Abstract
The micronutrient selenium is present in proteins as selenocysteine (Sec). In eukaryotes and archaea, Sec is formed in a tRNA-dependent conversion of O-phosphoserine (Sep) by O-phosphoseryl-tRNA:selenocysteinyl-tRNA synthase (SepSecS). Here, we present the crystal structure of Methanococcus maripaludis SepSecS complexed with PLP at 2.5 Å resolution. SepSecS, a member of the Fold Type I PLP enzyme family, forms an (α2)2 homotetramer through its N-terminal extension. The active site lies on the dimer interface with each monomer contributing essential residues. In contrast to other Fold Type I PLP enzymes, Asn247 in SepSecS replaces the conserved Asp in binding the pyridinium nitrogen of PLP. A structural comparison with Escherichia coli selenocysteine lyase allowed construction of a model of Sep binding to the SepSecS catalytic site. Mutations of three conserved active site arginines (Arg72, Arg94, Arg307), protruding from the neighboring subunit, led to loss of in vivo and in vitro activity. The lack of active site cysteines demonstrates that a perselenide is not involved in SepSecS-catalyzed Sec formation; instead, the conserved arginines may facilitate the selenation reaction. Structural phylogeny shows that SepSecS evolved early in the history of PLP enzymes, and indicates that tRNA-dependent Sec formation is a primordial process.
INTRODUCTION
The indirect tRNA-dependent pathways of aminoacyl-tRNA formation, in which a noncognate amino acid bound to tRNA is converted to the cognate one, are widely distributed in nature. In fact, the tRNA-dependent pathways for Gln and Asn formation are evolutionarily older than the corresponding direct aminoacylation route catalyzed by the aminoacyl-tRNA synthetases (1). Although selenocysteine occurs in organisms from all three domains of life (2,3), Sec-tRNA is synthesized solely by the indirect route; actually it is the only natural amino acid found in proteins for which a cognate aminoacyl-tRNA synthetase did not evolve. Seryl-tRNA synthetase (SerRS) forms Ser-tRNASec in bacteria (4), archaea (5,6) and eukaryotes (7). Using the selenium donor selenophosphate bacteria convert this misacylated aminoacyl-tRNA species to Sec-tRNASec by the action of the SelA protein, a PLP-dependent selenocysteine synthase (3). Some methanogenic archaea harbor a gene that was thought to encode a SelA homolog (e.g. MJ0158), but its product is unable to synthesize Sec-tRNASec in vitro (6).
On the other hand, eukaryotes and archaea require an additional phosphorylation step catalyzed by O-phosphoseryl-tRNASec kinase (PSTK) (8–10) and convert the resulting Sep-tRNASec to Sec-tRNASec by Sep-tRNA:Sec-tRNA synthase (SepSecS) (11,12). An unexpected and important property of the human SepSecS protein is the fact that it is the target antigen for soluble liver antigen/liver–pancreas (SLA/LP) autoantibodies (13–15) that are found in about a quarter of the patients with autoimmune hepatitis (16). The reactions catalyzed by PSTK and SepSecS are reminiscent of the indirect pathway of Cys-tRNACys synthesis in archaeal methanogens (17) where Sep-tRNACys is converted to Cys-tRNACys by Sep-tRNA:Cys-tRNA synthase (SepCysS), a PLP-dependent enzyme carrying out a β-replacement on tRNA-bound Sep. The crystal structure of Archaeoglobus fuldgidus SepCysS has been reported (18).
The initial characterization of SepSecS revealed that this protein is a PLP-dependent enzyme (11,12). In nature such enzymes are abundant; in some microbial genomes they represent as much as 1.5% of all genes (19). They have many diverse functions and are often involved in amino acid biosynthesis (20). The structures of three other PLP-dependent enzymes that use substrates (selenocysteine, cysteine) chemically similar to those of SepSecS have been solved. One is the A. fulgidus SepCysS (AFSepCysS) (18), while another one is Escherichia coli selenocysteine lyase (ECCsdB) (21), an enzyme that converts selenocysteine to alanine and elemental selenium (22). The last enzyme is the E. coli cysteine desulfurase (ECIscS) that catalyzes the desulfuration of cysteine (23). The reactions catalyzed by these enzymes are illustrated in Scheme 1; SepSecS and SepCysS carry out β-replacements on tRNA-bound Sep, while selenocysteine lyase removes the β-substituent of Sec to form elemental selenium, and cysteine desulfurase catalyzes the fragmentation of cysteine to alanine and elemental sulfur. Biochemical data on SepSecS and SepCysS currently do not exist.
Scheme 1.
Graphic representation of the reaction schemes. (A) MMPSepSecS catalyzes the conversion of tRNASec-bound Sep to Sec. (B) AFSepCysS mediates the tRNACys-dependent transformation of Sep to Cys. (C) ECCsdB converts selenocysteine to alanine and elemental selenium. (D) ECIscS converts cysteine to alanine and elemental sulfur.
Here, we report the crystal structure of Methanococcus maripaludis SepSecS (MMPSepSecS) at 2.5 Å resolution and perform a structural comparison with ECCsdB and AFSepCysS. We propose active site residues important for the enzymatic function of MMPSepSecS by employing a combination of mutational in vivo and in vitro activity analyses. Finally, we present a structural phylogeny of the Fold Type I family of PLP-dependent enzymes that documents the evolutionary history of SepSecS.
MATERIALS AND METHODS
General
Oligonucleotide synthesis and DNA sequencing was performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. [14C]Serine (163 mCi/mmol) and [α-32P]ATP (10 mmol/μCi) were obtained from Amersham Pharmacia Biosciences (Piscataway, NJ, USA). The E. coli BL21-CodonPlus (DE3)-RIL strain and the pUC18 vector were from Stratagene (LaJolla, CA, USA). The pET15b and the pACYC184 vectors were from Novagen (San Diego, CA, USA). Nickel-nitriloacetic acid agarose was from Qiagen (Valencia, CA, USA). Nickel-sepharose and Resource PHE were from GE Healthcare Bio-Sciences KK (Tokyo, Japan).
Bacterial strains and plasmids
Construction of the E. coli ΔselA deletion strain JS1 (DE3), cloning of the M. maripaludis SepSecS gene (MMP0595) and the E. coli SelD gene into the pET15b vector, of the M. maripaludis tRNASec gene into the pUC18 vector, and of the M. jannaschii PSTK gene into the pACYC184 vector were described previously (11). The M. maripaludis SepSecS mutants R72A, R72Q, R72K, R94A, R94Q, H166A, H166F, H166Q, R307A, R307Q, R307K, Q102A, K278A, N247A, D277A, and K278A were generated using the QuikChange site-directed mutagenesis kit (Stratagene) and cloned into the pET15b vector with an N-terminal His-tag. An N-terminal Δ1–34SepSecS deletion mutant was constructed by PCR using the primers 5′-CCGCTCGAGCATCGGAAAATTCCTGAAAACGGAATTGATGACG-3′ and 5′-GCTAGTTATTGCTCAGCGGTGGCAGC-3′ and the pET15b-sepsecS plasmid DNA as the template. The resulting DNA fragment was digested with BamHI and XhoI and re-inserted into the pET15b vector.
Protein expression and purification
Expression and purification of the M. maripaludis wild-type and mutant SepSecS proteins and the E. coli SelD (used for biochemical experiments) was done as described previously using Ni-NTA column chromatography (11). Purification of the wild-type SepSecS used for crystallization involved two chromatographic steps. Briefly, pET15b-sepsecS was transformed into the E. coli BL21-CodonPlus (DE3)-RIL strain, cells were grown to A600 = 0.6 and gene expression was induced with 0.4 mM IPTG. After growth for 17 h at 20°C, cells were harvested, resuspended in 50 mM Tris–HCl (pH 7.0), 300 mM NaCl, 10 μM PLP, 5 mM 2-mercaptoethanol, 10% glycerol, 1 mM PMSF and gently sonicated. After centrifugation at 14 000g for 30 min, the supernatant was collected and purified by sequential passage through a Ni-Sepharose and a Resource PHE chromatography column.
Gel filtration of MMPSepSecS
A 0.5 ml sample of a 1.5 mg/ml purified solution of selenomethionine-labeled MMPSepSecS was loaded onto a HiPrep 16/60 Sephacryl S-300 HR column (GE Healthcare). The column was run at 0.5 ml/min in the same buffer as crystallization, containing 20 mM HEPES (pH 7.0), 300 mM NaCl, 10 μM PLP and 5 mM DTT. The elution volume of MMPSepSecS was compared to the elution volumes of other oligomeric proteins according to the GE healthcare web site (http://www.gelifesciences.co.jp/catalog/pdf_attach/18106088AC.pdf).
Crystallization, structure determination and refinement
The purified SepSecS was dialyzed against crystallization buffer, containing 20 mM HEPES–NaOH (pH 7.0), 10 μM PLP and 5 mM DTT, 300 mM NaCl and was concentrated. Crystals of M. maripaludis SepSecS were grown within a day at 20°C by the sitting-drop vapor diffusion method. Drops were prepared by mixing equal volumes of the 6 mg/ml SepSecS solution and the reservoir solution, containing 45 mM HEPES–NaOH (pH 7.0), 10 mM MES–HCl (pH 6.5), 90 mM KCl, 9 mM CaCl2, 160 mM MgSO4 and 10% PEG550MME. Selenomethionine-labeled SepSecS was prepared by the conventional method and was purified in the same manner as the wild-type.
The SepSecS crystals were flash-cooled in a nitrogen stream at 100 K. All diffraction data sets were collected at the BL41XU at SPring-8 (Harima, Japan), and were processed with the HKL2000 suite. The crystals belong to the primitive monoclinic space group P21, with unit-cell parameters a = 75.7, b = 108.1, c = 110.4 Å, β = 97°. There are four SepSecS molecules in the asymmetric unit. A multiwavelength anomalous dispersion (MAD) data set of the selenomethionine-substituted crystals was collected, and was used to search for the locations of the selenium atoms by using the program SnB (24). Subsequent phase refinements were performed with the program SHARP (25), and the model was manually built into the electron density maps by using the program O (26). The model was refined against reflections up to 2.5 Å resolution by using the program CNS (27). The backbones of all residues were clearly defined in the final 2Fo−Fc electron density maps. Graphic representations were prepared with CueMol (http://www.cuemol.org).
Statistics on data collection, phasing and refinement are shown in Tables 1 and 2.
Table 1.
Data collection and phasing statistics
| X-ray source | SPring-8 BL41XU | ||||
|---|---|---|---|---|---|
| Peak | Edge | Reml | Remh | ||
| Data collection statistics | SeMet | ||||
| Wavelength (Å) | 0.9791 | 0.9793 | 0.9820 | 0.9770 | |
| Resolution (Å) | 50–2.5 (2.54–2.5) | 50–2.5 (2.54–2.5) | 50–2.5 (2.54–2.5) | 50–2.5 (2.54–2.5) | |
| Unique reflections | 59 938 | 59 274 | 59 463 | 59 123 | |
| Redundancy | 5.0 (2.4) | 4.9 (2.3) | 4.8 (2.2) | 4.8 (2.0) | |
| Completeness (%) | 97.9 (86.1) | 97.6 (86.1) | 97.0 (81.2) | 96.6 (79.3) | |
| I/s(I) | 17.0 (2.9) | 16.4 (2.7) | 16.3 (2.6) | 15.3 (2.4) | |
| Rsym | 0.135 (0.258) | 0.127 (0.272) | 0.125 (0.261) | 0.127 (0.272) | |
| Phasing statistics | |||||
| No. of Se sites | 31 | 31 | 31 | 31 | |
| Phasing power | |||||
| Iso (cen./acen.) | 0.633/0.587 | – | 1.006/0.879 | 0.706/0.653 | |
| Ano | 1.399 | 0.79 | 0.107 | 0.811 | |
| Rcullis | |||||
| Iso (cen./acen.) | 0.896/0.833 | – | 0.713/0.767 | 0.749/0.794 | |
| Ano | 0.754 | 0.89 | 0.987 | 0.89 | |
| Mean FOM | |||||
| Cen./Acen. | 0.406/0.461 | ||||
The numbers in parentheses are for the last shell.

Table 2.
Structure refinement statistics
| Refinement statistics | Se-Met |
|---|---|
| Resolution (Å) | 50–2.5 |
| No. of atoms | |
| Protein | 13 552 |
| Water | 146 |
| PLP | 160 |
| SO4 | 10 |
| Luzzati coordinate error (Å) | 0.3 |
| Cross-validated Luzzati coordinate error (Å) | 0.39 |
| RMSD of | |
| Bond length (Å) | 0.007 |
| Bond angle (°) | 1.39 |
| Dihedral angle (°) | 22.2 |
| Improper angle (°) | 0.89 |
| Average B factor (Å2) | 38.5 |
| Ramachandran plot | |
| Core region (%) | 88.2 |
| Additionally allowed region (%) | 11.3 |
| Generously allowed region (%) | 0.3 |
| Disallowed region (%) | 0.2 |
| Rwork/Rfree | 0.208/0.269 |
Rwork = Σ|Fo−Fc|/ΣFo for reflections of work set.
Rfree = Σ|Fo−Fc|/ΣFo for reflections of test set (10% of total reflections).
In vivo SepSecS assay
The M. maripaludis wild-type and mutant SepSecS genes were transformed into the ΔselA E. coli JS1 strain with or without the M. jannaschii PSTK gene. Aerobic overnight cultures were streaked in aerobic conditions on LB-agar plates supplemented with 0.01 mM IPTG, 1 μM Na2MoO4, 1 μM Na2SeO3 and 50 mM sodium formate. The plates were placed in an anaerobic incubation jar that was flushed with an N2: CO2: H2 (90 : 5 : 5) gas mix three times to give an anaerobic atmosphere and then grown for 16 h at 37°C and 36 h at 30°C. The plates were then overlaid with agar containing 1 mg/ml benzyl viologen (BV), 0.25 M sodium formate and 25 mM KH2PO4 adjusted to pH 7.0. The appearance of a blue/purple color is the indication of active formate dehydrogenase H (FDHH).
Preparation and purification of tRNA gene transcripts
The M. maripaludis tRNASec used as substrate in the in vitro assays was synthesized by in vitro T7 RNA polymerase run-off transcription as described (28). The tRNASec gene together with the T7 promoter was constructed from overlapping chemically synthesized oligonucleotides, cloned into the pUC18 plasmid and purified from E.coli DH5α transformants using a MaxiPrep plasmid purification kit (Qiagen). The purified plasmid was digested with BstN1 at 55°C for 16 h. The in vitro transcription reaction was performed at 37°C for 5 h in buffer containing 40 mM Tris–HCl (pH 8), 22 mM MgCl2, 25 mM DTT, 2 mM spermidine, 50 μg/ml BSA, 0.1 mg/ml pyrophosphatase, 4 mM of each nucleoside triphosphate, BstNI-digested vector containing the tRNASec gene (60 µg/ml) and 1 mM T7 RNA polymerase. The tRNASec transcript was purified by electrophoresis on a 12% denaturing polyacrylamide gel. Full-length tRNA was eluted and desalted on Sephadex G25 Microspin columns (Amersham). The tRNA transcripts were refolded by heating for 5 min at 70°C in buffer containing 10mM Tris–HCl (pH 7.0), followed by addition of 5 mM MgCl2 and immediate cooling on ice (5).
Preparation of 32P-labeled Sep-tRNASec
Refolded tRNASec transcript was 32P-labeled on the 3′ terminus by using the E. coli CCA-adding enzyme and [α-32P]AMP (Amersham) as previously described with some modifications (29). Briefly, 6μg of tRNASec transcript was incubated with the CCA-adding enzyme and [α-32P]ATP (50 μCi) for 1 h at room temperature in buffer containing 50 mM Tris–HCl (pH 8.0), 20 mM MgCl2, 5 mM DTT and 50 μM sodium pyrophosphate. After phenol/chloroform extraction the sample was passed over a Sephadex G25 Microspin column (Amersham) to remove excess ATP (30).
The recovered [32P]-labeled tRNASec was serylated and phosphorylated by M. maripaludis SerRS (5 μM) and M. jannaschii PSTK (1 μM) for 75 min at 37°C in buffer containing 50 mM HEPES (pH 7.5), 10 mM MgCl2, 20 mM KCl, 1 mM DTT, 1 mM serine and 10 mM ATP. After phenol/chloroform extraction aminoacylated Sep-tRNASec was ethanol precipitated at −20°C for 45 min and collected as a pellet by centrifugation at 10 000 g at 4°C for 30 min. After washing the pellet with 70% ethanol, it was allowed to dry on ice in order to avoid deacylation.
To check levels of serylation and Ser→Sep conversion 1 μl aliquots at the start and end of the reaction were quenched on ice with 3 μl of 100 mM sodium citrate (pH 4.75) and 0.66 mg/ml of nuclease P1 (Sigma). Following nuclease P1 digestion at room temperature for 1 h, 1.5 μl of the sample was spotted onto polyethyleneimine (PEI) cellulose 20 cm × 20 cm thin layer chromatography (TLC) plates (Merck). To separate the Sep-[32P]AMP spot from [32P]AMP and any remaining Ser-[32P]AMP the plates were developed for 75 min in buffer containing 100 mM ammonium acetate, 5% acetic acid. The plates were exposed on an imaging plate (FujiFilms) for 14 h, scanned using a Molecular Dynamics Storm 860 scanner and quantified using the ImageQuant densitometry software. The amount of Sep-tRNASec formed can be calculated by dividing the intensity of the Sep-[32P]AMP spot by the sum of the intensities of all spots (Sep-[32P]AMP, [32P]AMP and Ser-[32P]AMP).
In vitro conversion of Sep-tRNASec to Cys-tRNASec
Purified wild-type or mutant MMPSepSecS (1 μM) was incubated with 1 μM 32P-labeled Sep-tRNASec in buffer containing 50 mM HEPES (pH 7.0), 20 mM KCl, 10 mM MgCl2, 5 mM DTT, 2 μM PLP and 500 μM sodium thiophosphate. Reactions were carried out anaerobically at 37°C over 40 min. At each time point taken, 1 μl reaction aliquots were quenched on ice with 3 μl of 100 mM sodium citrate (pH 4.75) and 0.66 mg/ml of nuclease P1 (Sigma). Following nuclease P1 digestion, 1.5 μl of the sample was spotted onto PEI cellulose TLC plates that were developed, scanned and quantified as described above. To separate the Cys-[32P]AMP spot from the Sep-[32P]AMP, the [32P]AMP and any remaining Ser-[32P]AMP spots the plates were developed for 75 min in buffer 100 mM ammonium acetate, 5% acetic acid. The plates were exposed on an imaging plate (FujiFilms) for 14 h, scanned using a Molecular Dynamics Storm 860 scanner and quantified using the ImageQuant densitometry software. The amount of Cys-[32P]AMP formed was calculated by dividing the intensity of the Cys-[32P]AMP spot by the sum of the intensities of all spots (Cys-[32P]AMP, Sep-[32P]AMP, [32P]AMP and Ser-[32P]AMP). The added elevated concentration of PLP was used to assure that the mutant enzymes were saturated with the cofactor.
In vitro conversion of Sep-tRNASec to Sec-tRNASec
The Sep-to-Sec conversion reaction was carried out as described before (11). Briefly, purified tRNASec (10 μM) was incubated with M. maripaludis SerRS (6 μM) and M. jannaschii PSTK (3 μM) in reaction buffer containing 100 μM [14C]Ser, 100 mM HEPES (pH 7.0), 10 mM KCl, 10 mM magnesium acetate, 1 mM DTT and 0.1 mg/ml BSA at 37°C for 1 h. The aminoacylated Sep-tRNASec products were purified by phenol extraction followed by passage over a Sephadex G25 Microspin column (Amersham) and ethanol precipitation. Purified wild-type SepSecS or the R72Q mutant was incubated with 10 μM Sep-tRNASec and 100 μM purified E. coli SelD in reaction buffer containing 100 mM HEPES pH 7.0, 300 mM KCl, 10 mM MgCl2, 1 mM DTT and 250 μM Na2SeO3. All buffers were prepared anaerobically and the reaction was carried out in an anaerobic chamber at 37°C. After 30 min incubation the reaction was stopped by phenol extraction and the tRNAs were purified by application on a Sephadex G25 Microspin column (Amersham) and ethanol precipitation. Purified tRNA products were deacylated in 20 mM NaOH at room temperature for 10 min. The released amino acids were oxidized with performic acid and spotted onto silica gel 60 TLC aluminium sheets (Merck) that were subsequently developed in 85% ethanol.
Alignment and phylogeny
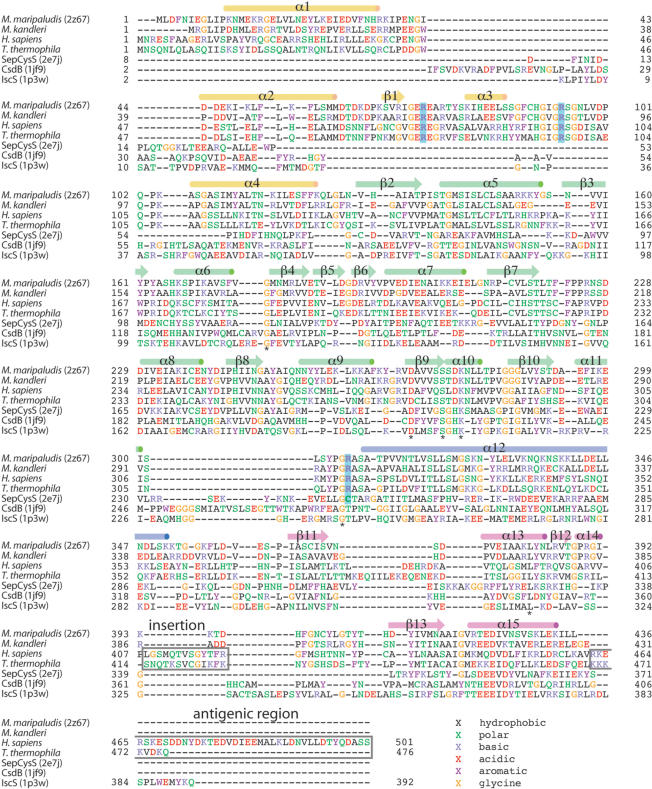
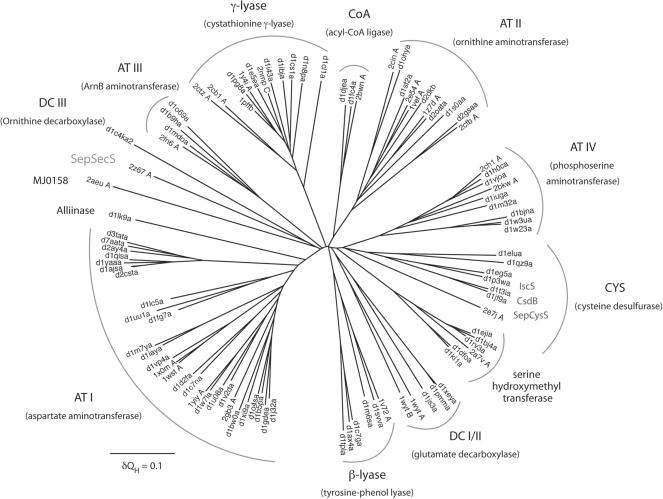
The STAMP (31) structural superposition algorithm in the Multiseq 2.0 module of VMD 1.8.6 (32) was used to establish a structure-based alignment between SepSecS and the other members of the fold type I PLP-dependent family. The structural similarity measure QH (33) was used to determine evolutionary distances between members of the fold type I group for the structural phylogeny shown in Figure 8. The tree was drawn using the programs NEIGHBOR and DRAWTREE in the Phylip 3.66 package (34). A similar structure-based alignment was used for the structure-based sequence alignment shown in Figure 2. First, SepSecS was structurally aligned to SepCysS, IscS and CsdB. This structure-based alignment was then supplemented with three additional SepSecS sequences, which had been previously aligned to MMPSepSecS with CLUSTAL (35). Some alignment ambiguities were corrected by manual adjustment to the structure-based sequence alignment.
Figure 8.
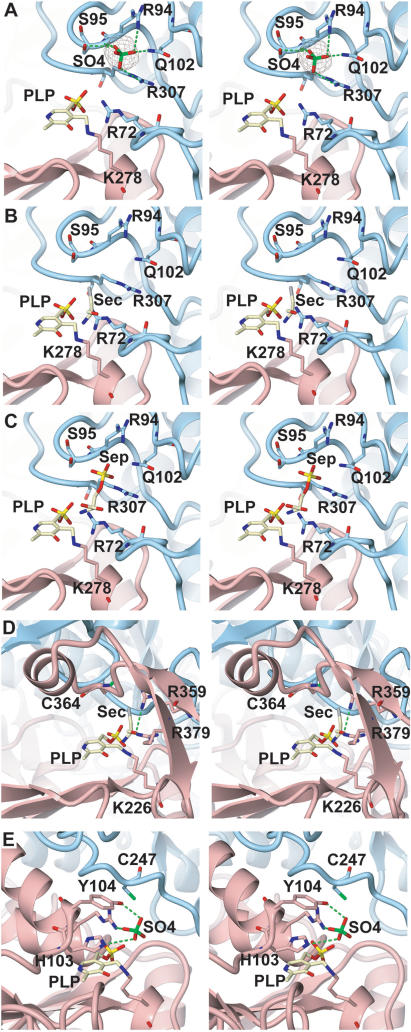
The active site. Close-up stereo views of the active site regions in MMPSepSecS, ECCsdB, and AFSepCysS. Chains A and B are colored pink and blue, respectively. The PLP molecule (shown as a ball and stick model) is covalently bound to chain A. (A) The recognition manner of the sulfate ion in the active site of MMPSepSecS. The sulfate ion is shown as a ball-and-stick model. The Fo−Fc omit map of the sulfate ion, contoured at 4.5 σ, is shown. (B) Sec docking model of MMPSepSecS, based on the structural comparison with ECCsdB. (C) Sep binding model of MMPSepSecS, based on the locations of the sulfate ion and Sep. (D) Close-up view of the active site region in ECCsdB. The PLP molecule is covalently bound to Lys226 of chain A. The orientation of the PLP molecule is the same as that in (A). The chain A residues that recognize PLP are shown. The PLP molecule is covered by the βαβ structural motif. (E) Close-up view of the active site region in AFSepCysS. The PLP molecule is covalently bound to Lys209 of chain A. The orientation of PLP is the same as that in (A). The chain A residues that recognize PLP and sulfate are shown.
Figure 2.
Structure-based amino acid sequence alignment. The alignment includes representative SepSecS sequences from two archaea (M. maripaludis and M. kandleri) and two eukaryotes (H. sapiens and T. thermophila) as well as A. fulgidus SepCysS, E. coli IscS and E. coli CsdB. Amino acids are colored according to residue types, and strictly conserved residues are marked (*). The secondary structure of MMPSepSecS is illustrated above the alignment, and the active site arginines are highlighted in blue. The autoimmune antigenic region of human SepSecS (37) and a eukaryotic specific insertion are shown in gray boxes.
Accession numbers
The Protein Data Bank (http://www.rscb.org/pdb) accession number for the coordinates of MMPSepSecS conjugated with PLP is 2Z67.
RESULTS
Experimental outline
The MMPSepSecS protein was overproduced in E. coli, then purified by two column chromatographic steps, and crystallized. Using selenomethionine-labeled protein the structure was solved by the MAD method. The complex of SepSecS with covalently-bound PLP was refined to an Rfree of 26.9% at 2.5 Å resolution.
In this first characterization of M. maripaludis SepSecS (MMPSepSecS) activity, we used the genetic complementation of an E. coli ΔselA deletion strain as an in vivo test. When grown anaerobically, E. coli produces the selenium-dependent formate dehydrogenase FDHH. Its activity enables the cells to reduce benzyl viologen (BV) in the presence of formate; this is usually observed by a blue/purple color in agar overlay plates under anaerobic conditions (36). Furthermore, we employed two in vitro tests. The first detects the enzyme's final reaction product, Sec-tRNASec, as determined by TLC of Sec released from tRNASec (11). Given the difficulty in working with selenophosphate (availability and oxygen sensitivity) we used thiophosphate as a surrogate substrate to measure the time course of Cys-tRNASec formation by SepSecS. In this assay, 32P-labeled Sep-tRNASec was incubated anaerobically with wild-type or mutant SepSecS proteins and thiophosphate. After nuclease P1 digestion of the tRNA in the reaction mixture, the product Cys-[32P]AMP was separated from Sep-[32P]AMP by TLC and quantitated.
Overall structure of M. maripaludis SepSecS
The MMPSepSecS protein adopts an L-shaped structure consisting of the N-terminal extension domain (1–130), a catalytic domain (131–309) and a C-terminal domain (353–436) (Figure 1A). Long, kinked helices (310–352) connect the catalytic and C-terminal domains. The PLP molecule is covalently bound to the conserved Lys residue K278 (Figure 2) at the active site. As can be seen in Figure 1, the overall architecture of MMPSepSecS is similar to the Fold Type I (20) PLP enzymes AFSepCysS (18) and ECCsdB (21) (Figure 1). These enzymes consist of a catalytic domain similar to that of MMPSepSecS, and a ‘small domain’, formed by the N-terminal polypeptide and C-terminal polypeptide, which also resembles the MMPSepSecS C-terminal domain (Figure 1B and C) (18,21). We constructed a structure-based sequence alignment (Figure 2) in order to accurately compare MMPSepSecS to three PLP enzymes, AFSepCysS, ECCsdB and ECIscS, that act upon chemically similar substrates. According to structural similarity measures SepCysS, CsdB and IscS are significantly more closely related to each other than they are to SepSecS (Figure 3). Sequence relationships show a similar trend with SepCysS, CsdB and IscS sharing 24 identical residues while these proteins have only 8 residues in common with SepSecS (Figure 2). The alignment also shows that the archaeal SepSecS proteins lack the C-terminal extension that has been identified as the major antigenic region for the SLA/LP autoantibodies (37).
Figure 1.
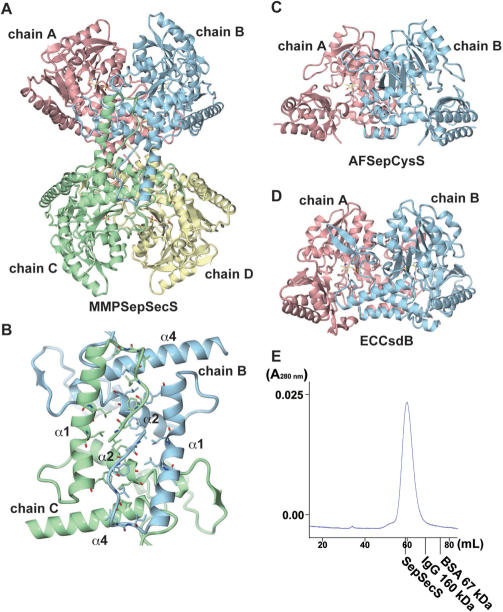
The overall architectures of SepSecS and related enzymes. (A) Stereo view of the MMPSepSecS structure. The N-terminal domain (residues 1–130), the catalytic domain (residues 131–309), the linker helix (310–352) and the C-terminal domain (residues 353–434) are colored yellow, green, blue and pink, respectively. The carboxy-terminal two residues are disordered. The PLP molecule bound to the active site is shown as a ball and stick model. (B) Stereo view of the AFSepCysS structure in the same orientation as (A) (18). The small domain, the linker helices and the catalytic domain are colored pink, blue and green, respectively. (C) Stereo view of the ECCsdB structure in the same orientation as (A) (21). The coloring scheme is the same as in (B).
Figure 3.
Structural phylogeny of the Fold Type I PLP-dependent enzyme family. The phylogenetic tree depicts the evolutionary history of the Fold Type I family. Branch lengths in the tree are proportional to structural differences between the members of the family according to the structural similarity measure QH (see ‘Materials and methods’ section). The subfamily clusters are denoted by arced lines and under each subfamily name (e.g. AT I) a representative enzyme name (e.g. aspartate aminotransferase) is given in parentheses. Isofunctional subfamilies are labeled only by their corresponding enzyme names. Protein structures used to build the tree are noted according to PDB (46) or SCOP (47) codes.
In solution, SepSecS is a tetramer as revealed by gel filtration (Figure 4E). The asymmetric unit of the crystal contains four SepSecS molecules (chains A, B, C and D) related by non-crystallographic symmetry, suggesting that SepSecS forms a homotetramer (dimer of homodimers) (Figure 4A). The four molecules are mostly identical, with root mean square (RMS) deviations of about 0.5 Å between the subunits. The N-terminal extension domain of each subunit plays a pivotal role in the tetrameric organization of SepSecS. These domains interact with each other to form a hydrophobic core (chain A with chain D, and chain B with chain C), which involves inward-facing hydrophobic residues protruding from the helices, α1, α2 and α4 (Figure 4B). Thus, the N-terminal extension facilitates tetramer formation and its deletion is predicted to produce a dimeric SepSecS. Interestingly, AFSepCysS (18) and ECCsdB (21) lack the N-terminal extension domain and form dimers (Figure 4C and D). We investigated whether SepSecS would be active in a homodimeric form by assaying the enzymatic activity of an MMPSepSecS protein that lacks the N-terminal extension residues 1–34. The Δ1–34SepSecS enzyme did not form Sec-tRNASec in vivo (Figure 5). It was not able to rescue selenoprotein biosynthesis in our in vivo complementation test of the E.coli ΔselA deletion strain. Since the active site of SepSecS, like other PLP enzymes, is formed at the dimer interface it is clear that SepSecS would not function as a monomer. Why the tetrameric organization of SepSecS is critical for function remains unclear, but it is possible that the quaternary structure of SepSecS is important for tRNA recognition. In such a scenario, when a tRNA acceptor stem is bound to the active site of one dimer, the other dimer in the tetramer could be interacting with other regions of the tRNA.
Figure 4.
The oligomeric states of SepSecS and related enzymes. (A) The overall architecture of the MMPSepSecS tetramer. Chains A and B, and chains C and D form dimers, respectively. The PLP molecules bound to each subunit are shown as ball and stick models. Chain A, chain B, chain C and chain D are colored pink, blue, green and yellow, respectively. (B) The N-terminal extension domains of chains B and C form a hydrophobic core that stabilizes the tetrameric state of MMPSepSecS. Three α-helices (α1,α2,α4) are labeled. Chains B and C are colored blue and green, respectively. (C) Stereo view of the AFSepCysS dimer in the same orientation as (A) (18). Chains A and B are colored pink and blue, respectively. The PLP molecules are shown as ball and stick models. (D) Stereo view of the ECCsdB dimer in the same orientation as (A) (21). The coloring scheme and the PLP representation are the same as in (C). (E) Gel filtration of selenomethionine-labeled SepSecS on Sephacryl S-300. Absorbance at 280 nm is shown as a blue line. The elution volumes of other oligomeric proteins are indicated in the chromatogram. The molecular weight of the MMPSepSecS monomer is 50 kDa. MMPSepSecS eluted at the size expected for a tetrameric species.
Figure 5.
In vivo assays of SepSecS mutants. Formation of Sec-tRNASec in vivo is assayed by the ability of the wild-type MMPSepSecS and its mutant variants (N-terminal deletion Δ1-34, R72A, R72Q, R72K, R94A, R94Q, H166A, H166F, H166Q, R307A, R307Q, R307K, Q102A, K278A, N247A, D277A, K278A) to restore the BV reducing activity of the selenoprotein FDHH in the E.coli selA deletion strain JS1. Cotransformation of the PSTK gene (indicated with +) from M. jannaschii is required for the formation of the Sep-tRNASec intermediate.
Recognition of PLP
As in other Fold Type I PLP enzymes the active sites of MMPSepSecS lie on the dimer interface with each monomer contributing essential residues (20). The active site of chain A is formed by chains A and B that both recognize the PLP molecule (Figure 6). The catalytic domain harbors a seven-stranded β–sheet, with only the sixth β–strand being antiparallel; this is a common feature of Fold Type I PLP enzymes (20,38) including AFSepCysS (18) and ECCsdB (21). In the MMPSepSecS structure, PLP is covalently bound via a Schiff base to the strictly conserved Lys278 (chain A) (Figure 2), which is located between the sixth and seventh β strand in the active site (Figure 6A). Indeed, a Lys278Ala mutation abolishes MMPSepSecS catalytic activity as shown both in vivo by the lack of BV reduction by FDHH (Figure 5), and in vitro by the inability of the Lys278Ala mutant to form Cys-tRNASec (Figure 7).
Figure 6.
PLP recognition. Stereo views of (A) Ribbon representation of the active site of MMPSepSecS in the dimer interface between chains A and B that are colored pink and blue, respectively. The PLP molecule is covalently bound to Lys278 of chain A. The Fo−Fc omit map of the PLP molecule, contoured at 3.5 σ, is shown. (B) The amino acid residues that recognize the PLP molecule. The residues of chains A and B are colored pink and blue, respectively. The Fo−Fc omit map (contoured at 3.5 σ) of all of the residues and PLP is shown.
Figure 7.
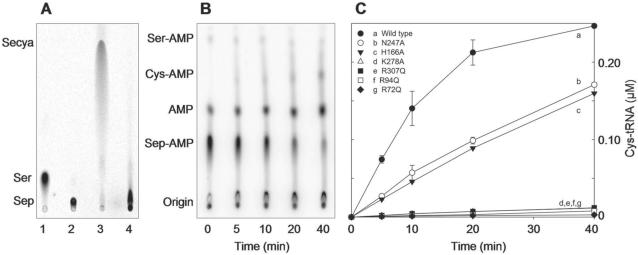
In vitro conversion of Sep-tRNASec to Sec-tRNASec or Cys-tRNASec. (A) Phosphorimages of TLC separation of [14C]Sep and [14C]Sec recovered from the aa-tRNAs of the SepSecS activity assay (see ‘Materials and methods’ section). Sec was analyzed in its oxidized form as selenocysteic acid (Secya). Lane 1, Ser marker; lane 2, Sep marker; lane 3, Sep-tRNASec with wild-type MMPSepSecS; lane 4, Sep-tRNASec with the R72Q MMPSepSecS mutant. (B) Representative phosphorimage for the H166A SepSecS mutant of the separation of Ser-[32P]AMP, Cys-[32P]AMP, [32P]AMP and Sep-[32P]AMP. At the indicated time points aliquots of the SepSecS reaction were quenched, digested with nuclease P1 and spotted onto PEI-cellulose TLC plates as described in the ‘Materials and methods’ section. (C) Plot of Cys-tRNASec formed versus time with 1 μM of wild-type and mutant SepSecS enzymes using Sep-tRNASec (1 μM) and thiophosphate (500 μM) as substrates. Following quantification of the intensities of Ser-[32P]AMP, Cys-[32P]AMP, [32P]AMP and Sep-[32P]AMP using ImageQuant, the concentration of Cys-tRNASec formed at each time point was calculated by dividing the intensity of the Cys-[32P]AMP spot by the total intensity. The experiment was carried out in duplicate.
All known Fold Type I PLP enzymes possess a critical aspartate that contacts the N1 atom of the pyridine ring (39), which is thought to further increase the electron sink character of the PLP cofactor. SepSecS is an exception to this paradigm, Asn247 is found in the corresponding position, and it forms a hydrogen bond with the pyridinium nitrogen (Figure 6). The other ‘nonconforming’ enzyme is SepCysS where the structure also reveals a similar Asn contact with PLP (18). The replacement of Asn247 with the uncharged Ala247 yields a partially active enzyme (Figures 5 and 7). The phosphate moiety of Sep appears to be such a good leaving group that the presence of an Asp or Asn residue is not required for the catalytic activity of SepSecS.
The aromatic pyridine ring of PLP is sandwiched between His166 (chain A) and Ala249 (chain A) through hydrophobic interactions at the bottom of the catalytic site (Figure 6). This recognition mode is commonly observed in the structures of Fold Type I PLP enzymes where the His residue increases the electron sink character of PLP's pyridine ring through the stacking interactions with it and may also play a role in substrate activation and acid base catalysis. In the latter case, the His residue makes a direct hydrogen bond with the critical aspartate that contacts the N1 atom of the coenzyme pyridine ring in all Fold Type I PLP enzymes (see above discussion) and is thus thought to assist in the dissipation of the negative charge generated around PLP's ring during catalysis (40). Furthermore, the phosphate moiety hydrogen bonds with the main-chain amide and carbonyl groups of Gly140 and Ser168, respectively (Figure 6B). The MMPSepSecS mutant His166Ala was partially active in forming Sec-tRNASec in vivo (Figure 5). In vitro, the His166Ala MMPSepSecS mutant was partially active in forming Cys-tRNASec as can be seen by the increase in intensity of the Cys-[32P]AMP spot and the concomitant decrease of the Sep-[32P]AMP spot during the course of the reaction (Figure 7B). Obviously, the mutant enzyme with Ala166 has PLP still in a partially functional position. A mutation of the corresponding His residue (position 143) in E. coli aspartate aminotransferase also led to a functional enzyme (40). In contrast, both the His166Gln and His166Phe mutants were inactive in vivo (Figure 5). The lack of enzymatic activity of the His166Phe mutant implicates His166 in the catalytic mechanism and not only in structural ring stacking interactions with the pyridine ring of PLP. It is interesting to note that with the exception of Plasmodium all known eukaryotic SepSecS proteins contain Gln at a homologous position to 166 in their sequences (Figure 2 and data not shown).
Since the catalytic pocket is formed in the dimerization interface, chain B also contributes residues that participate in PLP recognition. In particular, the guanidinium group of Arg72 and the main-chain amide group of Arg307 (from chain B) hydrogen bond to the phosphate moiety of PLP (Figure 6B). Mutations of Arg72 and Arg307 to Ala, Gln or Lys resulted in MMPSepSecS mutants that were significantly less active in Sec-tRNASec formation in vivo (Figure 5) and Cys-tRNASec formation in vitro (Figure 7C). We also show that the Arg72Gln mutant enzyme is unable to form Sec-tRNASec in vitro (Figure 7A).
Such PLP recognition differs from that of AFSepCysS (18) and ECCsdB (21) (Figure 8). Unlike in the MMPSepSecS structure, the adjacent subunit of ECCsdB does not come close to the active site. In ECCsdB, an additional βαβ structural motif covers the active site, presumably to stabilize the PLP and Sec substrates inside the pocket. This motif also precludes interaction between residues from the neighboring subunit and the active site (Figure 8D). Although the active site of AFSepCysS is formed by residues from chains A and B, the chain B amino acids are located too distant for PLP recognition (Figure 8E). The active sites of MMPSepSecS and AFSepCysS are spacious enough to accommodate the O-phosphoseryl-CCA end of the tRNASec or tRNACys species, respectively.
Phosphoserine binding model
Our crystallization solution contained 10 mM magnesium sulfate. In the present structure, a strong electron density (4.5σ), presumably corresponding to a sulfate ion, was observed adjacent to the PLP molecule of chains A and B (Figure 8A). The sulfate ion is recognized by Arg94, Ser95, Gln102 and Arg307 of chain B (Figure 8A). The structure of ECCsdB with Sec bound to the active site (Figure 8D) was reported (21). Using the CE program (41) we superposed the active site structure of MMPSepSecS onto that of ECCsdB. This allowed modeling of Sec from CsdB into the MMPSepSecS active site (Figure 8B). The distance between the selenium atom of the modeled Sec and the sulfur atom of the bound sulfate is 3.74 Å. We overlaid a Sep molecule on the position of the modeled Sec in the active site. The phosphate moiety of this modeled Sep overlapped with the bound sulfate, suggesting that this sulfate mimics the phosphate moiety of Sep attached to tRNASec. This allowed us to construct the Sep-binding model of MMPSepSecS (Figure 8C). Mutations of Arg94, Gln102 and Arg307 residues that according to our Sep-binding model recognize the phosphate moiety of Sep significantly decreased the catalytic activity of MMPSepSecS both in vivo (Figure 5) and in vitro (Figure 7). While the distance of the Schiff base linkage between the C4α atom of PLP and the Nζ atom of Lys278 is 1.74 Å, the PLP C4α atom is 2.19 Å away from the amino group of the modeled Sep; thus, the amino group of Sep forms a Schiff base with PLP as a reaction intermediate.
The active site
We should first note that the active site of MMPSepSecS lacks a Cys residue. In contrast, the crystal structures of ECCsdB (21) and of AFSepCysS (18) each possess a cysteine-active site residue; Cys364 in ECCsdB and Cys247 in AFSepCysS (Figure 8). As revealed by a structural study (21), Cys364 of ECCsdB recognizes Sec and withdraws its selenium to form perselenide as an intermediate. Cys364 of ECCsdB resides in the βαβ structural motif which does not exist in MMPSepSecS. On the other hand, Cys247 (from the neighboring subunit) of AFSepCysS is the candidate residue that forms a persulfide that is the sulfur source for enzyme-catalyzed Cys-tRNACys formation (18). The absence of such a Cys active site residue indicates that the reaction mechanisms and chemistries of SepSecS are fundamentally different from those of CsdB and SepCysS. Interestingly, SepSecS has an active site arginine (Arg307) in a homologous position to Cys247 in SepCysS (Figure 2).
The MMPSepSecS structure reveals three conserved arginines (Arg72, Arg94 and Arg307) that are located in proximity of each other and close to the active site; they recognize the phosphate groups of PLP and presumably of Sep acylated to tRNASec (Figures 2 and 8A–C). Mutations of Arg72, Arg94 and Arg307 to Ala, Gln or Lys yielded MMPSepSecS enzymes that were unable to form Sec-tRNASec in vivo (Figure 5). Asp277 interacts electrostatically with Arg72, and the Asp277Ala enzyme was also inactive in vivo (Figure 5). The Arg72Gln, Arg94Gln and Arg307Gln MMPSepSecS mutants were inactive in forming Cys-tRNASec in vitro (Figure 7). In addition to the PLP-conjugated Lys278, Sep-binding arginines may also facilitate general acid/base catalysis as was shown for the tRNA modification enzyme TrmH where an arginine activated by a phosphate group acts as a general base (42).
DISCUSSION
The mechanism
SepSecS is a PLP enzyme catalyzing a β-replacement (Scheme 1), leading to the exchange of a phosphate group for a selenol moiety. We were interested to see if SepSecS might employ a perselenide intermediate during catalysis, which would require the presence of an active site cysteine. There are four moderately-to-highly conserved cysteine residues in the SepSecS sequences, which are found in 35–97% of the SepSecSs. Three of these cysteines (Cys146, Cys214 and Cys237 in MMPSepSecS) are distant from the active site and have no chance to contribute to catalysis. The least conserved of these residues is a strictly conserved proline (Pro169) in the archaeal sequences, while the residue is conserved as a cysteine in 78% of the eukaryotic sequences. Interestingly, Pro169 in MMPSepSecS is in the active site and in close contact with the PLP moiety (3.77 Å at closest approach), which would then mean that most eukaryotic SepSecSs have a cysteine adjacent to the PLP. It is unclear whether the eukaryotic sequences make use of this cysteine in catalysis; nevertheless, this position is an intriguing candidate for mutagenesis in the eukaryotic context.
Concerning the archaeal SepSecSs, there are no cysteine residues that could be involved in the formation of a catalytically important perselenide intermediate. In contrast, SepCysS is proposed to use a persulfide mechanism (18). Thus, SepSecS and SepCysS, two related PLP enzymes that perform chemically analogous tRNA-dependent transformations of Sep to Sec or Cys, respectively, proceed with different selenium and sulfur transfer mechanisms. Based on the observed active site residues we propose that the arginine residues, that bind the phosphate groups of PLP and of Sep, also recruit the selenium donor selenophosphate. In this context, Arg307 in the present structure resides at a similar position to that of Cys364 in ECCsdB and of Cys247 in AFSepCysS.
Structural phylogeny of Fold Type I PLP enzymes
SepSecS belongs to the largest and most diverse family of PLP-dependent enzymes found in nature. The evolutionary history of the Fold Type I PLP-dependent enzymes, also referred to as the α-family, has been studied in detail with sequence-based phylogenetic methods (43). By applying the family profile analysis (FPA) technique (44), the authors were able to use sequence-based phylogenetics to partially capture the distant evolutionary events recorded in the sequences of the members of this family, some of whom share only 5% sequence identity. However, the low level of sequence similarity may lead to artifacts in the tree reconstruction process. Since crystal structures for most members of this protein family exist, we applied the technique of structural phylogeny (45) to explore the history of the Fold Type I family. This technique derives phylogenetic information directly from three-dimensional structures and thus allows an accurate reconstruction of the most distantly detectable evolutionary events. Our structural phylogeny (Figure 3) is largely in agreement with the previous sequence-based work, but some significant rearrangements in the tree can be seen. For example, the DC I and DC II subfamilies coalesce into a single subfamily according to structural similarity, and the γ-lyase and CYS groups are not one but two evolutionary distinct subfamilies.
There are two kinds of subfamilies in the Fold Type I group. The first type includes several proteins with distinct but related functions, which may have descended from a progenitor with a promiscuous enzymatic function. Examples of these progenitor enzymes include the four distinct aminotransferases (AT I–AT IV), an amino acid decarboxylase (DC I/II), a persulfide forming cysteine desulfurase/sulfhydrylase and finally separate β- and γ-amino acid lyases. Subsequent evolution in each of these subfamilies ultimately produced the specific enzymatic functions observed in modern PLP-dependent enzymes.
The second kind of subfamilies are those that include a set of isofunctional enzymes. The modern enzymes in these subfamilies trace back to ancestors that had already evolved their modern enzymatic specificity in the initial evolutionary radiation of the Fold Type I family. These isofunctional subfamilies include serine hydroxymethyl transferase, alliinase and an archaeal SelA-like protein (MJ0158), which is of unknown function.
Importantly, the structural phylogeny allows the accurate placement of SepSecS within its family tree and reveals how this enzyme came into being. SepSecS is also a founding member of the Fold Type I family. It shows no specific relationship to any of the other subfamilies and emerges near the root of its family tree. While SepCysS, CsdB and IscS are part of a multifunctional subfamily (CYS in Figure 8) and these proteins have chemically similar substrates to SepSecS, there is no special relationship between SepSecS and the CYS subfamily. This result indicates that SepSecS is truly an ancient enzyme, and thus tRNA-dependent selenocysteine biosynthesis is a primordial process.
ACKNOWLEDGEMENTS
We thank the beam-line staff at BL41XU of SPring-8 (Harima, Japan) for technical help in data collection. Dan Su, Markus Englert, Kelly Sheppard, Michael Hohn, Hee-Sung Park, Juan Salazar, Motoyuki Hattori, Kotaro Nakanishi and Tomoyuki Numata participated in many discussions on this topic.
This work was supported by a SORST Program grant from Japan Science and Technology (to O.N.), by grants from the Ministry of Education, Culture, Sports, Science and Technology (to R.I. and O.N.), by a National Project on Protein Structural and Functional Analyses grant from the Ministry of Education, Culture, Sports, Science and Technology (to O.N.) and by grants from the Department of Energy, the National Institute of General Medical Sciences and the National Science Foundation (to D.S.). S.P. holds a fellowship of the Yale University School of Medicine MD/PhD Program. R.L.S. is the recipient of a Ruth L. Kirschstein National Research Service Award from the National Institute of General Medical Sciences. P.O’D. holds a National Science Foundation Postdoctoral Fellowship in Biological Informatics. Funding to pay the Open Access publication charges for this article was provided by GM22854 (to D.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev. 2004;18:731–738. doi: 10.1101/gad.1187404. [DOI] [PubMed] [Google Scholar]
- 2.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol. Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böck A, Thanbichler M, Rother M, Resch A. Selenocysteine. In: Ibba M, Francklyn CS, Cusack S, editors. Aminoacyl-tRNA Synthetases. Georgetown, TX: Landes Bioscience; 2005. pp. 320–327. [Google Scholar]
- 4.Leinfelder W, Zehelein E, Mandrand-Berthelot MA, Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- 5.Bilokapic S, Korencic D, Söll D, Weygand-Durasevic I. The unusual methanogenic seryl-tRNA synthetase recognizes tRNASer species from all three kingdoms of life. Eur. J. Biochem. 2004;271:694–702. doi: 10.1111/j.1432-1033.2003.03971.x. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser JT, Gromadski K, Rother M, Engelhardt H, Rodnina MV, Wahl MC. Structural and functional investigation of a putative archaeal selenocysteine synthase. Biochemistry. 2005;44:13315–13327. doi: 10.1021/bi051110r. [DOI] [PubMed] [Google Scholar]
- 7.Ohama T, Yang DCH, Hatfield DL. Selenocysteine tRNA and serine tRNA are aminoacylated by the same synthetase, but may manifest different identities with respect to the long extra arm. Arch. Biochem. Biophys. 1994;315:293–301. doi: 10.1006/abbi.1994.1503. [DOI] [PubMed] [Google Scholar]
- 8.Mäenpää PH, Bernfield MR. A specific hepatic transfer RNA for phosphoserine. Proc. Natl Acad. Sci. USA. 1970;67:688–695. doi: 10.1073/pnas.67.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharp SJ, Stewart TS. The characterization of phosphoseryl tRNA from lactating bovine mammary gland. Nucleic Acids Res. 1977;4:2123–2136. doi: 10.1093/nar/4.7.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc. Natl Acad. Sci. USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan J, Palioura S, Salazar JC, Su D, O’Donoghue P, Hohn MJ, Cardoso AM, Whitman WB, Söll D. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl Acad. Sci. USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. doi:10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelpi C, Sontheimer EJ, Rodriguez-Sanchez JL. Autoantibodies against a serine tRNA-protein complex implicated in cotranslational selenocysteine insertion. Proc. Natl Acad. Sci. USA. 1992;89:9739–9743. doi: 10.1073/pnas.89.20.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa M, Rodriguez-Sanchez JL, Czaja AJ, Gelpi C. Isolation and characterization of cDNA encoding the antigenic protein of the human tRNP(Ser)Sec complex recognized by autoantibodies from patients with type-1 autoimmune hepatitis. Clin. Exp. Immunol. 2000;121:364–374. doi: 10.1046/j.1365-2249.2000.01280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wies I, Brunner S, Henninger J, Herkel J, Kanzler S, Meyer zum Büschenfelde KH, Lohse AW. Identification of target antigen for SLA/LP autoantibodies in autoimmune hepatitis. Lancet. 2000;355:1510–1515. doi: 10.1016/s0140-6736(00)02166-8. [DOI] [PubMed] [Google Scholar]
- 16.Herkel J, Manns MP, Lohse AW. Selenocysteine, soluble liver antigen/liver-pancreas, and autoimmune hepatitis. Hepatology. 2007;46:275–277. doi: 10.1002/hep.21807. [DOI] [PubMed] [Google Scholar]
- 17.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, III, Ibba M, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 18.Fukunaga R, Yokoyama S. Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus. J. Mol. Biol. 2007;370:128–141. doi: 10.1016/j.jmb.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 19.Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003;4:850–854. doi: 10.1038/sj.embor.embor914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 21.Lima CD. Analysis of the E. coli NifS CsdB protein at 2.0 A reveals the structural basis for perselenide and persulfide intermediate formation. J. Mol. Biol. 2002;315:1199–1208. doi: 10.1006/jmbi.2001.5308. [DOI] [PubMed] [Google Scholar]
- 22.Mihara H, Maeda M, Fujii T, Kurihara T, Hata Y, Esaki N. A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase. Gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J. Biol. Chem. 1999;274:14768–14772. doi: 10.1074/jbc.274.21.14768. [DOI] [PubMed] [Google Scholar]
- 23.Cupp-Vickery JR, Urbina H, Vickery LE. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 2003;330:1049–1059. doi: 10.1016/s0022-2836(03)00690-9. [DOI] [PubMed] [Google Scholar]
- 24.Weeks CM, Miller R. The design and implementation of SnB version 2.0. J. Appl. Cryst. 1999;32:120–124. [Google Scholar]
- 25.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 26.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 30.Bullock TL, Uter N, Nissan TA, Perona JJ. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J. Mol. Biol. 2003;328:395–408. doi: 10.1016/s0022-2836(03)00305-x. [DOI] [PubMed] [Google Scholar]
- 31.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 32.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Donoghue P, Luthey-Schulten Z. Evolutionary profiles derived from the QR factorization of multiple structural alignments gives an economy of information. J. Mol. Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 34.Felsenstein J. PHYLIP - Phylogeny Interference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 35.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacourciere GM, Levine RL, Stadtman TC. Direct detection of potential selenium delivery proteins by using an Escherichia coli strain unable to incorporate selenium from selenite into proteins. Proc. Natl Acad. Sci. USA. 2002;99:9150–9153. doi: 10.1073/pnas.142291199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herkel J, Heidrich B, Nieraad N, Wies I, Rother M, Lohse AW. Fine specificity of autoantibodies to soluble liver antigen and liver/pancreas. Hepatology. 2002;35:403–408. doi: 10.1053/jhep.2002.30699. [DOI] [PubMed] [Google Scholar]
- 38.Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 1998;8:759–769. doi: 10.1016/s0959-440x(98)80096-1. [DOI] [PubMed] [Google Scholar]
- 39.Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000;8:R1–R6. doi: 10.1016/s0969-2126(00)00085-x. [DOI] [PubMed] [Google Scholar]
- 40.Yano T, Kuramitsu S, Tanase S, Morino Y, Hiromi K, Kagamiyama H. The role of His143 in the catalytic mechanism of Escherichia coli aspartate aminotransferase. J. Biol. Chem. 1991;266:6079–6085. [PubMed] [Google Scholar]
- 41.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 42.Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, Hori H, Yokoyama S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure. 2004;12:593–602. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Christen P, Mehta PK. From cofactor to enzymes. The molecular evolution of pyridoxal- 5'-phosphate-dependent enzymes. Chem. Rec. 2001;1:436–447. doi: 10.1002/tcr.10005. [DOI] [PubMed] [Google Scholar]
- 44.Mehta PK, Argos P, Barbour AD, Christen P. Recognizing very distant sequence relationships among proteins by family profile analysis. Proteins. 1999;35:387–400. [PubMed] [Google Scholar]
- 45.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol. Mol. Biol. Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]