Abstract
Humans lack the ability to synthesize vitamin C (ascorbate) due to the absence of gulonolactone oxidase, the last enzyme in the biosynthetic pathway in most other mammals. The corresponding oxidoreductase in trypanosomes therefore represents a target that may be therapeutically exploitable. This is reinforced by our observation that Trypanosoma cruzi, the causative agent of Chagas' disease, lacks the capacity to scavenge ascorbate from its environment and is therefore dependent on biosynthesis to maintain intracellular levels of this vitamin. Here, we show that T. cruzi galactonolactone oxidase (TcGAL) can utilize both L-galactono-γ-lactone and D-arabinono-γ-lactone as substrates for synthesis of vitamin C, in reactions that obey Michaelis–Menten kinetics. It is >20-fold more active than the analogous enzyme from the African trypanosome Trypanosoma brucei. FMN is an essential cofactor for enzyme activity and binds to TcGAL non-covalently. In other flavoproteins, a histidine residue located within the N-terminal flavin-binding motif has been shown to be crucial for cofactor binding. Using site-directed mutagenesis, we show that the corresponding residue in TcGAL (Lys-55) is not essential for this interaction. In contrast, we find that histidine and tryptophan residues (His-447 and Trp-448), localized within a C-terminal motif (HWXK) that is a feature of ascorbate-synthesizing enzymes, are necessary for the FMN association. The conserved lysine residue within this motif (Lys-450) is not required for cofactor binding, but its replacement by glycine renders the protein completely inactive.
Keywords: ascorbate, flavin mononucleotide (FMN), galactonolactone oxidase, trypanosome, vitamin C
Abbreviations: ALO, arabinonolactone oxidase; APX, ascorbate-dependent peroxidase; CHEFE, contour-clamped homogeneous electric field electrophoresis; GAL, galactonolactone oxidase; GALDH, galactonolactone dehydrogenase; GLO, gulonolactone oxidase; GULDH, gulonolactone dehydrogenase; IPTG, isopropyl β-D-thiogalactoside; NEM, N-ethylmaleimide; Ni-NTA, Ni2+-nitrilotriacetic acid; pCMB, p-chloromercuribenzoate; Tb, Trypanosoma brucei; Tc, Trypanosoma cruzi; TCA, trichloroacetic acid
INTRODUCTION
The insect-transmitted protozoan Trypanosoma cruzi is the aetiological agent of Chagas' disease. There are more than 10 million people infected with this parasite in 21 countries across Central and South America, with another 40 million at risk. Approx. 14000 deaths annually are attributed to Chagas' disease [1,2]. There is no immediate prospect of a vaccine and the development of improved chemotherapy is a priority. Nifurtimox and benznidazole are the only drugs currently available for T. cruzi infections. However, both have toxic side effects and are largely non-effective against the chronic stage of the disease, which can occur years after the initial infection. The activation of nifurtimox within T. cruzi can lead to the production of reactive oxygen species, a process that has been implicated in trypanocidal activity [3]. As a consequence, oxidative defence in trypanosomes has been a major focus of research.
In aerobic organisms, endogenous biochemical processes result in the generation of reactive oxygen species, which include superoxide anions, hydrogen peroxide and hydroxyl radicals. Complex enzymatic and non-enzymatic pathways have evolved to protect cells from these toxic oxygen metabolites. In trypanosomatid parasites, many aspects of oxidative defence are distinct from those in the mammalian hosts. For example, trypanosomes lack catalase, their superoxide dismutases use iron as a cofactor and peroxide metabolism involves a complex series of overlapping pathways linked to the unique thiol trypanothione, a glutathione–spermidine conjugate [4–8]. In the T. cruzi endoplasmic reticulum, detoxification of hydrogen peroxide is dependent on an unusual plant-like haemoperoxidase [TcAPX (T. cruzi ascorbate-dependent peroxidase)] [5]. The activity of this enzyme is linked to trypanothione, via ascorbate reduction, in a process that involves a non-enzymatic interaction, with ascorbate acting as an electron shuttle. APX genes are present in T. cruzi and the related protozoan Leishmania, both of which have an obligatory intracellular mammalian stage. However, the gene is absent from the African trypanosome, Trypanosoma brucei, which is exclusively extracellular. Ascorbate may therefore be more important to oxidative defence in T. cruzi and Leishmania, parasites which are exposed to reactive nitrogen/oxygen species when they invade host macrophages [9–12]. In line with this, deletion of both TcAPX genes has only been possible in parasites containing an episomal copy (M. C. Taylor, unpublished work).
Ascorbate is a water-soluble vitamin that has multiple cellular functions, both as a cofactor and as an antioxidant molecule [13]. Two major properties enable ascorbate to detoxify reactive oxygen species directly. First, the low reduction potential of ascorbate and the ascorbyl radical allows them to reduce almost all radicals and oxidants [14]. Second, the ascorbyl radical, formed when ascorbate scavenges reactive nitrogen or oxygen species, is very stable and has a low reactivity [15,16]. Most animals and plants are able to synthesize ascorbate. Exceptions include humans and other primates, guinea-pigs and several species of fish [17]. Humans rely entirely on their diet for ascorbate, and, as such, are susceptible to scurvy, a syndrome caused by vitamin C deficiency [18,19]. The final stage in ascorbate biosynthesis is common to all organisms where this pathway is active, and involves the conversion of an aldonolactone substrate into ascorbate. The flavin-dependent oxidoreductases responsible utilize a range of substrates in different cell types, and are classified accordingly: gulonolactone oxidase (GLO) in mammalian cells [20], arabinonolactone oxidase (ALO) in fungal cells [21], galactonolactone dehydrogenase (GALDH) in plant cells [22,23] and gulonolactone dehydrogenase (GULDH) in bacteria [24,25]. In most flavoproteins, the cofactor (FAD or FMN) is bound non-covalently. However, in mammalian GLO and fungal ALO [21,22], this interaction is covalent, with the cofactor attached to a histidine residue, which is conserved in many other oxidoreductases where this type of linkage has been identified. Covalent binding of flavin cofactors typically involves histidine, although there are instances where linkage to cysteine or tyrosine residues has been identified [26].
In humans, the terminal enzyme in ascorbate biosynthesis is lacking, although a pseudogene remains [27]. This absence raises the possibility that specific inhibitors targeted at this biosynthetic step could have potential as antimicrobial or antiparasitic agents, in situations where this pathway is essential for pathogen viability. However, inhibitor design has so far been restricted by the lack of a three-dimensional structure for this class of enzyme and the absence of detailed information on the precise mechanism of action. Recently, we demonstrated that T. brucei has an ascorbate biosynthetic capacity and that this reaction occurs in the glycosome, a kinetoplastid-specific organelle related to peroxisomes [28]. The enzyme which catalyses the final step in this pathway can use both galactonolactone and arabinonolactone as substrates. It has a slight preference for the latter and was initially designated TbALO. In the present paper, we describe the properties of the corresponding enzyme from T. cruzi. We show that activity of this plant-like enzyme is dependent on the non-covalent binding of an FMN cofactor. Furthermore, using site-directed mutagenesis, we have identified residues that are required for flavin-binding, and consequently for enzyme activity.
EXPERIMENTAL
Parasites
T. cruzi epimastigotes (MHOM/BR/78/Silvio X10/6) were cultured at 28 °C in supplemented RPMI 1640 medium (Sigma) [29]. Culture-derived amastigotes were obtained from Vero cells (A.T.C.C. CRL-1586) infected with metacyclic stationary-phase trypomastigotes. Genomic DNA was isolated from parasites using the phenol/chloroform method, and chromosomes for CHEFE (contour-clamped homogeneous electric field electrophoresis) analysis were extracted using an agarose-embedding technique [30] and separated using a Bio-Rad CHEFE mapper. Total RNA was extracted from parasites using the RNeasy® mini-kit (Qiagen).
Ascorbate uptake
L-[carboxyl-14C]Ascorbate (13 mCi·mmol−1) and D-[2-3H]glucose (14 Ci·mmol−1) were supplied by GE Healthcare. For assays of dehydroascorbate uptake, radiolabelled ascorbate was oxidized to dehydroascorbate by the addition of 0.5% (v/v) bromine. The resulting solution was vortex-mixed for 30 s, and then nitrogen was bubbled through for 10 min to remove the bromine. Aliquots of dehydroascorbate were prepared freshly on each occasion, and used immediately. Epimastigotes, in the exponential phase of growth, were pelleted and washed three times in assay buffer (90 mM Tris/HCl, pH 7.5, 3.1 mM KCl, 96.9 mM NaCl, 5 mM MgCl2, 2 mM Na2HPO4 and 2 mM glycerol), then resuspended at 2×108 cells·ml−1. To initiate uptake, 50 μl of parasite suspension was added to an equal volume of assay buffer containing radiolabelled ascorbate (0.625 μCi) or glucose (5 μCi), giving final concentrations of 480 μM and 3.6 μM respectively. The cell suspension was layered over a 150 μl cushion of oil (1-bromododecane) in a 1.5 ml Eppendorf tube. Reaction mixtures were incubated at 22 °C and terminated by the addition of 200 μl of 10 mM unlabelled substrate in assay buffer (quench buffer) at <0 °C. The cells were then separated from the uptake buffer by centrifugation at 13000 g for 30 s, and the tubes were flash-frozen using liquid nitrogen. The parasite pellet was cut from the bottom of the tube into a scintillation vial, and lysed in 250 μl of 2% (w/v) SDS. A 3 ml aliquot of scintillation fluid (Ecoscint A, National Diagnostics) was added to each vial, before incubation at room temperature (22 °C) overnight. Incorporated radioactivity was then measured using a Beckman liquid-scintillation spectrometer. Vero-cell derived amastigotes were treated in a similar manner, except that cells were not layered over oil, and the assay was stopped by the addition of 2% (v/v) paraformaldehyde, followed by pelleting of the cells as before. Pellets were washed three times in 1× PBS, followed by lysis in 250 μl of 2% (w/v) SDS in a scintillation vial. As a control, quench buffer was added to the assay prior to the cells. The tube was then immediately centrifuged at 13000 g for 5 min, and the pellet were treated as for the experimental samples.
Isolation of the T. cruzi galactonolactone oxidase (Tcgal) gene
Tcgal was amplified from T. cruzi (X10/6 clone) genomic DNA using primers based on the sequence of the corresponding gene from the genome reference clone CL Brener (GeneDB accession number Tc00.1047053509179.100). Primers 5′-AGAggtaccTTGTGACGTTTCCATGCGGCCC-3′ and 5′-GGGaagcttTTACAAATGACTATTGGTGCT-3′ were used for amplification. Lower-case letters indicate restriction sites for KpnI and HindIII respectively, incorporated to facilitate cloning of the resulting fragment. Amplifications were performed under the following conditions: 94 °C for 2 min, followed by 30 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 2 min. The 1.5 kb product was ligated into the Escherichia coli expression vector pTrcHis-C (Invitrogen).
Expression and purification of His-tagged TcGAL
E. coli BL-21+ cells were transformed with the plasmid pTrcHisCTcGAL. Optimal expression of the soluble protein was obtained by induction with 100 μM IPTG (isopropyl β-D-thiogalactoside) for 6 h at 37 °C. The induced cells were pelleted and lysed by sonication in 20 ml of buffer A (300 mM NaCl and 50 mM sodium phosphate buffer, pH 8.0) containing 10 mM imidazole. The lysate was centrifuged at 7000 g, and the supernatant was applied to a 2 ml Ni-NTA (Ni2+-nitrilotriacetate) column (Qiagen), pre-equilibrated in buffer A. Initially, the column was washed with 40 ml of buffer A containing 20 mM imidazole, and the recombinant protein then eluted in buffer A containing 250 mM imidazole. Each step was performed in the presence of protease inhibitors (Roche Molecular Biochemicals). Fractions were analysed by SDS/PAGE (10% gels), and protein concentrations were determined by the bicinchoninic acid protein assay system (Pierce). Approximately 3 mg of purified recombinant protein was recovered per litre of E. coli culture.
Site-directed mutagenesis
Oligonucleotide-directed in vitro mutagenesis was performed using the Stratagene QuikChange® mutagenesis kit, following the manufacturer's protocol. Briefly, amplifications were performed in 50 μl reaction volumes, with pTrcHisCTcGAL as template. The reaction consisted of one cycle of 95 °C for 30 s, then 16 cycles of 95 °C for 30 s, 55 °C for 1 min and 68 °C for 6 min. Following this, 10 units of DpnI was added to the reaction mixture to digest parental double-stranded DNA. A 1 μl aliquot of DpnI-digested PCR product was then used to transform E. coli XL1-Blue supercompetent cells. The following sense primers (along with their antisense counterparts) (Sigma Genosys) were used to generate each of the desired mutations. The relevant substitution sites, incorporating the required base change, are underlined. K55G, 5′-TGCCGGGGCCGGAGGAAGCCCCAATGC-3′; K55H, 5′-TGCCGGGGCCGGACATAGCCCCAATGC-3′; K55L, 5′-TGCCGGGGCCGGACTAAGCCCCAATGC-3′; H447G, 5′-GGGCGGTAGGCCGGGATGGGCGAAGTACTACACATGG-3′; W448G, 5′-TAGGCCGCACGGTGCGAAGTACTACACATGG-3′; K450G, 5′-GCACTGGGCGGGATACTACACATGGGG-3′.
Tcgal mutants were confirmed by sequencing using a BigDye® terminator cycle sequencing kit (Applied Biosystems) and an ABI Prism® 377 automated sequencer.
Enzyme activity
Activity was measured by monitoring ferricytochrome c reduction, using an absorbance of 550 nm. A standard reaction mixture (1 ml) contained 50 mM potassium phosphate buffer, pH 8.0, 1 mM EDTA, 100 μl of cytochrome c and 12.2 μg of purified TcGAL. The background rate of cytochrome c reduction was determined, and the reaction was initiated by the addition of D-arabinono-γ-lactone (Dextra Lab), L-gulono-γ-lactone (Acros) or L-galactono-γ-lactone (Sigma). Enzyme activity was calculated using a ϵ value of 17.3 mM−1, and assumes that two molecules of ferricytochrome c are reduced per molecule of ascorbate oxidized [31]. Data were analysed using GraphPad Prism software. Inhibitory compounds [21,32] were added to the reaction mixture before reading the background rate of cytochrome c reduction.
HPLC analysis
All HPLC equipment and software were from Dionex (UK). Separations were carried out as outlined by Birch et al. [33] utilizing an ODS Hypersil 5 μm column (250 mm×4.6 mm) (Thermo Electron Corp.), eluting with acetonitrile/water/10% (w/v) trifluoracetic acid/phosphoric acid (14:84:1.5:0.09, by vol.) passing through the fluorescence detector (Dionex model RF 2000) set at the excitation wavelength of 430 nm and emission wavelength of 525 nm, at a flow rate of 0.5 ml·min−1. Peak identity in the protein fraction was confirmed by measuring the retention time, and comparison with commercially available FMN and FAD. On an assumption of one molecule of flavin per molecule of TcGAL, 12 mg of purified protein/ml was used to generate 0.1 mg of cofactor/ml. TcGAL was trypsin-digested in the dark for 3 h at 37 °C before mixing with a solution of ice-cold 5% (w/v) TCA (trichloroacetic acid) in water, and the precipitated protein was removed by centrifugation at 13000 g for 5 min at room temperature. A 20 μl aliquot of the resulting supernatant was injected on to the column, and the chromatograms were compared with those from the standard samples of FMN and FAD.
RESULTS
Ascorbate-uptake capacity of T. cruzi
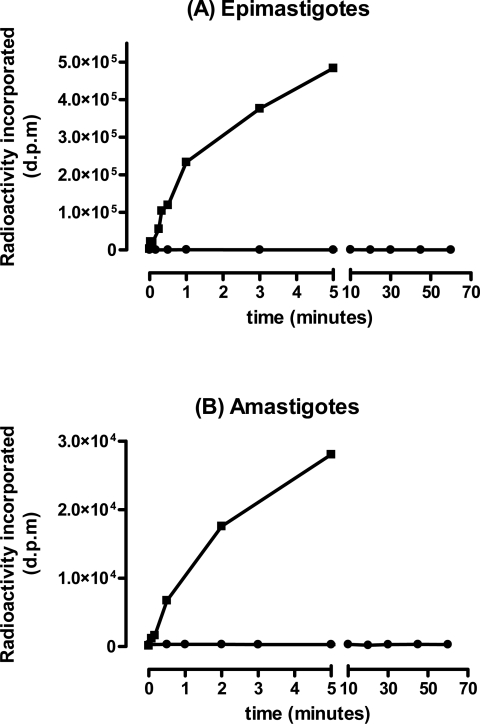
To determine whether T. cruzi epimastigotes and amastigotes can scavenge ascorbate from their environment, uptake assays were performed on both life-cycle stages. Parasites were incubated with radiolabelled ascorbate or glucose, and the extent of uptake was measured. Both parasite life-cycle stages took up glucose rapidly, reaching equilibrium within 5–10 min, indicating that they were competent for transport (Figure 1). Epimastigotes transported 20 times as much glucose as amastigotes (Vmax 10.95 and 0.49 pmol·min−1 per 107 cells respectively), partly reflecting their larger cell volume (2–3-fold [34]). In contrast, neither cell type displayed any capacity to take up ascorbate, even when followed for more than 1 h (Figure 1). Mammalian cells can also transport dehydroascorbate, the oxidized form of the vitamin [35]. Radiolabelled ascorbate was therefore converted into dehydroascorbate by the addition of bromine, and uptake assays were carried out as before. Neither cell type showed any capacity to take up dehydroascorbate over a 60 min time course (results not shown). These experiments imply that both the insect and intracellular mammalian stages of T. cruzi are incapable of obtaining ascorbate from their environment, and, as such, must acquire this vitamin by de novo synthesis.
Figure 1. T. cruzi lacks an ascorbate-uptake capacity.
Epimastigotes (A) and Vero cell-derived amastigotes (B) (107 parasites per assay) were incubated with 3H-labelled glucose (■) or 14C-labelled ascorbate (●) for the times indicated. Cells were prepared, and uptake of the corresponding radiolabel was measured by liquid-scintillation counting. The data are expressed in disintegrations per minute (d.p.m.). Similar results were obtained with dehydroascorbate.
Isolation and expression of TcGAL
A putative GAL/ALO gene was identified in the T. cruzi database [36]. DNA primers were designed and used to amplify the corresponding sequence from parasite genomic DNA (X10/6 clone). The 1.5 kb gene (designated Tcgal) has the potential to encode a 57 kDa protein (Figure 2) and is expressed as a 2.0 kb transcript (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/407/bj4070419add.htm). In CHEFE analysis, the Tcgal probe hybridized to a chromosome band of 1100 kb (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/407/bj4070419add.htm). With the genome reference strain CL Brener, bands of ∼3000, 1100 and 600 kb were detected. These differences reflect the hybrid nature of the CL Brener clone [37] and the fact that chromosome homologues in T. cruzi often vary considerably in size.
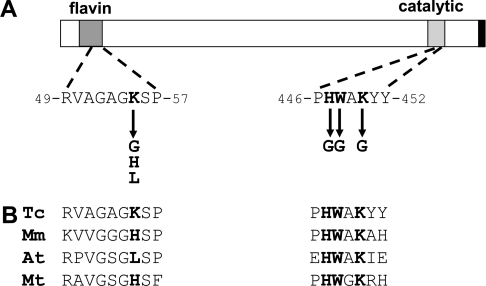
Figure 2. Organization of TcGAL.
(A) Schematic of TcGAL showing the flavin-binding motif (residues 22–57) (Pfam designation PF01565), catalytic region (residues 436–452) (Pfam designation PF04030) and glycosomal targeting signal (black bar) [28]. Residues selected for site-directed mutagenesis are highlighted in bold, with arrows indicating new amino acids that were generated. Numbers refer to the position of amino acids in the TcGAL sequence (GeneDB accession no. Tc00.1047053509179.100). (B) Alignment of selected regions of TcGAL (Tc) with the corresponding sequences from Mus musculus GLO (Mm) (GenBank® accession number AAH28828), Arabidopsis thaliana GALDH (At) (GenBank® accession number T06690), and Mycobacterium tuberculosis GULDH (Mt) (GenBank® accession number D70989).
Proteins that synthesize the final step in ascorbate biosynthesis do not display a high level of cross-species sequence conservation (∼15%). One feature they do share is an N-terminal flavin-binding motif (Figure 2A), which often contains a histidine residue implicated in the covalent attachment of FAD. Replacement of this residue with leucine in plant GALDHs has been interpreted as the reason that flavin binding occurs non-covalently. Unusually, in T. cruzi, the corresponding amino acid is a lysine residue (Lys-55). A second histidine residue, which is widely conserved in aldonolactone oxidases/dehydrogenases, has also been implicated in FAD binding in the flavoprotein vanillyl-alcohol oxidase [38]. In TcGAL, this residue is also a histidine (His-447) and is found within the motif HWXK (highlighted in Figure 2A), a feature that is completely conserved in ascorbate-synthesizing enzymes. The T. cruzi enzyme also contains the C-terminal tripeptide SHL (black bar in Figure 2A), which has been implicated in glycosomal targeting [28].
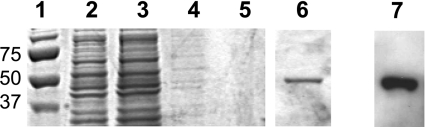
To investigate the biochemical properties of TcGAL, the gene was cloned into the vector pTrcHis-C and expressed in E. coli BL-21+ cells. In this system, recombinant TcGAL is tagged at its N-terminus with a histidine-rich sequence and an epitopetag detectable with the Anti-Xpress® monoclonal antibody (Invitrogen). After IPTG induction, the recombinant protein could be readily purified by one round of affinity chromatography on a Ni-NTA column, following washing with imidazole. SDS/PAGE gels and Western blotting using the Anti-Xpress® antibody confirmed the purification of the 55 kDa protein (Figure 3).
Figure 3. Purification of recombinant TcGAL.
Lanes 1–6, SDS/PAGE gel (10% polyacrylamide) stained with Coomassie Blue. Lane 1, size standards; lane 2, crude extract loaded on to a Ni-NTA column; lane 3, flowthrough. The column was washed extensively with 10 mM imidazole (lane 4, first wash; lane 5, last wash). Recombinant protein was eluted with 200 mM imidazole (lane 6). Lane 7, Western blot of samples from the 200 mM imidazole elution stage in TcGAL purification. Samples were Western-blotted and probed with Anti-Xpress® antibody to identify the recombinant protein. Sizes are indicated in kDa.
Characterization of TcGAL activity
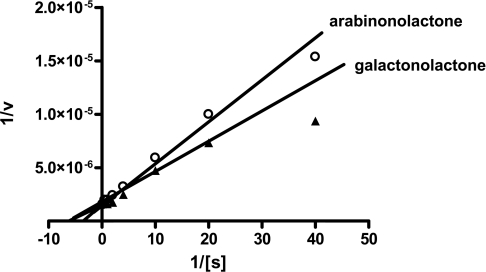
Assays were carried out using purified recombinant TcGAL (Figure 3) and, as a control, extracts of non-transformed E. coli BL-21+ that had been affinity-purified in parallel. Activity was assessed in the presence of various aldonolactone substrates. We found that recombinant TcGAL could synthesize vitamin C using the substrates L-galactono-γ-lactone and D-arabinono-γ-lactone, but no significant activity was detected with L-gulono-γ-lactone, the preferred substrate for the mammalian enzyme. Recombinant TcGAL was active across a wide range of temperatures, but was heat-inactivated above 50 °C. The optimal pH was between 7.5 and 8.0. Kinetic studies were carried out using L-galactono-γ-lactone and D-arabinono-γ-lactone as substrates, measuring the TcGAL-mediated ascorbate-dependent reduction of ferricytochrome c. Double-reciprocal plots of 1/TcGAL activity against 1/[substrate] were linear for substrate concentrations up to 4 mM (Figure 4), allowing kinetic constants for both metabolites to be calculated (Table 1a). On the basis of apparent Vmax values, TcGAL is >20-fold more active towards both substrates than the T. brucei homologue TbALO [28]. TcGAL exhibited a slightly higher turnover constant and catalytic efficiency value for L-galactono-γ-lactone compared with D-arabinono-γ-lactone, the normal substrates of the plant and fungal enzymes respectively (Table 1a). This, together with an analysis of the T. cruzi genome sequence [36], which suggests that L-galactono-γ-lactone is the normal physiological substrate (F. J. Logan, unpublished work), led us to designate the enzyme as TcGAL. The apparent Km value for L-galactono-γ-lactone (Table 1a) was in a similar range to that previously established for plant GALDHs (60–150 μM) (see Table 3).
Figure 4. Lineweaver–Burk plot of TcGAL activity.
The activity of TcGAL was assayed by following the ascorbate-dependent reduction of ferricytochrome c in the presence of 12.2 μg of TcALO and D-arabinono-γ-lactone (0.025–4 mM; ○) or L-galactono-γ-lactone (0.025–4 mM; ▲). Double-reciprocal plots of 1/TcGAL activity (nmol of ascorbate formed/min per mg) against 1/[galactonolactone] or 1/[arabinono-lactone] were linear for substrate concentrations up to 4 mM.
Table 1. Kinetic parameters of vitamin C formation and inhibition of TcGAL.
(a) Enzyme activity (Vmax) was calculated using an ϵ value of 17.3 mM−1 [31]. kcat assumes one catalytic site per 60 kDa monomer. Numbers in parentheses are for the corresponding T. brucei enzyme [28]. (b) Enzyme activity was determined at pH 8.0 and 21 °C, with 2 mM arabinonolactone. Inhibition was calculated as a percentage of activity of the untreated wild-type enzyme. Experiments were performed in triplicate and results are means±S.D.
| (a) | ||||
|---|---|---|---|---|
| Substrate | Michaelis–Menten constant (Km) (μM) | Activity (Vmax) (μmol of ascorbate formed/min per mg) | Turnover constant (kcat) (s−1) | Catalytic efficiency (kcat/Km) (M−1·s−1) |
| Arabinonolactone | 285±36 | 651±20 | 649 | 2.3×106 |
| (55±3) | (29.6±0.4) | (27.2) | (4.9×105) | |
| Galactonolactone | 161±24 | 675±22 | 673 | 4.2×106 |
| (154±24) | (22.8±0.8) | (20.9) | (1.4×105) | |
| (b) | ||||
| Inhibitor (1 mM) | Inhibition (%) | |||
| HgCl2 | 100±0 | |||
| PCMB | 100±5.0 | |||
| NEM | 77±2.4 | |||
| ZnSO4 | 63±3.2 |
Table 3. Comparison of the properties of the enzyme that mediates the final step in ascorbate (or erythroascorbate) biosynthesis across five kingdoms.
| Properties | GALDH (plants)* | GLO (mammals)* | ALO (fungi)* | GULDH (bacteria)† | GAL/ALO (trypanosomes)‡ |
|---|---|---|---|---|---|
| Product | L-Ascorbic acid | L-Ascorbic acid | D-Erythroascorbic acid | L-Ascorbic acid | Substrate-dependent |
| Cellular compartment | Mitochondria | Microsomes | Mitochondria | N/A | Glycosome |
| Molecular mass (kDa) | 56 | 51 | 56–60 | 61 | 57 (T. cruzi) 59 (T. brucei) |
| Cofactor Relative substrate specificity | Non-covalent FMN | Covalent FAD | Covalent FAD | Non-covalent flavin (proposed) | Non-covalent FMN |
| L-Galactonolactone | 100 | 87 | 87 | 0 | 100 (T. cruzi) 74 (T. brucei) |
| L-Gulonolactone | 0–20 | 100 | 24 | 100 | 3–9 |
| D-Arabinonolactone | − | − | 100 | − | 96 (T. cruzi) 100 (T. brucei) |
| Km (preferred substrate) | 0.12–3.3 mM | 0.066 mM (rat) 0.15 mM (goat) | 44.1 mM | 5.5 mM | 0.16 mM (T. cruzi) 0.05 mM (T. brucei) |
| Essential thiol groups | Yes | Yes | Yes | Yes | Yes |
It has previously been suggested that cysteine residues may play a role in aldonolactone oxidoreductase activity [21]. To establish whether this was the case for TcGAL, we carried out assays in the presence of pCMB (p-chloromercuribenzoate), NEM (N-ethylmaleimide), zinc and mercury ions. pCMB and NEM form thioether linkages with cysteine thiol groups, specifically inhibiting them, whereas zinc and mercury ions lead to toxicity, mainly because of thiol group inhibition [39]. The inhibition profile in each case suggested that cysteine residue(s) are essential for the activity of this enzyme (Table 1b). These results are similar to those seen with the Candida albicans ALO [21], which has several cysteine residues (Cys-27 and Cys-249) in common with TcGAL.
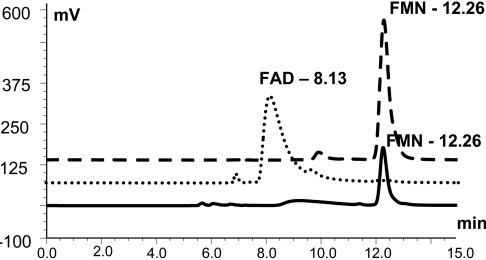
To determine the nature of the interaction between TcGAL and its flavin cofactor, we added 5% TCA to trypsin-digested recombinant enzyme and subjected the extract to HPLC analysis. This treatment resulted in the release of the flavin cofactor from the protein, indicating that it was non-covalently bound [40]. FAD and FMN standards were fractioned on the HPLC with retention times of 8.13 and 12.26 min respectively (Figure 5). The supernatant from the TcGAL extraction gave a retention time of 12.26 min for the flavin peak, identifying the cofactor as FMN. The T. brucei enzyme, which had previously been assumed to covalently bind FAD [28], also had a profile consistent with non-covalently bound FMN (results not shown). Therefore the trypanosome enzymes show a significant difference from the situation in the mammalian GLO and the fungal ALO, where the cofactor is covalently bound FAD [20,21], but similarity to plant GALDHs, which also non-covalently bind FMN [22]. In the case of bacterial GULDH, the cofactor has yet to be identified, but is a non-covalently bound flavin [25].
Figure 5. TcGAL non-covalently binds FMN.
HPLC analysis was performed as described in the Experimental section. Under these conditions, FAD (dotted line) and FMN standards (dashed line) could be separated with retention times of 8.13 and 12.26 min respectively. The supernatant from TCA-precipitated wild-type TcGAL (122 μg) had a retention time of 12.26 min (solid line), confirming FMN as the flavin cofactor.
Effects of mutated residues
The flavin-binding motif toward the N-terminus of trypanosome GAL/ALO (residues 22–57) (Figure 2) lacks the histidine residue which, in mammalian GLOs and fungal ALOs, covalently binds FAD via an 8-α-(N1-histidyl)-riboflavin linkage [20,26]. Instead, it contains a lysine residue (Lys-55) at this position. In plant GALDHs, where the corresponding residue is a leucine residue (Figure 2B), the flavin cofactor is FMN and it is bound non-covalently [22]. However, with bacterial GULDHs, where the flavin cofactor also binds non-covalently, there is a histidine residue at this position [25]. Therefore, rather than influencing the type of cofactor interaction, this residue could be a determinant of cofactor specificity, with the histidine residue being associated with FAD binding and the lysine/leucine residues with FMN binding. In the absence of information on the nature of the flavin cofactor in the bacterial enzyme, we therefore determined the functional importance of the corresponding residue in TcGAL.
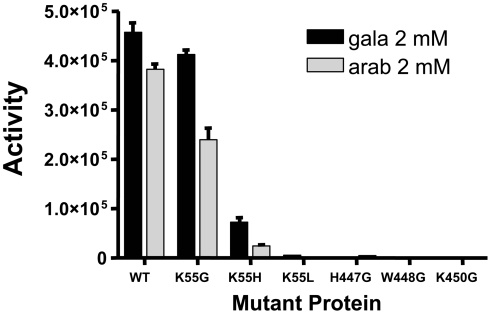
Using site-directed mutagenesis, we investigated the role of key residues (Lys-55, His-447, Trp-448 and Lys-450; Figure 2B) that are proposed to be involved in cofactor binding (Lys-55) and enzyme activity. We first converted the lysine in TcGAL into glycine (K55G). This had only a minor inhibitory effect (∼25%) on both FMN binding (Table 2) and biochemical activity (Figure 6). By inference, the lysine residue at this position in TcGAL does not have an essential role in either cofactor binding or enzyme activity. When the lysine residue was mutated to histidine (K55H), as in mammals, bacteria and fungi, FMN binding was dramatically reduced (>90%) and enzyme activity fell by a similar amount (Table 2, Figure 6). When the lysine was converted into leucine (K55L), as in plants, flavin was not detectable and the enzyme was inactive. It may be that the bulkier side chains in these amino acids provide steric hindrance that blocks FMN binding, with subsequent loss of enzyme activity. The mutation of Lys-55 to histidine, as in mammals, did not appear to alter the non-covalent nature of the residual FMN binding.
Table 2. FMN-binding properties of mutated TcGAL proteins.
Supernatants from TCA-precipitated TcGAL mutants (Figure 2) were analysed by HPLC. The resulting peaks had the same retention time as the FMN control (Figure 5). The extent of cofactor binding is expressed as a percentage of the peak height obtained with the same amount of wild-type (WT) enzyme (122 mg). Assays were performed in duplicate, and results are means±S.E.M.
| Mutant | FMN peak height (%WT) |
|---|---|
| K55G | 74.6±1.3 |
| K55H | 8.1±0.7 |
| K55L | 0 |
| H447G | 0 |
| W448G | 0 |
| K450G | 52.4±0.9 |
Figure 6. Activity of TcGAL mutant proteins.
Samples of 12.2 μg of TcGAL (WT) and six mutant proteins (Figure 2) were assayed for the production of ascorbate in the presence of 2 mM arabinonolactone (arab) or galactonolactone (gala). Activity is given in nmol of ascorbate formed/min per mg of protein. Assays were carried out in duplicate, and results are means±S.E.M.
The HWXK domain in the C-terminus of TcGAL (Figure 2B) is highly conserved among enzymes that synthesize ascorbate and is specific to this group. There are no detailed reports on the mechanism by which these enzymes function, and the role of this region has yet to be elucidated. When His-447 and Trp-448 in TcGAL were mutated individually to glycine, we found that the mutated enzyme was inactive (Figure 6) and FMN binding was not detectable (Table 2). Assuming that the amino acid changes that we introduced do not affect protein folding, these experiments therefore suggest that the histidine and tryptophan residues participate in non-covalent FMN binding, directly or indirectly. When the conserved lysine was replaced with glycine (K450G), this substitution resulted in an enzyme that retained an ability to bind FMN (∼50% of the wild-type enzyme), but which exhibited no activity. The result is consistent with Lys-450 having an important role in activity that is independent of flavin binding.
DISCUSSION
Humans do not have the capacity to synthesize ascorbate and rely on their diet to provide sufficient levels of this vitamin. In contrast, T. cruzi appears to lack the ability to scavenge ascorbate (Figure 1), and is dependent on biosynthesis to maintain intracellular levels. Ascorbate plays an important function in the T. cruzi oxidative defence system. In addition to its role as a free radical scavenger, it acts as an electron shuttle which transfers reducing equivalents from trypanothione to the endoplasmic-reticulum-localized haemoperoxidase TcAPX [5], an enzyme that appears to be essential for parasite survival (M. C. Taylor, unpublished work). In both T. brucei and T. cruzi, the enzyme responsible for the terminal step in ascorbate biosynthesis is a glycosomal flavin-dependent oxidoreductase (Table 3). Despite the widespread distribution of ascorbate-synthesizing enzymes, surprisingly little is known about their structure or the precise location of amino acids critical to function.
The mammalian and fungal ascorbate-synthesizing flavoproteins utilize covalently linked FAD as a cofactor [20,21]. In contrast, FMN is the cofactor found in plant enzymes, where it binds non-covalently [22]. In the present study, we have shown that the corresponding enzymes from T. cruzi and T. brucei resemble the plant enzymes with regard to flavin specificity and binding. Similarly to both the fungal ALOs and the plant GALDHs, the T. cruzi enzyme appears to require thiol groups for biochemical activity, as inhibition is observed in the presence of reagents which specifically inactivate these groups. The trypanosome enzymes, like those of fungi, can utilize either arabinonolactone or galactonolactone as substrates, compounds in which the hydroxy groups on the lactone ring are arranged in a trans-configuration. Whereas the T. brucei enzyme has a preference for arabinonolactone over galactonolactone (Km 55 and 154 μM respectively), TcGAL has a slightly higher affinity for galactonolactone over arabinonolactone (Km 161 and 285 μM respectively). In terms of specific activity, however, that of the T. cruzi enzyme is more than 20-fold higher than that of the T. brucei. This may reflect a greater requirement for ascorbate in a parasite that has an obligatory intracellular stage, and can be exposed to reactive oxygen and nitrogen species generated as part of the immune response. Consistent with this, T. cruzi, but not the extracellular parasite T. brucei, contains an endoplasmic-reticulum-localized APX [5].
Ascorbate-synthesizing enzymes, including those from trypanosomes, contain two conserved domains: the N-terminal flavin-binding region and the C-terminal HWXK motif. Since there is a paucity of structural data for this group of enzymes, we used site-directed mutagenesis to explore the functional significance of specific amino acids in these conserved regions. Within the N-terminal flavin-binding domain, residue 55 had been linked to cofactor binding. In the mammalian and fungal enzymes, this amino acid is a histidine residue which is covalently linked to FAD [20,21]. In trypanosomes, the corresponding residue is lysine, in plants it is leucine, and in bacteria it is histidine. Enzymes from each of these groups bind their cofactors non-covalently [21,25], although the identity of the flavin cofactor in bacteria is not yet known. One possibility is that the residue at this position could be a determinant of cofactor specificity, with histidine being associated with FAD and leucine/lysine with FMN. When Lys-55 in the T. cruzi enzyme was replaced with glycine, this had only a minor effect on enzyme activity and the extent of FMN interaction. In contrast, its replacement with histidine greatly reduced cofactor binding and enzyme activity, although there was no shift in the preference for FMN over FAD. Our results therefore show that, in TcGAL, Lys-55 does not have a direct influence on flavin specificity, although its replacement by residues with bulky side chains does reduce FMN binding, with a resultant loss of enzyme activity.
In the conserved C-terminal HWXK motif, we observed that mutations led to a loss of enzyme activity. In the case of mutants H447G and W448G, this was associated with an inability to bind FMN. With the K450G mutation, however, the enzyme retained a capacity to bind the flavin cofactor, although all activity was lost. These results highlight the significance of this conserved domain for ascorbate biosynthesis. In the flavoprotein vanillyl-alcohol oxidase [38], a C-terminal histidine residue covalently links the FAD cofactor, whereas an N-terminal histidine residue promotes this covalent linkage. If either histidine residue was mutated, however, the enzyme retained the ability to bind FAD non-covalently, although enzyme activity was reduced. It has been suggested, at least in the case of vanillyl-alcohol oxidase, that the C-terminal histidine residue could form part of a second flavin-binding domain, although a scenario where it might be involved in ‘docking’ the cofactor has been postulated. This has also been considered to be a possibility with TcGAL and TbALO [28]. Our findings indicate that, in the case of TcGAL, His-447 and Trp-448 appear to a have a role, directly or indirectly, in FMN binding, with a concomitant effect on enzyme activity. Furthermore, the observation that the K450G mutant retained a capacity for FMN binding in the absence of enzyme activity may point to this residue having an alternative role in the some other aspect of the biochemical mechanism, distinct from the cofactor interaction. Certainly the loss of activity associated with these mutations supports the hypothesis that the conserved HWXK motif plays an important role in the active site of the enzyme.
TcGAL shares many characteristics with the plant and fungal enzymes (Table 3), even though there is not a high degree of conservation at the level of primary sequence. The enzymes are of similar size and contain thiol groups that are necessary for full activity, as inferred from inhibition profiles. The trypanosomal enzyme shares with the plant enzyme a preference for non-covalently bound FMN as its prosthetic group. This contrasts with the fungal and mammalian enzymes which are reliant on covalently bound FAD. Although the trypanosomal and plant enzymes appear to be more closely related, they do not share all of the same characteristics. Both TcGAL and mammalian GLO are microsomally located, whereas the plant GALDH and fungal ALO are mitochondrial. The natural substrate of the T. cruzi enzyme is not yet known. TcGAL has the ability to use arabinonolactone or galactonolactone, which would give rise to D-erythroascorbic acid or L-ascorbic acid respectively. These products have indistinguishable biochemical properties. Fungi synthesize D-erythroascorbic acid from arabinose, whereas plants synthesize ascorbate in several ways, the best characterized pathway being from glucose via mannose and galactose to reach galactonolactone and then ascorbate. Many of the genes in this pathway, such as galacturonate reductase and GDP-mannose-3,5-epimerase, have been described previously [41]. The mammalian vitamin C biosynthetic pathway also starts with glucose, but from there it follows a different path, through glucuronate to GLO [42]. Putative homologues of each of the genes from the plant pathway can be identified in the T. cruzi genome, suggesting that ascorbate biosynthesis in trypanosomes could proceed by way of a plant-like pathway, via L-galactose and L-galactono-γ-lactone [28].
In summary, the present study shows that trypanosomes are unable to obtain ascorbate from the environment, and that the terminal step in vitamin C biosynthesis in T. cruzi involves an enzyme that is significantly different from the mammalian equivalent. Furthermore, we have demonstrated, for the first time, the importance of the HWXK motif to this reaction. Genetic-based approaches, aimed at resolving the biological importance of TcGAL, should now provide additional experimental evidence for the validation of this pathway as a candidate drug target.
Online data
Acknowledgments
We thank Dr Nick Shaw of Lonza, Visp, Switzerland, for advice on flavin analysis, and Dr Quinton Fivelman (London School of Hygiene and Tropical Medicine) for assistance with uptake assays. This work was supported by a Medical Research Council doctoral training grant to F. J. L. and the Wellcome Trust. The Bill and Melinda Gates Foundation provided support for the analytical facility through an award to the Gates Malaria Partnership at the London School of Hygiene and Tropical Medicine.
References
- 1.World Health Organization. The World Health Report 2004: Changing History. 2004 http://www.who.int/whr/2004/en/index.html.
- 2.Barrett M. P., Burchmore R. J. S., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S. The trypanosomiases. Lancet. 2003;362:1469–480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 3.Prathalingam S. R., Wilkinson S. R., Horn D., Kelly J. M. Deletion of the Trypanosoma brucei superoxide dismutase gene sodb1 increases sensitivity to nifurtimox and benznidazole. Antimicrob. Agents Chemother. 2007;51:755–758. doi: 10.1128/AAC.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carnieri E. G., Moreno S. N., Docampo R. Trypanothione-dependent peroxide metabolism in Trypanosoma cruzi different stages. Mol. Biochem. Parasitol. 1993;61:76–86. doi: 10.1016/0166-6851(93)90160-y. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson S. R., Obado S. O., Mauricio I. L., Kelly J. M. Trypanosoma cruzi expresses a plant-like ascorbate-dependent hemoperoxidase localized to the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13453–13458. doi: 10.1073/pnas.202422899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilkinson S. R., Meyer D. J., Taylor M. C., Bromley E. V., Miles M. A., Kelly J. M. The Trypanosoma cruzi enzyme TcGPXI is a glycosomal peroxidase and can be linked to trypanothione reduction by glutathione or tryparedoxin. J. Biol. Chem. 2002;277:17062–17071. doi: 10.1074/jbc.M111126200. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson S. R., Taylor M. C., Touitha S., Mauricio I. L., Meyer D. J., Kelly J. M. TcGPXII, a glutathione-dependent Trypanosoma cruzi peroxidase with substrate specificity restricted to fatty acid and phospholipid hydroperoxides, is located to the endoplasmic reticulum. Biochem. J. 2002;364:787–794. doi: 10.1042/BJ20020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson S. R., Temperton N. J., Mondragon A., Kelly J. M. Distinct mitochondrial and cytosolic enzymes mediate trypanothione-dependent peroxide metabolism in Trypanosoma cruzi. J. Biol. Chem. 2000;275:8220–8225. doi: 10.1074/jbc.275.11.8220. [DOI] [PubMed] [Google Scholar]
- 9.Clark D., Albrecht M., Arevalo J. Ascorbate variations and dehydroascorbate reductase activity in Trypanosoma cruzi epimastigotes and trypomastigotes. Mol. Biochem. Parasitol. 1994;66:143–145. doi: 10.1016/0166-6851(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 10.Krauth-Siegel R. L., Ludemann H. Reduction of dehydroascorbate by trypanothione. Mol. Biochem. Parasitol. 1996;80:203–208. doi: 10.1016/0166-6851(96)02689-8. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez M. N., Piacenza L., Irigoin F., Peluffo G., Radi R. Macrophage-derived peroxynitrite diffusion and toxicity to Trypanosoma cruzi. Arch. Biochem. Biophys. 2004;432:222–232. doi: 10.1016/j.abb.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Sousa-Franco J., Araujo-Mendes E., Silva-Jardim I., L.-Santos J., Faria D. R., Dutra W. O., Horta M. F. Infection-induced respiratory burst in BALB/c macrophages kills Leishmania guyanensis amastigotes through apoptosis: possible involvement in resistance to cutaneous leishmaniasis. Microb. Infect. 2006;8:390–400. doi: 10.1016/j.micinf.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Frei B., England L., Ames B. N. Ascorbate is an outstanding antioxidant in human blood plasma. Proc. Natl. Acad. Sci. U.S.A. 1989;86:6377–6381. doi: 10.1073/pnas.86.16.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turrens J. F. Oxidative stress and antioxidant defenses: a target for the treatment of diseases caused by parasitic protozoa. Mol. Aspects Med. 2004;25:211–220. doi: 10.1016/j.mam.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 15.Halliwell B. Vitamin C: antioxidant or pro-oxidant in vivo. Free Radical Res. 1996;25:439–454. doi: 10.3109/10715769609149066. [DOI] [PubMed] [Google Scholar]
- 16.Buettner G. R. The pecking order of free radicals and antioxidants: lipid peroxidation, α-tocopherol, and ascorbate. Arch. Biochem. Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- 17.Pauling L. Evolution and the need for ascorbic acid. Proc. Natl. Acad. Sci. U.S.A. 1970;67:1643–1648. doi: 10.1073/pnas.67.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carr A. C., Frei B. Toward a new recommended dietary allowance for vitamin C based on antioxidant and health effects in humans. Am. J. Clin. Nutr. 1999;69:1086–1107. doi: 10.1093/ajcn/69.6.1086. [DOI] [PubMed] [Google Scholar]
- 19.Levine M., Rumsey S. C., Daruwala R., Park J. B., Wang Y. Criteria and recommendations for vitamin C intake. JAMA, J. Am. Med. Assoc. 1999;281:1415–1423. doi: 10.1001/jama.281.15.1415. [DOI] [PubMed] [Google Scholar]
- 20.Brush J. S., May H. E. A kinetic study of the mechanism of action of L-gulononlactone oxidase. J. Biol. Chem. 1966;241:2907–2912. [PubMed] [Google Scholar]
- 21.Huh W.-K., Kim S.-T., Yang K.-S., Seok Y.-J., Hah Y. C., Kang S.-O. Characterisation of D-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur. J. Biochem. 1994;225:1073–1079. doi: 10.1111/j.1432-1033.1994.1073b.x. [DOI] [PubMed] [Google Scholar]
- 22.Mapson L. W., Breslow E. Biological synthesis of ascorbic acid: L-galactono-γ-lactone dehydrogenase. Biochem. J. 1958;68:395–406. doi: 10.1042/bj0680395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostergaard J., Persiau G., Davey M. W., Bauw G., Van Montagu M. Isolation of a cDNA coding for L-galactono-γ-lactone dehydrogenase, an enzyme involved in the biosynthesis of ascorbic acid in plants. J. Biol. Chem. 1997;272:30009–30016. doi: 10.1074/jbc.272.48.30009. [DOI] [PubMed] [Google Scholar]
- 24.Sugisawa T., Ojima S., Matzinger P. K., Hoshino T. Isolation and characterisation of a new vitamin C producing enzyme (L-gulononlactone dehydrogenase) of bacterial origin. Biosci. Biotech. Biochem. 1995;59:190–196. [Google Scholar]
- 25.Wolucka B. A., Communi D. Mycobacterium tuberculosis possesses a functional enzyme for the synthesis of vitamin C, L-gulono-1,4-lactone dehydrogenase. FEBS J. 2006;273:4435–4445. doi: 10.1111/j.1742-4658.2006.05443.x. [DOI] [PubMed] [Google Scholar]
- 26.Mewies M., McIntire W. S., Scrutton N. S. Covalent attachment of flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) to enzymes: the current state of affairs. Protein Sci. 1998;7:7–20. doi: 10.1002/pro.5560070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishikimi M., Fukuyama R., Minoshima S., Shimizu N., Yagi K. Cloning and chromosomal mapping of the human non-functional gene for L-gulonolactone oxidase, the enzyme for L-ascorbic acid biosynthesis missing in man. J. Biol. Chem. 1994;269:13685–13688. [PubMed] [Google Scholar]
- 28.Wilkinson S. R., Prathalingam S. R., Taylor M. C., Horn D., Kelly J. M. Vitamin C biosynthesis in trypanosomes: a role for the glycosome. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11645–11650. doi: 10.1073/pnas.0504251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kendall G., Wilderspin A. F., Ashall F., Miles M. A., Kelly J. M. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase does not conform to the ‘hotspot’ topogenic signal model. EMBO J. 1990;9:2751–2758. doi: 10.1002/j.1460-2075.1990.tb07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson W. C., Miles M. A. The karyotype and ploidy of Trypanosoma cruzi. EMBO J. 1986;5:1299–1305. doi: 10.1002/j.1460-2075.1986.tb04359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oba K., Ishikawa S., Nishikawa M., Mizuno H., Yamamoto T. Purification and properties of L-galactono-γ-lactone dehydrogenase, key enzyme for ascorbic acid biosynthesis, from sweet potato roots. J. Biochem. (Tokyo) 1995;117:120–124. doi: 10.1093/oxfordjournals.jbchem.a124697. [DOI] [PubMed] [Google Scholar]
- 32.Kim S.-T., Huh W.-K., Kim J.-Y., Hwang S.-W., Kang S.-O. D-Arabinose dehydrogenase and biosynthesis of erythroascorbate in Candida albicans. Biochim. Biophys. Acta. 1996;1297:1–8. doi: 10.1016/0167-4838(96)00077-5. [DOI] [PubMed] [Google Scholar]
- 33.Birch O. M., Hewitson K. S., Fuhrmann M., Burgdorf K., Baldwin J. E., Roach P. L., Shaw N. M. MioC is an FMN-binding protein that is essential for Escherichia coli biotin synthase activity in vitro. J. Biol. Chem. 2000;275:32277–32280. doi: 10.1074/jbc.M004497200. [DOI] [PubMed] [Google Scholar]
- 34.Rohloff P., Rodrigues C. O., Docampo R. Regulatory volume decrease in Trypanosoma cruzi involves amino acid efflux and changes in intracellular calcium. Mol. Biochem. Parasitol. 2003;126:219–230. doi: 10.1016/s0166-6851(02)00277-3. [DOI] [PubMed] [Google Scholar]
- 35.Vera J. C., Rivas C. I., Fischbarg J., Golde D. W. Mammalian facilitative hexose transporters mediate the transport of dehydroascorbic acid. Nature. 1993;364:79–82. doi: 10.1038/364079a0. [DOI] [PubMed] [Google Scholar]
- 36.El-Sayed N. M., Myler P. J., Bartholomeu D. C., Nilsson D., Aggarwal G., Tran A. N., Ghedin E., Worthey E. A., Delcher A. L., Blandin G., et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 37.Machado C. A., Ayala F. J. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. U.S.A. 2001;98:7396–7401. doi: 10.1073/pnas.121187198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraaije M. W., van den Heuvel R. H. H., van Berkel W. J. H., Mattevi A. Structural analysis of flavinylation in vanillyl-alcohol oxidase. J. Biol. Chem. 2000;275:38654–38658. doi: 10.1074/jbc.M004753200. [DOI] [PubMed] [Google Scholar]
- 39.Stohs S. J., Bagchi D. Oxidative mechanisms in the toxicity of metal ions. Free Radical Biol. Med. 1995;18:321–336. doi: 10.1016/0891-5849(94)00159-h. [DOI] [PubMed] [Google Scholar]
- 40.Cisneros R. J., Dunlap R. B. Development of a trichloroacetic acid precipitation assay for covalent adducts of thymidylate synthase. Anal. Biochem. 1990;186:202–208. doi: 10.1016/0003-2697(90)90067-j. [DOI] [PubMed] [Google Scholar]
- 41.Smirnoff N., Conklin P. L., Loewus F. A. Biosynthesis of ascorbic acid in plants: a renaissance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:437–467. doi: 10.1146/annurev.arplant.52.1.437. [DOI] [PubMed] [Google Scholar]
- 42.Linster C. L., Van Schaftingen E. Vitamin C biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274:1–22. doi: 10.1111/j.1742-4658.2006.05607.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.