Abstract
The lipid second messenger ceramide regulates the rate of β cleavage of the Alzheimer's disease APP (amyloid precursor protein) by affecting the molecular stability of the β secretase BACE1 (β-site APP cleaving enzyme 1). Such an event is stimulated in the brain by the normal process of aging, and is under the control of the general aging programme mediated by the insulin-like growth factor 1 receptor. In the present study we report that BACE1 is acetylated on seven lysine residues of the N-terminal portion of the nascent protein. This process involves lysine acetylation in the lumen of the ER (endoplasmic reticulum) and is followed by deacetylation in the lumen of the Golgi apparatus, once the protein is fully mature. We also show that specific enzymatic activities acetylate (in the ER) and deacetylate (in the Golgi apparatus) the lysine residues. This process requires carrier-mediated translocation of acetyl-CoA into the ER lumen and is stimulated by ceramide. Site-directed mutagenesis indicates that lysine acetylation is necessary for nascent BACE1 to leave the ER and move ahead in the secretory pathway, and for the molecular stabilization of the protein.
Keywords: Alzheimer's disease, β-site APP cleaving enzyme 1 (BACE1), ceramide, lysine acetylation, post-translational regulation
Abbreviations: APP, amyloid precursor protein; BACE1, β-site APP cleaving enzyme 1; CBP, cAMP-response-element-binding protein-binding protein; CHO, Chinese hamster ovary; DTT, dithiothreitol; Endo H, endoglycosidase H; ER, endoplasmic reticulum; ERAD, ER-associated degradation; HAT, histone acetyltransferase; IGF1-R, insulin-like growth factor 1 receptor; UGGT, UDP-glucose:glycoprotein glucosyltransferase
INTRODUCTION
Even though the native conformation of a protein lies encoded in its primary amino acid sequence, the efficiency of folding itself is greatly enhanced by the ER (endoplasmic reticulum) through different complex systems that require chaperones that protect nascent proteins and enzymes that modify, either temporally or definitively, the protein [1]. The first system includes BiP and calnexin, whereas the second includes OST (oligosaccharyltransferase) and UGGT (UDP-glucose:glycoprotein glucosyltransferase), which re-attaches one glucose residue to the improperly folded nascent glycoprotein and regulates its interaction with the lectin chaperone calnexin [2,3]. Once folding is completed, the nascent protein dissociates from the calnexin/calreticulin cycle and moves ahead in the secretory pathway. In contrast, proteins that have not successfully reached an appropriate folding status, are removed through ERAD (ER-associated degradation) [4]. Therefore the half-life of a membrane protein is highly affected by the ability to fold correctly and leave the ER successfully. The lack of the appropriate post-translational modification can make the protein fail quality control and be directed toward ERAD for final degradation [1].
Reversible acetylation of lysine residues was originally identified in histones and then extended to transcription factors [5,6]. During the last few years a growing number of cytosolic and nuclear proteins have been reported as targets of lysine acetylation [6]. Being a novel form of post-translational modification, the purpose of lysine acetylation is still mostly unknown. Recognized functions include regulation of activity, and molecular stabilization of a protein [6]. From the biochemical point of view, acetylation of a protein requires three components: (i) an appropriate acceptor [a given protein with the appropriate lysine residue(s)]; (ii) a donor of the acetyl group (acetyl-CoA); and (iii) an enzyme able to transfer the acetyl group from the donor to the acceptor [an acetyl-CoA:lysine acetyltransferase (or simply called acetyltransferase)]. The above components have so far been identified only in the cytoplasm and lysine acetylation has been regarded solely as a cytoplasmic event [5,6].
Our group has shown that the lipid second messenger ceramide regulates the rate of β cleavage of the APP (amyloid precursor protein) by affecting the molecular stability of the BACE1 (β-site APP cleaving enzyme 1) [7]. Such an event is stimulated in the brain by the normal process of aging [8] and is under the control of the general aging programme mediated by the IGF1-R (insulin-like growth factor 1 receptor) [9]. In the present study we report that BACE1 is acetylated on seven different lysine residues, all facing the N-terminal portion of the nascent protein. This process involves lysine acetylation in the lumen of the ER and is followed by deacetylation in the lumen of the Golgi apparatus once the nascent protein is fully mature. We also show that a dual-enzymatic machinery acts in the ER and Golgi apparatus respectively to acetylate and deacetylate the lysine residues. In addition, this process requires carrier-mediated translocation of acetyl-CoA into the lumen of the ER and is stimulated by the lipid second messenger ceramide. Finally, site-directed mutagenesis shows that the transient acetylation of BACE1 is required for the nascent protein to leave the ER and proceed toward the secretory pathway.
MATERIALS AND METHODS
Cell cultures and treatment, cell lysis and sample preparation, BACE1 immunoprecipitation, Endo H (endoglycosidase H) digestion, site-directed mutagenesis, mRNA quantitation, disorder-prediction analysis, protein digestion and LC-MS/MS analysis are described in Supplementary methods at http://www.BiochemJ.org/bj/407/bj4070383add.htm.
Antibodies and Western blot analysis
Western blotting was performed on 10% Bis/Tris, 4–12% Bis/Tris or 7% Tris/Acetate SDS/PAGE systems (NuPAGE; Invitrogen) as described previously [7–10]. The following antibodies were used in the present study: anti-BACE1 N-terminal (monoclonal; R&D Systems); anti-BACE1 C-terminal (polyclonal; Abcam); anti-pro-BACE1 (polyclonal; Abcam); anti-acetylated lysine (monoclonal; Abcam); anti-calreticulin (ER marker, polyclonal; Abcam); anti-58 K Golgi/formiminotransferase cyclodeaminase (Golgi marker, polyclonal; Abcam); anti-syntaxin (Golgi marker, monoclonal; Abcam); anti-EEA1 (early endosome antigen 1, endosomes, monoclonal; BD Transduction Laboratories); anti-actin (polyclonal; Cell Signaling). Secondary antibodies (Amersham) were used at a 1:6000 dilution. Binding was detected by chemiluminescence (LumiGLO kit; KPL). Only a representative blot of at least three different experiments is shown throughout the paper.
Pixel densities (for signal-area) of scanned images were calculated with Adobe Photoshop; densitometry (for signal-density) was analysed with the EpiChemi3 Darkroom™ (UVP Bioimaging Systems) using Labworks Image Acquisition and Analysis Software 4.5.
In vitro acetylation
BACE1–myc was purified from stably-transfected CHO (Chinese hamster ovary) cells with the ProFound c-Myc-Tag IP/Co-IP Kit (Pierce). BACE1–myc was incubated with the purified recombinant HAT (histone acetyltransferase) fragment of CBP (cAMP-response-element-binding protein-binding protein)/p300 (5 units; Upstate Biotechnology) in the presence of [3H]acetyl-CoA (1000 cpm/pmol) (200 mCi/mmol; American Radiolabeled Chemicals) for 1 h at 30 °C. The reaction was performed in acetylation buffer [50 mM Tris/HCl (pH 8.0), 0.1 mM EDTA, 1 mM DTT (dithiothreitol), 10% glycerol and 20 μM acetyl-CoA] and stopped by lowering the temperature to 0–4 °C. BACE1 was immunoprecipitated with an anti-BACE1 N-terminal monoclonal antibody and then counted on a liquid scintillation counter. As a control, affinity-purified BACE1 was also incubated in the absence of the enzyme (HAT) and in the presence of pre-boiled (denaturated) enzyme.
Purification of ER and Golgi intact vesicles
Intact vesicles from the ER and the Golgi apparatus were purified on a 10–34% Iodixanol (OptiPrep; Axis-Shield) continuous gradient as described previously [10]. The complete migration of subcellular markers in a typical gradient is shown in [9,10]. Fractions enriched in ER and Golgi markers (simply called ER or Golgi vesicles thereafter) were separated, pooled together and resuspended in isotonic/cryogenic buffer [0.25 M sucrose and 10 mM Tris/HCl (pH 7.4)] in the presence of protease inhibitors (Roche). Latency of ER and Golgi vesicles was determined with the glucose-6-phosphatase [11] and the sialyl-transferase [12] methods respectively. The purity of our preparation was further confirmed by assaying the above enzymatic activities, together with transport of CMP-sialic acid. Approx. >95% of vesicles were sealed and of the same membrane topographical orientation as in vivo [13].
Trypsin digestion of ER and Golgi vesicles
Vesicles were incubated for 60 min at 25 °C with trypsin (Sigma) at a final concentration of 1 μg of protease per μg of ER/Golgi proteins. Digestion was halted by the addition of anti-trypsin specific inhibitor (Sigma) and by lowering the temperature to 0–4 °C. The anti-trypsin inhibitor was used at a final concentration of 1 μg of inhibitor per μg of protease. As a control, 0.05% (v/v) Triton X-100 was added in some experiments in order to allow access of trypsin to the lumen of ER and Golgi vesicles.
Acetyl-CoA transport assay
Transport of acetyl-CoA into ER and Golgi vesicles was performed as previously described for nucleotide sugars [12,14] with some modification. Briefly, assays were performed in 100 μl final volume (isotonic/cryogenic buffer) using 25–50 μg of ER/Golgi vesicles protein. The rate of acetyl-CoA uptake at different concentrations was determined maintaining the amount of radioactive acetyl-CoA constant, whereas unlabelled acetyl-CoA ranged between 0.125 and 100 μM. After 5 min at 30 °C, reactions were stopped by lowering the temperature to 0–4 °C and by adding 100 μl of ice-cold isotonic/cryogenic buffer (as described above). Reaction mixtures were immediately spun at 25 p.s.i. (100000 g) for 15 min using an air-driven ultracentrifuge (Airfuge; Beckman). Pellets were then dissolved in 0.6 ml of 1 M NaOH; after neutralization with 0.2 ml of 4 M HCl, samples were counted by scintillation spectrometry. Transport of CMP-sialic acid and ATP was performed at two different concentrations (5 and 10 μM) of solute as described above and fully characterized in previous publications [12,14].
Membrane acetyltransferase/deacetylase activities
For the acetyltransferase activity from in vivo membranes, BACE1–myc was incubated with purified Golgi and ER membrane vesicles and [3H]acetyl-CoA (1000 cpm/pmol) for 1 h at 30 °C. The reaction was performed in 200 μl of acetylation buffer [50 mM Tris/HCl (pH 8.0), 0.1 mM EDTA, 1 mM DTT, 10% glycerol and 20 μM acetyl-CoA] in the presence or absence of 0.2% (v/v) Triton X-100. The reaction was stopped by adding 200 μl of ice-cold buffer and immediate immersion in ice; BACE1 was then immunoprecipitated and counted on a liquid scintillation counter. As a control, affinity-purified BACE1 was also incubated in the absence of ER vesicles (no enzyme) and in the presence of membranes that had been boiled prior to the assay.
For the deacetylase activity, BACE1–myc was first acetylated in vitro using the recombinant HAT fragment of CBP/p300 and [3H]acetyl-CoA (as described above) and then purified again with magnetic beads cross-linked to anti-BACE1 antibodies. BACE1 was eluted by lowering the pH to 2.0 and recovered by centrifuging at 604 g for 2 min. The pH was immediately neutralized by adding 10 μl of neutralizing buffer [1 M Tris (pH 9.5)] per 200 μl of low-pH elution buffer (Pierce). Acetylated BACE1 was then incubated in the presence of Golgi and ER membrane vesicles for 1 h at 30 °C. The reaction was performed in 200 μl of acetylation buffer (without EDTA) in the presence or absence of 0.2% (v/v) Triton X-100, and stopped as above. BACE1 was then immunoprecipitated and counted on a liquid scintillation counter. The deacetylation assay was performed in the presence of the same controls described above.
Protein labelling, pro-peptide maturation and cell-surface biotinylation
Confluent cells were starved for 1 h in methionine/cysteine free medium, labelled with 100 μCi [35S]methionine/cysteine (Trans35S-Label, >1000 Ci/mmol; Pierce) in starvation medium/dish and chased for 8, 16 and 24 h with DMEM (Dulbecco's modified Eagle's medium) supplemented with 5 mM non-radiolabelled methionine/cysteine. Cell extracts were pre-cleared with BioMag Protein A (Polysciences); beads were used at a concentration of 15 μl of the slurry per sample and separated with a magnetic holder. BACE1 was then immunoprecipitated with anti-BACE1 antibodies and recovered with BioMag Protein A as above. The immunoprecipitate–bead complexes were washed three times and boiled in sample buffer. Beads were then separated and the samples were analysed by reducing SDS/PAGE (on a 4–12% Bis/Tris NuPAGE system) and autoradiography.
For the analysis of pro-BACE1 maturation, cells were labelled and chased for 10 min and 2 h as above. Pro-BACE1 was immunoprecipitated from total cell lysates with an antibody specific for the pro-domain of BACE1 and then recovered with BioMag Protein A. Unbound BACE1 was further immunoprecipitated with anti-BACE1 antibodies. The immunoprecipitate–bead complexes were washed three times and boiled in NuPAGE sample buffer. Beads were then separated and the samples were analysed by reducing SDS/PAGE and autoradiography.
For cell-surface biotinylation, cells were labelled as above and chased for 1.5 h. Cells were then washed twice, resuspended in PBS containing EZ-Link Sulfo-NHS-LC-Biotin (Pierce) at a final concentration of 2 mg/ml, and incubated for 30 min at room temperature (25 °C). Biotinylated proteins were separated from cell extracts using ImmunoPure Immobilized Streptavidin (following the manufacturer's protocol; Pierce). Proteins were eluted and recovered as above; BACE1 was then immunopurified and analysed by reducing SDS/PAGE and autoradiography.
Statistical Analysis
Results are expressed as means±S.D. of the indicated number of determinations. The data were analysed by ANOVA and Student's t test comparison, using GraphPad InStat3 software. Statistical significance was reached at P<0.05.
RESULTS
BACE1 is acetylated in the lumen of the ER and deacetylated in the lumen of the Golgi apparatus
CHO cells expressing human BACE1 were treated with C6-ceramide (simply called ceramide thereafter) for different periods of time; BACE1 was then immunoprecipitated and analysed by Western blotting. As expected, we found a progressive increase in the steady-state levels of both immature and mature (fully glycosylated) BACE1 with an apparent plateau at day 4 (Figure 1A; see also [7]). Surprisingly, the lower band migrating as the immature form of BACE1 could also be detected with an antibody raised against acetylated lysine residues suggesting that this form, and not the mature form, was acetylated (Figure 1A; bottom panel). The pattern of acetylation would also suggest that the lysine modification occurs in the ER, where the immature form of the protein is synthesized and found. Therefore we isolated ER and Golgi fractions and analysed both the expression levels and acetylation status of BACE1. As expected, BACE1 was found to be enriched in the Golgi apparatus when compared with the ER, and to migrate slightly higher than the ER-form on gel electrophoresis (Figure 1B). However, analysis of immunoprecipitated BACE1 with an anti-acetylated lysine antibody revealed that BACE1 was acetylated in the ER compartment, but not in the Golgi apparatus (Figure 1B). The sensitivity to Endo H digestion of BACE1 purified from the ER (Figure 1C) confirmed that it represents an immature and only partially glycosylated form. In fact, resistance to Endo H is acquired in the cis/medial Golgi apparatus where the ‘nascent’ and incomplete oligosaccharide chain is processed and completed [15,16].
Figure 1. BACE1 is transiently acetylated in the lumen of the ER.
(A) CHO cells stably expressing BACE1 were treated with ceramide for the indicated time. BACE1 was immunoprecipitated from total cell lysates, separated on a 7% Tris/Acetate NuPAGE system, and then analysed by Western blotting using either anti-BACE1 or anti-acetylated lysine antibodies. Immature (im. BACE1) and mature (m. BACE1) BACE1 are indicated. (B) BACE1 was immunoprecipitated (IP) from ER and Golgi vesicles and then analysed with the indicated antibodies. ER (calreticulin) and Golgi (58 Golgi protein) markers are shown. For this experiment, samples were separated on a 4–12% Bis/Tris NuPAGE system, which allows a better resolution of total (mature and immature) BACE1. (C) BACE1 was immunoprecipitated from ER and Golgi vesicles as in (B) and then digested with Endo H prior to immunoblotting. (D) Intact ER vesicles were digested with trypsin for 30 min at 25 °C, in the presence or absence of 0.05% (v/v) Triton X-100. Digestion was halted using an anti-trypsin specific inhibitor. BACE1 was then immunoprecipitated using a monoclonal antibody against the N-terminal domain and analysed by Western blotting using the indicated antibodies. (E) BACE1–myc was purified using an anti-myc affinity column and incubated in the presence of HAT and [3H]acetyl-CoA for 1 h at 30 °C. BACE1 was then purified again and analysed on a scintillation liquid counter. *P<0.0005; n=4. (F) Primary neurons and human neuroblastoma (SH-SY5Y) cell lines were treated with ceramide for 6 days. The steady-state levels of BACE1 (top panel) were assessed by immunoblotting of total cell lysates, whereas the acetylation of BACE1 (bottom panel) was assessed following BACE1 immunoprecipitation. Electrophoresis was performed on a 4–12% Bis/Tris NuPAGE system. Neurons were prepared from wild-type mice as described in [8,9]. (G) Primary neurons were cultured in vitro up to 24 days as described previously [8,9] and then analysed for BACE1 acetylation with the indicated antibody. (H) BACE1 was immunoprecipitated from brain (cortex) extracts of 1-month-old wild-type and p44+/+ mice [9] and then analysed for lysine acetylation with the appropriate antibody. Ac. Lys., acetylated lysine; IP, immunoprecipitate; Wb, Western blot.
BACE1 is a type I membrane protein with one single transmembrane domain (∼17 residues) and a 24-amino-acid long cytosolic tail. The large N-terminal domain with 460 amino acids faces the lumen of the ER and Golgi apparatus while being synthesized and translocated to the plasma membrane. The primary sequence of BACE1 shows 16 lysine residues; however, only one is found in the cytosolic tail, being the very last residue of BACE1. In order to analyse whether BACE1 acetylation occurred on the endo-lumenal or cytosolic portion of the protein, we purified intact ER vesicles following ceramide treatment, and subjected them to trypsin digestion in the absence and presence of mild concentrations of Triton X-100. It is worth stressing that vesicles were sealed and of the same membrane topographical orientation as in vivo. In the absence of detergent, trypsin did not have access to the lumen of the vesicles and, therefore, could only digest the cytoplasmic tail of ER-membrane proteins (Figure 1D). Analysis of intact ER vesicles prior to trypsin digestion allowed detection of BACE1 with antibodies directed against both the N- and C-terminal domains, and against acetylated lysine residues (Figure 1D; control lane). Following proteolytic digestion in the absence of detergent, BACE1 was detected with antibodies against the N- but not the C-terminus, confirming successful removal of the cytosolic tail (Figure 1D). This is further indicated by the slight change in protein migration on gel electrophoresis following trypsin treatment. Surprisingly, BACE1 could still be detected when we used antibodies against acetylated lysine residues indicating that acetylation occurred in the lumen of the ER. Complete digestion of BACE1 in the presence of Triton X-100 served as a further control confirming successful trypsin digestion (Figure 1D). It is also worth stressing that both the Endo H sensitivity of the ER-form (Figure 1C) and the inability of trypsin to digest the N-terminal domain of the protein in the absence of detergent (Figure 1D) prove that nascent BACE1 is correctly inserted into the ER membrane.
We next used an in vitro system to assess whether BACE1 could serve as a substrate for lysine acetylation. A myc-tagged (at the C-terminus) version of BACE1 [7] was purified from CHO cells grown in the absence of the second messenger ceramide. Purified BACE1 was then incubated in the presence of radiolabelled acetyl-CoA, as donor of the acetyl group, and the HAT domain of recombinant CBP/p300, a well-characterized acetyl-CoA:lysine acetyltransferase localized in the cytosol [17]. Figure 1(E) shows that BACE1 can be acetylated in vitro, provided that it is incubated in the presence of an enzymatically active acetyltransferase.
Finally, we mapped the acetylation sites by comparing peptide masses (MS/MS) generated from tryptic digests of BACE1 purified from stably transfected cells grown in the absence or presence of ceramide (see Supplementary Table 2 at http://www.BiochemJ.org/bj/407/bj4070383add.htm). Analysis of mass spectra from several individual digests of BACE1 identified seven different lysine residues. Lys299 and Lys307 were found to be acetylated even in the absence of ceramide; all the other residues (Lys126, Lys275, Lys279, Lys285 and Lys300) were found to be acetylated only following ceramide treatment. No additional residues appeared to be acetylated or modified following fingerprinting analysis.
When taken together, the above results indicate that BACE1 can be acetylated both in vivo and in vitro on seven lysine residues situated in the N-terminal and globular part of the protein, which faces the lumen of the ER. The ability of BACE1 to undergo lysine acetylation in vivo was further confirmed with primary neurons and human neuroblastoma (SH-SY5Y) cells on an endogenous BACE1 background (Figure 1F). As we have demonstrated in a previous paper, primary neurons undergo spontaneous activation of p75NTR signalling, downstream of IGF1-R, that leads to a pronounced activation of endogenous ceramide and BACE1 levels [9]. We also demonstrated that p44+/+ transgenic animals, which display hyperactivation of IGF1-R signalling and an accelerated aging phenotype, have increased levels of ceramide in the brain when compared with age-matched controls [9]. In addition, the same animals have increased levels of BACE1 and increased production of amyloid β-peptide [9]. Therefore we decided to analyse the acetylation pattern of endogenous BACE1 under those situations and in the absence of exogenous treatment. Figure 1(G) shows that primary neurons in culture display a progressive increase in the levels of lysine acetylation of BACE1 that follows the same trend of ceramide activation described previously [9]. In addition, 1-month-old p44+/+ mice display increased acetylation of BACE1 in the cortex (Figure 1H), further confirming that the post-translational event described in the present study occurs in vivo, is not limited to transgenic BACE1, and does not require exogenous ceramide. Our results also suggest that acetylation is a transient event, most likely requiring deacetylation in the Golgi apparatus following full maturation of the protein. Finally, they indicate that ceramide treatment stimulates BACE1 acetylation by facilitating the efficiency of lysine acetylation; this could require changes either in the availability of the acetyl donor, the enzymatic activity of the acetyltransferase, or both.
Lysine acetylation is required for the ceramide-dependent stabilization of BACE1
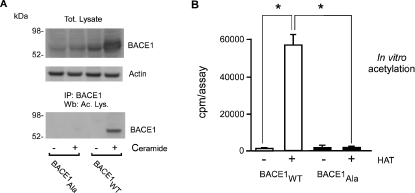
In order to analyse whether the lysine acetylation is required for the molecular stabilization of BACE1 induced by ceramide [7], we mutated the seven lysine residues described above to alanine residues. The lysine to alanine substitution precludes the acetylation of the protein and eliminates the negative charges introduced by the acetyl groups, therefore behaving as a ‘loss-of-function’. Next, we analysed the steady-state levels and acetylation of both the native (BACE1WT) and the mutated (BACE1Ala) form of BACE1 following ceramide treatment of stably transfected cells. Figure 2(A) (top panel) shows that, in contrast with BACE1WT, the steady-state levels of BACE1Ala were not affected by ceramide treatment. In addition, ceramide did not induce acetylation of BACE1Ala, indicating that this mutant form could not be acetylated in vivo (Figure 2A, bottom panel). Finally, incubation of purified BACE1 with the HAT domain of recombinant CBP/p300 in vitro resulted in acetylation of BACE1WT, but not BACE1Ala, further confirming that the mutated version of the protein could not be acetylated and that the remaining nine lysine residues of the protein did not act as a substrate for acetylation (Figure 2B).
Figure 2. Substitution of the lysine residues blocked the ability of BACE1 to be acetylated in vivo and in vitro.
(A) CHO cells stably expressing the native (BACE1WT) or mutated (BACE1Ala) form of BACE1 were grown in the presence of ceramide and then analysed by Western blotting using an anti-BACE1 antibody (top panel). Cell lysates were separated on a 4–12% Bis/Tris SDS/PAGE system prior to Western blotting. For BACE1 acetylation, BACE1 was immunoprecipitated from total cell lysates and then analysed using an anti-acetylated lysine antibody (bottom panel). (B) Both BACE1WT and BACE1Ala were purified from stably transfected cells using a specific anti-BACE1 antibody cross-linked to BioMag Protein A, eluted by lowering the pH, and then incubated in the presence of HAT and [3H]acetyl-CoA for 1 h at 30 °C. BACE1 was then purified again and analysed on a scintillation liquid counter. *P<0.0005; n=3. IP, immunoprecipitate; Wb, Western blot.
In contrast with alanine, glutamine is known to mimic the effect of lysine acetylation behaving as a bona fide ‘gain-of-function’ [18]. Therefore we mutated the seven lysine residues described above to glutamine (BACE1Gln) and analysed the maturation process of both BACE1Ala and BACE1Gln. For this purpose, we initially decided to investigate how efficiently the pro-domain was removed from newly synthesized BACE1. In fact, BACE1 contains a pro-peptide sequence at the N-terminus that is removed in the Golgi apparatus by a furin-like proprotein convertase [19,20]. Pulse-chase experiments showed that BACE1Ala displayed a marked delay in the removal of the pro-domain sequence when compared with either BACE1WT or BACE1Gln (Figure 3A; left-hand panel). This was also accompanied by a reduced and delayed appearance of the cleaved BACE1 product (Figure 3A; right-hand panel). Finally, the removal of the pro-domain occurred faster and more efficiently with BACE1Gln (Figure 3A), suggesting a more efficient ER-to-Golgi translocation. This conclusion was further confirmed when we analysed how efficiently the different mutant forms of BACE1 were able to reach the cell surface along the secretory compartment. Indeed, pulse-chase and cell-surface biotinylation experiments indicated that ∼9% of newly synthesized BACE1Ala was found at the cell-surface 1.5 h after the pulse, compared with ∼20% of the native counterpart and with ∼42% of BACE1Gln (Figure 3B). It is worth noting that ceramide treatment of BACE1WT-expressing cells produced a similar effect by increasing the translocation of newly synthesized BACE1 to the cell surface to levels almost similar to BACE1Gln (Figure 3B).
Figure 3. Transient lysine acetylation controls both translocation along the secretory pathway and molecular stability of the nascent BACE1 protein.
(A) BACE1WT, BACE1Ala, and BACE1Gln cells were labelled with a mixture of radioactive methionine/cysteine for 10 min and then chased for 2 h. Pro-BACE1 was immunoprecipitated from total cell lysates with a specific antibody against the pro-domain region of BACE1. Unbound BACE1 (without the pro-domain) was then immunoprecipitated with an antibody against the C-terminal domain of BACE1. Immunoprecipitates were analysed by SDS/PAGE and autoradiography. (B) BACE1WT, BACE1Ala and BACE1Gln cells were labelled with a mixture of radioactive methionine/cysteine for 10 min and then chased for 1.5 h. Cell surface proteins were biotinylated, separated with Immobilized Streptavidin and eluted by lowering the pH. BACE1 was then immunoprecipitated as above and analysed by reducing SDS/PAGE and autoradiography. The typical migration of cell-surface BACE1 on reducing SDS/PAGE is shown in Supplementary Figures 1A and 1B at http://www.BiochemJ.org/bj/407/bj4070383add.htm. Results are expressed as a percentage of total newly synthesized BACE1. *P<0.005; n=4. (C and D) BACE1WT, BACE1Ala and BACE1Gln cells were labelled with a mixture of radioactive methionine/cysteine for 30 min. and then chased for 8, 16 and 24 h. BACE1 was immunoprecipitated from total cell lysates and analysed by SDS/PAGE and autoradiography. The half-life of BACE1 from five different readings is shown in (C), whereas a representative radiogram is shown in (D). (E) Cell lysates from BACE1WT, BACE1Ala and BACE1Gln cells were separated either on a 7% Tris/Acetate (TA) or on a 4–12% Bis/Tris (BT) SDS/PAGE system, blotted on to a PVDF membrane, and probed with anti-BACE1 antibodies. Both immature (im. BACE1) and mature (m. BACE1) BACE1 are indicated. (F) Autoradiography of BACE1 immunoprecipitated from stably transfected cells immediately after a 30 min period of labelling with [35S]methionine/cysteine (top panel). Samples were separated on a 4–12% Bis/Tris SDS/PAGE system. BACE1 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; control lane) mRNA levels were quantified after total RNA extraction and RT (reverse transcriptase)-PCR (bottom panel). IP, immunoprecipitate; WT, wild-type.
In our previous studies we have shown that ceramide treatment increases the half-life of both newly synthesized and pre-formed BACE1 [7]. Therefore we decided to analyse the half-life of both the native and mutated forms of BACE1 following labelling with a radioactive mixture of methionine and cysteine. As shown previously [7,21], the half-life of native BACE1 was ∼16 h, but increased to ∼24 h following ceramide treatment (Figures 3C and 3D). However, BACE1Ala showed a significant reduction in the half-life with only ∼5% still detecTable 24 h after labelling, compared with ∼22% of BACE1WT and ∼55% of BACE1Gln (Figures 3C and 3D). Once again, BACE1WT under ceramide treatment behaved very similarly to BACE1Gln, further suggesting that neutralization of the positive charges of the lysine residues provided by the acetylation (and mimicked by glutamine) affected the molecular stability and/or conformational maturation of BACE1. Consistent with the above results, analysis of total cell lysates on a 7% Tris/Acetate electrophoresis system revealed a marked decrease in the steady-state levels of BACE1Ala (Figure 3E). The overall reduction in the levels of the mutant protein was also evident when the same samples were analysed on a 4–12% Bis/Tris gel, which resolves both mature and immature forms of BACE1 in one single band (Figure 3E). In contrast with BACE1Ala, BACE1Gln showed a marked increase in the steady-state levels when compared with BACE1WT. These results were observed in the absence of any difference in the levels of BACE1 mRNA (Figure 3F) or in the rate of incorporation of a radiolabelled methionine/cysteine mixture into the different BACE1 versions (Figure 3F) excluding any defect in either transcription or translation induced by the mutagenesis.
Even though the results obtained with the lysine to alanine substitution were supported by those obtained with the lysine to glutamine substitution, we cannot completely rule out the possibility that they were in part caused by the generation of an unstable protein that was not able to fold. Therefore we decided to mutate the same lysine residues to arginine (BACE1Arg), which is thought to mimic more closely the lysine structure and to be a less perturbing mutation than alanine [18]. However, analysis of both the steady-state levels (Figure 4A) and half-life (Figure 4B) of BACE1 with either the lysine to alanine or the lysine to arginine substitutions did not seem to show any apparent difference, both being below the levels observed with BACE1WT.
Figure 4. Both the lysine to alanine and lysine to arginine (loss-of-acetylation) substitutions generate an unstable BACE1 protein that is retained in the early secretory pathway and degraded by a proteasome-independent system.
(A) The steady-state levels of BACE1Arg were analysed as described in Figure 3(E). (B) The half-life of BACE1Arg was analysed as in Figure 3(C) and compared with BACE1Ala (n=3). (C) The cellular distribution of the wild-type and mutant forms of BACE1 was analysed by SDS/PAGE and immunoblotting after separation of intracellular membranes on a 10–24% discontinuous Nycodenz gradient (described in [9,10]). Membranes were developed in parallel on the same film. The appropriate subcellular markers are indicated: calreticulin (ER), syntaxin (Golgi apparatus) and EEA1 (endosomes). A longer exposure of BACE1WT and BACE1Gln gradients showing the relative distribution of mature and immature BACE1 is found in Supplementary Figure 1(C) at http://www.BiochemJ.org/bj/407/bj4070383add.htm. (D) BACE1Ala and BACE1WT-expressing CHO cells were treated with two different proteasome inhibitors for 10 h. BACE1 steady-state levels were assessed by immunoblotting of total cell lysates on a 4–12% Bis/Tris NuPAGE system. The effects of proteasome inhibition on wild-type BACE1 have been described previously [26]. WT, wild-type.
Therefore, when taken together, the above results indicate that the neutralization of the positive charges of the lysine residues provided by the acetyl groups (and mimicked by the lysine to glutamine substitution) is required for an efficient translocation of the nascent protein to the Golgi apparatus and to the cell surface along the secretory compartment. The retention of non-acetylated species of BACE1 in the early secretory pathway is further shown by the subcellular distribution pattern of the wild-type and mutated forms of BACE1 (Figure 4C). In fact, the ‘loss-of-acetylation’ mutants (BACE1Ala and BACE1Arg) were mostly observed in fractions corresponding to the ER and showed very low levels of mature BACE1. In contrast, the ‘gain-of-acetylation’ mutant (BACE1Gln) displayed a predominant localization in the late-Golgi and early-endosomal compartments with high levels of the mature form of the protein (visible as a double band in Figure 4C; see also Supplementary Figure 1C at http://www.BiochemJ.org/bj/407/bj4070383add.htm). This distribution is consistent with the previously reported localization of BACE1 and with the normal distribution of β secretase activity in the cell [22–25].
Even though the lysine to alanine substitution impaired the ability of the nascent BACE1 protein to leave the ER, we did not observe an accumulation of the protein, suggesting a rapid and efficient removal of the non-acetylated intermediates (Figure 3A). Inhibition of the proteasome machinery with either MG-132 or lactacystin successfully increased the steady-state levels of BACE1WT but not those of BACE1Ala (Figure 4D) indicating that an alternative system degrades the non-acetylated intermediates of nascent BACE1.
Acetyltransferase and deacetylase activities exist in the ER and Golgi apparatus respectively
When taken together, the above results indicate that the lysine acetylation of BACE1 is a transient event: the lysine residues are acetylated in the lumen of the ER and then deacetylated in the Golgi apparatus while the nascent protein moves ahead in the secretory pathway. This conclusion is supported by the fact that only the immature and ER-based form of BACE1 is acetylated (Figure 1) and by the fact that the gain-of-acetylation mutant (BACE1Gln) can move ahead in the secretory pathway, whereas the loss-of-acetylation mutants (BACE1Ala and BACE1Arg) are retained in the ER (Figures 3 and 4).
However, for the above events to occur, the ER must possess an acetyl-CoA:lysine acetyltransferase, whereas the Golgi apparatus must possess a lysine deacetylase. In both cases, the catalytic site of the enzyme is predicted to face the lumen of the organelles. Since the donor of the acetyl group, acetyl-CoA, is highly charged and unable to cross the ER membrane, our results also predict a membrane transporter responsible for the translocation of acetyl-CoA from the cytoplasm to the lumen of the ER, where it can then serve as a donor for the biochemical reaction.
We decided to test the above predictions by analysing each enzymatic step in vitro from highly purified intact ER and Golgi vesicles. Once again, vesicles were sealed and of the same membrane topographical orientation as in vivo. When ER and Golgi vesicles were incubated in the presence of increasing concentrations of acetyl-CoA, we found a progressive accumulation of acetyl-CoA in the lumen of ER-, but not Golgi-derived vesicles (Figures 5A–5C). Transport was saturable with an apparent Km of 14 μM and a Vmax of 824 pmol/5 min/mg of protein (Figures 5A and 5B). Finally, transport was temperature-dependent (Figure 5C), further suggesting that acetyl-CoA was translocated into the ER lumen in a carrier-mediated manner. Analysis of ER vesicles from ceramide-treated cells revealed a ∼30% increase in the Vmax (1086±92 compared with 824±75 pmol/5 min/mg of protein; P<0.005), whereas the Km was comparable with untreated cells as demonstrated by the double reciprocal plot (Lineweaver–Burk transformation; Figure 5B). Analysis of CMP-sialic acid transport into ER and Golgi vesicles confirmed the purity of our in vitro system (Figure 5D). Indeed CMP-sialic acid transport only occurs in the Golgi apparatus, where it serves for sialylation of terminal galactose residues on both glycolipids and glycoproteins [27]. Finally, analysis of ATP transport into ER vesicles purified from control and ceramide-treated cells showed no apparent difference (Figure 5E) indicating that the increased translocation of acetyl-CoA described in Figure 5(A) is not caused by generalized changes in membrane permeability or transport activities of the ER following ceramide treatment.
Figure 5. The ER membrane has an acetyl-CoA transport activity.
(A and B) Intact ER vesicles were incubated for 5 min at 30 °C with increasing concentrations of acetyl-CoA while maintaining [3H]acetyl-CoA constant. Translocation was measured as described in the Materials and methods section. The points of the double reciprocal plot (Lineweaver–Burk transformation (B) of the data shown in (A) were fitted by linear regression analysis to give a Km of 14 μM and a Vmax of 824 pmol/mg/5 min (control) (n=6). (C) ER and Golgi vesicles were assayed for transport of acetyl-CoA as described above. *P<0.0005; n=3 (D) Transport of CMP-sialic acid into ER and Golgi intact vesicles was measured as described in (A). The assay was performed at two different concentrations (5 and 10 μM) of solute. *P<0.0005; n=3 (E) Transport of ATP into ER intact vesicles obtained from control and ceramide-treated CHO cells was measured as described in (A). The assay was performed at two different concentrations (5 and 10 μM) of solute.
Incubation of purified BACE1 with radiolabelled acetyl-CoA and ER vesicles revealed the existence of an acetyl-CoA:lysine acetyltransferase activity in ER vesicles that could not be observed in the absence of mild concentrations of Triton X-100, indicating that the catalytic site of the enzyme faced the lumen of the ER (Figure 6A). In addition, the acetyltransferase activity was not detected in Golgi-purified vesicles confirming that the enzyme was an ER-resident protein (Figure 6A).
Figure 6. Acetyltransferase and deacetylase activities exist in the lumen of ER and Golgi apparatus respectively.
(A) BACE1–myc was purified with an anti-myc affinity column, and then incubated with [3H]acetyl-CoA and ER or Golgi vesicles in the presence/absence of 0.2% (v/v) Triton X-100 for 1 h at 30 °C. Reaction was stopped by lowering the temperature; BACE1 was then purified again and analysed on a scintillation liquid counter. As control, BACE1 was also incubated with [3H]acetyl-CoA in the absence of ER/Golgi vesicles and with ER vesicles that had been boiled for 10 min prior to the reaction. *P<0.0005; n=4 (B) Purified BACE1–myc was first acetylated in vitro as described in Figure 1(E) and then purified again in order to eliminate unbound acetyl-CoA. For the in vitro deacetylation, acetylated BACE1 was incubated with ER or Golgi intact vesicles in the presence/absence of 0.2% (v/v) Triton X-100 for 1 h at 30 °C. Reaction was stopped by lowering the temperature; BACE1 was then immunoprecipitated and analysed on a liquid scintillation counter. As control, BACE1 was also incubated with Golgi vesicles that had been boiled for 10 min prior to the reaction in order to inactivate any enzymatic activity. *P<0.005; n=4. (C) Both the in vitro acetylation and deacetylation of BACE1 were repeated by using intact ER (for acetylation) or Golgi (for deacetylation) vesicles prepared from control and ceramide-treated CHO cells. The assays were performed at 30 °C and in the presence of 0.2% (v/v) Triton X-100, as described in (A) (for acetylation) or (B) (for deacetylation). *P<0.0005; n=4.
We next incubated BACE1 with radiolabelled acetyl-CoA and the HAT domain of CBP/p300. The in vitro acetylated BACE1 was then purified again and incubated with either ER or Golgi vesicles in the presence or absence of Triton X-100. Incubation with permeabilized Golgi vesicles resulted in a dramatic decrease in the levels of acetylation of BACE1 (Figure 6B); such an effect was not observed when the incubation was performed in the presence of ER or pre-boiled Golgi vesicles (Figure 6B). Finally, deacetylation of BACE1 was only observed when the Golgi vesicles were permeabilized with Triton X-100 (Figure 6B) indicating that the active site of the deacetylase faces the lumen of the Golgi apparatus.
We did not detect any apparent difference when the in vitro acetylation assay was performed using ER membrane vesicles purified from either control or ceramide-treated cells (Figure 6C; left-hand panel), suggesting that ceramide did not act directly on the acetyltransferase, at least in vitro. However, we did observe increased deacetylation of BACE1 when the in vitro deacetylation assay was performed with Golgi vesicles purified from ceramide-treated cells (Figure 6C; right-hand panel). When taken together, the above results indicate that the ER possesses both an acetyl-CoA membrane transporter and an acetyltransferase activity, whereas the Golgi apparatus possesses a deacetylase activity. Finally, they also indicate that the efficiency of both acetyl-CoA translocation across the ER membrane and deacetylation in the Golgi apparatus is stimulated by the second messenger ceramide.
DISCUSSION
In the present study we show that BACE1 undergoes transient acetylation on seven different lysine residues, all facing the N-terminal portion of the nascent protein. This process involves lysine acetylation in the lumen of the ER and is followed by deacetylation in the lumen of the Golgi apparatus. We also show that a dual-enzymatic machinery acts in the ER and Golgi apparatus respectively to acetylate and deacetylate the lysine residues (Figure 7). In addition, this process requires carrier-mediated translocation of acetyl-CoA into the lumen of the ER and is stimulated by the lipid second messenger ceramide. Finally, we show that the transient/reversible lysine acetylation is required for nascent BACE1 to leave the ER and move ahead in the secretory pathway, and for the molecular stabilization of the protein.
Figure 7. Schematic model of BACE1 transient lysine acetylation/deacetylation.
During translation/translocation across the ER membrane (1), BACE1 undergoes acetylation in seven different lysine residues facing the lumen of the organelle. The reaction requires transfer of acetyl-CoA from the cytoplasm, where it is generated, to the lumen of the ER. The acetyl-CoA will then serve as donor of the acetyl group in the reaction of acetylation. This process is favoured by the second messenger ceramide, thereby increasing the concentration of acetyl-CoA in the ER. The enzyme that carries out the reaction, the acetyl-CoA:lysine acetyltransferase, is also an ER-resident protein with the catalytic site facing the lumen of the organelle. The acetylation of BACE1 provides conformational stability to the nascent protein (2). Once this has been achieved, BACE1 moves to the Golgi apparatus, where a Golgi-resident deacetylase will remove the acetyl groups (3). This event is most likely necessary in order to decrease the electron density of the globular domain of the protein (4) and allow conformational flexibility when the mature and active protein needs to shift from the ligand-free to the ligand-bound (and vice versa). In analogy with the many ER and Golgi resident glucosyltransferases already identified, both the acetyltransferase and the deacetylase are depicted here as membrane proteins. However, further studies are required to confirm this assumption.
Even though the native conformation of a protein lies encoded in its primary amino acid sequence, the efficiency of folding itself is greatly enhanced by the ER through different complex systems that require, among others, enzymes that modify, either temporally or definitively, the protein [1]. The typical example of transient modification of a nascent protein that controls the efficiency of folding is the UGGT/calnexin system, which re-monoglucosylates nascent glycoproteins and regulates their interaction with the lectin chaperone calnexin [2,3]. Once folding is completed, the protein is released from the calnexin cycle and left free to move ahead in the secretory pathway.
The acetylation/deacetylation system seems to work in the same way. Only immature BACE1 was found acetylated, thus suggesting that the process is transient. Indeed, our biochemical analysis identified both acetylase and deacetylase machineries that are physically separated: the acetylase machinery (acetyltransferase and acetyl-CoA transporter) was found only in the ER, whereas the deacetylase machinery was found only in the Golgi apparatus. Substitution of all the lysine residues that can act as acceptors for the acetyl group for alanine residues generated a BACE1 protein (BACE1Ala) that could not be acetylated in vivo or in vitro, and that was not affected by ceramide treatment. Finally, this mutated version of BACE1 showed a decreased efficiency in the translocation to the Golgi apparatus and to the plasma membrane, suggesting physical retention in the ER, and a reduced half-life. In contrast, when the same lysine residues were substituted with glutamine residues to mimic constitutive acetylation, the new protein (BACE1Gln) showed a longer half-life, together with increased efficiency in its translocation to the Golgi apparatus and the plasma membrane along the secretory pathway. The fact that the lysine to alanine substitution was also mimicked by the lysine to arginine substitution strengthens our results and seems to exclude possible artifacts produced by the mutagenesis. Finally, the results obtained with the lysine to glutamine substitution mimicked very closely those obtained with wild-type BACE1 under ceramide treatment.
It is worth noting that disorder prediction of BACE1 revealed that five of the acetylated lysine residues were either in (Lys275, Lys279 and Lys285) or close to (Lys126 and Lys307) weak electron density areas (Figure 8A). Further comparison with the structural determinants and temperature factor (B-factors) distribution of BACE1 [28] indicated that, with the exception of Lys126, all of the acetylated lysine residues are clustered in a highly-disordered region of the globular part of the C lobe of the protein (Figure 8B), which seems to be required for the conformational flexibility of the protein when shifting from the ligand-free to the ligand-bound (and vice versa) state [28–30].
Figure 8. The acetylated lysine residues are clustered in a disordered region of BACE1.
(A) Disorder prediction of BACE1 protein sequence. The prediction was performed using DRIP-PRED analysis (http://www.sbc.su.se/∼maccallr/disorder/) of the Stockholm Bioinformatics Center, Stocholm University, Sweden. The colours used are from blue to red through light green and yellow, where dark blue indicates highly ordered regions. Underlined regions scored >0.5 in the prediction algorithm and are probably disordered (with the probability being higher for red regions). The transmembrane domain is indicated by a box and appears highly ordered; the acetylated lysine residues are circled. (B) Three-dimensional view of BACE1 with the acetylated lysine residues. The structure information, together with the graphic representation, was prepared using the Entrez Molecular Modeling Database (MMDB) available at http://www.ncbi.nlm.nih.gov/Structure/. The acetylated lysine residues are indicated. The white dots correspond to Lys299 and Lys307, which can also be found acetylated prior to ceramide treatment; the other lysine residues are shown in red. The arrowheads indicate the two catalytic aspartic acid groups of the enzyme. (C) Schematic view of the lysine residues that undergo acetylation in BACE1 and p53.
Intrinsically unstructured protein domains are important for the activity of many proteins with very diverse functions, which include regulation of transcription and translation, signal transduction and chaperone-assisted unfolding of kinetically trapped folding intermediates [31]. The disordered region provides structure flexibility and allows complex molecular interactions that require movement, transition-state or coupled folding and binding to a certain substrate. Incorrect folding of this region can ultimately interfere with the functional activity of the protein. Acetylation neutralizes the positive charges of the lysine residues, and can function as an electrostatic mechanism to help the globular domain of the protein to assume the correct folding state. Consistent with this prediction, long-time molecular dynamic simulation of both ligand-free and ligand-bound BACE1 indicates that the disordered regions of the globular domain of BACE1 allow for structural fluctuation of the enzyme, which is probably necessary for correct substrate binding.
The above assumptions seem supported by the post-translational marking of the histone N-terminal tails (also referred to as the ‘histone code’) [32,33]. These are intrinsically disordered regions that are subject to post-translational modification by acetylation, methylation and phosphorylation, and that are required to fluctuate in order to allow access to DNA. Another example is offered by the regulatory domain of p53, which shows intrinsically disordered regions that provide flexibility for the various conformational requirements of the protein [34,35]. Similar to BACE1, p53 has seven lysine residues that are acetylated [36], six of which are concentrated in disordered regions of the protein (Figure 8C). In the case of p53, lysine acetylation increases with cellular senescence [37], is required for its transcriptional activities and induces molecular stability of the protein [38]. However, in contrast with p53, which is cytoplasmic, BACE1 is a membrane protein and the transitory acetylation occurs in the lumen of the ER and Golgi apparatus.
In addition to the acceptor substrate, the reaction of lysine acetylation requires a donor of the acetyl group, acetyl-CoA, and an enzyme, acetyltransferase, that transfers the acetyl group from the donor to the acceptor. In order for the reaction to occur in the lumen of the ER, both the enzyme and the donor must be made available. The present study shows that the ER membrane has an enzymatic activity that acts as acetyl-CoA:lysine acetyltransferase both in vivo and in vitro. It also indicates that the active site of the acetyltransferase faces the lumen of the organelle, and that the ER membrane is able to translocate the donor (normally found in the cytoplasm) inside the ER. Both the transferase and the transporter were found to act only in the ER and not in the Golgi apparatus, thus providing biochemical confirmation of the model shown in Figure 7. Conversely, an enzymatic activity able to function as a lysine deacetylase in vitro was identified in highly purified intact Golgi vesicles, and was shown to be absent in the ER.
The identity of the acetyltransferase, acetyl-CoA transporter, and deacetylase is still unknown; therefore further studies will be required for their identification. However, a putative acetyl-CoA transporter has been identified and found to localize in the ER system [39]. This protein has homologues in lower organisms, including Saccharomyces cerevisiae, Caenorhabditis elegans and Drosphila melanogaster [40] suggesting a fundamental and conserved function. Finally, it is up-regulated as a result of induced ER-stress, suggesting a possible role during the unfolded protein response [41].
Interestingly, ceramide was shown to stimulate the efficiency of acetyl-CoA transport across the ER membrane. This fact alone could potentially explain the increased acetylation of BACE1 observed following ceramide treatment. Indeed, an increased concentration of acetyl-CoA in the lumen of the ER would favour the kinetic of the reaction (lysine acetylation) leading to more protein and/or more lysine residues being acetylated per unit of time. Similar effects have already been described in patients affected by leucocyte adhesion deficiency type II, where a selective reduction in the Vmax (but not in the Km) of GDP-fucose transport into the Golgi lumen leads to a generalized defect in fucosylation of major glycoconjugates [42]. It is worth stressing the fact that only two lysine residues were found acetylated prior to ceramide treatment; this number increased to seven following ceramide treatment. These results appear consistent with the kinetics of acetyl-CoA translocation across the ER membrane. An increase in the concentration of the donor is expected to facilitate the reaction and allow the acetylation of all lysine residues. Obviously, this also seems to implicate that the kinetics of acetylation might be different for the different lysine residues, with the Lys299 and Lys307 occurring at lower concentrations of the donor. In addition to the transport of acetyl-CoA across the ER membrane, ceramide was also able to stimulate the rate of deacetylation in the lumen of the Golgi apparatus, therefore suggesting a common regulatory mechanism that controls the reversible/transient acetylation of ER/Golgi transiting membrane proteins.
Even though the lysine to alanine substitution impaired the ability of the nascent BACE1 protein to leave the ER, we did not observe an accumulation of the protein intermediate. Interestingly enough, the steady-state levels of BACE1Ala were not ‘normalized’ by inhibiting the proteasomes, therefore suggesting that an alternative system degrades the pool of BACE1Ala that is not able to complete maturation and translocation along the secretory compartment. Future work is required to analyse whether the disposal of non-acetylated BACE1Ala could require a specific chaperone and/or protease. Indeed, the lysine acetylation might serve as a recognition or binding site for proteins that mediate structural organization or disposal of unfolded and kinetically-trapped folding intermediates. Such a mechanism has been proposed to explain the effects produced by lysine methylation of histones and could also explain, at least in part, some of the effects induced by the acetylation [43]. Consistent with this hypothesis, several proteins containing a conserved acetyl-lysine-binding domain (the ‘bromodomain module’) that serve as transcriptional regulators have been identified [2,44]. However, once again they appear to be situated in the cytoplasm and not in the lumen of the ER.
While this manuscript was under preparation, a new study has revealed the existence of a reversible form of lysine acetylation in the mitochondrial matrix that regulates the activity of mitochondria-based enzymes [45]. Therefore this transient form of post-translational modification, previously thought to be limited to histones and cytoplasmic proteins, appears to play a more general function for the ‘physiology’ of different organelles. The fact that the events described in the present study act downstream of IGF1-R, the common regulator of lifespan [46–48], and neurotrophin signalling to regulate the rate of amyloid β-peptide generation during aging [7–9], suggests that they might provide a novel target to prevent the Alzheimer's disease risk associated with aging.
Online data
Acknowledgments
We thank Dr Carlos B. Hirschberg for helpful discussion and Dr Heidi Scrable for critical reading of an early version of this manuscript. We are grateful to Dr Amy Harms and Jim Brown for their constant support with the MS. This work was supported by P. H. S. grant NS045669 from the National Institute of Neurological Disorders and Stroke (to L. P.). C. C. was enrolled in the Programme of ‘Molecular and Cellular Biology and Pathology’ (Department of Pathology, University of Verona, Italy) and partially supported by a Fellowship from the University of Verona, Italy.
References
- 1.Trombetta E. S., Parodi A. J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 2003;344:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 2.Dempski R. E., Jr, Imperiali B. Oligosaccharyl transferase: gatekeeper to the secretory pathway. Curr. Opin. Chem. Biol. 2002;6:844–850. doi: 10.1016/s1367-5931(02)00390-3. [DOI] [PubMed] [Google Scholar]
- 3.Kleizen B., Braakman I. Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 2004;16:343–349. doi: 10.1016/j.ceb.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 5.Wade P. A., Pruss D., Wolffe A. P. Histone acetylation: chromatin in action. Trends Biochem. Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 6.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puglielli L., Ellis B. C., Saunders A. J., Kovacs D. M. Ceramide stabilizes β-site amyloid precursor protein-cleaving enzyme 1 and promotes amyloid β-peptide biogenesis. J. Biol. Chem. 2003;278:19777–19783. doi: 10.1074/jbc.M300466200. [DOI] [PubMed] [Google Scholar]
- 8.Costantini C., Weindruch R., Della Valle G., Puglielli L. A TrkA-to-p75NTR molecular switch activates amyloid β-peptide generation during aging. Biochem. J. 2005;391:59–67. doi: 10.1042/BJ20050700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costantini C., Scrable H., Puglielli L. An aging pathway controls the TrkA to p75(NTR) receptor switch and amyloid β-peptide generation. EMBO J. 2006;25:1997–2006. doi: 10.1038/sj.emboj.7601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puglielli L., Konopka G., Pack-Chung E., Ingano L. A., Berezovska O., Hyman B. T., Chang T. Y., Tanzi R. E., Kovacs D. M. Acyl-coenzyme A: cholesterol acyltransferase modulates the generation of the amyloid β-peptide. Nat. Cell. Biol. 2001;3:905–912. doi: 10.1038/ncb1001-905. [DOI] [PubMed] [Google Scholar]
- 11.Clairmont C. A., De Maio A., Hirschberg C. B. Translocation of ATP into the lumen of rough endoplasmic reticulum-derived vesicles and its binding to luminal proteins including BiP (GRP 78) and GRP 94. J. Biol. Chem. 1992;267:3983–3990. [PubMed] [Google Scholar]
- 12.Puglielli L., Hirschberg C. B. Reconstitution, identification, and purification of the rat liver Golgi membrane GDP-fucose transporter. J. Biol. Chem. 1999;274:35596–35600. doi: 10.1074/jbc.274.50.35596. [DOI] [PubMed] [Google Scholar]
- 13.Carey D. J., Hirschberg C. B. Topography of sialoglycoproteins and sialyltransferases in mouse and rat liver Golgi. J. Biol. Chem. 1981;256:989–993. [PubMed] [Google Scholar]
- 14.Puglielli L., Mandon E. C., Hirschberg C. B. Identification, purification, and characterization of the rat liver golgi membrane ATP transporter. J. Biol. Chem. 1999;274:12665–12669. doi: 10.1074/jbc.274.18.12665. [DOI] [PubMed] [Google Scholar]
- 15.Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 16.Nohturfft A., Brown M. S., Goldstein J. L. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 18.Masumoto H., Hawke D., Kobayashi R., Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 19.Bennett B. D., Denis P., Haniu M., Teplow D. B., Kahn S., Louis J. C., Citron M., Vassar R. A furin-like convertase mediates propeptide cleavage of BACE, the Alzheimer's β-secretase. J. Biol. Chem. 2000;275:37712–37717. doi: 10.1074/jbc.M005339200. [DOI] [PubMed] [Google Scholar]
- 20.Capell A., Steiner H., Willem M., Kaiser H., Meyer C., Walter J., Lammich S., Multhaup G., Haass C. Maturation and pro-peptide cleavage of β-secretase. J. Biol. Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- 21.Huse J. T., Pijak D. S., Leslie G. J., Lee V. M., Doms R. W. Maturation and endosomal targeting of β-site amyloid precursor protein-cleaving enzyme. The Alzheimer's disease β-secretase. J. Biol. Chem. 2000;275:33729–33737. doi: 10.1074/jbc.M004175200. [DOI] [PubMed] [Google Scholar]
- 22.Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., et al. β-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 23.Hussain I., Powell D., Howlett D. R., Tew D. G., Meek T. D., Chapman C., Gloger I. S., Murphy K. E., Southan C. D., Ryan D. M., et al. Identification of a novel aspartic protease (Asp 2) as β-secretase. Mol. Cell. Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 24.Yan R., Bienkowski M. J., Shuck M. E., Miao H., Tory M. C., Pauley A. M., Brashier J. R., Stratman N. C., Mathews W. R., Buhl A. E., et al. Membrane-anchored aspartyl protease with Alzheimer's disease β-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 25.Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., et al. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 26.Qing H., Zhou W., Christensen M. A., Sun X., Tong Y., Song W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- 27.Deutscher S. L., Nuwayhid N., Stanley P., Briles E. I., Hirschberg C. B. Translocation across Golgi vesicle membranes: a CHO glycosylation mutant deficient in CMP-sialic acid transport. Cell. 1984;39:295–299. doi: 10.1016/0092-8674(84)90007-2. [DOI] [PubMed] [Google Scholar]
- 28.Patel S., Vuillard L., Cleasby A., Murray C. W., Yon J. Apo and inhibitor complex structures of BACE (β-secretase) J. Mol. Biol. 2004;343:407–416. doi: 10.1016/j.jmb.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 29.Xiong B., Huang X. Q., Shen L. L., Shen J. H., Luo X. M., Shen X., Jiang H. L., Chen K. X. Conformational flexibility of β-secretase: molecular dynamics simulation and essential dynamics analysis. Acta Pharmacol. Sin. 2004;25:705–713. [PubMed] [Google Scholar]
- 30.Hong L., Tang J. Flap position of free memapsin 2 (β-secretase), a model for flap opening in aspartic protease catalysis. Biochemistry. 2004;43:4689–4695. doi: 10.1021/bi0498252. [DOI] [PubMed] [Google Scholar]
- 31.Dyson H. J., Wright P. E. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 32.Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 33.Berger S. L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 34.Bell S., Klein C., Muller L., Hansen S., Buchner J. p53 contains large unstructured regions in its native state. J. Mol. Biol. 2002;322:917–927. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 35.Ayed A., Mulder F. A., Yi G. S., Lu Y., Kay L. E., Arrowsmith C. H. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 36.Scrable H., Sasaki T., Maier B. ΔNp53 or p44: priming the p53 pump. Int. J. Biochem. Cell. Biol. 2005;37:913–919. doi: 10.1016/j.biocel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P. P., Pelicci P. G. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 2000;406:207–210. doi: 10.1038/35018127. [DOI] [PubMed] [Google Scholar]
- 38.Brooks C. L., Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 39.Kanamori A., Nakayama J., Fukuda M. N., Stallcup W. B., Sasaki K., Fukuda M., Hirabayashi Y. Expression cloning and characterization of a cDNA encoding a novel membrane protein required for the formation of O-acetylated ganglioside: a putative acetyl-CoA transporter. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2897–2902. doi: 10.1073/pnas.94.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirabayashi Y., Kanamori A., Nomura K. H., Nomura K. The acetyl-CoA transporter family SLC33. Pflugers Arch. 2004;447:760–762. doi: 10.1007/s00424-003-1071-6. [DOI] [PubMed] [Google Scholar]
- 41.Shaffer A. L., Shapiro-Shelef M., Iwakoshi N. N., Lee A. H., Qian S. B., Zhao H., Yu X., Yang L., Tan B. K., Rosenwald A., et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Sturla L., Puglielli L., Tonetti M., Berninsone P., Hirschberg C. B., De Flora A., Etzioni A. Impairment of the Golgi GDP-L-fucose transport and unresponsiveness to fucose replacement therapy in LAD II patients. Pediatr. Res. 2001;49:537–542. doi: 10.1203/00006450-200104000-00016. [DOI] [PubMed] [Google Scholar]
- 43.Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 44.Jacobson R. H., Ladurner A. G., King D. S., Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 45.Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Longo V. D., Finch C. E. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- 47.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 48.Puglielli L. Aging of the brain, neurotrophin signaling, and Alzheimer's disease: Is IGF1-R the common culprit? Neurobiol. Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.01.010. doi: 10.1016/j.neurobiolaging.2007.01.010. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.