouse line (Supplementary Figure S6A–D) (Mangiarini et al, 1996; Davies et al, 1997). Thus, the reduction of NF-Y binding to DNA as well as reduced expression of HSP70 protein was generally observed in different stages and different lines of mutant Htt transgenic mice.
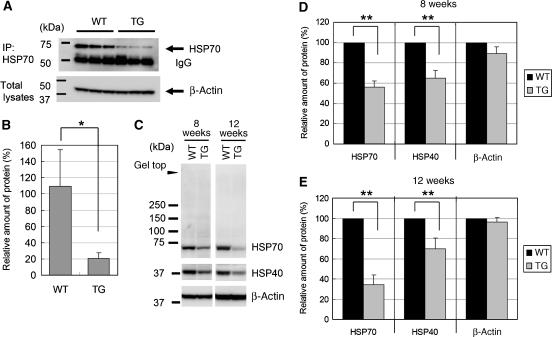
Figure 7.
Expression of HSP70 protein was decreased in 8- or 12-week-old R6/2 mouse brain. (A) Cerebrums from 12-week-old R6/2 or control mice were subjected to immunoprecipitation with anti-HSP70 (SPA-801), and precipitated HSP70 was detected with the same antibody. β-Actin proteins in total lysates are also shown. (B) Quantification of the amount of immunoprecipitated HSP70 protein. (C) Total lysates of cortexes from 8- or 12-week-old R6/2 or control mice were subjected to western blot analysis using antibodies against HSP70 (sc-24), HSP40 or β-actin. (D, E) Quantification of the amount of each protein of 8-week-old (D) or 12-week-old (E) mouse brain cortexes. Values are means±s.d. of three independent experiments (*P<0.05, **P<0.01).
Expression of HSP70 mRNA was decreased in HD model mouse brain
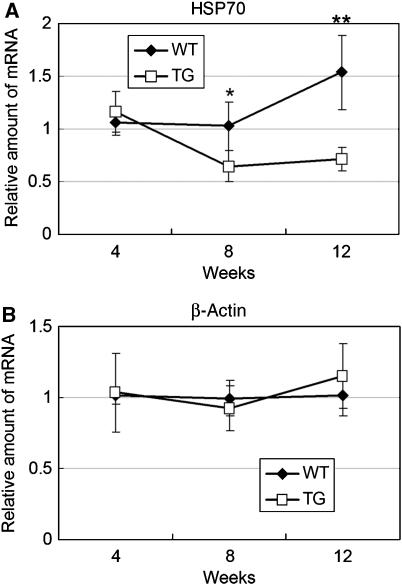
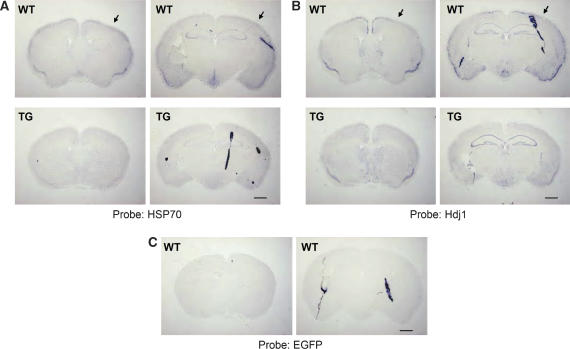
Immunoprecipitation analysis using anti-HSP70 antibody showed the reduction of soluble HSP70 protein in the brain of 12-week-old R6/2 mice (Figure 7A and B). By western blot analysis using another HSP70 antibody, we noticed that reduction of HSP70 in 8- and 12-week-old R6/2 mouse brain cortex was not accompanied by appearance of gel top band for this protein (Figure 7C–E), suggesting that reduction of HSP70 is not caused by its insolubilization. To examine whether the reduction of HSP70 protein is caused by reduction of HSP70 transcription, we performed RT–PCR using 4-, 8- or 12-week-old R6/2 or control cerebrums. The mRNA expression of HSP70 started to decrease at 8 weeks after birth and remained this level until 12 weeks, whereas its expression increased at 12 weeks in control mice (Figure 8A). In contrast, the mRNA levels of β-actin were not changed (Figure 8B). To confirm the data further, we performed in situ hybridization using a mouse HSP70 antisense probe. HSP70 mRNA was densely detected at cortical regions of control mouse brain, which were severely reduced in R6/2 mouse brain (Figure 9A). These signals were not observed if we used EGFP antisense probe, which was useful for in situ hybridization of mouse brain section (Kotliarova et al, 2005), as a negative control (Figure 9C). Taken together, these results indicate that the expression of HSP70 mRNA, which is highly detected in cortical regions, was severely suppressed in R6/2 mouse.
Figure 8.
Expression of HSP70 mRNA was decreased in 8- or 12-week-old R6/2 mouse brain. Quantitative RT–PCR analysis of HSP70 (A) or β-actin (B) in 4, 8 or 12 week-old cerebrum of R6/2 (TG) or control mice (WT). Values are means±s.d. of four independent experiments (*P<0.05, **P<0.01).
Figure 9.
Cortical expression of HSP70 or Hdj1 mRNA is reduced in 12-week-old R6/2 mouse. In situ hybridization of brain sections from 12-week-old R6/2 (TG) or control (WT) mouse using antisense probe for HSP70 (A), Hdj1 (B) or EGFP (C). Strong expression of HSP70 or Hdj1 mRNA at cortical regions (arrows) in WT mouse was severely reduced in TG mouse. No clear signals were detected by EGFP probe. Scale bar: 1 mm.
Because the promoter region of Hdj1, one of the HSP40 isoforms, is also reported to contain NF-Y-binding sites (Hata and Ohtsuka, 1998; Imbriano et al, 2001), and its protein expression was reduced in R6/2 mouse brain (Hay et al, 2004), we also checked Hdj1mRNA expression by in situ hybridization using mouse Hdj1 antisense probe. Hdj1 mRNA is expressing in cortical region similarly to HSP70, and is partially suppressed in R6/2 mice (Figure 9B). The reduction of Hdj1 protein expression was also observed in R6/2 and R6/1 mouse brain cortex (Figure 7C–E; Supplementary Figure S6D).
Importance of NF-Y binding to HSP70 promoter region on its transcription in neuronal cells
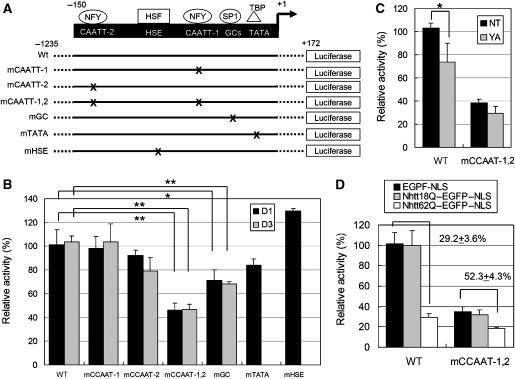
Finally, we examined the requirement of NF-Y for HSP70 transcription in neurons by reporter gene assay. We first constructed reporter gene vectors that contain the human HSP70 promoter (−1235 to +172) with or without mutation(s) in the transcriptional factor-binding site (Figure 10A). These reporter genes were introduced into cultured cortical neurons and luciferase activity was measured 1 or 3 days after transfection. In our experimental condition, two-thirds of transfected cells were positive for the neuronal cell marker NeuN (data not shown). Luciferase activity was markedly reduced (2–3% of that of wild-type) when we used a reporter vector without the HSP70 promoter region (data not shown), meaning that the HSP70 promoter region used here has transcriptional activity in the transfected cells. Interestingly, mutations in both CCAAT regions (mCCAAT-1,2) significantly reduced reporter activity at day 1 or 3 after transfection (Figure 10B). Mutations in the SP-1-binding site also slightly reduced reporter activity, whereas mutations in the TBP- or HSF1-binding site did not (Figure 10B).
Figure 10.
Importance of NF-Y-binding sites on promoter activity of human HSP70 in primary cultured cortical neurons. (A) Reporter gene constructs containing −1235 to +172 of human HSP70 promoter fused with luciferase gene. WT, wild type with no mutation; mCCAAT-1, single mutation in proximal NF-Y-binding site; mCCAAT-2, single mutation in distal NF-Y-binding site; mCCAAT-1,2, double mutation in both NF-Y-binding sites; mGC, single mutation in SP1-binding site; mTATA, single mutation in TBP-binding site; mHSE, single mutation in HSF1-binding site. (B) In vitro-cultured cortical neurons were transfected with reporter gene construct together with a CMV-LacZ vector. At 1 (D1) or 3 days (D3) after transfection, luciferase assay and β-galactosidase assay were performed. (C) In vitro-cultured cortical neurons were transfected with siRNA oligos for NF-YA (YA) or control oligo (NT) together with reporter gene construct and CMV-LacZ vector. At 3 days after transfection, luciferase assay and β-galactosidase assay were performed. (D) In vitro-cultured cortical neurons were transfected with indicated expression vectors together with reporter gene construct and CMV-LacZ vector. At 6 days after transfection, luciferase assay and β-galactosidase assay were performed. The values were luciferase activities normalized by β-galactosidase activities obtained from more than three independent experiments (*P<0.05, **P<0.01).
We further performed knockdown of NF-YA using siRNA oligos, which could effectively knock down endogenous NF-YA when introduced into neuro2a cells (data not shown). We found that NF-YA RNAi could suppress promoter activity compared with non-targeting (NT) control (Figure 10C). The effect of NF-YA RNAi was not obvious if we used an mCCAAT-1,2 construct (Figure 10C). Taken together, these data indicate that NF-Y binding to the HSP70 promoter region is important for its transcription in cortical neurons.
Importantly, overexpression of Nhtt62Q–EGFP–NLS, but not Nhtt18Q–EGFP–NLS, reduced HSP70 promoter activity to 29.2±3.6% compared with control (EGFP–NLS) (Figure 10D). This reduction is comparable to that observed by mutation in mCCAAT-1,2. Furthermore, the sensitivity to mutant Htt was partly reduced by mutations in NF-Y-binding sites (52.3±4.3% compared with control) (Figure 10D). On the contrary, mutation in TBP-binding site did not affect mutant Htt-mediated repression of HSP70 promoter activity (Supplementary Figure S7). Thus, NF-Y-binding sites are responsible for repression of HSP70 promoter activity by mutant Htt in primary cultured cortical neurons.
Discussion
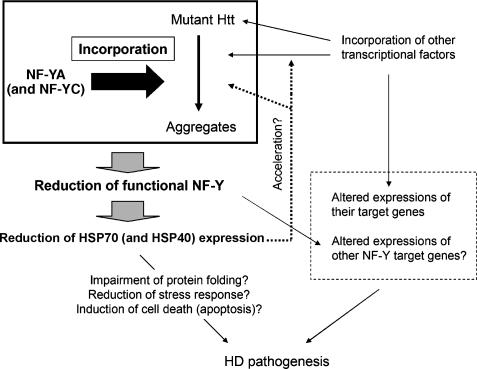
Previous studies have suggested the involvement of transcriptional dysregulation in HD pathogenesis (Cha, 2000; Harjes and Wanker, 2003; Sugars and Rubinsztein, 2003; Li and Li, 2004), although the precise mechanism remains obscure. In the present study, we identified NF-Y components (NF-YA and NF-YC) as novel mutant Htt aggregates-interacting proteins in vitro and in vivo. EMSA analysis using the HSP70 promoter region as a probe showed that NF-Y binding of HSP70 promoter was reduced in the cortex of HD model mice. Furthermore, expression of HSP70 mRNA was reduced in the cortex of these mice. Finally, a reporter gene assay indicated the importance of NF-Y on HSP70 transcription in cortical neurons. These data indicate that the NF-Y complex loses its function due to sequestration of NF-YA to mutant Htt aggregates, resulting in the reduction of HSP70 gene transcription (Figure 11).
Figure 11.
Hypothetical model. Incorporation of NF-YA (and probably with NF-YC) into mutant Htt aggregates causes reduction of functional NF-Y, resulting in suppression of HSP70/40 expression. Reduced expression of HSP70/40 would lead to acceleration of mutant Htt aggregation. This would also induce incorporation of some transcriptional factors into aggregates, which results in alteration of expression of their target genes. Expressions of NF-Y target genes other than HSP70 might be also affected. The reduction of HSP70/40 may result in reduction of their physiological functions, accelerating the aggregation formation and other pathological processes. These phenomenon might be cooperating together to modulate HD progression.
We further showed that overexpression of mutant Htt in cultured cortical neurons suppressed HSP70 promoter activity and the sensitivity to mutant Htt was reduced by mutations in NF-Y-binding sites, although its promoter activity was still sensitive to mutant Htt (Figure 10D). Thus, at least in in vitro condition, there would be additional target(s) of mutant Htt, in addition to NF-Y, for suppression of HSP70 promoter activity. TBP and HSF1 seem to be not involved in this process, because mutation in TBP- or HSF1-binding site did not show clear reduction of HSP70 promoter activity (Figure 10B), and mutation in TBP did not affect mutant Htt effect (Supplementary Figure S7). On the contrary, mutation in SP1-binding site showed some reduction of HSP70 promoter activity in cultured neurons (Figure 10B). Because suppression of SP1 activity by mutant Htt has been reported (Dunah et al, 2002; Li et al, 2002), SP1 could be another target of mutant Htt to induce the suppression of HSP70 promoter activity at least in cultured neurons. However, in R6/2 and R6/1 mice brains, SP1 binding to DNA was not reduced (Supplementary Figure S3–6). Further study is necessary to clarify the role of SP1 on HSP70 repression in HD in vivo. Recently, Tagawa et al (2007) used primary cultured neurons to show that mutant Htt induces HSP70 expression in cerebellar granule cells, which are insensitive to mutant Htt-induced degeneration, but not in cortical neurons, which are highly sensitive to it. They also found that p53 is involved in this cell-type-specific expression and suggest that this is one of mechanisms underlying vulnerabilities to mutant Htt among neuronal cell types. Considering our findings, this mechanism might be additively involved in the reduction of HSP70 expression in cortical neurons of HD model mice.
Because of suppressive roles of HSP70 and/or HSP40 on the pathological process in HD models (Sakahira et al, 2002; Barral et al, 2004), it seems that reduction of HSP70/40 enhances the HD pathological process by several mechanisms. One possible mechanism is acceleration of aggregation of mutant Htt, because HSP70/40 has been reported to suppress aggregation of mutant Htt (Figure 11; Muchowski et al, 2000; Wacker et al, 2004). Furthermore, HSP70/40 has been reported to suppress incorporation of some transcriptional factors into aggregates (Schaffar et al, 2004), thus the reduction of HSPs may also enhance sequestration of these transcriptional factors, leading to changes the expression of their target genes (Figure 11). This may also accelerate co-aggregation of NF-Y components, which would make a positive feedback loop on Htt aggregation. Another possible mechanism is the reduction of normal HSP70/40 functions, including protein folding, stress response, antiapoptotic function and regulation of signal transduction pathways (Figure 11; Nollen and Morimoto, 2002; Guzhova and Margulis, 2006). Consistent with this notion, Gidalevitz and co-workers have shown that expression of expanded polyglutamine proteins disrupts the global balance of protein folding control in Caenorhabditis elegans (Gidalevitz et al, 2006). Suppressive roles of HSP70 on mutant Htt-induced activation of cell death-related pathways have been also reported (Zhou et al, 2001; Merienne et al, 2003).
The NF-Y-binding motif, CCAAT, is one of the common promoter elements present in the proximal promoter regions of numerous mammalian genes transcribed by RNA polymerase II (Maity and de Crombrugghe, 1998; Mantovani, 1999). By re-analysing GeneChip data previously reported (Kotliarova et al, 2005), we found that the expressions of 147 genes was decreased in 8-week-old R6/2 brain cerebrum (data not shown) and 36 genes contain NF-Y-binding sites (CCAAT box) in putative promoter regions those were predicted by the UCSC Genome Browser (http://genome.ucsc.edu/) (Supplementary Table III). Interestingly, 12 of these genes contain more than two CCAAT sequences in their putative promoter regions like HSP70 (Supplementary Table III), implying that reductions of their expression in R6/2 mouse brain might be caused by NF-YA sequestration (Figure 11). These include preproenkephalin (PPE); however, chromatin-immunoprecipitation assay showed that NF-Y binding to PPE promoter region was not reduced in R6/2 mice brain (Chen-Plotkin et al, 2006), which seems to contradict our results using HSP70 promoter. One possible reason is that reduction of NF-Y binding of DNA by mutant Htt is promoter specific. Such promoter specificity has been reported in SCA17 model mouse brain (Friedman et al, 2007). Alternatively, the difference in brain regions used for assay could produce different results, because Chen-Plotkin and co-workers used whole brain, whereas we used highly affected regions including cortex/striatum in R6/2 mouse brain. Further studies will be needed to understand the relationship between the NF-Y function and the altered other gene expressions in HD models.
In summary, we identified NF-YA and NF-YC, as new aggregate-binding proteins and probable modulators of the HD pathological process. Further studies of the role of NF-Y in HD pathology would reveal novel aspect of neuronal degeneration.
Materials and methods
Mice
Heterozygous htt exon 1 transgenic male mice of the R6/2 strain and R6/1 strain were obtained from Jackson Laboratory (Bar Harbor, ME). In this paper, only male of R6/2 (over 120 CAG repeats), R6/1 and control mice were used for experiments.
Antibodies
Polyclonal antibody NF-YA (YA1-3) or NF-YC (YC5-3 or YC6-3) was generated against the C-terminal 14 aa of mouse NF-YA or NF-YC, respectively. Each antibody was purified by respective antigen peptide conjugated to SulfoLink Coupling Gel (PIERCE Biotechnology, Rockford, IL). Anti-htt (MAB5374) and anti-ubiquitin (MAB1510) were from Chemicon International (Temecula, CA); anti-NF-YA (sc-10779), anti-NF-YB (sc-13045), anti-HSP70 (sc-24) and anti-TBP (sc-204) were from Santa Cruz Biotechnology Inc.; anti-SP1 (07–645) was from Upstate; anti-HSP40 (Hdj1) (SPA-400), anti-HSF1 (SPA-901), anti-HSP70 (SPA-801) were from Stressgen; anti-V5 (R960-25) was from Invitrogen; anti-β-actin (A5441) was from Sigma-Aldrich; anti-ubiqutin (Z0458) was from DAKO.
Expression vectors
The cDNAs for NF-YA, NF-YB and NF-YC were kindly provided as FANTOM3 clones (Carninci et al, 2005). cDNAs were subcloned into the pcDNA3.1/V5–His for expression in mammalian cells (Invitrogen), or pET-15b for expression in Escherichia coli. cDNAs for N-terminal Htt (exon 1) containing Nhtt18Q–EGFP–NLS, Nhtt62Q–EGFP–NLS or Nhtt150Q–EGFP–NLS were subcloned into pcDNA3.1 vector (Invitrogen) as described previously (Wang et al, 2000; Doi et al, 2004). cDNAs for N-terminal Htt containing 18Q, 42Q or 62Q were subcloned into pGEX-6P vector for expression in E. coli.
Cell culture
Neuro2a cells or stably transfected neuro2a cells, which inducibly express Nhtt–EGFP–NLS containing 18 or 150Q under the control of ponasterone A were cultured as described previously (Doi et al, 2004). Method for in vitro culture of mouse cortical neurons was precisely described in Supplementary data. Transfection was performed by Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol.
Immunofluorescence microscopy and immunohistochemistry
Neuro2a cells or frozen brain sections (10 μm) were fixed with 4% paraformaldehyde/PBS for 10 min and stained as described previously (Wang et al, 2000; Doi et al, 2004). Immunohistochemistry was performed as described previously (Oyama et al, 2006).
Immunoprecipitation and western blotting
R6/2 or control mice cerebrums were homogenized with a Teflon homogenizer in lysis buffer containing 20 mM HEPES, pH 7.2, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% NP-40, 1 × Complete Protease Inhibitor (Roche). After sonication and centrifugation at 15 000 r.p.m. for 30 min, the supernatants were incubated with antibody-conjugated protein A Sepharose for 60 min at 4°C, after which the Sepharoses were washed twice with the lysis buffer. Immunoprecipitates were eluted with SDS-sample buffer and subjected to SDS–PAGE and western blotting as described previously (Yamanaka et al, 2006). Chemiluminescent signals were quantified using LAS-1000 (Fuji film).
TaqMan RT–PCR and in situ hybridization
The methods for TaqMan RT–PCR and in situ hybridization were described in Supplementary data.
Protein purification and in vitro binding assay
Methods for purification of GST–Nhtt and His–NF-YA and in vitro binding assay were described in detail in Supplementary data.
Electrophoretic mobility shift assay
EMSA was performed as described previously (Mosser et al, 1988) with slight modifications. The methods were described in Supplementary data.
Reporter gene assay
Human HSP70 (hspa1a) promoter (−1235 to +172) was amplified from human genomic DNA (Clontech) by PCR and subcloned into a pGL-3 vector (Promega). Mutations in transcriptional binding sites were generated using oligonucleotides listed in Supplementary Table II by a PCR-based method. Cortical neurons seeded on 12-well plates were transfected with these reporter vectors and pCMV-LacZ with or without Nhtt expression vector by Lipofectamine2000. After 1, 3 or 6 days, cells were lysed with Passive Lysis Buffer (Promega), and subjected to a luciferase assay (Promega) and β-galactosidase assay (Clontech). For RNAi, 20 pmol of oligos for NF-YA siRNA (M-065522-00) or NT control (D-001210-01-05) purchased from Dharmacon were co-introduced into cortical neurons with reporter vector and pCMV-LacZ by Lipofectamine2000. After 3 days, a luciferase assay and β-galactosidase assay were performed as above.
Supplementary Material
Supplementary data
Acknowledgments
We thank the staff at the Research Resource Center (RIKEN Brain Science Institute) for DNA sequencing analysis, peptide synthesis and antibody preparation; Gen Matsumoto for critical suggestions and technical help; Kumi Kaneko, Misako Okuno, Hideaki Shimizu and Yoshiaki Furukawa for technical help; and all the members of our laboratory (Laboratory for Structural Neuropathology, RIKEN Brain Science Institute) for helpful comments. This work was supported by Grant-in-Aid from Ministry of Education, Culture, Sports, Science and Technology of Japan (17025044, 18700354), the Ministry of Health, Welfare and Labour, Japan, and RIKEN Special Postdoctoral Researchers Program.
References
- Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH, Montell C, Ross CA, Snyder SH, Sawa A (2005) p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron 47: 29–41 [DOI] [PubMed] [Google Scholar]
- Barral JM, Broadley SA, Schaffar G, Hartl FU (2004) Roles of molecular chaperones in protein misfolding diseases. Semin Cell Dev Biol 15: 17–29 [DOI] [PubMed] [Google Scholar]
- Bevilacqua A, Fiorenza MT, Mangia F (1997) Developmental activation of an episomic hsp70 gene promoter in two-cell mouse embryos by transcription factor Sp1. Nucleic Acids Res 25: 1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Engemann S, Lurz R, Okazawa H, Lehrach H, Wanker EE (2003) Mutant huntingtin promotes the fibrillogenesis of wild-type huntingtin: a potential mechanism for loss of huntingtin function in Huntington's disease. J Biol Chem 278: 41452–41461 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, et al. , The FANTOM Consortium (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Cha JH (2000) Transcriptional dysregulation in Huntington's disease. Trends Neurosci 23: 387–392 [DOI] [PubMed] [Google Scholar]
- Chan EY, Luthi-Carter R, Strand A, Solano SM, Hanson SA, DeJohn MM, Kooperberg C, Chase KO, DiFiglia M, Young AB, Leavitt BR, Cha JH, Aronin N, Hayden MR, Olson JM (2002) Increased huntingtin protein length reduces the number of polyglutamine-induced gene expression changes in mouse models of Huntington's disease. Hum Mol Genet 11: 1939–1951 [DOI] [PubMed] [Google Scholar]
- Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM (2000) Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet 9: 2811–2820 [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Sadri-Vakili G, Yohrling GJ, Braveman MW, Benn CL, Glajch KE, DiRocco DP, Farrell LA, Krainc D, Gines S, MacDonald ME, Cha JH (2006) Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol Dis 22: 233–241 [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90: 537–548 [DOI] [PubMed] [Google Scholar]
- Doi H, Mitsui K, Kurosawa M, Machida Y, Kuroiwa Y, Nukina N (2004) Identification of ubiquitin-interacting proteins in purified polyglutamine aggregates. FEBS Lett 571: 171–176 [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D (2002) Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296: 2238–2243 [DOI] [PubMed] [Google Scholar]
- Friedman MJ, Shah AG, Fang ZH, Ward EG, Warren ST, Li S, Li XJ (2007) Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat Neurosci 10: 1519–1528 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A (2005) Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem 280: 34908–34916 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI (2006) Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474 [DOI] [PubMed] [Google Scholar]
- Greene JM, Larin Z, Taylor IC, Prentice H, Gwinn KA, Kingston RE (1987) Multiple basal elements of a human hsp70 promoter function differently in human and rodent cell lines. Mol Cell Biol 7: 3646–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzhova I, Margulis B (2006) Hsp70 chaperone as a survival factor in cell pathology. Int Rev Cytol 254: 101–149 [DOI] [PubMed] [Google Scholar]
- Harjes P, Wanker EE (2003) The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci 28: 425–433 [DOI] [PubMed] [Google Scholar]
- Hata M, Ohtsuka K (1998) Characterization of HSE sequences in human Hsp40 gene: structural and promoter analysis. Biochim Biophys Acta 1397: 43–55 [DOI] [PubMed] [Google Scholar]
- Hay DG, Sathasivam K, Tobaben S, Stahl B, Marber M, Mestril R, Mahal A, Smith DL, Woodman B, Bates GP (2004) Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet 13: 1389–1405 [DOI] [PubMed] [Google Scholar]
- Holbert S, Denghien I, Kiechle T, Rosenblatt A, Wellington C, Hayden MR, Margolis RL, Ross CA, Dausset J, Ferrante RJ, Neri C (2001) The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington's disease pathogenesis. Proc Natl Acad Sci USA 98: 1811–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbriano C, Bolognese F, Gurtner A, Piaggio G, Mantovani R (2001) HSP-CBF is an NF-Y-dependent coactivator of the heat shock promoters CCAAT boxes. J Biol Chem 276: 26332–26339 [DOI] [PubMed] [Google Scholar]
- Jana NR, Tanaka M, Wang G, Nukina N (2000) Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet 9: 2009–2018 [DOI] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P, Benzer S (2000) Genetic suppression of polyglutamine toxicity in Drosophila. Science 287: 1837–1840 [DOI] [PubMed] [Google Scholar]
- Kotliarova S, Jana NR, Sakamoto N, Kurosawa M, Miyazaki H, Nekooki M, Doi H, Machida Y, Wong HK, Suzuki T, Uchikawa C, Kotliarov Y, Uchida K, Nagao Y, Nagaoka U, Tamaoka A, Oyanagi K, Oyama F, Nukina N (2005) Decreased expression of hypothalamic neuropeptides in Huntington disease transgenic mice with expanded polyglutamine-EGFP fluorescent aggregates. J Neurochem 93: 641–653 [DOI] [PubMed] [Google Scholar]
- Kuhn A, Goldstein DR, Hodges A, Strand AD, Sengstag T, Kooperberg C, Becanovic K, Pouladi MA, Sathasivam K, Cha JH, Hannan AJ, Hayden MR, Leavitt BR, Dunnett SB, Ferrante RJ, Albin R, Shelbourne P, Delorenzi M, Augood SJ, Faull RL et al. (2007) Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet 16: 1845–1861 [DOI] [PubMed] [Google Scholar]
- Landles C, Bates GP (2004) Huntingtin and the molecular pathogenesis of Huntington's disease. Fourth in molecular medicine review series. EMBO Rep 5: 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP (1998) Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J 17: 6300–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Cheng AL, Zhou H, Lam S, Rao M, Li H, Li XJ (2002) Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol 22: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Li XJ (2004) Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet 20: 146–154 [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, Woods AM, Chan EY, Kooperberg C, Krainc D, Young AB, Tapscott SJ, Olson JM (2002) Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet 11: 1911–1926 [DOI] [PubMed] [Google Scholar]
- Maity SN, de Crombrugghe B (1998) Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci 23: 174–178 [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP (1996) Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87: 493–506 [DOI] [PubMed] [Google Scholar]
- Mantovani R (1999) The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27 [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Kim S, Morimoto RI (2006) Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. J Biol Chem 281: 4477–4485 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Bukau B (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merienne K, Helmlinger D, Perkin GR, Devys D, Trottier Y (2003) Polyglutamine expansion induces a protein-damaging stress connecting heat shock protein 70 to the JNK pathway. J Biol Chem 278: 16957–16967 [DOI] [PubMed] [Google Scholar]
- Morgan WD (1989) Transcription factor Sp1 binds to and activates a human hsp70 gene promoter. Mol Cell Biol 9: 4099–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan WD, Williams GT, Morimoto RI, Greene J, Kingston RE, Tjian R (1987) Two transcriptional activators, CCAAT-box-binding transcription factor and heat shock transcription factor, interact with a human hsp70 gene promoter. Mol Cell Biol 7: 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser DD, Theodorakis NG, Morimoto RI (1988) Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol 8: 4736–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Schaffar G, Sittler A, Wanker EE, Hayer-Hartl MK, Hartl FU (2000) Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA 97: 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Morimoto RI (2002) Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock' proteins. J Cell Sci 115: 2809–2816 [DOI] [PubMed] [Google Scholar]
- Nucifora FC Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA (2001) Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291: 2423–2428 [DOI] [PubMed] [Google Scholar]
- Obrietan K, Hoyt KR (2004) CRE-mediated transcription is increased in Huntington's disease transgenic mice. J Neurosci 24: 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama F, Miyazaki H, Sakamoto N, Becquet C, Machida Y, Kaneko K, Uchikawa C, Suzuki T, Kurosawa M, Ikeda T, Tamaoka A, Sakurai T, Nukina N (2006) Sodium channel beta4 subunit: down-regulation and possible involvement in neuritic degeneration in Huntington's disease transgenic mice. J Neurochem 98: 518–529 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Norflus F, Singh B, Swindell MK, Buzescu R, Bejarano M, Chopra R, Zucker B, Benn CL, DiRocco DP, Cha JH, Ferrante RJ, Hersch SM (2006) Sp1 is up-regulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J Biol Chem 281: 16672–16680 [DOI] [PubMed] [Google Scholar]
- Sakahira H, Breuer P, Hayer-Hartl MK, Hartl FU (2002) Molecular chaperones as modulators of polyglutamine protein aggregation and toxicity. Proc Natl Acad Sci USA 99 (Suppl 4): 16412–16418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, Sakahira H, Siegers K, Hayer-Hartl M, Hartl FU (2004) Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell 15: 95–105 [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90: 549–558 [DOI] [PubMed] [Google Scholar]
- Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker EE, Bates GP, Housman DE, Thompson LM (2000) The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA 97: 6763–6768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugars KL, Rubinsztein DC (2003) Transcriptional abnormalities in Huntington disease. Trends Genet 19: 233–238 [DOI] [PubMed] [Google Scholar]
- Suhr ST, Senut MC, Whitelegge JP, Faull KF, Cuizon DB, Gage FH (2001) Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J Cell Biol 153: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa K, Marubuchi S, Qi ML, Enokido Y, Tamura T, Inagaki R, Murata M, Kanazawa I, Wanker EE, Okazawa H (2007) The induction levels of heat shock protein 70 differentiate the vulnerabilities to mutant huntingtin among neuronal subtypes. J Neurosci 27: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983 [DOI] [PubMed] [Google Scholar]
- van Roon-Mom WM, Reid SJ, Jones AL, MacDonald ME, Faull RL, Snell RG (2002) Insoluble TATA-binding protein accumulation in Huntington's disease cortex. Brain Res Mol Brain Res 109: 1–10 [DOI] [PubMed] [Google Scholar]
- Wacker JL, Zareie MH, Fong H, Sarikaya M, Muchowski PJ (2004) Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat Struct Mol Biol 11: 1215–1222 [DOI] [PubMed] [Google Scholar]
- Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N (2000) Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet 9: 1795–1803 [DOI] [PubMed] [Google Scholar]
- Wu BJ, Kingston RE, Morimoto RI (1986) Human HSP70 promoter contains at least two distinct regulatory domains. Proc Natl Acad Sci USA 83: 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka T, Horikoshi Y, Izumi N, Suzuki A, Mizuno K, Ohno S (2006) Lgl mediates apical domain disassembly by suppressing the PAR-3–aPKC–PAR-6 complex to orient apical membrane polarity. J Cell Sci 119: 2107–2118 [DOI] [PubMed] [Google Scholar]
- Yu ZX, Li SH, Nguyen HP, Li XJ (2002) Huntingtin inclusions do not deplete polyglutamine-containing transcription factors in HD mice. Hum Mol Genet 11: 905–914 [DOI] [PubMed] [Google Scholar]
- Zhou H, Li SH, Li XJ (2001) Chaperone suppression of cellular toxicity of huntingtin is independent of polyglutamine aggregation. J Biol Chem 276: 48417–48424 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data