Abstract
Interferon regulatory factor 3 (IRF3) is an essential transcriptional regulator of the interferon genes. IRF3 is constitutively present in a latent conformation in the cell cytoplasm. In cells infected by Sendai virus, IRF3 becomes phosphorylated, homodimerizes, translocates to the nucleus, binds to target genes and activates transcription by interacting with CBP/p300 co-activators. In this study, we report that in non-infected cells IRF3 is post-translationally modified by S-glutathionylation. Upon viral-infection, it undergoes a deglutathionylation step that is controlled by the cytoplasmic enzyme glutaredoxin-1 (GRX-1). In virus-infected GRX-1 knockdown cells, phosphorylation, homodimerization and nuclear translocation of IRF3 were not affected, but the transcriptional activity of IRF3 and the expression of interferon-β (IFNβ), were severely reduced. We show that deglutathionylation of IRF3 is necessary for efficient interaction of IRF3 with CBP, an event essential for transcriptional activation of the interferon genes. Taken together, these findings reveal a crucial role for S-glutathionylation and GRX-1 in controlling the activation of IRF3 and IFNβ gene expression.
Keywords: glutaredoxin, interferon beta, IRF3, S-glutathionylation
Introduction
Reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) are formed and degraded by all aerobic organisms. Generation of excessive amounts of ROS into cells, called oxidative stress, alters their oxidant/redox status. Although ROS, in general, damage macromolecules including DNA and proteins, they can also function as regulators or secondary messengers of intracellular signal transduction pathways (Thannickal and Fanburg, 2000). Among the most susceptible oxidant-sensitive targets are thiol groups on proteins that can be reversibly oxidized to sulphenic acid (-SOH). Reversible oxidation is considered to be a homeostatic mechanism that protects proteins from irreversible oxidation (Thomas et al, 1995) and also constitutes a critical post-translational modification mechanism that can regulate or alter protein function (Cotgreave and Gerdes, 1998; Klatt and Lamas, 2000). The sulphenic acid moiety is very unstable and readily reacts with other thiols to form intra- or intermolecular disulphides. Reaction with glutathione (GSH) results in the formation of mixed disulphides with GSH known as S-glutathionylation (protein-SSG formation). This modification has been shown to be essential for the function of several proteins such as NF1, c-jun, p50/NF-kB, actin and IkB kinase subunit β (IKKβ) (Bandyopadhyay et al, 1998; Klatt et al, 1999; Pineda-Molina et al, 2001; Wang et al, 2001; Reynaert et al, 2006). Glutaredoxin-1 (GRX-1) is a cytoplasmic enzyme that catalyzes the reversible reduction of the protein-glutathionyl-mixed disulphides to free sulphhydryl groups through a monothiol mechanism (Gravina and Mieyal, 1993; Yang et al, 1998). During this process, GRX-1 becomes S-glutathionylated at Cys-23 of its conserved catalytical motif Cys-X-X-Cys. Reduction of GRX-1–SSG is accomplished by GSH and glutathione disulphide reductase at the expense of NADPH to NADP transformation (Yang et al, 1998). As mammalian GRX-1 functions in a substrate-specific manner towards S-glutathionylated proteins (Gravina and Mieyal, 1993; Fernandes and Holmgren, 2004) and because some of them are crucial components of many signalling pathways, GRX-1 is believed to have an important function in signal transduction (Ghezzi, 2005; Shelton et al, 2005).
Interferons (IFNs) are cytokines involved in diverse biological functions including antiviral response, inhibition of proliferation, induction of differentiation, modulation of immune system and inhibition of angiogenenesis (Uddin and Platanias, 2004). Initially, IFNs were discovered through their anti-viral activities. Type I IFNs (IFNα and IFNβ) are produced by virus-infected host cells and constitute the primary response against virus infection. All IFNs bind to specific receptors present on the cell surface and by doing so activate the JAK–STAT signalling pathway eliciting their effects through the transcriptional activation of target genes bearing specific DNA-binding recognition sites (ISREs) within their promoters (Darnell et al, 1994; Ihle, 1996; Stark et al, 1998). Rapid induction of type I IFNs following virus infection requires post-translational modification of latent transcription factors involved in immunomodulation, including NF-κB, ATF-2/c-Jun and interferon regulatory factor 3 (IRF3) (Thanos and Maniatis, 1995a, 1995b; Juang et al, 1998; Sato et al, 2000). IRF3 is a 55 kD protein constitutively expressed in a variety of tissues and maintained in a latent conformation in the cytoplasm (Nguyen et al, 1997). Virus-induced C-terminal phosphorylation of IRF3 represents a key post-translational modification, leading to dimerization, cytoplasmic-to-nuclear translocation, association with CBP/p300 co-activators, binding to ISREs of target genes, including the PRDIII-I regions of the IFNβ gene promoter, and transcriptional activation (Fujita et al, 1988; Miyamoto et al, 1988; Lin et al, 1998; Sato et al, 1998; Weaver et al, 1998; Yoneyama et al, 1998).
In this study, we investigated the role of GRX-1 in the signal transduction pathways leading to IFNβ production in virus-infected cells. Our data demonstrated that, in GRX-1 knockdown (KD) cells infected with Sendai virus, the expression of IFNβ was significantly reduced, suggesting a critical role for GRX-1 in the IFNβ gene activation pathway. More specifically, we found that GRX-1 was essential for the transcriptional activity of IRF3. We showed that IRF3 is S-glutathionylated in non-infected cells, but upon viral induction, IRF3 undergoes deglutathionylation catalyzed by GRX-1. Our experiments revealed that, in GRX-1 KD cells, phosphorylation, dimerization and nuclear translocation of IRF3 were not altered, but its transactivation ability was reduced due to the inefficient recruitment of the CBP transcription co-activator. Collectively, this study uncovered an essential role for GRX-1 in the IRF3 activation and the signalling regulation of the IFNβ pathway. Our work also underscores the role of S-glutathionylation as a crucial post-translational modification that is able to regulate signalling pathways.
Results
GRX-1 is essential for the IRF activation and IFN β production
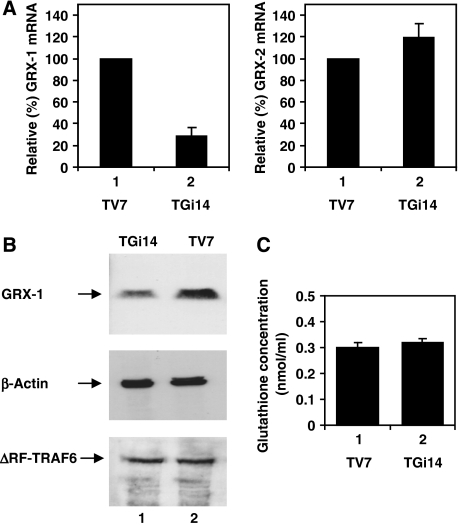
To study the role of GRX-1 on the IFNβ pathway activation, we generated a HEK293 stable cell line that underexpresses GRX-1 due to RNA interference, namely TGi14, and a control cell line, TV7, transfected with the empty vector. GRX-1 mRNA levels in TGi14 and TV7 cells were estimated by quantitative real-time PCR. TGi14 cells showed a 71% reduction of GRX-1 mRNA compared with control cells (Figure 1A). The RNAi specificity for GRX-1 was confirmed by real-time PCR for GRX-2, another glutaredoxin located in mitochondria (Figure 1A). The endogenous GRX-1 protein levels in TV7 cells were below the detection limit for the two different anti-GRX-1 antibodies used; thus we analysed the protein levels of GRX-1 in TV7 and TGi14 cells co-transfected with pIRES–GRX-1 and a ΔRF–TRAF6 expression plasmid as transfection efficiency marker. GRX-1 was reduced in TGi14 cells compared with TV7 cells (Figure 1B). Reduction of the GRX-1 levels did not affect the glutathione concentration in TGi14 cells compared to the control cell line (Figure 1C).
Figure 1.
(A) RNAi-mediated knockdown of GRX-1. Real-time PCR was performed in cDNA from TV7 and TGi14 cells to estimate the mRNA levels of GRX-1 (left panel) and GRX-2 (right panel). Values have been corrected by the respective values of GAPDH mRNA for each sample. Mean values from three independent experiments are presented. (B) TV7 and TGi14 cells were transfected with 0.5 μg of pIRES-GRX-1 along with 0.5 μg cDNA3 Flag–ΔRF–TRAF6 as marker for transfection efficiency. Whole-cell extracts were separated on SDS gel and blotted with antibodies to GRX-1, Flag epitope and β-actin as loading control. (C) Glutathione concentration was measured in TV7 and TGi14 cells. Average values from three independent experiments, expressed as nmol ml−1 of cell extracts, are presented.
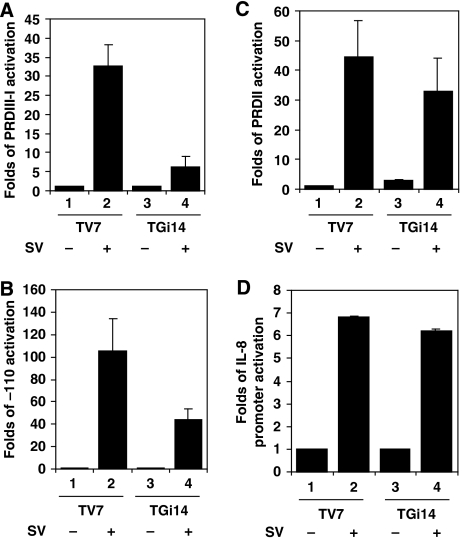
To investigate whether GRX-1 was involved in the IFNβ gene activation pathway, TV7 and TGi14 cells were transfected with either PRDIII–I3-luc (12 IRF-binding sites) or −110-luc (IFNβ promoter) reporter plasmids and 18 h later were infected with Sendai virus for 9 h. As shown in Figure 2A (compare lanes 2 and 4), reduction of the GRX1 levels resulted in a strong decrease of IRF-dependant activation after viral infection. These results suggest that GRX-1 is required for IRF-dependant transcription upon viral infection. Moreover, GRX-1 underexpression resulted in almost 60% lower activation of the entire IFNβ promoter upon viral infection (Figure 2B). We have also analysed the effect of GRX-1 underexpression on NF-κB activation, using the PRDII4-luc (four NF-κB-binding sites) reporter plasmid, as well as the IL-8 promoter. Both IL-8 and -110 promoters contain NF-κB- and AP-1-binding sites, but the IL-8 promoter has a C/EBP-binding site instead of IRF-binding sites. NF-κB activation in virally induced TGi14 cells was slightly affected (Figure 2C), whereas there was almost no effect on the activation of the IL-8 promoter (Figure 2D). These results suggest that GRX-1 primarily affects the IRF-dependant transcription activation pathways. Similar results were also obtained by the respective transfection experiments in a HeLa stable cell line that underexpresses GRX-1 (HGR7) and a control cell line (HV7) (Supplementary Figure S2 and S3).
Figure 2.
Activation of PRDIII-I, PRDII, –110 and IL-8 reporters in TV7 and TGi14 cell lines. Cells were transfected with the indicated plasmids (0.3 μg per well) and 18 h later were infected with Sendai virus (SV) for 9 h. (A) activation of the PRDIII–I3-luc; (B) activation of the -110-luc; (C) activation of the PRDII4–luc; (D) activation of the IL-8-luc. Luciferase values have been normalized by the respective β-galactosidase values (0.1 μg pGK-β-gal reporter per well co-transfected) in all experiments. Data are expressed as means±s.d. from a minimum of three independent experiments.
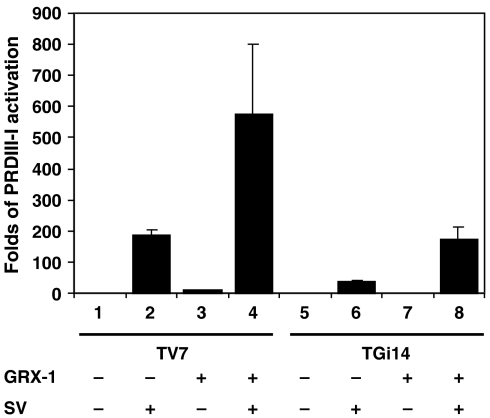
To examine whether restoration of GRX-1 in TGi14 cells could reverse the loss of IRF activation, TV7 and TGi14 cells were co-transfected with PRDIII–I3-luc reporter along with empty vector or pIRES-GRX-1 and infected with Sendai virus as described above. As shown in Figure 3, control cells transfected with GRX-1 and infected with Sendai virus could activate transcription more efficiently when compared with cells infected only with virus indicating synergistic interactions (compare bars 2, 3 and 4). GRX-1 expression in TGi14 cells resulted in restoration of IRF activation upon viral induction, reaching levels similar to control cells transfected with empty vector and infected with virus (compare bars 2 and 8).
Figure 3.
Activation of PRDIII-I reporter in TV7 and TGi14 cells transfected with GRX-1 and induced with Sendai virus. Cells were transfected with PRDIII-I3-luc (0.3 μg per well) and GRX-1 (0.4 μg per well) and 18 h later were infected with Sendai virus for 9 h. Luciferase values have been normalized by β-galactosidase values (0.1 μg pGK–β-gal reporter per well).
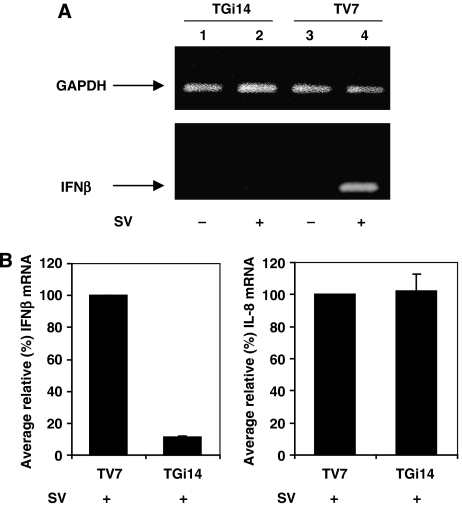
To estimate the physiological impulse of GRX-1 to the anti-viral response, TV7 and TGi14 cells were infected with Sendai virus, and after 9 h, total RNA was isolated and used for cDNA production in reverse transcriptase (RT) reaction. cDNA was used as template in semi-quantitative PCR assay for IFNβ and GAPDH as control (Figure 4A). Quantitative real-time PCR for IFNβ and IL-8 was also performed (Figure 4B). Both analysis showed that in TGi14 cells the transcription of IFNβ gene upon viral induction was diminished, whereas IL-8 production was not affected, indicating a key role for GRX-1 in the primary anti-viral response of the cells.
Figure 4.
Underexpression of GRX-1 drastically reduces IFNβ gene transcription upon viral infection. (A) Semi-quantitative RT–PCR for IFNβ expression in TV7 and TGi14 cells mock-induced or induced with virus for 9 h. Upper panel shows the respective expression of GAPDH in those cells. (B) Real-time PCR for IFNβ (left panel) and IL-8 (right panel) gene expression, in TV7 and TGi14 cells infected with virus for 9 h. Both experiments were normalized with the respective real-time PCR values for GAPDH, and data are presented as means of % relative mRNA production±s.d. from three independent experiments.
IRF3 is S-glutathionylated and GRX-1 is needed for its deglutathionylation upon viral infection
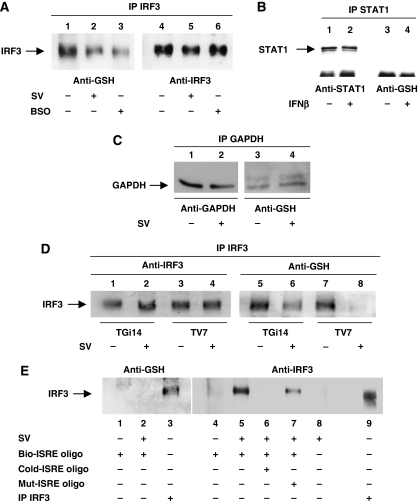
GRX-1 is a constitutively active cytoplasmic enzyme, known to control S-glutathionylation of target proteins (Gravina and Mieyal, 1993; Yang et al, 1998). As IRF3 is a transcription factor located in a latent form in the cytoplasm of uninfected cells (Nguyen et al, 1997), we tested whether IRF3 was S-glutathionylated and whether GRX-1 controlled this modification. TV7 cells were either mock- or virus-infected for 9 h or cultured in the presence of 200 mM L-buthionine-(S,R)-sulphoximine (BSO) (an inhibitor of GSH synthesis) (Anderson, 1997) for 4 days. Cytoplasmic cell extracts were prepared and used in immunoprecipitation experiments with anti-IRF3 antibody followed by western blot analysis using anti-IRF3 and anti-GSH antibodies (Figure 5A). We found that IRF3 was S-glutathionylated in its inactive form (lane 1) and its S-glutathionylation was significantly reduced upon viral infection (lane 2). The amount of IRF3 S-glutathionylation upon viral infection resembles the one observed in non-infected cells cultured with BSO (lane 3). Immunoprecipitation with anti-STAT1 antibody was also performed in cytoplasmic extracts from TV7 cells either mock-induced or induced with recombinant IFNβ, as control experiment for S-glutathionylation specificity. As shown in Figure 5B, no S-glutathionylation was observed on STAT1 under normal conditions or upon induction with IFNβ. To determine the specificity of IRF3 deglutathionylation upon virus infection, we analysed the extent of deglutathionylation of GAPDH, a protein that has been reported to be S-glutathionylated (Lind et al, 1998). GAPDH was immunoprecipitated from cytoplasmic extracts of TV7 cells either mock-induced or induced with virus for 9 h and western blotted for GSH. No difference in the S-glutathionylation of GAPDH was observed upon viral induction (Figure 5C), suggesting that the effect of Sendai virus induction on the S-glutathionylation levels of IRF3 is specific. GRX-1 was responsible for deglutathionylation of IRF3, as the overall amount of S-glutathionylated IRF3 that became deglutathionylated upon viral infection in TGi14 cells was much less, compared with control cells (Figure 5D).
Figure 5.
IRF3 is S-glutathionylated in its inactive form and its glutathionylation status is altered upon viral infection. (A) IRF3 was immunoprecipitated from cytoplasmic extracts (300 μg) of TV7 control cells either mock-induced or induced with Sendai virus for 9 h or cultured with 200 mM BSO for 4 days. Proteins were separated on SDS–PAGE and probed with anti-IRF3 and anti-GSH antibodies. (B) Cytoplasmic extracts (300 μg) of TV7 cells either mock-induced or induced with IFNβ (500 IU ml−1) for 30 min were used for STAT1 immunoprecipitation. Proteins were separated on SDS–PAGE and probed with anti-STAT1 and anti-GSH antibodies. (C) GAPDH was immunoprecipitated from cytoplasmic extracts (300 μg) of TV7 cells either mock-induced or induced with Sendai virus for 9 h. Proteins were western blotted with anti-GAPDH and anti-GSH antibodies. (D) IRF3 was immunoprecipitated from cytoplasmic extracts (300 μg) of TV7 and TGi14 cells either mock-induced or induced with Sendai virus for 9 h. The proteins were separated on SDS–PAGE and probed with anti-IRF3 and anti-GSH antibodies. (E) Double-stranded biotinylated-ISRE oligo was bound to magnetic streptavidin beads. Equal amounts of nuclear extracts (100 μg) from TV7 cells mock-induced or induced with virus for 4 h were mixed with Bio-ISRE beads in the presence or absence of mutated ISRE or wild-type ISRE, incubated at room temperature for 20 min, washed and subjected to SDS–PAGE. The proteins were probed with anti-IRF3 and anti-GSH antibodies. Immunoprecipitated IRF3 from mock TV7 cytoplasmic extracts was used as positive control (lanes 3 and 9).
Next, we analysed the glutathionylation status of IRF3 bound to Bio-ISRE (oligo-binding IRF3) in nuclear extracts prepared from mock- and virus-infected TV7 cells. Binding of IRF3 to the Bio-ISRE oligo was observed in nuclear extracts from virus-infected cells (Figure 5E, compare lanes 4 and 5). However, no S-glutathionylation was observed in IRF3 when bound to DNA (lane 2). Immunoprecipitated IRF3 from mock cytoplasmic extracts of TV7 cells was used as positive control for S-glutathionylation (Figure 5E, lane 3). These results suggest that activated IRF3 that can bind DNA is deglutathionylated; however, from these experiments, we cannot conclude that glutathionylation affects the DNA binding of IRF3.
GRX-1 underexpression does not affect the dimerization and nuclear translocation of IRF3
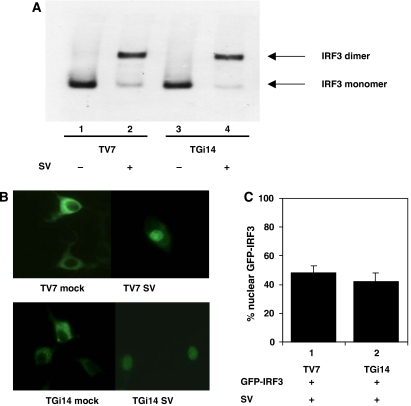
Viral infection activates IRF3 through a biochemical cascade leading to phosphorylation and subsequent dimerization of the molecule. The activated IRF3 enters the nucleus, associates with transcription co-activators and promotes transcription of target genes (Lin et al, 1998; Sato et al, 1998; Weaver et al, 1998; Yoneyama et al, 1998). To investigate whether GRX-1 has an important function in IRF3 dimerization, whole-cell extracts from TV7 and TGi14 cells, mock-infected or infected with Sendai virus for 9 h, were prepared. Extracts were separated in a 7.5% native gel and blotted with anti-IRF3 antibody (Figure 6A). Underexpression of GRX1 in TGi14 cells did not alter the formation of the IRF3 homodimer upon viral induction (compare lanes 2 and 4). Moreover, nuclear translocation experiments performed in TV7 and TGi14 cells, transfected with GFP–IRF3 and then infected with Sendai virus revealed no significant differences in IRF3 translocation between the two cell lines (Figure 6B and C).
Figure 6.
GRX-1 underexpression does not affect dimerization and nuclear translocation of IRF3 upon viral infection. (A) Western blot analysis of IRF3 in TV7 and TGi14 cells either mock-induced or induced with virus for 9 h. Whole-cell extracts (20 μg) have been separated on a 7.5% acrylamide native gel, and the active IRF3 dimer is visible. (B) TV7 and TGi14 cells were transfected with GFP–IRF3 (0.5 μg per well) and mock-stimulated or stimulated with virus for 16 h. Nuclear translocation of GFP–IRF3 was measured by microscopy observation of 250 living cells in each well. (C) The percentage of cells with nuclear translocation of GFP–IRF3 induced by virus is shown.
GRX-1 is essential for IRF3 ability to promote transcription
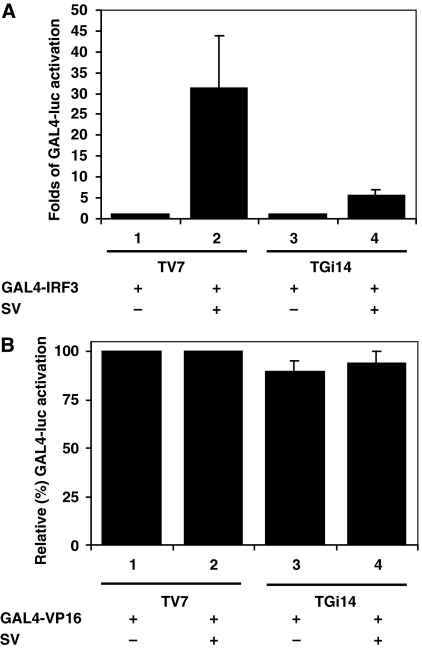
As IRF3 could be activated and translocated in the nucleus when GRX-1 was underexpressed, we tested whether deglutathionylation was essential for IRF3 to promote transcription. We transfected both cell lines with a GAL4-luc reporter together with the GAL4–IRF3 DN133 construct in which the IRF3 DNA-binding domain was replaced with the GAL4 DNA-binding domain, ensuring its binding to the GAL4 luciferase reporter. Moreover, the construct bears its own nuclear localization signal (NLS) to guarantee the entry of the factor into the nucleus. As shown in Figure 7A, upon viral infection, transcriptional activation of GAL4 luciferase was reduced by 80% in TGi14 cells compared with control cells (lanes 2 and 4), suggesting that GRX-1 underexpression affects the ability of the factor to promote transcription through its transactivation domain. To ensure that binding ability of GAL4 constructs was not altered by GRX-1 underexpression, we performed similar experiments using GAL4-VP16, a potent transcription activator. No significant differences in GAL4-luciferase reporter activation between TV7 and TGi14 cell lines were observed (Figure 7B).
Figure 7.
GRX-1 affects the ability of IRF3 to promote transcription through its transactivation domain. (A) Average activation of GAL4-luc reporter (0.5 μg per well) in TV7 and TGi14 cells, co-transfected with GAL4–IRF3 DN133 (0.2 μg per well) and either mock-induced or induced with virus for 9 h. Luciferase values have been normalized by β-galactosidase values (0.1 μg pGK–βgal reporter per well). (B) Average relative transcriptional activation (%) of GAL4-luc reporter (0.5 μg per well) in TV7 and TGi14 cells transfected with GAL4-VP16 (0.2 μg per well) either mock-induced or induced with virus for 9 h. Data are expressed as means±s.d. from a minimum of three independent experiments.
Deglutathionylation of IRF3 catalyzed by GRX-1 is required for IRF3–CBP interaction
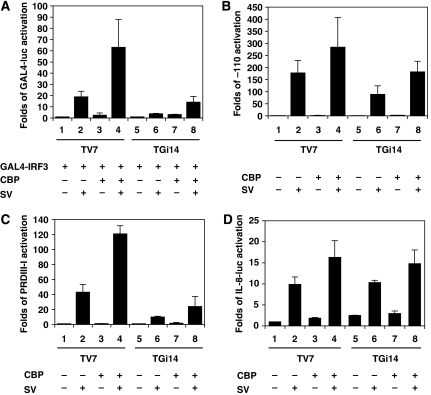
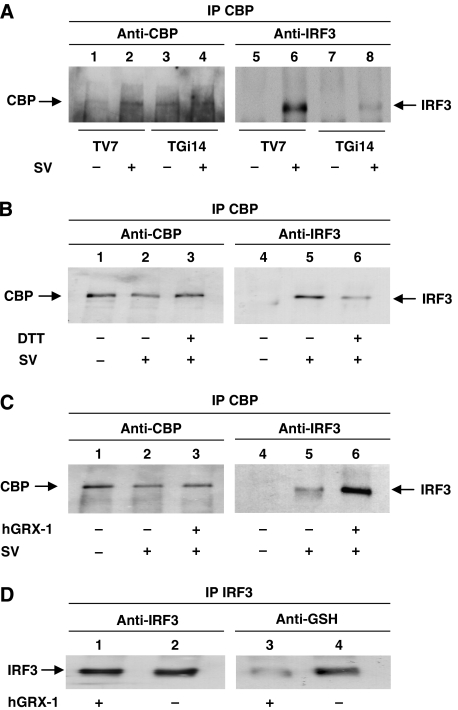
IRF3 bears a hydrophobic surface, exposed upon dimer formation, that recruits CBP/p300 co-activator and functions as a transactivation domain (Qin et al, 2003). As GRX-1 was essential for transcriptional activation by IRF3, the possibility that GRX-1 influences IRF3–CBP interaction was investigated. To address whether CBP overexpression can restore the loss of IRF3 transcription activity when GRX-1 is underexpressed, we conducted the transfection experiments described in Figure 8. As expected, CBP overexpression improved the transcriptional activation of all reporters used (GAL4-luc, PRDIII–I3-luc and –110-luc), but could not restore the transactivation ability of IRF3 in TGi14 cells (compare lanes 4 and 8 in A, B and C). These results indicate that IRF3 could not interact efficiently with CBP when IRF3 was not deglutathionylated. In contrast, transcriptional activation of the IL-8 reporter in TGi14 cells was not affected, indicating that GRX-1 does not alter the ability of CBP to activate transcription (Figure 8D, compare lanes 4 and 8). To further investigate the regulatory role of GRX-1 in the IRF3–CBP interaction, nuclear extracts from TV7 and TGi14 cells induced for 9 h with Sendai virus were prepared, subjected to immunoprecipitation with anti-CBP antibody and blotted for CBP and IRF3. As shown in Figure 9A, there was much less IRF3 co-precipitated with CBP in virus-infected TGi14 cells than in respective TV7 cells, although the amounts of immunoprecipitated CBP were similar (compare lanes 6 and 8). These results showed that GRX-1 was required for efficient interaction of CBP with the activated IRF3 in virus-infected cells. To confirm the observation that S-glutathionylated IRF3 does not bind CBP, we treated nuclear extracts from TGi14 cells either mock- or virus-infected with DTT (a non-specific reducer of disulphide bonds) or purified hGRX-1 before CBP immunoprecipitation. As shown in Figure 9B, DTT treatment resulted in even less co-precipitation of IRF3 with CBP (compare lanes 5 and 6), suggesting that certain disulphide bonds are needed for efficient interaction between CBP and activated IRF3. In contrast, after GRX-1 treatment, the co-precipitated IRF3 was strongly increased in the viral-induced nuclear extracts (Figure 9C, compare lanes 5 and 6). These results strongly suggest that deglutathionylation of IRF3 is required for its efficient interaction with CBP. As control, we showed that hGRX-1 can deglutathionylate IRF3 under experimental conditions (Figure 9D).
Figure 8.
Activation of GAL4-luc, PRDIII-I and -110 reporters in TV7 and TGi14 cells upon CBP overexpression. Average activation of reporter plasmids in TV7 and TGi14 cells co-transfected with RSV–CBP (0.5 μg per well) and either mock-induced or induced with virus for 9 h. (A) GAL4-luc (0.5 μg per well) and GAL-4–IRF3 DN133 (0.2 μg per well) (B) -110-luc (0.3 μg per well) (C) PRDIII–I3-luc (0.3 μg per well) (D) IL-8-luc (0.3 μg per well). In all four experiments, luciferase values have been normalized by total protein amount of lysates, and data are expressed as means±s.d. from a minimum of three independent experiments.
Figure 9.
GRX-1 underexpression affects the IRF3–CBP interaction. (A) Immunoprecipitation of CBP from nuclear extracts (150 μg) of TV7 and TGi14 cells either mock-induced or induced with virus for 9 h. Proteins were separated on SDS–PAGE and probed with anti-IRF3 and anti-CBP antibodies. (B) Immunoprecipitation of CBP from nuclear extracts (300 μg) untreated or pretreated with DTT (10 mM for 15 min at 25°C) of TGi14 cells either mock-induced or induced with virus for 9 h. Proteins were western blotted with anti-IRF3 and anti-CBP antibodies. (C) Immunoprecipitation of CBP from nuclear extracts (300 μg) untreated or pretreated with hGRX-1 (400 μg ml−1 for 15 min at 25°C) of TGi14 cells either mock-induced or induced with virus for 9 h. Proteins were western blotted with anti-IRF3 and anti-CBP antibodies. (D) Immunoprecipitation of IRF3 from cytoplasmic extracts (300 μg) untreated or pretreated with hGRX-1 (400 μg ml−1 for 15 min at 25°C) of mock TV7 cells. Proteins were western blotted with anti-IRF3 and anti-GSH antibodies.
Discussion
S-glutathionylation is a post-translational modification having an important function in the role of several proteins as well as in the pathways these proteins may be involved in (Bandyopadhyay et al, 1998; Klatt et al, 1999; Pineda-Molina et al, 2001; Wang et al, 2001; Reynaert et al, 2006). Our interest was to investigate a possible role of GRX-1 in the IFNβ pathway. To accomplish this, we generated a stable HEK 293 GRX-1 knockdown cell line, TGi14. According to our data, TGi14 cells were at least 50% less efficient in supporting transactivation of the IFNβ promoter compared to control cells, when induced with Sendai virus. This reduction was specific for the IFNβ promoter, as the IL-8 promoter, which also contains NF-κB- and AP1-binding sites, but a C/EBP site instead of the IRF-binding sites, was fully active in GRX-1-underexpressing cells. It was shown recently by Reynaert et al (2006) that GRX-1 regulates the activation of IKKβ upon TNFα stimulation. Their findings revealed that IKKβ is S-glutathionylated and needs to be deglutathionylated by GRX-1 to be fully catalytically active. We observed a 25% loss of the PRDII promoter activation in GRX-1 KD cells, indicating a small reduction in the NF-κB activation, probably due to inefficient IKKβ activation. This low reduction of NF-κB activation in the GRX-1 KD cells was also reflected in the 9% reduction of the IL-8 promoter activation observed in these cells. Strikingly, the PRDIII-I promoter, which binds IRFs, was 85% less active in GRX-1-underexpressing cells compared with control cells upon viral induction, suggesting that fall of the -110 promoter activation in GRX-1 KD cells was mainly due to the IRF proteins. To demonstrate the general relevance of our observation, we have repeated these experiments with similar results using a stable HeLa GRX-1 knockdown cell line.
The GRX-1 restoration experiments revealed that the decreased activation by IRFs was specifically correlated with the underexpression of GRX-1 in the knockdown cells. An interesting finding was that overexpression of GRX-1 combined with virus infection in control cells resulted in ‘super' activation of PRDIII-I promoter in a synergistic manner, indicating that GRX-1 may be a limiting factor for the activation of IRF. However, virus infection had no effect on gene expression of GRX-1 (data not shown). More importantly, real-time PCR experiments for IFNβ expression showed that GRX-1 underexpression dramatically reduced the IFNβ transcription upon viral induction, suggesting that GRX-1 is an essential regulator of the IFNβ pathway.
It has been previously reported that Sendai virus induces IRF3 activation (Lin et al, 1998; Yoneyama et al, 1998; Hiscott et al, 1999) and this activation was independent of both interferon production and virus replication (Collins et al, 2004). Furthermore, IRF3, but not IRF1, IRF7 or IRF9, was critical for the initial response to virus particle entry (Collins et al, 2004). Therefore, we investigated the possibility that GRX-1 regulates the IRF3 activation pathway. Immunoprecipitation experiments revealed that IRF3 is S-glutathionylated under normal conditions and that GRX-1 is responsible for its deglutathionylation upon viral infection. Moreover, a DNA-binding assay for IRF3 revealed that the activated factor binds to DNA in its deglutathionylated form. Upon virus induction, IRF3 is phosphorylated at its C-term region, and it dimerizes and enters the nucleus to promote transcription of specific promoters bearing ISRE-binding elements. Our data revealed that formation of IRF3 homodimers, a requirement for the factor's activation process, was not altered in GRX-1 KD cells, suggesting that deglutathionylation of IRF3 is not essential for the phosphorylation and dimerization of the factor in the cytoplasm. In addition, nuclear translocation of GFP-IRF3 constructs upon viral infection was not affected by GRX-1 underexpression.
We used the IRF3-GAL4 fusion to determine the transactivation ability of IRF3 upon induction with virus in control and GRX-1 KD cells. GRX-1 underexpression severely reduced GAL4-luc activation, indicating that the transactivation ability of IRF3 is regulated by GRX-1 and that deglutathionylation is required for its ability to transactivate transcription. CBP/p300 is an essential co-activator for several transcription factors and its amount in vivo is not excessive, but rather limiting (Tanaka et al, 1997). IRF3 interacts with the CBP/p300 co-activators to initiate transcription (Yoneyama et al, 1998). Our initial hypothesis was that deglutathionylation of IRF3 may be required for efficient interaction with CBP. Overexpression of CBP in cells raised the transcriptional activity of IRF3, but failed to restore the loss of transcriptional activity of IRF3 in GRX-1 KD cells, suggesting that S-glutathionylated IRF3 cannot interact with CBP. Immunoprecipitation of CBP in nuclear extracts of control and GRX-1 KD cells showed dramatically decreased amount of co-precipitated IRF3 in virally induced GRX-1-underexpressing cells, indicating that deglutathionylation of IRF3 is needed for efficient interaction between CBP and the activated IRF3. The role of the deglutathionylation of IRF3 for its interaction with CBP was also demonstrated by the increased amount of IRF3 that was co-precipitated with CBP in virally induced GRX-1 KD nuclear extracts pretreated with hGRX-1. In addition, treatment of virally infected nuclear extracts with DTT diminished the CBP-IRF3 co-precipitation, indicating a requirement for thiol bonds for this interaction. Cysteines participating in thiol bonds are crucial for the function of IRF3 (Qin et al, 2003) and some of them might be protected from non-reversible oxidation by being S-glutathionylated, although the factor remains inactivated in the cytoplasm. Sendai virus induction results in deglutathionylation of these cysteines through GRX-1; thus, the activated IRF3 may form the essential thiol bonds needed to establish interaction with CBP.
The role of the cysteines in the transactivation ability of IRF3 was investigated by transfection experiments using mutant forms of IRF3 bearing cysteine-to-serine substitutions. We constructed six single-point mutants, one for each cysteine of the IRF3 molecule (C167S, C222S, C267S, C289S, C347S and C371S) and analysed the glutathionylation status of these mutated proteins in immunoprecipitation experiments from transfected TV7 cells. All of them were S-glutathionylated, suggesting that there are more than one glutathionylated cysteines in the molecule (data not shown). We also tested their transactivation ability in transfection experiments in TV7 and TGi14 cells. In TV7 cells, mutants C164S and C267S have similar transactivation ability as the wild-type IRF3. Mutation of cysteines 222 or 347 resulted in 25 and 30% lower activation, respectively. Mutation of cysteines 289 and 371 resulted in 50% loss of the transactivation ability of the molecule, respectively (Supplementary data, Supplementary Figure S4). These findings indicate that more than one cysteine residues are involved in the transactivation ability of IRF3, probably through a network of thiol bonds. Therefore, these thiol bonds cannot be formed when one or both of the participating cysteines are glutathionylated. Interestingly, mutation of these cysteines could affect not only their glutathionylation state but also their ability to participate in disulphides with other cysteines in cis or trans. Thus, elimination of a specific cysteine would not necessarily result in hyperactivation of IRF3, but it could lower its transactivation ability. Indeed, we found that some of these mutations decreased the ability of IRF3 to activate transcription. To identify the cysteines that are glutathionylated, we immunoprecipitated IRF3 and subjected the protein to mass spectrometry analysis. Despite our intense efforts, standard MS methods failed to identify IRF3 (not shown) probably due to lack of peptide ionization. We should point out that no IRF3 MS identifications have been reported.
GRX-1 is constantly active and able to perform deglutathionylation of S-glutathionylated proteins when modified cysteines can be accessed by the enzyme (Supplementary data, Supplementary Figure S1) (Fernandes and Holmgren, 2004; Ghezzi, 2005). Phosphorylation of IRF3, leading to exposure of its transactivation domain, might also be the critical event needed for GRX-1 to approach and deglutathionylate the factor. Sendai virus does not induce GRX-1 expression, so we believe that it makes IRF3 available, as substrate for GRX-1.
Inactive IRF3 constitutively shuttles in and out of the nucleus, with its export signal more active than its NLS, favouring the factor's presence to the cytoplasm (Reich, 2002). We have shown that S-glutathionylation of IRF3 inhibits its transactivation ability, thus the modification may also act as an extra ‘secure lock' preventing any unnecessary or random accidental activation of IRF3 and subsequently of IFNβ.
Collectively, our findings demonstrate a novel regulatory role for GRX-1 in the IFNβ pathway. We propose that IRF3 is S-glutathionylated and upon viral infection it becomes deglutathionylated by the action of GRX-1. This modification is prerequested for IRF3, to associate with the CBP co-activator and become fully transcriptionally active. Our findings support, along with previous reports, an essential role of S-glutathionylation as a crucial post-translational protein modification mechanism that may control cell signal pathways associated with viral infections.
Materials and methods
Plasmids
PRDII4–luc, PRDIII–I3-luc and –110-luc reporter plasmids were constructed by subcloning four PRDII, seven PRDIII-I sites and the IFNβ promoter (−1 to −110 bp upstream the initiation codon of ifnβ gene) respectively, to pGL3 basic vector (Promega). These plasmids were used for assays of NF-κB, IRF and IFNβ promoter activation. GFP-IRF3 plasmid was constructed by cloning the entire IRF3 ORF into the pEGFP-C2 mammalian expression vector (BD Biosciences) and was used for IRF3 nuclear translocation assay. pIRES–GRX-1 was constructed by cloning the entire ORF of GRX-1 into the pIRES-neo mammalian expression vector (CLONTECH) and was used for GRX-1 overexpression. Plasmid GAL4-luc (5xGAL4-TATA-luc) (Sun et al, 1994), GAL4–IRF3 DN133 (Senger et al, 2000), GAL4–VP16 (White et al, 1992), IL-8 promoter-luc (Agelopoulos and Thanos, 2006), pRSV–CBP (Merika et al, 1998) and pGK–β-gal (Luftig et al, 2003) have been described. PCDNA3–Flag–ΔRF–TRAF6 was kindly provided by G Mosialos (Aristotle University of Thessaloniki, Greece). Vector pSuperior-puro siRNA system was purchased from Oligo Engine and used for generating stable control and siRNA cell lines. All constructs were verified by DNA sequencing.
Cell cultures, transfections, viral infections, IFN β induction, reporter gene and glutathione assays
HEK293 and HeLa cells (ATCC) were cultivated in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 10% heat-inactivated FBS (GIBCO/BRL), 2 mM L-glutamine (Sigma) and 80 μg ml−1 gentamycin (GIBCO/BRL). For BSO experiments, cells were cultured for 4 days with 200 mM BSO (Sigma). For transfection experiments 3 × 105 cells per well onto 12-well culture plates were transfected with LF 2000 (Invitrogen). After 4 h of incubation, transfection mixture was replaced with culture medium. Cell lysates were obtained 20–24 h post-transfection. Luciferase reporter assays were normalized for transfection efficiency by co-transfection of β-galactosidase expression plasmid (pGK-β-gal). β-gal activity values were measured with the Galacton-Plus substrate system (Tropix, Bedford, MA). In experiments where CBP was overexpressed, luciferase assays were normalized with the amount of total protein of the samples, measured by Bradford protein assay (Sigma).
For GFP-IRF3 nuclear translocation assays, cells were transfected with 0.5 μg GFP-IRF3 and 18 h later were either mock-infected or infected with Sendai virus. Sixteen hours post-infection, living cells were observed under a fluorescent microscope to determine the percentage of cells with GFP-IRF3 nuclear translocation.
Viral infections were performed with 50 hemagglutinating units ml−1 culture media of Sendai virus (Cantell strain; Charles River laboratories) for 9 or 16 h in antibiotic free medium.
Inductions with recombinant IFNβ (ImmunoTools) were performed with 500 IU ml−1 in antibiotic free medium for 30 min.
Glutathione concentrations in TV7 and TGi14 cells were measured according to Akerboom and Sies (1981).
siRNA
Stable siRNA cell lines were produced by the pSuperior-puro vector system for inducible expression of siRNA (OligoEngine). The following oligonucleotides encoding the desired siRNA strands for GRX-1 mRNA were annealed and cloned to pSuperior-puro vector: 5′-GTACCCCCCATCAAACAAGGGCTTCTTTCAAGAG AAGAAGCCCTTGTTTGATGGGGC-3′ (sense); 5′-TCGAGCCCCATCAAACAAGGGCTTCTTCTCTTGA AAGAAGCCCTTGTTTGATGGGGG-3′ (anti-sense).
pSuperior-puro vector (control) or pSuperior-puro vector where the oligonucleotide was cloned were transfected to HEK 293 or HeLa cells. Forty-eight hours post-transfection, puromycin (Sigma) was added to the cultures to a final concentration of 2 μg ml−1. After 7 days in puromycin, colonies of stably transfected cells were collected. Control and stable siRNA lines were tested for GRX-1 expression with real-time PCR.
Immunoassays, western blot analysis and antibodies
Cells were washed with PBS (137 mM NaCl/3 mM KCl/6.5 mM Na2HPO4/0.5 mM KH2PO4) (pH 7.4), collected by scraping and centrifuged at 3000 g for 1 min. Cytoplasmic protein extracts were prepared by fractionation of cells with a Dounce homogenizer under hypotonic conditions (buffer A: 10 mM HEPES–KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM Na3VO4, 0.5% Nonidet P-40, 1 mM phenylmethylsulphonyl fluoride (PMSF) and 1 × protease inhibitor mixture (Roche Diagnostics)). Nuclei were precipitated by centrifugation at 12 000 g for 20 s at 4°C. For nuclear protein extracts preparation, sucrose was added to the lysates to a 0.25 M final concentration and nuclei were precipitated by centrifugation (1000 g) in a swing rotor. Nuclei pellet was resuspended in buffer A with 0.25 M sucrose and precipitated again by centrifugation. The pellet was washed five times with hypotonic buffer without sucrose, resuspended in hypertonic buffer (buffer B: 25 mM HEPES (pH 7.9), 420 mM NaCl, 0.5% Nonidet P-40, 1.5 mM MgCl2, 1 mM Na3VO4, 2 mM PMSF and 1 × protease inhibitor mixture) and incubated for 20 min under shaking at 4°C. Insoluble material was removed by centrifugation (12 000 g, 20 min, 4°C). Protein yields were measured with the Bradford protein assay kit. Whole-cell protein extracts were prepared by heating cells suspended in 1 × SDS-loading buffer (Roth) at 100°C for 5 min.
For immunoprecipitation experiments, equal protein amounts of cytoplasmic or nuclear extracts were incubated with protein A-agarose beads (Gibco/BRL) crosslinked with pre-immune rabbit or mouse serum for 2 h at 4°C (pre-clear). Crosslinking of the beads with the pre-immune serum was performed by DSS (Pierce) according to the manufacturer's protocol. Following pre-clearing, extracts were incubated with an excess of antibody overnight at 4°C followed by incubation with protein A-agarose beads for 2 h at 4°C, washed with buffer A, suspended in 1 × SDS buffer and heated for 5 min at 80°C.
Proteins were separated by SDS–PAGE transferred to nitrocellulose membranes (Protran) by wet transfer at 250 mA for 2–14 h at 4°C. Membranes were blocked with 5% nonfat dry milk in PBS-0.1%Tween-20 (PBST) at room temperature for 1 h. Native gel electrophoresis and protein transfer to nitrocellulose membrane were performed as described previously (Seth et al, 2005). Whole-cell extracts for native gel electrophoresis were prepared by combining cytoplasmic and nuclear extract preparations.
Treatment of nuclear extracts with DTT was carried out by addition of freshly prepared DTT to 300 μg virally induced TGi14 nuclear extracts to a final concentration of 10 mM. Extracts were incubated at room temperature for 15 min and then used for immunoprecipitation. Treatment of nuclear extracts with purified hGRX-1 was carried out as follows: 300 μg of virally induced TGi14 nuclear extracts were incubated at room temperature for 15 min with 0.5 mM GSH (Sigma), 1.5 U glutathione reductase from bakers yeast (Sigma), 400 μg ml−1 NADPH (Sigma) with or without 400 μg ml−1 hGRX-1 (IMCO, Sweden). Extracts were then subjected to immunoprecipitation with CBP antibody.
Mouse Abs for GSH moieties on proteins (101-A, Virogen, Watertown, MA), GRX-1 (ABNOVA), β-Actin (Sigma), GAPDH (Chemicon) and Flag epitope (Santa Cruz) were used at 1:1000, 1:500, 1:5000, 1:2000 and 1:300 dilutions in PBST, respectively, overnight at 4°C. Rabbit polyclonal Abs for IRF3, STAT1 and CBP (Santa Cruz) were used in 1:300 dilution in PBST for 3 h at room temperature. Anti-rabbit and anti-mouse-HRP secondary antibodies were from Jackson Immunoresearch Lab Inc.
Total RNA isolation, reverse transcription and real-time PCR
Total RNA was isolated from cells using Trizol reagent (Gibco BRL). RNA quality was checked electrophoretically, and quantification was done spectrophotometrically. RNA samples were treated with DNAse (Promega). First-strand cDNAs were obtained by RT reaction, performed with 2 μg RNA, 50 pmol random primer (6-mer), 10 mM dNTPs and 25 units of StrataScript Reverse Transcriptase (Stratagene) at 42°C for 1 h.
Quantitative real-time PCR (qRT–PCR) was performed using a Chromo4 Real-Time System (Bio-Rad), selected gene primers, cDNA template and Platinum Cyber Green qPCR Supermix UTG (Invitrogen, Carlsbad, CA) under the following conditions: 52°C for 2 min, 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, 58°C for 35 s and 72°C for 5 s. Data were obtained using Opticon Monitor Version 3.1 software. All reactions were performed in triplicate, together with blank (no template) controls. Melting curves for each PCR reaction were generated to ensure the purity of the amplified products. No primer dimers were detected when denaturation curves performed. For normalization, respective GAPDH mRNA quantities for each cDNA sample were measured. Changes in gene expression level were calculated by the 2ΔΔ Ct method (Livak and Schmittgen, 2001). Data are presented as means±s.d. from a minimum of three independent experiments.
The following primer pairs were used for qRT–PCR:
- GRX-1 forward:
CACAGCCACCAACCACACTA
GCCGCGTCAGCAGTTCCCC
CTTTGGAGAATTTAGCGACGGC
CTGGTTTCCATATTCAAGCAGGT
CACGACAGCTCTTTCCATGA
AGCCAGTGCTCGATGAATCT
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- GRX-1 backward:
GCCGCGTCAGCAGTTCCCC
CTTTGGAGAATTTAGCGACGGC
CTGGTTTCCATATTCAAGCAGGT
CACGACAGCTCTTTCCATGA
AGCCAGTGCTCGATGAATCT
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- GRX-2 forward:
CTTTGGAGAATTTAGCGACGGC
CTGGTTTCCATATTCAAGCAGGT
CACGACAGCTCTTTCCATGA
AGCCAGTGCTCGATGAATCT
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- GRX-2 backward:
CTGGTTTCCATATTCAAGCAGGT
CACGACAGCTCTTTCCATGA
AGCCAGTGCTCGATGAATCT
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- IFNβ forward:
CACGACAGCTCTTTCCATGA
AGCCAGTGCTCGATGAATCT
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- IFNβ backward:
AGCCAGTGCTCGATGAATCT
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- IL8 forward:
TAGGACAAGAGCCAGGAAGAAAC
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- IL8 backward:
GGAGTATGTCTTTATGCACTG
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- GAPDH forward:
CATGAGAAGTATGACAACAGCCT
AGTCCTTCCACGATACCAAAGT
- GAPDH backward:
AGTCCTTCCACGATACCAAAGT
IRF3 DNA binding assay
Nuclear extracts from TV7 and TGi14 cells either mock-induced or induced with Sendai virus for 4 h were prepared as described above. Biotinylated-ISRE ds-oligo (Bio-ISRE) from the ISG15 promoter (Biotin-CATGCCTCGGGAAAGGGAAACCGAAACTGAAGCC) (Lin et al, 1998) was incubated with equilibrated magnetic steptavidin beads (Dynal) in binding buffer A (1 M NaCl, 5 mM Tris–HCl pH 7.5, 0.5 mM EDTA pH 8.0) for 20 min under rotation at room temperature. Equal protein amounts of extracts were not incubated or pre-incubated in DNA binding buffer B (100 mM Tris–HCl pH 7.5, 10 mM EDTA pH 8.0, 50% glycerol, 2 μg ml−1 poly dI/dC) with 50 times more non-biotinylated ISRE ds-oligo (wt-ISRE) (CATGCCTCGGGAAAGGGAAACCGAAACTGAAGCC) or non-biotinylated ISRE mutated ds-oligo (Mut-ISRE) (GGCGGGATCGGGACACCGACACTGAA) for 10 min on ice.
Equal amount of streptavidin beads bound to Bio-ISRE was added to all samples, but control sample where empty streptavidin beads were added, and incubated at room temperature for 20 min. Beads were washed five times with binding buffer B, diluted in 1 × SDS-loading buffer, heated at 100°C for 5 min and subjected to SDS–PAGE electrophoresis. Proteins were transferred to nitrocellulose membrane and western blotted with anti-IRF3 and anti-GSH antibodies.
Supplementary Material
Supplementary data
Acknowledgments
We thank, Alexandros Gouveris, Maria Bessa, Marios Aggelopoulos and Alexios Vlamis-Gardikas for their technical assistance and helpful discussions throughout this work. We thank Paschalis Sideras for supporting our work and providing us with critical materials. We thank George Panayotou and Martina Samiotaki for proteomics analysis (BSRC Fleming). We thank Spyros Garbis and Theodoros Roumeliotis for proteomics analysis (BRFAA). We thank George Panayotou and George Mosialos for critical reading of the manuscript. This work was financially supported by the Biomedical Research Foundation of the Academy of Athens to GS and from the March of Dimes and Phillip Morris Inc USA to DT.
References
- Agelopoulos M, Thanos D (2006) Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J 25: 4843–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerboom TP, Sies H (1981) Assay of glutathione, glutathione disulfide, and glutathione mixed disulfides in biological samples. Methods Enzymol 77: 373–382 [DOI] [PubMed] [Google Scholar]
- Anderson ME (1997) Glutathione and glutathione delivery compounds. Adv Pharmacol 38: 65–78 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Starke DW, Mieyal JJ, Gronostajski RM (1998) Thioltransferase (glutaredoxin) reactivates the DNA-binding activity of oxidation-inactivated nuclear factor I. J Biol Chem 273: 392–397 [DOI] [PubMed] [Google Scholar]
- Collins SE, Noyce RS, Mossman KL (2004) Innate cellular response to virus particle entry requires IRF-3 but not virus replication. J Virol 78: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotgreave IA, Gerdes RG (1998) Glutathione–protein interactions: a molecular link between oxidative stress and cell proliferation? Biochem Biophys Res Commun 242: 1–9 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr, Kerr IM, Stark GR (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264: 1415–1421 [DOI] [PubMed] [Google Scholar]
- Fernandes AP, Holmgren A (2004) Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal 6: 63–74 [DOI] [PubMed] [Google Scholar]
- Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T (1988) Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-b gene regulatory elements. EMBO J 7: 3397–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P (2005) Regulation of protein function by glutathionylation. Free Radic Res 39: 573–580 [DOI] [PubMed] [Google Scholar]
- Gravina SA, Mieyal JJ (1993) Thioltransferase is a specific glutathionyl mixed disulfide oxidoreductase. Biochemistry 32: 3368–3376 [DOI] [PubMed] [Google Scholar]
- Hiscott J, Pitha P, Genin P, Nguyen H, Heylbroeck C, Mamane Y, Algarte M, Lin R (1999) Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res 19: 1–13 [DOI] [PubMed] [Google Scholar]
- Ihle JN (1996) STATs: signal transducers and activators of transcription. Cell 84: 331–334 [DOI] [PubMed] [Google Scholar]
- Juang YT, Lowther W, Kellum M, Au WC, Lin R, Hiscott J, Pitha PM (1998) Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA 95: 9837–9842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt P, Lamas S (2000) Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem 267: 4928–4944 [DOI] [PubMed] [Google Scholar]
- Klatt P, Molina EP, De Lacoba MG, Padilla CA, Martinez-Galesteo E, Barcena JA, Lamas S (1999) Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB J 13: 1481–1490 [DOI] [PubMed] [Google Scholar]
- Lin R, Heylbroeck C, Pitha PM, Hiscott J (1998) Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol 18: 2986–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind C, Gerdes R, Schuppe-Koistinen I, Cotgreave IA (1998) Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: catalysis by glutaredoxin. Biochem Biophys Res Commun 247: 481–486 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue J, Nakano H, Mak TW, Yeh WC, Li X, Akira S, Suzuki N, Suzuki S, Mosialos G, Kieff E (2003) Epstein-Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci USA 100: 15595–15600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merika M, Williams AJ, Chen G, Collins T, Thanos D (1998) Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell 1: 277–287 [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Fujita Ô, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T (1988) Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to the IFN-b gene regulatory elements. Cell 54: 903–913 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Hiscott J, Pitha PM (1997) The growing family of interferon regulatory factors. Cyt Growth Fact Rev 8: 293–312 [DOI] [PubMed] [Google Scholar]
- Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, Perez-Sala D, Lamas S (2001) Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry 40: 14134–14142 [DOI] [PubMed] [Google Scholar]
- Qin BY, Liu C, Lam SS, Srinath H, Delston R, Correia JJ, Derynck R, Lin K (2003) Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol 10: 913–921 [DOI] [PubMed] [Google Scholar]
- Reich NC (2002) Nuclear/cytoplasmic localization of IRFs in response to viral infection or interferon stimulation. J Interferon Cytokine Res 22: 103–109 [DOI] [PubMed] [Google Scholar]
- Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, Heintz NH, Heim J, Ho YS, Matthews DE, Wouters EF, Janssen-Heininger YM (2006) Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci USA 103: 13086–13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13: 539–548 [DOI] [PubMed] [Google Scholar]
- Sato M, Tanaka N, Hata N, Oda E, Taniguchi T (1998) Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett 425: 112–116 [DOI] [PubMed] [Google Scholar]
- Senger K, Merika M, Agalioti T, Yie J, Escalante CR, Chen G, Aggarwal AK, Thanos D (2000) Gene repression by coactivator repulsion. Mol Cell 6: 931–937 [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Shelton MD, Chock PB, Mieyal JJ (2005) Glutaredoxin: role in reversible protein S-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal 7: 348–366 [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD (1998) How cells respond to interferons. Annu Rev Biochem 67: 227–264 [DOI] [PubMed] [Google Scholar]
- Sun P, Enslen H, Myung P, Maurer RA (1994) Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev 8: 2527–2539 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S (1997) Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA 94: 10215–10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal VJ, Fanburg BL (2000) Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physio 279: L1005–L1028 [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T (1995a) Identification of the rel family members required for virus induction of the human beta interferon gene. Mol Cell Biol 15: 152–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, Maniatis T (1995b) Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83: 1091–1100 [DOI] [PubMed] [Google Scholar]
- Thomas JA, Poland B, Honzatko R (1995) Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys 319: 1–9 [DOI] [PubMed] [Google Scholar]
- Uddin S, Platanias LC (2004) Mechanisms of type-I interferon signal transduction. J Biochem Mol Biol 37: 635–641 [DOI] [PubMed] [Google Scholar]
- Wang J, Boja ES, Tan W, Tekle E, Fales HM, English S, Mieyal JJ, Chock PB (2001) Reversible glutathionylation regulates actin polymerization in A431 cells. J Biol Chem 276: 47763–47766 [DOI] [PubMed] [Google Scholar]
- Weaver BK, Kumar KP, Reich NC (1998) Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol 18: 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P (1992) The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J 11: 2229–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Jao S, Nanduri S, Starke DW, Mieyal JJ, Qin J (1998) Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity. Biochemistry 37: 17145–17156 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T (1998) Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 17: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data