Abstract
The activity of Rb (retinoblastoma protein) is regulated by phosphorylation and acetylation events. Active Rb is hypophosphorylated and acetylated on multiple residues. Inactivation of Rb involves concerted hyper-phosphorylation by cyclin–CDK (cyclin-dependent kinase) complexes combined with deacetylation of appropriate lysine residues within Rb. In the present study, using in vivo co-immunoprecipitation experiments, we identified mammalian SIRT1 (sirtuin 1) as a binding partner for Rb and its family members p107 and p130. Formation of Rb–SIRT1 complexes required the pocket domain of Rb. p300 catalysed the acetylation of Rb, and SIRT1 was a potent deacetylase for Rb. The ability of SIRT1 to catalyse the deacetylation of Rb was dependent on NAD and was inhibited by the SIRT1 inhibitor nicotinamide. Deacetylated lysine residues within Rb formed a domain similar to the SIRT1-targeted domain of the p53 tumour suppressor protein. Cultures of arrested cells, via contact inhibition or DNA damage, exhibited decreased Rb phosphorylation and increased Rb acetylation. Overexpression of SIRT1 in either confluent or etoposide-treated cells resulted in a significant reduction in Rb acetylation, which was restored with nicotinamide. Gene knockdown of SIRT1 by siRNA (short interfering RNA) produced an accumulation of acetylated Rb. This increase was augmented further when siRNA against SIRT1 was used in conjunction with nicotinamide. In conclusion, our results demonstrate that SIRT1 is an in vitro and in vivo deacetylase for the Rb tumour suppressor protein.
Keywords: acetylation, deacetylase, p300, p53, retinoblastoma protein (Rb), sirtuin 1 (SIRT1), silent information regulator 2 (Sir2)
Abbreviations: CBP, cAMP-response-element-binding protein-binding protein; CDK, cyclin-dependent kinase; DMEM, Dulbecco's modified Eagle's medium; E1A, early region 1A; HAT, histone acetyltransferase; HDAC, histone deacetylase; NP40, Nonidet P40; PCAF, p300/CBP-associated factor; Rb, retinoblastoma protein; RNAi, RNA interference; Sir2, silent information regulator 2; siRNA, short interfering RNA; SIRT1, sirtuin 1; TBST, Tris-buffered saline containing Tween 20; TSA, trichostatin A
INTRODUCTION
The tumour suppressor Rb (retinoblastoma protein) is a nuclear protein that functions to regulate the G1/S transition of the cell cycle through its interaction with the E2F family of transcription factors [1]. Not only does Rb inhibit E2F transcriptional activity, but it also actively represses transcription by recruiting chromatin-remodelling enzymes, such as HDACs (histone deacetylases), components of the human SWI2/SNF2 complex and methyltransferases, to E2F target gene promoters [2,3]. Promoters of cell-cycle genes, such as cyclins and CDKs (cyclin-dependent kinases), as well as proteins involved in DNA replication and synthesis, contain E2F sites and repression of these genes during growth suppression has been shown to be Rb-mediated [4].
Transient phosphorylation of Rb by G1-specific CDKs is the main mechanism by which Rb activity is regulated [5,6]. Hypo-phosphorylated Rb predominates in resting and early G1 cells. In response to mitogenic signals, CDK4/6, together with the D family of cyclins, triggers phosphorylation of Rb. Cyclin E, expressed in late G1, forms an active kinase with CDK2 to further phosphorylate Rb near the end of G1. Hyper-phosphorylated Rb causes dissociation of Rb–E2F complexes, enabling E2F-dependent transcription of genes that mediate S-phase entry.
Previous studies [7–10] indicate that Rb also undergoes acetylation as an additional mechanism of regulating Rb activity. Mutation of the lysine residues in the central B domain of Rb, which is conserved in the Rb family proteins, results in the loss of acetylation [7]. It is unknown what biological function acetylation of this series of lysine residues serves. Rb can also be acetylated at the C-terminus by p300, a member of the p300/CBP (cAMP-response-element-binding protein-binding protein) family of HAT (histone acetyltransferase) transcriptional co-activators [8]. This acetylation obstructs efficient phosphorylation of Rb, leading to the maintenance of Rb in its active hypo-phosphorylated form. As a result, cells are maintained in a growth-arrested state. Although the mechanism behind Rb acetylation is not fully understood, the viral oncoprotein E1A (early region 1A) can potentiate the process [8]. However, in cells that normally do not express E1A, there may be a similar cellular protein that functions to bridge p300 and Rb into a multiprotein complex to facilitate Rb acetylation. Studies into the identity of a cellular cofactor led to the discovery of PCAF (p300/CBP-associated factor), as an in vivo Rb-acetylating protein [9]. PCAF acetylates Rb to induce Rb-mediated terminal cell-cycle exit and expression of late myogenic genes [9]. In addition to PCAF, Leduc et al. [10] have shown that Tip 60, a MYST-related HAT, catalyses Rb acetylation to control Rb expression levels. Thus in vivo post-translational modification of Rb by acetylation reveals a new level of Rb regulation.
Protein acetylation is a reversible reaction, subject to deacetylation by numerous enzymes within the cell. One of these deacetylation enzymes, Sir2 (silent information regulator 2), was initially identified in yeast for the repression of mating type loci, telomeres and rDNA (ribosomal DNA) [11–14]. Sir2 is a class III HDAC that can deacetylate specific lysine residues of core histone proteins H3 and H4 to promote heterochromatin silencing [15]. Unlike the classic class I and II HDACs, Sir2 is regulated by the cofactor NAD+. In the absence of NAD+, Sir2 displays no deacetylase activity [15]. Sir2 is highly conserved, as homologues of Sir2 have been identified in organisms ranging from bacteria to humans [16]. Interestingly, mammalian Sir2, or SIRT1 (sirtuin 1), not only deacetylates histones [17], but also a number of non-histone proteins, including p53 [18–20], TAFI68 (TATA-box-binding protein-associated factor I 68) [21], PCAF/MyoD (myogenic differentiation) [22], Foxo (forkhead box O) [23,24], NF-κB (nuclear factor κB) [25] and PGC-1α (peroxisome-proliferator-activated receptor γ co-activator 1α) [26,27]. It is becoming increasingly clear that mammalian SIRT1 has roles in diverse biological processes involving heterochromatin silencing, differentiation, cell survival and metabolism.
In the present study, we investigated the role of SIRT1 in regulating Rb acetylation, and report that a functional interaction exists between SIRT1 and Rb. This interaction appeared to be conserved among all three Rb family members (i.e. Rb, p107 and p130). We confirmed in an in vitro assay that Rb is acetylated by p300. Using our Rb acetylation assay, we determined that Rb is a substrate for deacetylation by both mouse and human SIRT1. We observed a striking similarity in the p53 and Rb acetylation domains upon amino-acid-sequence analysis and demonstrated that an anti-(acetylated p53) antibody is able to detect acetylated Rb. Furthermore, we found that acetylated Rb increases in response to contact inhibition, and overexpression of SIRT1 reduced the levels of Rb acetylation in vivo. Additionally, overexpression of SIRT1 also diminished levels of DNA-damage-induced accumulation of acetylated Rb. To target endogenous cellular SIRT1, we performed knockdown experiments using siRNA (short interfering RNA). Silencing of SIRT1 by RNAi (RNA interference) caused an increase in Rb acetylation. Knockdown of both SIRT1 expression by SIRT1 siRNA and SIRT1 activity by nicotinamide resulted in an even greater level of Rb acetylation. These results lead us to propose a model in which active Rb is both acetylated and hypo-phosphorylated, and that SIRT1-mediated deacetylation is required to deactivate Rb.
EXPERIMENTAL
Cell culture
C33A (Rb-null) cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) fetal bovine serum, 2 mM glutamine, 0.1 mM non-essential amino acids and 1 mM sodium pyruvate at 37 °C in 6% CO2. BJT cells (BJ fibroblasts immortalized by ectopic expression of telomerase) and BJThSir2WT cells stably overexpressing human SIRT1 (hereafter referred to as BJTSIRT1 cells), generously provided by Dr Shin-ichiro Imai (Division of Biology and Biological Sciences, Washington University School of Medicine, St Louis, MO, U.S.A.), were grown in DMEM supplemented with 10% (v/v) fetal bovine serum at 37 °C in 5% CO2.
BJT and BJTSIRT1 cells were seeded in culture dishes and allowed to grow for 1 day beyond confluence for density-dependent growth inhibition. Cells growing asynchronously were collected from subconfluent cultures.
To induce DNA damage, BJT and BJTSIRT1 cells were treated with either DMSO (vehicle) or 70 μM etoposide for 18 h before they were harvested for experiments. For inhibition of SIRT1, BJT and BJTSIRT1 cells were treated with 20 μM etoposide and 5 mM nicotinamide for 18 h before they were harvested. DMSO, etoposide and nicotinamide were added directly to the tissue culture medium of BJT and BJTSIRT1 cells.
Plasmids
Mouse SIRT1 constructs pcDNA3mSir2wt and pcDNA3mSir2H355A (hereafter referred to FLAG–SIRT1 and FLAG–SIRT1H355A respectively) were generously provided by Professor Wei Gu (Institute for Cancer Genetics, Columbia University, New York, NY, U.S.A.) [18]. Full-length Rb plasmid was kindly provided by Professor J. William Harbour (Department of Ophthalmology and Visual Sciences, Washington University School of Medicine, St Louis, MO, U.S.A.). Full-length p107 and p130 plasmids were gifts from Dr Gustavo Leone (Department of Molecular Genetics, The Ohio State University, Columbus, OH, U.S.A.). Rb mutant constructs were provided by Dr Douglas C. Dean (Department of Ophthalmology, University of Louisville Health Sciences Center, Louisville, KY, U.S.A.).
Generation of the SIRT1 antibody
The SIRT1 antibody was generated by injection of a synthesized peptide representing the C-terminus of mouse SIRT1 protein into chickens (Aves Labs).
Transfections and immunoprecipitations
C33A cells were transfected by the calcium phosphate DNA precipitation method and lysed 48 h after transfection in lysis buffer [50 mM Hepes (pH 7.2), 5 mM EDTA, 0.5% NP40 (Nonidet P40) and 250 mM NaCl, or 50 mM Tris/HCl (pH 8.0), 5 mM EDTA, 0.5% NP40 and 150 mM NaCl] in the presence of a protease inhibitor cocktail (Roche). Rb, p107 and p130 were immunoprecipitated with anti-Rb (IF8; Santa Cruz Biotechnology), anti-p107 (C-18; Santa Cruz Biotechnology) and anti-p130 (3F327; USBiological) respectively. FLAG-tagged SIRT1 was immunoprecipitated with anti-FLAG M2 agarose-affinity gel (Sigma). GAL4-tagged large pocket Rb and pocket mutant proteins were immunoprecipitated with anti-GAL4 (RK5C1; Santa Cruz Biotechnology). Following sonication, samples were immunoprecipitated overnight at 4 °C and washed in lysis buffer. The protein concentration of cell lysates was determined using the Bradford assay (Bio-Rad Laboratories).
Western blot analysis
Collected immune complexes and cell lysates were resolved using SDS/PAGE [6, 8 or 10% (w/v) polyacrylamide gels or gradient 4–20% (w/v) Precise (Pierce) pre-cast polyacrylamide gels] and transferred on to nitrocellulose membranes (Osmonics). Membranes were blocked with 5% (w/v) non-fat dried milk in TBST [Tris-buffered saline containing Tween 20; 25 mM Tris/HCl (pH 8.0), 125 mM NaCl and 0.05% Tween 20, or 20 mM Tris/HCl (pH 7.6), 137 mM NaCl and 0.1% Tween 20] and were probed with the following antibodies: anti-SIRT1 (Aves Labs; also provided by Professor Wei Gu [18]), anti-Rb [C-15 (IF8; Santa Cruz Biotechnology); XZ55 (BD PharMingen) or G3-245 (BD PharMingen)], anti-p107 (C-18; Santa Cruz Biotechnology), anti-p130 [C-20 (Santa Cruz Biotechnology) or 3F327 (USBiological)], anti-(acetylated lysine residue) (Ac-K-103; Cell Signaling Technology), anti-[acetylated p53 (Lys382)] (Cell Signaling Technology), anti-p27 (C-19; Santa Cruz Biotechnology), anti-p53 (DO-1; Santa Cruz Biotechnology), anti-p21 (C-19; Santa Cruz Biotechnology) or anti-γ-tubulin (C-20; Santa Cruz Biotechnology) antibodies. HRP (horseradish peroxidase)-coupled goat anti-(chicken IgY) (Aves Labs), rabbit anti-(mouse IgG) (Sigma), donkey anti-(mouse IgG) (Jackson ImmunoResearch Laboratories), goat anti-(rabbit IgG) (Jackson ImmunoResearch Laboratories) or rabbit anti-(goat IgG) (Zymed) were used as secondary antibodies. Protein bands were visualized by autoradiography using ECL® Plus Western blotting detection reagents (Amersham Biosciences). Densitometry was performed using either Image J (National Institutes of Health) or ImageQuant TL (Amersham Biosciences) software.
Expression and purification of His6-tagged mouse SIRT1
Bacterial cells expressing His6-tagged full-length mouse SIRT1 as well as deacetylase-compromised SIRT1, kindly provided by Dr Shin-Ichiro Imai, were used to inoculate cultures containing 75 μg/ml kanamycin and 37 μg/ml chloramphenicol. IPTG (isopropyl β-D-thiogalactoside) was added to a final concentration of 0.1 mM to induce the expression of mouse SIRT1. Cells were resuspended in lysis buffer [20 mM Tris/HCl (pH 7.0), 50 mM NaCl, 1 mM EDTA, 1 mM imidazole, 1 mM DTT (dithiothreitol), 1 mM PMSF and protease inhibitor cocktail (Roche)], sonicated and incubated on ice in the presence of 1% (v/v) Triton X-100. Clarified lysates were incubated with Ni-NTA (Ni2+-nitrilotriacetate) Superflow resin (Qiagen). The resin was washed with buffer containing 20 mM Tris/HCl (pH 8.0), 300 mM NaCl, 10% (v/v) glycerol, 0.1% Triton X-100 and 1 mM imidazole. Proteins were eluted in wash buffer containing 350 mM imidazole. Protein concentration was determined by the Bradford assay (Bio-Rad Laboratories).
In vitro Rb acetylation/deacetylation assays
For Rb acetylation, 1 μg of Rb (unless otherwise indicated) and 0.3 μg of p300 HAT domain recombinant proteins (Upstate) were incubated together with unlabelled acetyl-CoA (0.33 mM; Sigma) and HAT assay buffer [25 mM Tris/HCl (pH 8.0), 2.5% (v/v) glycerol, 0.05 mM EDTA and 25 mM KCl] at 30 °C for 20, 40, 60 or 80 min, or overnight, in a final reaction volume of 30 μl. Reactions were stopped by adding SDS/PAGE sample buffer. Proteins were separated by SDS/PAGE on 10% (w/v) polyacrylamide gels and immunoblotted as described above. For Rb deacetylation, Rb acetylation reactions were allowed to proceed for the first 20 min, after which recombinant mouse SIRT1 or recombinant human SIRT1 (Biomol) was added (1, 10 or 20 μg of mouse SIRT1, and 0.05, 0.5 or 5 units of human SIRT1) in the presence or absence of 1 mM NAD (Sigma). TSA (trichostatin A; 5 μM; Upstate) was added to all of the reactions at this time as well as 50 mM nicotinamide (Sigma), where indicated. Reactions were then incubated at 30 °C for another 45 min before the addition of SDS/PAGE sample buffer.
SIRT1 siRNA nucleofections
BJT cells were nucleofected with either 3 μg of siRNA against human SIRT1 (Hs_SIRT1_3_HP; Qiagen) or control siRNA against luciferase (Dharmacon), according to the manufacturer's instructions (Amaxa Biosystems). Nucleofected cells were replated and harvested at subconfluency 24 h after incubation. For SIRT1-knockdown experiments with nicotinamide, 5 mM nicotinamide was added to the culture medium 5 h after nucleofected BJT cells were reseeded. Cells were also visually checked for adherence by standard microscopy prior to the addition of nicotinamide. Cells were harvested 19 h after nicotinamide treatment for a total of 24 h post-nucleofection.
RESULTS
SIRT1 interacts with Rb pocket proteins in vivo
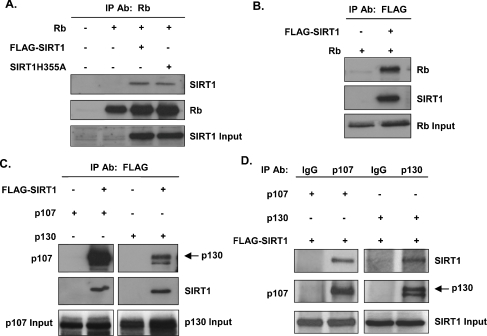
In order to identify a protein capable of deacetylating Rb in vitro and in vivo, we first sought to define interactions between Rb and known deacetylases. We began our studies with SIRT1, given the recent finding that Rb and SIRT1 interacted in a GST (glutathione transferase) pull-down assay (W. Gu, personal communication). Co-immunoprecipitation assays were done to determine whether Rb and murine SIRT1 interact in vivo. Rb-deficient C33A cells were co-transfected with Rb and FLAG–SIRT1 expression vectors. Cellular extracts from the transfected cells were immunoprecipitated with the anti-Rb antibody and the resulting immune complexes were collected and analysed by immunoblotting with the anti-SIRT1 antibody. Immunoprecipitation of Rb resulted in co-precipitation of FLAG–SIRT1 (Figure 1A). SIRT1 containing a point mutation that weakens its deacetylase activity also interacted with Rb, suggesting that catalytically active SIRT1 is not a requisite for the interaction (Figure 1A). Similarly, when the immunoprecipitation was performed with the anti-FLAG antibody, Rb was detected in the FLAG immunoprecipitate (Figure 1B), indicating that the interaction between Rb and SIRT1 can be detected by precipitating with antibodies against either protein.
Figure 1. Interaction of SIRT1 with all three Rb family proteins in vivo.
(A) Whole-cell lysates from transfected C33A cells were immunoprecipitated (IP) with the anti-Rb antibody. Complexes were resolved by SDS/PAGE, transferred on to nitrocellulose and immunoblotted using the anti-SIRT1 antibody (top panel). The blot was stripped and reprobed with the anti-Rb antibody for the presence of immunoprecipitated Rb (middle panel). Input protein representing 10% of the cell lysate is shown in the bottom panel. (B) The experiment was conducted as described in (A), except that immunoprecipitations were carried out using the anti-FLAG antibody and Western blotting with the anti-Rb antibody. (C) The experiment was conducted as described in (A and B), except that immunoprecipitations were carried out using the anti-FLAG antibody and Western blotting with either the anti-107 or anti-p130 antibody. (D) The experiment was conducted as described in (A), except that immunoprecipitations were carried out using control IgG, the anti-p107 antibody or the anti-p130 antibody and Western blotting with the anti-SIRT1 antibody. Expression of empty vector was used as a negative control for (A)–(C).
Rb-related proteins p107 and p130, together with Rb, make up the family of pocket proteins [28]. Rb, p107 and p130 share structural and biochemical similarities and have extensive overlapping functions. To determine whether the interaction between Rb and SIRT1 is conserved in p107 and p130 or unique to Rb, additional co-immunoprecipitations were performed. Cell lysates from p107- and p130-transfected C33A cells were immunoprecipitated with the anti-FLAG antibody and immunoblotted with the anti-p107 or anti-p130 antibody. As shown in Figure 1(C), SIRT1 interacted with both p107 and p130 in vivo. We also detected these interactions reciprocally by using the appropriate Rb family member antibody for the immunoprecipitations and the anti-SIRT1 antibody for Western blot analyses (Figure 1D).
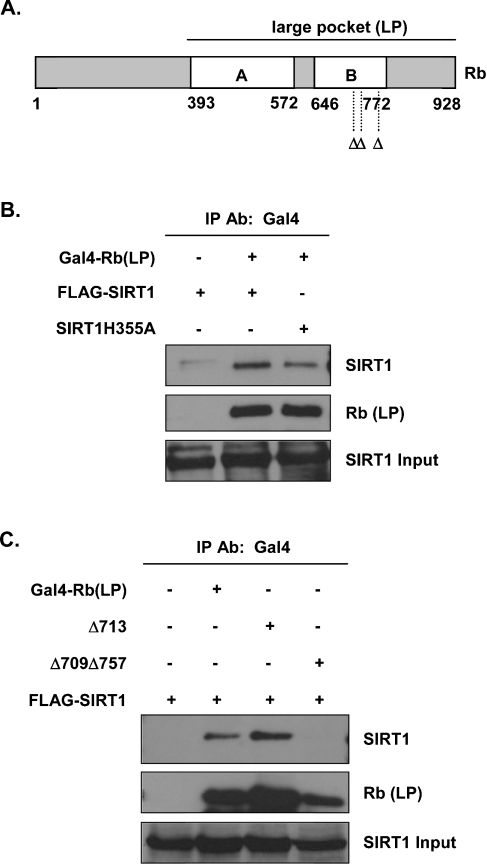
Rb contains three distinctive structural domains: the N-terminus, the central A/B pocket, which is conserved in p107 and p130, and the C-terminus [28] (Figure 2A). As the A/B pocket is required for the interaction of Rb with many of its associated proteins and for carrying out the repressor functions of Rb, we tested whether the A/B pocket domain was important for the interaction between Rb and SIRT1. Co-immunoprecipitation assays were performed using a GAL4-tagged large-pocket Rb expression vector (amino acids 379–928 of Rb) (Figure 2A). As shown in Figure 2(B), SIRT1 interacted with large-pocket Rb, suggesting that the N-terminal region of Rb is dispensable for the interaction. Similarly, mutant SIRT1 also interacted with large-pocket Rb (Figure 2B), reinforcing the idea that the interaction of SIRT1 with Rb does not require active SIRT1.
Figure 2. Interaction of SIRT1 with the pocket region of Rb in vivo.
(A) Schematic representation of large-pocket Rb and mutations in the pocket region of Rb. Δ indicates mutations in the LXCXE Rb-binding site (residues 709, 713 and 757). (B) Whole-cell lysates from transfected C33A cells were immunoprecipitated (IP) with the anti-GAL4 antibody. Complexes were resolved by SDS/PAGE, transferred on to nitrocellulose and immunoblotted using the anti-SIRT1 antibody (top panel). The blot was stripped and reprobed with the anti-Rb antibody for the presence of immunoprecipitated Rb (middle panel). Input protein representing 10% of the cell lysate is shown in the bottom panel. (C) C33A cells were transfected with the indicated expression vectors. The experiment was performed as described in (B).
Many Rb-binding proteins harbour a critical LXCXE (LeuXaaCysXaaGlu) motif. This motif makes important contacts with several conserved amino acid residues in the A/B pocket of Rb as determined by crystal structure [29] and, as such, mutation of either the LXCXE motif in target proteins or in the A/B pocket of Rb results in the loss of Rb–target interaction. When constructs containing mutations in the LXCXE Rb-binding site (RbΔ713) were used in co-immunoprecipitation assays, we found no decrease in the formation of Rb–SIRT1 complexes (Figure 2C). However, a double mutation in the pocket, RbΔ709/Δ757, completely abolished this interaction (Figure 2C), indicating that a functional pocket domain of Rb is required for its interaction with SIRT1. Of note, we were unable to locate an appropriate LXCXE or LXCXE-like sequence within the coding region of either mouse or human SIRT1, suggesting that the interaction of SIRT1 with the pocket domain of Rb is unique in that it does not involve a classic Rb-binding site in SIRT1. Nonetheless, our results show that SIRT1 interacts with all three Rb family members in vivo and implicate Rb as a potential SIRT1 target for deacetylating activity.
Rb is acetylated by p300 in vitro
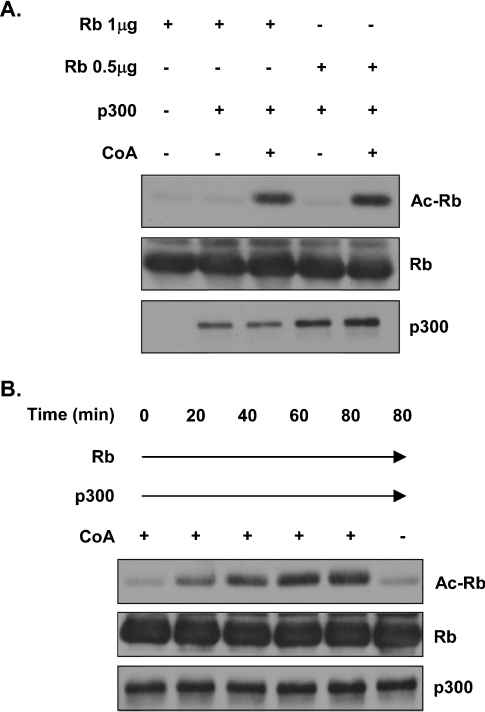
In order to study the deacetylation of Rb, we first developed a non-radioactive in vitro Rb acetylation assay. A recombinant C-terminal fragment of Rb containing the main sites of acetylation [8] was incubated with recombinant p300 and unlabelled acetyl-CoA. After incubation, the reactions were resolved by SDS/PAGE and immunoblotted with an anti-(acetylated lysine residue) antibody. Results of these in vitro experiments showed that Rb was indeed acetylated by p300 and that the amount of recombinant Rb was not limiting in the reaction (Figure 3A). Additionally, we established a time course of Rb acetylation by p300. Recombinant Rb was acetylated by p300 in vitro in increasing amounts over 60 min and remained unchanged up to 80 min, indicating that acetylation of Rb in this reaction is maximal at 60 min (Figure 3B).
Figure 3. Rb acetylation by p300 in vitro.
(A) Recombinant Rb and p300 were incubated together in the presence or absence of unlabelled acetyl-CoA (CoA). After incubation, proteins were separated by SDS/PAGE and analysed by Western blotting. Acetylation of Rb (Ac-Rb) was detected using an anti-(acetylated lysine residue) antibody (top panel). The blot was stripped and reprobed with the anti-Rb antibody to verify the presence of an overlapping Rb band (middle panel). (B) Acetylation of Rb by p300 is time-dependent. Rb acetylation reactions were carried out as described in (A), but with increasing incubation times, as indicated. Acetylated Rb and Rb were detected as described in (A).
Sequence homology is present at the acetylation sites of Rb and p53
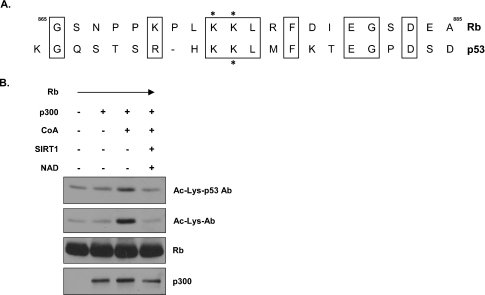
Mouse and human SIRT1 bind and deacetylate the tumour suppressor protein p53 [18–20]. In particular, SIRT1 preferentially deacetylates Lys382 of p53 thereby reducing the transcriptional activity of p53 [19]. Through in vitro acetylation studies of mutant derivatives, acetylation of Rb was mapped to a 87-residue region at the C-terminus [8]. Further analysis identified Lys873 and Lys874 to be acetylated and important in blocking Rb phosphorylation [8]. Given the ability of both Rb and p53 to act as gatekeepers of the cell cycle, we hypothesized that their acetylation, and by inference their deacetylation, might somehow be regulated in a similar fashion. In fact, Rb and p53 are both acetylated by p300 [8,30–32]. We speculated that Rb and p53 might share a common target consensus sequence for SIRT1-mediated deacetylation. To address this possibility, we performed a sequence alignment of amino acid residues flanking the acetylation sites of Rb and p53. Indeed, Rb and p53 share a surprising amount of similarity in their acetylation domains (Figure 4A). We reasoned that the amount of conservation seen in this acetylation motif might be observed experimentally through the use of antibodies recognizing acetylated moieties. Recombinant Rb was used as a substrate for p300-directed acetylation, and acetylated Rb was visualized with antibodies recognizing either acetylated lysine residues or those raised against acetylated Lys382 on p53. Using the antibody recognizing acetylated Lys382 on p53, we were able to detect acetylated recombinant Rb (Figure 4B, top panel), demonstrating that the p53-specific antibody cross-reacts with acetylated Rb. Blots were stripped and reprobed with antibodies recognizing acetylated lysine residues to confirm the acetylation of recombinant Rb (Figure 4B, upper-middle panel). Given that SIRT1 has been shown to deacetylate this conserved lysine residue on p53 proteins, we added recombinant SIRT1 proteins to our Rb acetylation reactions. Consistent with the ability of SIRT1 to deacetylate p53 at its C-terminal acetylation site, we found that human SIRT1 could also deacetylate Rb, presumably at this homologous lysine motif, as indicated by the loss of signal seen with the antibody recognizing acetylated Lys382 on p53 (Figure 4B, lane 4).
Figure 4. Similarity in p53 and Rb acetylation domains.
(A) Sequence homology is present at the acetylation sites of p53 and Rb. Formatted alignment between Rb and p53 at their acetylation sites was carried out, with similar identities between the two proteins being boxed. Acetylated lysine residues in Rb and the acetylated lysine residue in p53 that is deacetylated by SIRT1 are identified by asterisks. (B) The anti-[acetylated p53 (Lys382)] antibody recognizes acetylated Rb in vitro. Recombinant Rb and p300 were incubated together in the presence or absence of unlabelled acetyl-CoA (CoA). After incubation, proteins were separated by SDS/PAGE and analysed by Western blotting. Acetylated Rb was detected using the anti-[acetylated p53 (Lys382)] antibody (Ac-Lys-p53 Ab) first, after which the blot was stripped and reprobed with an anti-(acetylayed lysine residue) antibody (Ac-Lys-Ab). The blot was then stripped again and reprobed with the anti-Rb antibody to detect the presence of an overlapping Rb band.
Rb is a substrate for deacetylation by SIRT1 in vitro
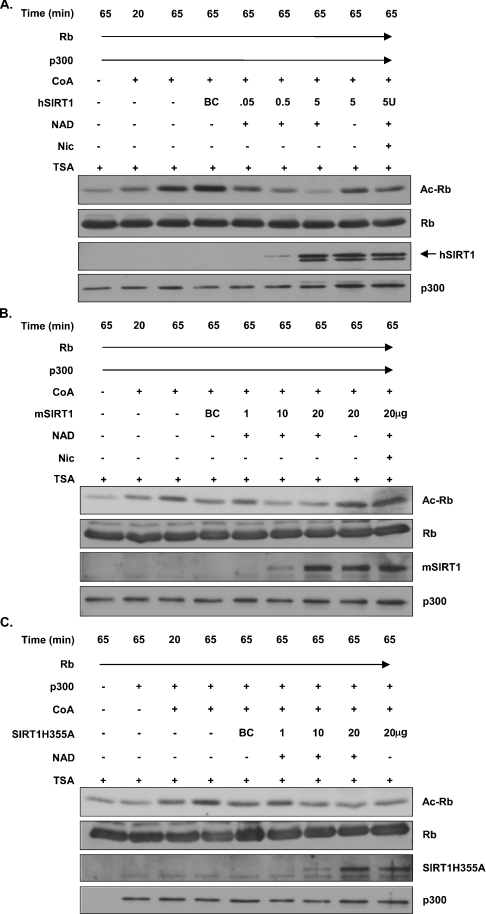
To address the specificity and dynamics of Rb deacetylation by SIRT1, we performed additional in vitro Rb acetylation assays under various conditions that affect SIRT1 activation. First, we acetylated Rb using recombinant p300 together with acetyl CoA before adding recombinant mouse or human SIRT1 to the acetylation reactions. We included three controls to ensure that the deacetylation was SIRT1-dependent. (i) Reactions were carried out in the presence or absence of NAD, since the deacetylase activity of Sir2 is tightly coupled to NAD hydrolysis [33–35]. (ii) Reactions were performed in the presence of TSA, a potent HDAC inhibitor [36]. The deacetylase activity associated with Sir2 is reportedly TSA-insensitive [15]. (iii) Reactions were treated with nicotinamide, a product of Sir2-mediated NAD-dependent deacetylation that significantly compromises the deacetylase activity of Sir2 [18,19]. Rb acetylation was gradually reduced with increasing amounts of either human or mouse SIRT1 respectively (Figures 5A and 5B). This reduction in Rb acetylation was dependent upon the presence of NAD and was not sensitive to TSA (Figures 5A and 5B). In the presence of the SIRT1 inhibitor nicotinamide, the Rb acetylation signal was enhanced (Figures 5A and 5B), demonstrating a requirement for active SIRT1 in the in vitro deacetylation of Rb. As an additional control, similar in vitro Rb acetylation/deacetylation assays were run using a mutant form of SIRT1 known to have compromised deacetylase activity [18]. We observed a slight reduction in the Rb acetylation signal when mutant SIRT1 was added (Figure 5C), which is consistent with the idea that, although this mutant is mostly deficient, it still possesses approx. 10% of wild-type deacetylase activity in vitro (S.-i. Imai, personal communication).
Figure 5. NAD-dependent deacetylation of Rb by SIRT1 in vitro.
(A) Rb acetylation reactions were allowed to proceed for the first 20 min, after which either buffer or recombinant human SIRT1 was added in the presence or absence of NAD. TSA was added to all reactions at this time as well as nicotinamide (Nic), where indicated. Samples were then incubated for another 45 min before they were resolved by SDS/PAGE and analysed by Western blotting. BC, buffer control; U, unit. (B) Reactions were carried out as described in (A), except that recombinant mouse SIRT1 was used. (C) Reactions were carried out as described in (A), except that recombinant mouse SIRT1 with a mutation in the catalytic domain (SIRT1H355A) was used.
Rb is a substrate for SIRT1-mediated deacetylation in vivo
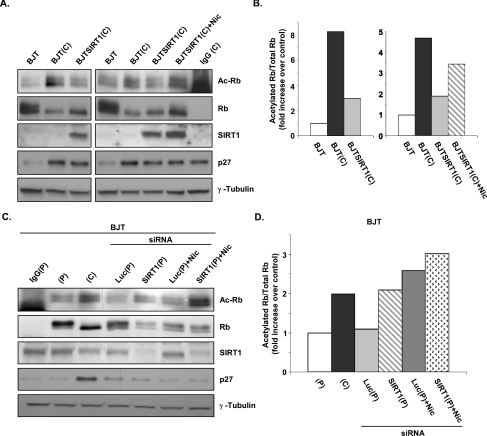
We next investigated whether SIRT1 mediates deacetylation of Rb in vivo. As active repression by the Rb–E2F complex has been shown to mediate G1 arrest triggered by contact inhibition [37], we hypothesized that Rb acetylation might be regulated in this context. In this setting, Rb is maintained in a hypo-phosphorylated state, and acetylation of Rb might serve to further activate Rb molecules. Cells from subconfluent and densely populated cultures were harvested, immunoprecipitated with anti-Rb antibodies and immunoblotted with antibodies recognizing acetylated lysine residues. We observed a significant increase in acetylated Rb after cells were allowed to grow beyond confluence, when hypo-phosphorylated Rb was prevalent (Figures 6A and 6B). Expression of p27, a cell-cycle factor connected to density-dependent inhibition of growth [38], was up-regulated in contact-inhibited cells (Figure 6A). Consistent with our in vitro findings, the levels of acetylated Rb were markedly lower in cells stably overexpressing human SIRT1 (Figures 6A and 6B, lanes 3 and 6), demonstrating the ability of SIRT1 to deacetylate Rb in vivo. Inhibition of SIRT1 by nicotinamide in BJTSIRT1 cells returned acetylated Rb levels to nearly those seen in confluent cultures (Figures 6A and 6B, lane 7). Rb remained in a hypo-phosphorylated state regardless of SIRT1 activity due to the lack of cyclin–CDK activity in confluent quiescent cultures [39,40]. These observations indicate that, in response to density-dependent cell-cycle arrest, Rb is acetylated and that the expression of SIRT1 can reverse contact-induced Rb acetylation.
Figure 6. Deacetylation of Rb by SIRT1 in vivo following density-dependent growth inhibition.
(A) Proliferating and contact-inhibited BJT and BJTSIRT1 cells were harvested and lysed, and proteins were immunoprecipitated with the anti-Rb antibody. Complexes were resolved by SDS/PAGE and analysed by Western blotting using the appropriate antibodies. Acetylated Rb was detected using an anti-(acetylated lysine residue) antibody. The blots were stripped and reprobed with the anti-Rb antibody for the presence of an overlapping Rb band (top two panels). (B) Densitometry of acetylated Rb and total Rb bands from (A) was conducted using Image J software. The ratios of acetylated Rb to total Rb were calculated and the fold increase over control (proliferating BJT cells) is shown. (C) SIRT1 knockdown by RNAi. BJT cells were nucleofected with either SIRT1 siRNA or control luciferase siRNA. Nicotinamide (Nic.) was added, where indicated, 5 h after cells were nucleofected with siRNA and replated. Cells were harvested for Rb immunoprecipitations 24 h post-nucleofection and Western blotted using the same antibodies described in (A). (P), proliferating; (C), contact-inhibited. (D) Densitometry of acetylated Rb and total Rb bands from (C) was performed using ImageQuant TL software. The ratios of acetylated Rb to total Rb were calculated and the fold increase over control (proliferating BJT cells) is shown.
As the in vivo data generated involved SIRT1 overexpression, we performed knockdown experiments using siRNA to target endogenous cellular SIRT1. We initially tested different pre-designed SIRT1 siRNA duplexes in BJTSIRT1 cells and identified one that induced SIRT1 knockdown as early as 24 h after nucleofection (results not shown). As expected, control siRNA against luciferase did not alter SIRT1 expression levels (results not shown). To determine whether SIRT1 is a deacetylase for Rb at endogenous protein levels, we nucleofected BJT cells with either SIRT1 siRNA or control luciferase siRNA and harvested them while they were growing asynchronously. We chose to assess acetylation of Rb in cycling cells rather than in contact-inhibited cells. As the signal for acetylated Rb is typically not robust, we reasoned that our chances of detecting a change were better in proliferating cells where the levels of acetylated Rb are still within the linear range. In contrast, acetylated Rb levels would already be high in cells that are contact-inhibited and mostly probably be close to, or outside of, the linear range. As such, a change where an even greater quantity of acetylated Rb is expected from the SIRT1 knockdown would be difficult to observe. As shown in Figures 6(C) and 6(D), luciferase siRNA had no effect on Rb acetylation, whereas introduction of SIRT1 siRNA resulted in an enhancement of acetylated Rb. Treatment with nicotinamide alone likewise increased the levels of acetylated Rb. However, in combination with SIRT1 siRNA, a higher level of acetylated Rb was achieved than with either SIRT1 RNAi or nicotinamide alone. These results demonstrate that, under physiological conditions, blocking SIRT1 protein expression and activity increases the pool of acetylated Rb.
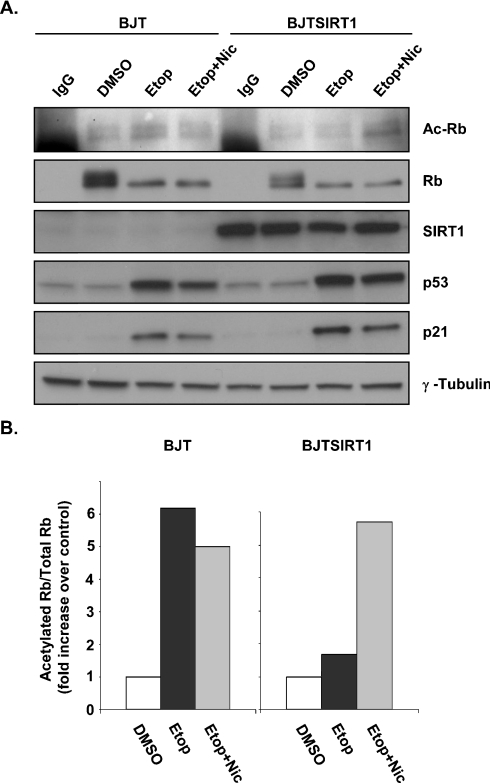
A recent study has shown that Rb is acetylated in response to DNA damage [41]. NIH-3T3 fibroblasts treated with etoposide, a chemotherapeutic drug that generates DNA double-strand breaks, lead to an accumulation of acetylated Rb. Therefore we decided to evaluate Rb deacetylation under DNA-damaging conditions. As expected, treating BJT cells with etoposide yielded elevated levels of acetylated Rb (Figure 7A, lane 3, and Figure 7B, lane 2). In contrast, etoposide-treated BJTSIRT1 cells did not exhibit an increase in Rb acetylation, suggesting that Rb was undergoing continual SIRT1-mediated deacetylation in these cells (Figure 7A, lane 7, and Figure 7B, lane 5). Although there was a slight reduction in acetylated Rb levels when etoposide was added in combination with nicotinamide, an accumulation of acetylated Rb in BJTSIRT1 cells treated with both agents was observed (Figure 7A, lanes 4 and 8, and Figure 7B, lanes 3 and 6). These results suggest that acetylated Rb induced by DNA damage can be deacetylated by SIRT1.
Figure 7. Deacetylation of Rb by SIRT1 in vivo during DNA damage.
(A) BJT and BJTSIRT1 cells were treated with etoposide (Etop) alone or together with nicotinamide (Nic) for 18 h and analysed as described in Figure 6(A). (B) Densitometry results for (A) were calculated as described in Figure 6(B) and represent the fold increase over control (DMSO).
DISCUSSION
Mammalian SIRT1 is a protein deacetylase that metabolizes NAD to mediate its physiological functions. Unlike yeast Sir2, not all targets of SIRT1 are histone proteins, as a growing number of studies reveal that SIRT1 regulates non-histone protein deacetylation. In the present study, we have identified SIRT1 as an Rb deacetylase and provide evidence for SIRT1 being a novel link between the Rb and p53 pathways in response to cell-cycle arrest cues. Rb is acetylated by p300 HAT [8], and we have confirmed this modification via an in vitro acetylation assay in which Rb is acetylated only in the presence of acetyl-CoA. We utilized this Rb acetylation assay to demonstrate that mouse and human SIRT1 can mediate Rb deacetylation. A gradual reduction in acetylated Rb was seen with increasing levels of SIRT1. Several supplementary observations indicate that SIRT1 is a bona fide deacetylase for Rb. First, SIRT1 is unable to reduce levels of Rb acetylation when NAD is omitted. Secondly, exposure to TSA, a specific inhibitor of HDAC, does not affect the Rb deacetylation activity of SIRT1. Thirdly, addition of nicotinamide inhibits SIRT1 deacetylation of Rb. Combined, these results argue that Rb is a substrate for a NAD-dependent and nicotinamide-sensitive deacetylase. Our present findings appear to mirror those seen in the regulation of p53 deacetylation. In fact, we found significant sequence homology at the acetylation sites of Rb and p53. Given that the p53 acetylation site has been shown to be deacetylated by SIRT1 in a NAD-dependent and nicotinamide-sensitive manner, our findings suggest that SIRT1 might recognize this conserved motif on both proteins to orchestrate their deacetylation. Recognition of acetylated Rb by a specific antibody that detects acetylated Lys382 of p53 strengthens this possibility.
We have also shown that Rb and SIRT1 interact in vivo and have found that SIRT1 interacts with additional Rb family members, p107 and p130. However, the N-terminus of Rb is not required for SIRT1 binding. SIRT1 appears to interface with the classical binding site of Rb (utilized by LXCXE-containing proteins), as the double-pocket mutant RbΔ709/Δ757 failed to bind SIRT1. Unexpectedly, we did not find an LXCXE-binding motif in the amino acid sequences of either mouse or human SIRT1, although not all Rb-binding proteins require an LXCXE-binding motif. E2F transcription factors and HDAC3, for example, lack an LXCXE site, yet bind Rb [42–44]. Our results suggest that SIRT1 uses a different structural motif that requires multiple contacts to the pocket domain of Rb.
In order to determine whether SIRT1 deacetylated Rb in vivo, we needed a physiological setting whereby Rb is acetylated. Rb has been shown to be acetylated under several conditions [8,9,41]. Accumulation of hypo-phosphorylated Rb coincides with Rb acetylation in these studies, implying that activation of Rb might consist of both a lack of Rb phosphorylation and increased acetylation events. It has been argued that acetylation of Rb prevents the hyper-phosphorylation of Rb by cyclin–CDK complexes [8]. If this model is correct, then a stimulus that increases Rb activity (hypo-phosphorylation) should also exhibit increased Rb acetylation. In the present study, we found increased Rb acetylation following density-dependent growth inhibition, a stimulus known to elicit active Rb [45]. This provided us with a means by which to study the effect of SIRT1 on Rb acetylation in vivo. Our results indicate that Rb is a substrate for SIRT1-mediated deacetylation in vivo. In SIRT1-overexpressing cells, we observed that the levels of acetylated Rb were diminished after contact-induced cell-cycle arrest. In contrast, there was a noticeable increase in acetylated Rb when cells were treated with the SIRT1 inhibitor nicotinamide, demonstrating a requirement for SIRT1 activity in the deacetylation of Rb in vivo. The fact that Rb remains in a hypo-phosphorylated state following deacetylation by SIRT1 indicates that SIRT1 expression is unable to stimulate cell-cycle progression or, more specifically, CDK activation in quiescent cells.
In corroboration of our findings, experiments utilizing SIRT1 siRNA show there was more acetylated Rb in cells lacking endogenous SIRT1. In the presence of SIRT1 siRNA and the SIRT1 antagonist nicotinamide, the increase in the pool of acetylated Rb was augmented further, suggesting that deacetylation of Rb in vivo requires translated SIRT1 proteins that are catalytically competent.
DNA-damage-induced Rb acetylation also presented us with another opportunity to assess the effect of SIRT1 on Rb acetylation in vivo. Our results in this setting recapitulated what we observed in contact-inhibited cells, lending credence to the notion that SIRT1 could concurrently regulate both Rb and p53 pathways.
When the immunoblots were probed with the anti-(acetylated lysine residue) antibody, we observed a doublet that overlapped with the hyper- and hypo-phosphorylated forms of Rb, suggesting that Rb can be both hyper-phosphorylated and acetylated. Although the significance of this is not currently understood, it does appear to weaken the notion that hyper-phosphorylated Rb is more resistant to acetylation. The existence of other Rb acetylases that target different lysine residues, the acetylation of which elicits discrete biological outcomes, may also account for this observation. Tip60, a member of the MYST family of HATs, acetylates Rb and stimulates its proteasomal degradation [10]. Since both contact-inhibition and DNA damage require the stabilization of active hypo-phosphorylated Rb, it might be proposed that any excess hyper-phosphorylated Rb undergoes Tip60-mediated acetylation and subsequent degradation by the proteasome. Additionally, it is possible that this population of Rb triggers a separate cellular function yet to be identified.
The G1 arrest caused by cell–cell contact inhibition induces an accumulation of hypo-phosphorylated Rb [38] and requires the Rb–E2F complex to actively repress the transcription of cell-cycle genes [37]. Rb is also required in imposing G1 arrest in response to DNA damage [46–48]. Our present results suggest that Rb is not only hypo-phosphorylated in establishing G1 arrest, but is also acetylated. We propose that once growth arrest is reversed, when cells are released from density-dependent growth arrest or DNA damage is repaired, acetylated Rb is deacetylated by SIRT1, ensuring that the cell cycle recommences. Specifically, deacetylated Rb can now be phosphorylated to relieve Rb-mediated repression of E2F-regulated cell-cycle genes. In this model, SIRT1 would negatively regulate Rb and act as a switch to restart the cell cycle in combination with CDK activities.
Acknowledgments
We thank Wei Gu, Douglas Dean, J. William Harbour, Gustavo Leone and Shin-ichiro Imai for providing reagents. We are grateful to Helen Piwnica-Worms, Shin-ichiro Imai, J. William Harbour, Howard McLeod and Sheila Stewart for their numerous helpful suggestions, and members of the Weber laboratory for advice. This work was supported by the Pew Scholars Program in Biomedical Sciences (to J. D. W.).
References
- 1.Dimova D. K., Dyson N. J. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 2.Harbour J. W., Dean D. C. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 3.Zhu L. Tumour suppressor retinoblastoma protein Rb: a transcriptional regulator. Eur. J. Cancer. 2005;41:2415–2427. doi: 10.1016/j.ejca.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg R. A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 6.Mittnacht S. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 7.Brown V. D., Gallie B. L. The B-domain lysine patch of pRB is required for binding to large T antigen and release of E2F by phosphorylation. Mol. Cell. Biol. 2002;22:1390–1401. doi: 10.1128/mcb.22.5.1390-1401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan H. M., Krstic-Demonacos M., Smith L., Demonacos C., La Thangue N. B. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 2001;3:667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen D. X., Baglia L. A., Huang S. M., Baker C. M., McCance D. J. Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. EMBO J. 2004;23:1609–1618. doi: 10.1038/sj.emboj.7600176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leduc C., Claverie P., Eymin B., Col E., Khochbin S., Brambilla E., Gazzeri S. p14ARF promotes RB accumulation through inhibition of its Tip60-dependent acetylation. Oncogene. 2006;25:4147–4154. doi: 10.1038/sj.onc.1209446. [DOI] [PubMed] [Google Scholar]
- 11.Rine J., Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 13.Bryk M., Banerjee M., Murphy M., Knudsen K. E., Garfinkel D. J., Curcio M. J. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- 14.Smith J. S., Boeke J. D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 15.Imai S., Armstrong C. M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 16.Brachmann C. B., Sherman J. M., Devine S. E., Cameron E. E., Pillus L., Boeke J. D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 17.Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 19.Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 20.Langley E., Pearson M., Faretta M., Bauer U. M., Frye R. A., Minucci S., Pelicci P. G., Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muth V., Nadaud S., Grummt I., Voit R. Acetylation of TAFI68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fulco M., Schiltz R. L., Iezzi S., King M. T., Zhao P., Kashiwaya Y., Hoffman E., Veech R. L., Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 23.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 24.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 25.Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoto S., Fergusson M. M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 27.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 28.Classon M., Dyson N. p107 and p130: versatile proteins with interesting pockets. Exp. Cell Res. 2001;264:135–147. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- 29.Lee J. O., Russo A. A., Pavletich N. P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 30.Gu W., Roeder R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 31.Sakaguchi K., Herrera J. E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C. W., Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraham J., Kelly J., Thibault P., Benchimol S. Post-translational modification of p53 protein in response to ionizing radiation analyzed by mass spectrometry. J. Mol. Biol. 2000;295:853–864. doi: 10.1006/jmbi.1999.3415. [DOI] [PubMed] [Google Scholar]
- 33.Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner K. G., Landry J., Sternglanz R., Denu J. M. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanny J. C., Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. U.S.A. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida M., Kijima M., Akita M., Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 37.Zhang H. S., Postigo A. A., Dean D. C. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFβ, and contact inhibition. Cell. 1999;97:53–61. doi: 10.1016/s0092-8674(00)80714-x. [DOI] [PubMed] [Google Scholar]
- 38.Polyak K., Kato J. Y., Solomon M. J., Sherr C. J., Massague J., Roberts J. M., Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 39.Sherr C. J. The ins and outs of RB: coupling gene expression to the cell cycle clock. Trends Cell Biol. 1994;4:15–18. doi: 10.1016/0962-8924(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 40.Weber J. D., Raben D. M., Phillips P. J., Baldassare J. J. Sustained activation of extracellular-signal-regulated kinase 1 (ERK1) is required for the continued expression of cyclin D1 in G1 phase. Biochem. J. 1997;326:61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markham D., Munro S., Soloway J., O'Connor D. P., La Thangue N. B. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;7:192–198. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helin K., Lees J. A., Vidal M., Dyson N., Harlow E., Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 43.Shan B., Durfee T., Lee W. H. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 1996;93:679–684. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emiliani S., Fischle W., Van Lint C., Al-Abed Y., Verdin E. Characterization of a human RPD3 ortholog, HDAC3. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2795–2800. doi: 10.1073/pnas.95.6.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sherr L. Adherence: sticking to the evidence. AIDS Care. 2000;12:373–375. doi: 10.1080/09540120050123765. [DOI] [PubMed] [Google Scholar]
- 46.Harrington E. A., Bruce J. L., Harlow E., Dyson N. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11945–11950. doi: 10.1073/pnas.95.20.11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shapiro G. I., Edwards C. D., Ewen M. E., Rollins B. J. p16INK4A participates in a G1 arrest checkpoint in response to DNA damage. Mol. Cell. Biol. 1998;18:378–387. doi: 10.1128/mcb.18.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brugarolas J., Moberg K., Boyd S. D., Taya Y., Jacks T., Lees J. A. Inhibition of cyclin-dependent kinase 2 by p21 is necessary for retinoblastoma protein-mediated G1 arrest after γ-irradiation. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1002–1007. doi: 10.1073/pnas.96.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]