Abstract
Yeast LAG1 was one of the first longevity genes found. Subsequent analysis showed that it encodes a component of ceramide synthase. Homologs of LAG1 have been identified in all eukaryotes examined for their presence, and multiple homologs are the norm. In human and mouse, the LAG1 counterpart is called LASS1. The involvement of this gene in determining yeast replicative life span led us to ask whether longevity effects could be found in C. elegans. Extended longevity was seen when we used RNAi to decrease expression of the worm homolog of LAG1, termed hyl-1, for Homolog of Yeast Longevity gene. In contrast, neither deletion of the gene nor overexpression resulted in life extension. There was no evidence that hyl-1 interacts with the insulin/IGF-1 like signaling pathway to specify longevity or dauer formation, nor were effects on stress resistance detected. Gene expression of hyl-1 homologs was altered in the deletion mutant and by RNAi, showing distinct evidence for compensation at the transcript level. These regulatory changes may explain the subtle phenotypic effects found under the conditions studied here.
Keywords: Aging, Ceramide synthase, Compensation, IIL pathway, RNAi, Stress resistance
Introduction
LAG1 was one of the first longevity genes to be isolated. It was found among genes that were differentially expressed over the replicative life span in the budding yeast, Saccharomyces cerevisiae (Egilmez et al. 1989; D’mello et al. 1994). A deletion in LAG1 was shown to result in a 50% increase in mean and maximum life span, whereas overexpression strains had an increase of replicative life span of as much as 27%. However, the extent of the effect on longevity is dependent on level of gene expression, displaying a maximum beyond which substantial curtailment of longevity occurs (Jiang et al. 2004). Homologs from both human and Caenorhabditis elegans are able to functionally complement the yeast gene LAG1 (Jiang et al. 1998). S. cerevisiae contains a close homolog of LAG1, termed LAC1; a deletion of both genes is lethal (Jiang et al. 1998). These genes encode proteins of the endoplasmic reticulum which are involved in ceramide biosynthesis (Guillas et al. 2001; Schorling et al. 2001). Ceramide signaling has been shown to affect growth, differentiation, stress resistance, apoptosis, cell senescence, and insulin action, any of which could have an effect on life span (Cutler and Mattson 2001; Jazwinski and Conzelmann 2002; Obeid and Hannun 2003; Stratford et al. 2004). LAG1 is the founding member of a large family of genes (Venkataraman and Futerman 2002). All LAG1/LASS1 proteins, and in particular those shown to possess ceramide synthase activity (Jazwinski and Conzelmann 2002), have a common sequence termed the Lag1p motif (Jiang et al. 1998). Specific amino acids in this motif are required for ceramide synthase activity (Spassieva et al. 2006).
The nematode C. elegans has long been a model in which genes specifying aging and longevity could be identified (Johnson and Wood 1982; Kenyon 2005). Currently, more than 200 genes leading to life extension have been found. In this study, we asked whether hyl-1 (Homolog of Yeast Longevity gene) specifies similar functions to those in yeast, where it is involved in ceramide metabolism and also affects replicative life span. We also looked at other worm homologs of LAG1 and asked how these genes affect life span. In addition, we saw that the effects of manipulation of the hyl-1 gene are subtle and apparently need to be fine-tuned to achieve a long-life phenotype. Studies of gene expression also indicated that changes and compensation are taking place both in deletion mutants and after RNAi treatment again suggesting subtle effects of genetic manipulation.
Materials and methods
Strains
Worms were grown on NGM plates spotted with E. coli strain OP50 using standard conditions (Brenner 1974), unless otherwise stated. Worm strains used in these experiments were: N2 (WT ancestral strain CGCb), CH1035 [rol-6(su1006) II], RB1036 [hyl-1(ok976) IV], TJ1090 and TJ1091 [hyl-1(ok976) backcrossed 6X and 10X to N2], TJ1052 [age-1(hx546) II], TJ356 [daf-16::GFP(zIs356)], VC334 [hyl-1(gk203) IV]. RNAi constructs were obtained from the Ahringer collection, the Andy Fire collection, Sam Henderson, or were newly constructed (Table 1). All constructs are carried by E. coli strain HT115.
Table 1.
RNAi constructs
| Gene (name) | Sequence | RNAi strain | Description |
|---|---|---|---|
| age-1 | B0334.8 | II-7J02 | Ahringer library |
| daf-2 | Y55D5A.5 | pSH510 | From S. Henderson |
| daf-16 | R13H8.1 | pSH508 | From S. Henderson |
| gfp | Green fluorescent protein | pPD128.110 | From A. Fire |
| ev | None | pPD129.36 | Empty vector (A. Fire) |
| hyl-2 | K02G10.6 | X-2D18 | Ahringer library |
| lagr-1 | Y6B3B.10 | I-6P22 | Ahringer library |
| tram-1 | C24F3.1 | IV-5K16 | Ahringer library |
| hyl-1 | C09G4.1 | pJW9 | hyl-1 seq spanning Lag1p motif* |
| pUC18 | None | pUC18 | Classic pUC18, control for pJW9 |
*hyl-1 nucleotides 486–639 between T7 and T3 promoters in pUC18 plasmid
RNAi
RNAi feeding was performed as described by Kamath and Ahringer (2003). Briefly, RNAi feeding bacteria were grown overnight at 37°C in LB media plus 50 μg/ml ampicillin. Small NGM plates containing 1 mM IPTG and 50 μg/ml ampicillin were spotted with the RNAi feeding bacteria and grown overnight at room temp (~23°C). Plates were stored at 2°C and used within 1–2 weeks. Young adult worms were placed onto RNAi plates and a staged egg lay was performed. Assays were carried out as usual on RNAi plates using staged young adult animals. Plates were checked to see that animals were not starved or contaminated. A number of variables can affect assays, including temperature, freshness of bacteria, gene of interest, and RNAi construct used.
Biological assays
Survival assays were performed as previously described and survival curves were compared via log rank tests (Johnson and Wood 1982). Dauer formation was assessed by initiating a staged egg lay at 20°C on NGM plates spotted with OP50 or an HT115 RNAi bacterial strain. Eggs were shifted to 27°C for 3 days and then scored visually for dauer formation. If dauers looked atypical, a 1% SDS treatment for 30 min to 1 h was performed and survivors were scored as dauer. Fertility was assessed using three to five single-worm replicates. These were placed onto NGM plates at 20°C and transferred daily; progeny reaching adulthood were scored as total brood.
Stress Resistance Assays-heat, juglone, UV, oxygen, and nuclear localization of DAF-16
For all stress assays, eggs were laid at 20°C onto NGM plates spotted with OP50 unless otherwise specified. On day 3 of life, animals were exposed to the appropriate stressor. For heat stress, plates were put at 35°C and followed until all animals were dead. For juglone treatment, NGM plates with 240 μM juglone were prepared (de Castro et al. 2004); animals were placed at 20°C and followed for survival until all were dead. For UV treatment, worms were transferred to NGM plates without OP50, irradiated with 254 nm light at 2,000 J/m2 in a UV Stratalinker 2400 (Stratagene), removed to NGM plates with OP50 at 20°C and scored for survival at 24-h intervals until all were dead (Murakami and Johnson 1996). For oxygen stress, animals were put into an oxygen chamber at 99% oxygen, 40 PSI for 24 h at 20°C. Plates were then removed and assayed for survival at intervals up to 24 h later. Survival means, p values, and other statistics were performed with Statistica 99 (StatSoft) software. For nuclear localization, eggs from strain TJ356 were laid onto plates seeded with various RNAi strains and adults were scored for presence or absence of nuclear localization of DAF-16 using a Leica MZFL III Fluorescence Stereomicroscope.
Real time-reverse transcriptase PCR (RT-PCR) for hyl-1, hyl-2, lagr-1, and tram-1
- Primers
Sequence
- ama-1 (control gene): ama-1F:
5′-ATAGGAATGGATCCACAATTCGC-3′
- ama-1R:
5′-TTCAGTCGCTGCTTGATCGATT-3′
- hyl-1 hyl-1F:
5′-AGTCGGAACCTTGATTCTTC-3′
- hyl-1R:
5′-CCCAAAGTATGTAATCTGGC-3′
- hyl-2 hyl-2F:
5′-TCGTCGTCGCAACATCTCCTA-3′
- hyl-2R:
5′-CCCAATGTTTGCGCAGACA-3′
- lagr-1 lagr-1F:
5′-TCGAATGATCTGGCGCGTA-3′
- lagr-1R:
5′-TCGTCGAGCTTCTGCTGATTG-3′
- tram-1 tram-1F:
5′-TTGCTGGATCGTCGTGCA-3′
- tram-1R:
5′-GGTGGAATGACTCGCCAAATT-3′
For real time RT-PCR, 2 μg of worm RNA extracted by Trizone (Invitrogen) was used for reverse transcription. The resulting cDNA, equivalent to 0.01 μg of total RNA, was used as template for PCR in a dilution series. SYBR-Green PCR Master Mix (ABI) was employed, and PCR carried out on the ABI 7000 Sequence Detection system. Results were normalized to the transcript of ama-1, as a control. The significance of differences in RNA was assessed using a two-tailed, unpaired t test with unequal variances. The results are presented as relative values for comparison, by setting the expression levels for N2 and puc18 at 1.0 for each determination.
Results
Transgenic strains
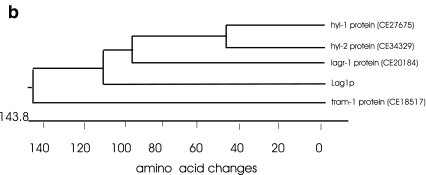
C. elegans has four homologs of yeast LAG1, called hyl-1 (Homolog of Yeast Longevity gene), hyl-2, lagr-1 (LAG Related), and tram-1 (Fig. 1). hyl-1 and hyl-2 are most closely related and, along with lagr-1, are all equally related to yeast LAG1 (Fig. 1b). The fourth homolog, tram-1, is a distant relative belonging to a larger gene family. The tram-1 protein lacks the Lag1p motif (Fig. 1a), and there are several amino acids required for ceramide synthase activity missing from the corresponding region of tram-1 protein (Spassieva et al. 2006). Overexpression of LAG-1 in yeast results in life extension (Jiang et al. 2004). We first asked if overexpression of hyl-1, the best nematode homolog to LAG1, would result in increased life span. Transgenic strains were constructed using standard worm methods. Specifically, an N2 strain was injected with either pRF4, a plasmid carrying rol-6(su1006) or pRF4 and pJJ2, a plasmid carrying the complete genomic sequence of hyl-1. Some strains were maintained as extra chromosomal lines (designated zEX) and some were integrated using gamma irradiation (designated zIs). All strains were backcrossed 7–10 times to N2, our reference wild-type strain. Three independent transgenic lines were isolated and tested, two containing hyl-1 and rol-6 and one containing only rol-6. Additional controls included CH1035 [rol-6(su1006)] and TJ1052 [age-1(hx546)], which served as a positive control for longevity increases.
Fig. 1.
Protein sequence alignment of yeast Lag1 and four worm homologs, hyl-1, hyl-2, lagr-1, and tram-1. a Alignment using CLUSTER METHOD of DNASTAR program. Shaded residues are identical. The region spanning amino acids 246–297 (yeast Lag1p coordinates) shows the Lag1p motif. Another highly homologous region, amino acids 356–409 (Lag1p coordinates) in the C-terminus, was also identified. b Phylogenetic tree of Lag1p homologs
Transgenic strains carrying extra copies of hyl-1 did not give a consistent long-life phenotype at 20°C or 25.5°C (data not shown). Transgenic strains were also tested for resistance to heat, juglone, and UV, and no significant differences were seen, compared to controls (data not shown). We checked transgenic strains for dauer formation at 27°C and none were constitutive dauer formers (Daf-c phenotype).
Deletion mutants
Two deletion mutants in hyl-1 are currently available. The first, VC334 [hyl-1(gk203)], contains a 197 bp deletion in hyl-1. Survival experiments showed no increase in the longevity of this strain compared to wild type. The second, RB1036 [hyl-1(ok976)], contains an 863 bp deletion in hyl-1 and initially showed an increased life span compared to N2. We backcrossed the hyl-1(ok976) deletion with N2 and generated the strains TJ1090 and TJ1091, backcrossed six and ten times respectively. The presence of the deletion was confirmed by PCR. Survival assays were performed, and we found that TJ1090 and TJ1091 were not longer-lived than N2. However, in the same experiment, the mean life span of the original unbackcrossed strain RB1036 was 21% longer than N2. These data suggested that backcrossing the strain had removed an unlinked locus responsible for increased life span in the original deletion strain, RB1036.
RNAi mediated survival
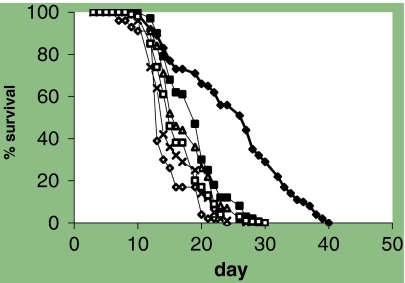
Despite the lack of success with the deletion mutants, we decided to try an RNAi approach to partially reduce the level of hyl-1 and also to target its homologs. Studies of yeast LAG1 suggested that this might be a useful approach (Jiang et al. 2004). A hyl-1 RNAi vector was constructed to encompass the Lag1p motif sequence (Fig. 1a and Table 1). The Lag1p motif sequence is a consensus sequence that was first identified by comparing several LAG1 homologs, including hyl-1 and hyl-2 (Jiang et al. 1998). The wild-type strain, N2, was put onto various RNAi strains and followed for survival at 20°C and 25°C. We found that the animals grown on the Lag1p motif vector (pJW9) had a greater mean life span than any of the negative control vectors (Fig. 2 and Table 2). Increases in mean life span at 20°C ranged from 14% to 31%, all with p < 0.00001. The increase in life span when compared to the RNAi bacterial strain HT115 (ht) carrying no vector was 8% with a p value of 0.01068. In these experiments, we used several negative control vectors including puc18, ev (HT115 with an empty vector), and gfp (HT115 carrying a green fluorescent protein sequence; Table 1). Since all of these negative controls were shorter lived, the longevity effect of the pJW9 vector was convincing. As expected, our positive RNAi control vector, daf-2, allowed longer life than all other strains and gave us confidence that the assay was working.
Fig. 2.
Combined survival data for pJW9 at 20°C. Wild-type animals were put onto various RNAi strains and followed for survival. Strains were daf-2 (♦), gfp (⋄), ht (△), ev (×), pJW9 (▪), and pUC18 (□)
Table 2.
RNAi mediated survival
| RNAi Strain | n | Rep | Mean (days) | SEM | p vs pJW9 | Temp |
|---|---|---|---|---|---|---|
| pJW9 | 192 | 3 | 18.9 | 0.3 | 20 | |
| pUC18 | 175 | 3 | 16.5 | 0.3 | 0.00001 | 20 |
| Ht* | 137 | 2 | 17.5 | 0.4 | 0.01068 | 20 |
| ev | 85 | 2 | 15.6 | 0.4 | 0.00001 | 20 |
| gfp | 46 | 1 | 14.3 | 0.5 | 0.00001 | 20 |
| daf-2 | 103 | 3 | 25.0 | 0.8 | 0.00001 | 20 |
| pJW9 | 68 | 1 | 11.0 | 0.2 | 25 | |
| pUC18 | 60 | 1 | 10.1 | 0.3 | 0.03380 | 25 |
| daf-2 | 23 | 1 | 18.7 | 0.9 | 0.00001 | 25 |
Wild-type animals were put onto indicated RNAi strains and followed for survival at 20°C or 25°C
*Ht is the E.coli strain HT115 with no RNAi vector
We repeated the above experiments at 25°C to see if increasing the temperature changed the phenotype (Table 2). At 25°C, we still saw a life span increase in worms grown on the pJW9 vector but the effect was smaller. The mean life span increase was now 9% with a p value of 0.034. We used one negative control vector for these experiments, pUC18, and the positive RNAi control vector, daf-2.
In many cases, RNAi can be more efficacious for longevity extension at lower levels of interference (Ventura and Rea 2007), so we reduced the amount of the pJW9 RNAi vector to 10% and 1% by diluting the pJW9 RNAi strain with the empty vector strain puc18. Neither of these conditions gave a long-life phenotype (data not shown), indicating that the undiluted RNAi vector pJW9 was the best choice for inducing longevity. We also looked at the effect of growing wild-type worms for more than one generation on pJW9. In these experiments, increased life span was only seen with first generation RNAi worms and not with worms grown for two or three generations on pJW9.
hyl-1 and other genes
hyl-1 has three homologs (Table 3), and it may be that more than one is involved in ceramide synthesis and specification of longevity. Therefore, we looked at these and a few other genes implicated in the aging process using RNAi mediated survival assays (Table 4). N2 animals treated with RNAi against hyl-2, lagr-1, or tram-1 did not have increased longevity, compared to empty vector. In fact, each of these vectors produced a decrease in mean life span, which reached significant p values for lagr-1 with a 10% decrease. In these experiments, daf-2 and age-1 served as long-lived controls and showed an increase in longevity of 77% and 56% with p values <0.00001.
Table 3.
hyl-1 homologs
| Protein | Gene | Descript | Blast score | % Alignment | AA | Alleles |
|---|---|---|---|---|---|---|
| CE27675 | C09G4.1 | hyl-1 | 368 | 2 | ||
| CE34329 | K02G10.6 | hyl-2 | 2.1 × e-65 | 98.5% | 329 | 2 |
| CE20184 | Y6B3B.10 | lagr-1 | 8.1 × e-18 | 63.6% | 360 | 4 |
| CE18517 | C24F3.1 | tram-1 | 6.3 × e-5 | 66.8% | 371 | 0 |
Data from WormBase Version WS174
Table 4.
RNAi mediated survival using hyl-1 homologs
| RNAi Strain | n | Mean fife span (days) | SEM (days) | p vs ev | Ratio/ev |
|---|---|---|---|---|---|
| ev | 50 | 17.2 | 0.5 | ||
| hyl-2 | 38 | 15.4 | 0.6 | 0.05200 | 0.90 |
| daf-2 | 41 | 30.5 | 1.4 | 0.00001 | 1.77 |
| ev | 40 | 15.8 | 0.5 | ||
| lagr-1 | 38 | 14.1 | 0.4 | 0.00001 | 0.90 |
| tram-1 | 47 | 15 | 0.4 | 0.16376 | 0.95 |
| age-1 | 33 | 24.7 | 1.5 | 0.00001 | 1.56 |
N2 worms were put onto RNAi strains and followed for survival at 20°C
RT-PCR analysis
Given the results obtained with RNAi mediated survivals, we decided to look at gene expression levels using RT-PCR. RT-PCR analysis showed significant changes in gene expression for hyl-1 and its homologs (Table 5). In the deletion strain TJ1091, levels of hyl-1 were undetectable but, surprisingly, we saw more than a 50% increase in levels of hyl-2 mRNA. We also observed decreased levels for lagr-1 (0.76-fold), and increased levels of tram-1 of 1.29-fold. All of these changes were significant with p values of 0.004 or less, but did not lead to increased life span in the deletion mutant. By contrast, treatment with the pJW9 RNAi vector resulted in a significant decrease in both hyl-1 and hyl-2, compared to the control vector pUC18. N2 worms on pJW9 had hyl-1 and hyl-2 levels of 0.7 and 0.68, respectively, compared to the pUC18 vector (p = 0.0002 and 0.025), but there were no significant changes in lagr-1 or tram-1 expression levels. This combination of expression level changes is apparently sufficient to produce a long-lived response.
Table 5.
Relative expression for hyl-1 homologs
| Gene | Deletion mutant | N2 on RNAi | ||
|---|---|---|---|---|
| N2 | TJ1091 | pUC18 | pJW9 | |
| hyl-1 | 1 | undetected | 1 | 0.7 (p = 0.0002) |
| hyl-2 | 1 | 1.66 (p = 0.002) | 1 | 0.68 (p = 0.025) |
| lagr-1 | 1 | 0.76 (p = 0.0037) | 1 | 1.02 (p = 0.76) |
| tram-1 | 1 | 1.29 (p = 0.0015) | 1 | 1.003 (p = 0.97) |
p values were calculated using a two-tailed t test for unpaired samples with unequal variances
Results are presented as relative values for comparison, by setting the expression levels for N2 and pUC18 at 1.0 for each determination
hyl-1 and the daf-16 pathway
We wanted to look at the interaction of hyl-1 with the daf-16 insulin/IGF-1 like (IIL) signaling pathway and in particular with the age-1 gene. To do this, we put TJ1052 [age-1(hx546)] onto the RNAi vectors pJW9, four negative controls, and one positive control, and followed these for survival at 20°C (Table 6). We did not see an increase in life span with the pJW9 vector, when compared with any of the four controls. However, we did see a large increase in mean life span when TJ1052 was placed onto the RNAi vector for daf-2. These data show that reducing hyl-1 expression does not further extend the life span of age-1 animals. However, reducing expression of daf-2 doubles the life span of age-1.
Table 6.
Survivals with TJ1052 [age-1 (hx546)]
| RNAi strain | n | Mean (d) | SEM | p vs pJW9 |
|---|---|---|---|---|
| pJW9 | 26 | 22.6 | 1.1 | |
| pJW9 (uninduced)* | 22 | 24.2 | 1.1 | 0.4334 |
| pUC18 | 29 | 22.9 | 1.0 | 0.89175 |
| ev | 28 | 20.9 | 0.8 | 0.22907 |
| gfp | 22 | 20.5 | 0.7 | 0.13677 |
| daf-2 | 16 | 41.7 | 2.6 | 0.00001 |
TJ1052 worms were placed onto RNAi strains and followed for survival at 20°C
*No IPTG was added to these RNAi feeding plates which served as a negative control
Because TJ1052 has been backcrossed three times to a wild-type strain, and because it can respond to daf-2 RNAi, we believe the lack of response to pJW9 is real and not due to background or strain effects.
Another experiment was performed to address the question of whether the hyl-1 longevity increase is mediated via DAF-16. Several long-lived mutants in the daf-16 pathway form constitutive dauers at 27°C. We looked to see if hyl-1 produced dauers at 27°C or if it affected dauer formation of age-1. We placed N2 and TJ1052 onto the RNAi vector pJW9 and looked for dauer formation at 27°C. N2 showed no dauers and TJ1052 was 100% dauer suggesting that pJW9 has no effect on dauer formation for these strains.
We then asked if hyl-1 causes DAF-16 to localize to the nucleus as is seen when RNAi treatment to daf-2 is implemented (Henderson and Johnson 2001). TJ356, an integrated DAF-16::GFP roller strain, was placed onto RNAi plates containing the hyl-1 Lag1p motif sequence (pJW9) or the empty vector control (pUC18) at 20°C. A limited lay was performed and animals were checked for DAF-16 nuclear localization as staged young adults under a GFP fluorescence dissecting microscope. The animals were then allowed to starve on the plates and checked again as a mixed population. We saw no localization of DAF-16 on unstarved plates with either the pJW9 RNAi vector or the pUC18 control vector. On starved plates, both RNAi vectors showed some animals with nuclear localization of DAF-16. We concluded that pJW9 does not cause nuclear localization in TJ356 but neither does it interfere with starvation-induced localization. Therefore, it appears likely that reducing hyl-1 expression via RNAi does not activate or inhibit daf-16 function.
Other assessments of hyl-1 worms
Young adult wild-type animals raised on RNAi plates at 20°C were put at 35°C and followed until death. Data from two experiments showed no significant resistance to heat for animals on the RNAi vector pJW9 as compared to our negative control vector puc18. However, our positive control vector, daf-2, showed significant resistance to heat compared to vector pUC18 with a p value of 0.00001 (data not shown).
Fertility was assessed at 20°C on wild-type animals grown on RNAi vectors pJW9, pUC18, ev, ht, gfp, and daf-2. We saw no significant difference between pJW9 and any of the other vectors suggesting that pJW9 does not affect fertility (data not shown).
Discussion
We have characterized several types of mutations in hyl-1, the C. elegans homolog of yeast LAG1, to determine if hyl-1 is a longevity gene in worms, as it is for yeast replicative life span. Overexpression did not lead to increased longevity or stress resistance. Since overexpression leads to about a 20% increase in longevity in yeast, it may be that the effect is smaller or more variable in worms and therefore undetectable. Moreover, the effect in yeast is on replicative and not chronological life span, although replicative life span correlates well with total yeast life span (Minois et al. 2005). When hyl-1 deletion mutants became available in worms, we assessed their longevity, since the longevity effect in yeast LAG1 deletions is on the order of 50%. Neither hyl-1(ok976) nor hyl-1(gk203) showed extended life span. Therefore, we switched to an RNAi feeding approach, which is currently a powerful method for blocking gene function in C. elegans (Fire et al. 1998; Kamath and Ahringer 2003), reasoning that partial reduction in expression of hyl-1 might affect life span as suggested by studies of LAG1 in yeast (Jiang et al. 2004).
Using an RNAi vector spanning the Lag1p motif in hyl-1 (pJW9), we saw a longevity effect of about 15%. This effect was small but consistent and statistically significant in three out of four experiments at 20°C. The Lag1p motif is a consensus sequence found across species in LAG1 homologs including hyl-1 in C. elegans. It consists of a highly conserved stretch of 52 amino acids and is necessary for ceramide synthase activity (Kageyama-Yahara and Riezman 2006; Spassieva et al. 2006). Ceramide signaling has been shown to affect growth, differentiation, stress resistance, apoptosis, cell senescence, and insulin action, any of which could have an effect on life span (Cutler and Mattson 2001; Jazwinski and Conzelmann 2002; Obeid and Hannun 2003; Stratford et al. 2004).
To further investigate this system, we used RNAi vectors to other members of the hyl-1 family in C. elegans. None of the homologs of hyl-1 showed a longevity effect; in fact, hyl-2 and lagr-1 showed shorter life spans. We also examined the IIL pathway, which is known to play a part in the long-life phenotype. Inhibition of this pathway leads to longevity in many different organisms including worms, fruit flies, and mice (Clancy et al. 2001; Henderson and Johnson 2001; Holzenberger et al. 2003; Kenyon 2005). We looked at the interaction of this pathway with hyl-1 in several different ways. We found no evidence for synergism in prolonging life. However, since there is evidence for ceramide modulation of the insulin signaling pathway (Burow et al. 2000; Teruel et al. 2001; Gorski et al. 2002; Straczkowski et al. 2004), we performed a few more tests. Most long-lived mutations in the insulin-like pathway have increased dauer formation, but we observed no effect of hyl-1 on dauer formation. We also concluded that hyl-1 does not promote or interfere with DAF-16 nuclear localization. Under the conditions tested, there was no evidence for interaction between hyl-1 and the IIL pathway.
We performed several additional assays to determine if hyl-1 acts via other known mechanisms of aging. We looked at the fertility of Lag1p RNAi animals and saw no deficit. We also did not detect heat resistance of wild-type animals on Lag1p RNAi even though heat resistance is highly correlated with longevity (Walker et al. 2001; Olsen et al. 2006). Mass spectrometry showed changes in ceramide species for both the deletion mutant and the RNAi vector pJW9 compared to controls (L. Obeid, personal communication). However, where the longevity effects were consistent, RNAi mass spec profiles were not. Presumably the RNAi treatments are somewhat variable and this is reflected in the mass spec profiles. There are numerous reports (Simmer et al. 2003; Echeverri et al. 2006) suggesting that RNAi is quite variable, and this is perhaps a quantitative example of that variability.
RT-PCR proved to be quite informative, and we saw significant changes in expression of hyl-1 and its homologs. In the hyl-1 deletion strain TJ1091, we observed significant changes for all homologs. hyl-1 was undetectable, hyl-2 and tram-1 were increased, and lagr-1 was decreased. However, the deletion mutant does not have an increased life span. RT-PCR analysis of worms treated with Lag1p RNAi also showed significant changes, but here hyl-1 and hyl-2 were reduced to about 70% each and the other two homologs were unaffected. This combination of expression levels appears to be sufficient for increased longevity, but there may be other combinations that would confer the same or a more extreme effect. It should be noted that there are multiple LAG1/LASS1 encoded ceramide synthases in cells, and they display different tissue and acyl-CoA specificities (Mizutani et al. 2005); we may have uncovered a regulated series of interactions among these family members at the transcript level. Moreover, it may be that both hyl-1and hyl-2 need to be down-regulated simultaneously. We saw compensation by hyl-2 in the hyl-1 deletion, and this may explain why no longevity effect was seen there. In contrast, the RNAi treatment with the Lag1p motif, which down-regulated both transcripts, did give a longevity response. The level of gene expression that leads to increased mean life span is apparently quite sensitive and is best seen with one generation on the undiluted hyl-1 RNAi vector.
In conclusion, hyl-1 under certain conditions has proven to be a longevity gene in C. elegans. The mechanism of its effect is yet to be elucidated, but does not appear to be related to fertility, heat resistance or the IIL pathway. It is possible that hyl-1, a worm homolog to yeast LAG1, represents a novel mechanism for affecting life span in C. elegans.
Acknowledgments
This work was supported by grants from the National Institutes of Health (NIH), AG16219 to T.E.J. and AG006168 to S.M.J. Some nematode strains were provided by the Caenorhabditis Genetics Center which is funded by the National Center for Research Resources (NCRR) of the NIH. We would like to thank Heather Greer, Sarah Hegi, Ibett Hynek, and Chris Link for technical help and Lina Obeid for permission to cite her unpublished work. Address correspondence to Thomas E. Johnson, PhD, Institute for Behavioral Genetics, Campus Box 447, University of Colorado, Boulder, CO 80309.
Abbreviations
- IIL
insulin/IGF-1 like
- IGF
insulin-like growth factor 1
- RNAi
RNA interference
- PCR
poymerase chain reaction
References
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77(1):71–94 [DOI] [PMC free article] [PubMed]
- Burow ME, Weldon CB, Collins-Burow BM, Ramsey N, McKee A, Klippel A, McLachlan JA, Clejan S, Beckman BS (2000) Cross-talk between phosphatidylinositol 3-kinase and sphingomyelinase pathways as a mechanism for cell survival/death decisions. J Biol Chem 275(13):9628–9635 [DOI] [PubMed]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L (2001) Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292:104–106 [DOI] [PubMed]
- Cutler RG, Mattson MP (2001) Sphingomyelin and ceramide as regulators of development and lifespan. Mech Ageing Dev 122(9):895–908 [DOI] [PubMed]
- de Castro E, Hegi de Castro S, Johnson TE (2004) Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic Biol Med 37(2):139–145 [DOI] [PubMed]
- D’mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM (1994) Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J Biol Chem 269:15451–15459 [PubMed]
- Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R (2006) Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods 3(10):777–779 [DOI] [PubMed]
- Egilmez NK, Chen JB, Jazwinski SM (1989) Specific alterations in transcript prevalence during the yeast life span. J Biol Chem 264:14312–14431 [PubMed]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed]
- Gorski J, Dobryn A, Zendzian-Piotrowska M (2002) The sphingomyelin-signaling pathway in skeletal muscles and its role in regulation of glucose uptake. Ann NY Acad Sci 967:236–248 [DOI] [PubMed]
- Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A (2001) C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J 20(11):2655–2665 [DOI] [PMC free article] [PubMed]
- Henderson ST, Johnson TE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11(24):1975–1980 [DOI] [PubMed]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y (2003) IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421:182–187 [DOI] [PubMed]
- Jazwinski SM, Conzelmann A (2002) LAG1 puts the focus on ceramide signaling. Int J Biochem Cell Biol 34(11):1491–1495 [DOI] [PubMed]
- Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM (1998) Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res 8:1259–1272 [DOI] [PubMed]
- Jiang JC, Kirchman PA, Allen M, Jazwinski SM (2004) Suppressor analysis points to the subtle role of the LAG1 ceramide synthase gene in determining yeast longevity. Exp Gerontol 39(7):999–1009 [DOI] [PubMed]
- Johnson TE, Wood WB (1982) Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci USA 79(21):6603–6607 [DOI] [PMC free article] [PubMed]
- Kageyama-Yahara N, Riezman H (2006) Transmembrane topology of ceramide synthase in yeast. Biochem J 398:585–593 [DOI] [PMC free article] [PubMed]
- Kamath RS, Ahringer J (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30(4):313–321 [DOI] [PubMed]
- Kenyon C (2005) The plasticity of aging: insights from long-lived mutants. Cell 120(4):449–460 [DOI] [PubMed]
- Minois N, Frajnt M, Wilson C, Vaupel JW (2005) Advances in measuring lifespan in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 102:402–406 [DOI] [PMC free article] [PubMed]
- Mizutani Y, Kihara A, Igarashi Y (2005) Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J 390:263–271 [DOI] [PMC free article] [PubMed]
- Murakami S, Johnson TE (1996) A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143(3):1207–1218 [DOI] [PMC free article] [PubMed]
- Obeid LM, Hannun YA (2003) Ceramide, stress, and a “LAG” in aging. Sci Aging Knowledge Environ 39:PE27 [DOI] [PubMed]
- Olsen A, Vantipalli MC, Lithgow GJ (2006) Lifespan extension of Caenorhabditis elegans following repeated mild hormetic heat treatments. Biogerontology 7(4):221–230 [DOI] [PubMed]
- Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D (2001) Lag1p and Lac1p are essential for the Acyl-CoA dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol Biol Cell 12:3417–3427 [DOI] [PMC free article] [PubMed]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH (2003) Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol 1(1):E12 [DOI] [PMC free article] [PubMed]
- Spassieva S, Seo JG, Jiang JC, Bielawdki J, Alvarez F, Jazwinski SM, Hannun YA, Obeid LM (2006) Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J Biol Chem 281(45):33931–33938 [DOI] [PubMed]
- Straczkowski M, Kowalska I, Nikolajuk A, Dzienis-Straczkowska S, Kinalska I, Baranowski M, Zendzian-Piotrowska M, Brzezinska Z, Gorski J (2004) Relationship between insulin sensitivity and sphingomyelin signaling pathway in human skeletal muscle. Diabetes 53(5):1215–1221 [DOI] [PubMed]
- Stratford S, Hoehn KL, Liu F, Summers SA (2004) Regulation of insulin action by ceramide: dual mechanisms linking ceramide accumulation to the inhibition of Akt/protein kinase B. J Biol Chem 279(35):36608–36615 [DOI] [PubMed]
- Teruel T, Hernandez R, Lorenzo M (2001) Ceramide mediates insulin resistance by tumor necrosis factor-alpha in brown adipocytes by maintaining Akt in an inactive dephosphorylated state. Diabetes 50(11):2563–2571 [DOI] [PubMed]
- Venkataraman K, Futerman AH (2002) Do longevity assurance genes containing HOX domains regulate cell development via ceramide synthesis. FEBS Lett 528:3–4 [DOI] [PubMed]
- Ventura N, Rea SL (2007) Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnol J 2(5):584–595 [DOI] [PubMed]
- Walker GA, White TM, McColl G, Jenkins NL, Babich S, Candido EP, Johnson TE, Lithgow GJ (2001) Heat shock protein accumulation is upregulated in a long-lived mutant of Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci 56(7):B281–B287 [DOI] [PubMed]