Abstract
RNA–RNA recombination is an important pathway in virus evolution and has been described for many viruses. However, the factors driving recombination or promoting the selection of recombinants are still unclear. Here, we show that the small movement protein (2b) was able to promote selection of RNA 1/2–RNA 3 recombinants within a chimeric virus having RNAs 1 and 2 from cucumber mosaic virus, and RNA 3 from the related tomato aspermy virus, along with heterologous 2b genes. The source of the 2b also determined the selection of the acceptor RNA and the crossover site, as well as affecting the rate of selection of the recombinant RNAs. The nature of the RNA 3 also influenced the selection of the recombinant RNAs. A 163-nt tandem repeat in RNA 3 significantly affected the rate of selection of the recombinant RNA, while a single nucleotide within the repeat affected the crossover site. The recombination occurred in a non-random manner, involved no intermediates and probably was generated via a copy-choice mechanism during (+) strand RNA synthesis.
INTRODUCTION
RNA–RNA recombination is one of the most important pathways in virus evolution. It was first discovered in the early 1960s in poliovirus (1,2) and has been documented in various genera of animal viruses, plant viruses and bacterial viruses group. Studies conducted on viruses during the last 40 years indicated that RNA–RNA recombination could occur between RNAs of the same or different strains of one species (3,4), of different species (5) or between viral and host cellular RNAs (6,7). RNA hairpins or mutations in a replicase domain have been implicated to play an important role in promoting RNA–RNA recombination (8–11). While none of those studies demonstrated a role for non-replicase proteins in viral RNA–RNA recombination or selection of recombinants, recombination without replication has been described (12,13).
RNA recombination has been studied extensively in the plant virus genus Cucumovirus. Those studies have involved two of the three recognized viral species in this genus, Cucumber mosaic virus (CMV) and Tomato aspermy virus (TAV). These viruses both contain a single-stranded and positive-sense RNA genome divided into three species designated RNAs 1, 2 and 3 (14,15). RNA 1 encodes the 1a protein involved in virus replication (16,17). RNA 2 encodes two proteins: the 2a, also involved in virus replication (16,17) and the 2b protein, involved in virus movement and the suppression of RNA silencing (18–21). RNA 3 also encodes two proteins, the 3a movement protein and the capsid protein, both of which are also involved in virus movement (22–24). Protein 2b is translated from a subgenomic RNA of RNA 2, RNA 4A (25,26), while the capsid protein is translated from a subgenomic RNA of RNA 3, RNA 4 (27). The 3′ terminal 305–310-nt non-translated region (NTR) is nearly identical in each of the three genomic RNAs of each virus and represents promoters for (−) viral RNA synthesis during viral replication (23,28,29). The location of most of the recombinants described involving CMV and TAV is within these 3′ NTR sequences.
RNA–RNA recombination was first described in cucumoviruses isolated from tobacco plants after multiple passage of a pseudorecombinant (reassortant) virus containing RNAs 1 and 2 from CMV, and RNA 3 from TAV (30). The progeny RNA 3 consisted mostly of TAV RNA 3 but with the 3′ terminal sequence from CMV RNA 2. In addition, in an earlier passage, there was an intermediate RNA 3 present containing a duplicated 21-nt sequence at the crossover site (30). Subsequently, several other cucumoviral recombinant RNAs were also discovered in plants infected with a so-called quadripartite hybrid virus having RNAs 2 and 3 from CMV, and RNAs 1 and 2 from TAV (31), with a mixture of wild-type (wt) CMV and wt TAV (32,33), with mixed CMV isolates (34–36), or within a single wt CMV (37). These characterized recombinant RNAs had non-template-derived nucleotides at the crossover site (31) and had CMV sequences at one end of the crossover site and TAV sequence at the other end (32) or had the same 3′-terminal NTR sequence for all three genomic RNAs (37). Some of the recombinants had crossover sites localized within a short region of high sequence similarity (32). Others were predicted to occur in regions of high secondary structure (33,34,38). TAV RNA 3 itself has a direct repeat of 163 nt (differing by one nucleotide) in the 3′ NTR, which is not essential for infection (39) and in some cases, was deleted during long-term incubation (40). While some of the above studies examined recombinants formed in the directly inoculated leaves and thus avoided detecting only those replicable recombinants that might be selected (32,33), other studies examined those recombinants detected after slow selection (31,38) and found that some recombinants could co-exist in the same plants (38). However, what factors may have led to the selection of particular recombinants has not previously been examined.
Here we show that infection and passage of a chimeric virus having RNAs 1 and 2 from CMV, with RNA 3 and the 2b gene from TAV consistently resulted in the slow selection of recombinant viruses. We demonstrate that the 2b gene plays a key role in promoting this selection.
MATERIALS AND METHODS
Plasmid construction
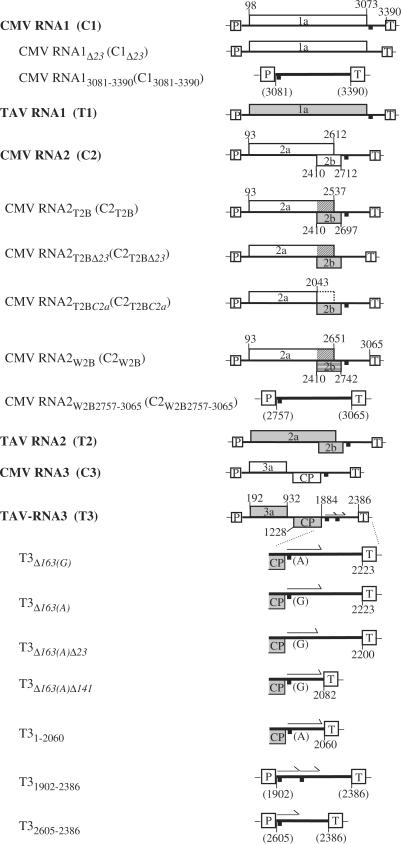
Plasmid cDNA clones, with transcription in planta controlled by the cauliflower mosaic virus (CaMV) 35S promoter and terminator, were used for plant inoculation. A pGEX2T-based plasmid (Amersham Pharmacia Biotech), pGEX-T2b, in which the gene encoding the TAV 2b protein was fused with sequences encoding GST, was used to express GST/TAV2b for RNA-binding assay. Various plasmids used in this study are shown in Figure 1. Plasmid cDNA clones C1 (previously named pQCD1), C2 (pQCD2), C2T2B (pQCD2qt), C2T2BC2a (pQCD2qt1), C2W2B (pQCD2qw), C3 (pQCD3), T1 (pCass1T1), T2 (pCass1T2), T3 (pCass1T3) and T3Δ163(A) (pCass1T3Δ163(A)) have all been previously described (26,39,41–43). The other plasmid cDNA clones were constructed as follows.
Figure 1.
Schematic representation of constructs used in this article. The constructs regulated from the 35S promoter and 35S terminator are indicated. The open reading frames encoding various cucumoviral proteins on the constructs are shown. The 23 nt of sequence identity between all genomic RNAs is indicated by a short thick bar. The tandem repeat of 163 nt is indicated by an arrow. The crosshatch lines represent the amino acid sequences different from the native sequence for either 2a or 2b.
Plasmid cDNA clones C1Δ23, C2T2BΔ23 and T3Δ163(A)Δ23 were constructed through three steps: first, amplifying two fragments with appropriate primer pairs (see Supplementary Table 1 for detailed primers), then, digesting the amplified fragments with appropriate restriction endonucleases; i.e. BstXI/BamHI for C1Δ23, Asp718/BamHI for C2T2BΔ23 and NheI/AvrII for T3Δ163(A)Δ23; and finally, by ligating the two digested fragments into the appropriate sites created by digestion with the same enzymes. Plasmid cDNA clones C13081–3390, C2W2B2757–3065, T3Δ163(A)Δ141, T31–2060, T31–2223, T31902–2386 and T32065–2386 were constructed via two steps: first amplifying appropriate fragments, and then ligating these fragments into the pCass1 vector (43). Plasmid cDNA clones C2W2BC2a and T3Δ163(G) were constructed with a QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instruction. All the amplified fragments as well as their joined regions in the plasmids were sequenced to ensure whether they were correct.
Plant inoculation and RNA analyses
Two plant species, Nicotiana clevelandii and Nicotiana glutinosa, were chosen for inoculation of the plasmid cDNA clones. Unless specified otherwise, all inocula used were a mixture of the three genomic cDNA clones (corresponding to RNAs 1, 2 and 3), each at an equal concentration of 10 μg/10 μl per plant. For each test, at least five, four-leaf plants of each of the above two species were inoculated mechanically after a 24-h dark treatment. Each test was repeated at least once.
Total plant RNAs were extracted from infected plants and analysed by northern blot hybridization as described previously (26). The nylon membranes were hybridized with a 32P-labelled transcript probe, complementary to the 3′ terminal sequences of either TAV RNA 3 (nucleotide 2287–2386) or RNA 2 of the Q-strain of CMV (Q-CMV; nucleotide 2871–3035). These two probes would hybridize to all the genomic and subgenomic RNAs of the corresponding viruses (39). An additional probe complementary to nucleotide 1–1884 of TAV RNA 3 was specifically used to hybridize to the 5′-terminal region of TAV RNA 3.
RT-PCR and sequence determination
Viral particles and virion RNAs were purified according to the method of Peden and Symons (44). Individual viral RNAs were purified from the virion RNAs first via an agarose gel and then via a polyacrylamide gel (45). The purified, single, viral RNAs were polyadenylated with Escherichia coli poly(A) polymerase and then RT-PCR was done using an oligo dT primer as described previously (43). The amplified products were cloned into the pBluescript SK+ vector and sequenced from both orientations.
The purified, single, recombinant RNAs, total RNAs extracted from plants and total virion RNAs were also reverse transcribed using primers T3′ and C3′, respectively. T3′ is complementary to the 3′-terminal sequence of TAV, while C3′ is complementary to the 3′-terminal sequence of Q-CMV (see Supplementary Table 1 for details). The synthesized first-strand cDNAs from the templates were amplified with different pairs of primers, C3′/T3-1187, C3′/C2-980, C3′/C1-2032, T3′/T3-1187, T3′/C2-980 and T3′/C1-2032 (see Supplementary Table 1 for details), using the thermocycler program: 1 min at 94°C, 1 min at 52°C and 2 min at 72°C for 30 cycles. The amplified products were also cloned into the pBluescript SK+ vector and sequenced from both orientations. At least five clones were sequenced from each PCR product.
DNA sequencing was carried out using 4 pmol of a specific primer and 0.1 μg of a cDNA clone. The sequence reaction was incubated at 37°C for 10 min and then analysed via a manual sequencing gel system. RNA sequencing was carried out under the same conditions as the DNA sequencing except that 10 μg of the total plant RNAs, 2 μg of the virion RNAs or 1 μg of a single RNA were used as a template and that the sequence reaction was incubated at 42°C for 30 min.
Purification of the TAV 2b protein from Escherichia coli
The plasmid pGEX-T2b was constructed via two steps: amplifying the PCR fragments first and then cloning the PCR fragments into appropriate vectors (as above). PGEX-T2b was transformed into E. coli BL21 (DE3). Protein expression was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Purification of the protein was carried out using glutathione beads. The cleavage of GST/TAV2b protein was carried out using thrombin (Novagen) as recommended by the manufacturer. The purity of the purified protein was analysed by SDS–PAGE. The amount of purified protein was measured using the Bio-Rad protein assay.
Preparation of full-length TAV RNA 3 transcripts
DNA templates representing a full-length RNA 3 of TAV and including T7 or T3 promoter were generated by PCR using the infectious cDNA clone of TAV RNA 3 (T3) as a template and primer pairs T3′ and T7-TR3(+) (a primer used for transcription of the sense RNA), or T5′ (nucleotides 1–27 of TAV RNA 3) and T3-TR3(−) (a primer used for transcription of the anti-sense RNA; see Supplementary Table 1 for the primer sequences). Labelled transcripts from the DNA templates were obtained using [32P]-α-UTP and T7 or T3 DNA-dependent RNA polymerase. The free nucleotides in the transcription reaction were removed using P-30 micro Bio-Spin columns (Bio-Rad), while the DNA templates were removed by incubation with DNase I. The labelled TAV RNA 3 transcripts were purified through a 4% denaturing polyacrylamide gel and quantified by UV spectrophotometry (Beckman).
Protein-RNA-binding assay
Three different amounts (150, 300 and 450 ng) of the TAV 2b protein purified from E. coli were each incubated with 4 ng of each of the [32P]-labelled TAV RNA 3 transcripts in a binding buffer [50 mM Tris–HCl (pH 8.2), 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 200 ng of yeast tRNA (Sigma), and 2 U of RNase inhibitor (Promega)] at 25°C for 30 min (46). After incubation, potential protein-RNA complexes were analysed via electrophoresis on a 4% non-denaturing polyacrylamide gel and detected by autoradiography.
RESULTS AND DISCUSSION
Identification and characterization of recombinant RNAs in passaged 2b-chimeric cucumovirus
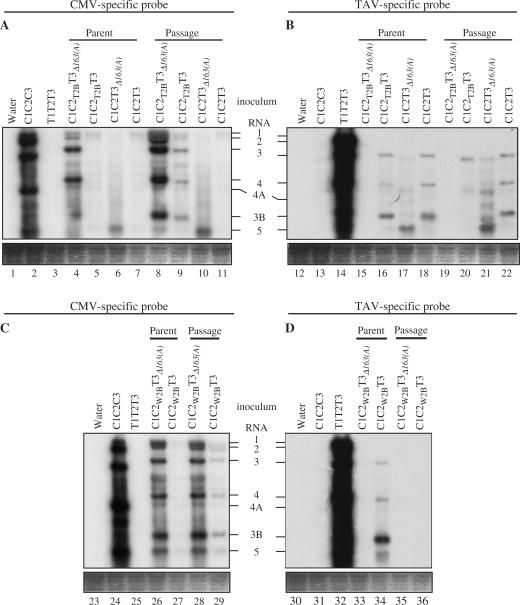
Pseudorecombinant viruses formed between RNAs 1 and 2 of CMV and RNA 3 of TAV have been shown gradually to yield stable, recombinant viruses after multiple passage (30,31,38). However, the effects of the various viral genes on the selection of these recombinant viruses have not been examined. Previously, using interspecies hybrid viruses, we examined the roles of different 2b genes on virulence and virus accumulation and showed that while the nature of the 2b gene influenced virulence, the RNA 3 (and therefore the two genes encoded by RNA 3; the 3a movement protein and the capsid protein) did not (21,47). Therefore, the hybrid viruses generated previously, C1C2T2BC3 and C1C2T2BT3, where C indicates Q-CMV, T indicates TAV, 1, 2 and 3 indicate RNAs 1, 2 and 3, and the subscript T2B indicates that the 2b gene of Q-CMV was replaced precisely with that of TAV (Figure 1), were subjected to long-term, multiple passages in N. glutinosa to determine whether the 2b gene had any effect on the selection of recombinant viruses. The progeny viral RNAs were examined by northern blot hybridization, with probes specific to the 3′ terminal NTR sequences of either the three Q-CMV RNAs or the three TAV RNAs (Figure 1). This analysis indicated that in the multiple passage plants TAV RNA 3 derived from C1C2T2BT3 had recombined with CMV RNA, as the progeny RNA 3 was detected by both the Q-CMV-specific and TAV-specific probes (Figure 2A and B, compare lanes 9 and 20). By contrast, RNA 3 derived from the same virus maintained for several weeks in the plasmid-inoculated plants was not a recombinant RNA, as it was only detected by the TAV-specific probe, but not by the Q-CMV-specific probe (Figure 2A and B, compare lanes 5 and 16). The specificity of the CMV-specific and TAV-specific probes was demonstrated in that both probes only detected their own specific viral RNAs (Figure 2A and B, compare lanes 2 and 3, and lanes 13 and 14). The other RNAs of C1C2T2BT3 were still detected by their specific probes in both the inoculated plants and the multiple passage plants (Figure 2A and B).
Figure 2.
Northern hybridization of accumulated viral RNAs in N. glutinosa. Total RNAs extracted from inoculated and passage N. glutinosa plants were fractionated in 1.2% of agarose gel containing 1.1% formaldehyde, and then blotted to membranes. The hybridization of the membranes was done with probes specific for the 3′ NTRs of the Q-CMV or TAV RNAs. The positions of RNAs 1, 2, 3, 4, 4A, 3B and 5 are indicated. CMV RNA 5 is 307–310 nt and represents the 3′ NTR of the genomic RNAs 1–3. TAV RNA 5 is 323 nt, while TAV RNA 3B is 486 nt; both are derived from the 3′ NTR of TAV RNA 3.
To confirm that RNA 3 was a recombinant, the RNA 3 derived from C1C2T2BT3 was gel-purified, reverse transcribed with primers C3′ and T3′, respectively, and then amplified with different pairs of primers as described in the Materials and Methods section. Only the primer pair C3′/T3-1187 was able to generate a specific RT-PCR product (data not shown). T3-1187 corresponded to sequences ending with nucleotide position 1187 of TAV RNA 3, while C3′ was complementary to the 3′-terminal sequence of Q-CMV. This indicates that the RNA 3 was indeed a recombinant, having a TAV RNA 3 sequence in the capsid protein region and a CMV RNA sequence at the 3′ terminus.
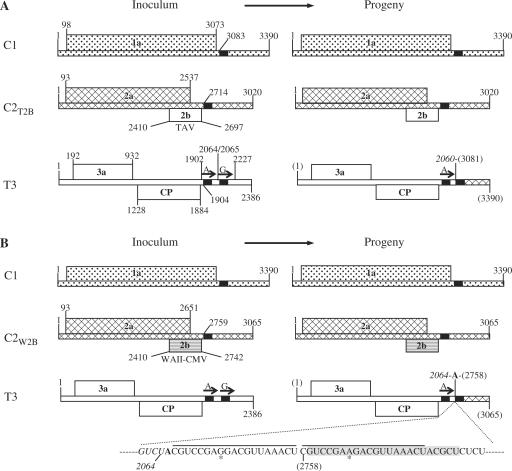
To determine the nature of the recombinant RNAs derived from C1C2T2BT3, the recombinant RNAs derived from this virus were purified from infected plants, subjected to RT-PCR and the RT-PCR products were then cloned into the pBluescript SK+ vector for sequencing. The sequencing results from five independent clones showed that there was one type of RNA 3 recombinant derived from C1C2T2BT3, containing 2370 nt, of which the 5′ terminal 2060 nt was derived from the 5′ terminal 2060 nt of TAV RNA 3 (Figure 3A right, open bar), while the 3′ terminal 310 nt was identical to the 3′ terminal 310 nt of Q-CMV RNA 1 (Figure 3A right, stippled bar). The crossover site on the TAV RNA 3 sequence was 7 nt 5′ of a homologous sequence of 23 nt conserved in all three genomic RNAs, and 3 nt 5′ of this conserved 23 nt sequence in the Q-CMV RNA 1 sequence.
Figure 3.
Genome organization of C1C2T2BT3, C1C2W2BT3 and their derived recombinant viruses. The genomic RNAs or 2b genes of each virus are indicated by different pattern fills. The short black bars on the RNAs represent the 23 nt of sequence identity. The arrows on RNA 3 represent the tandem repeats (first repeat, A, or second repeat, G). The junction site of each RNA 3 recombinant is indicated. The nucleotide sequence in the junction site of the RNA 3 recombinant derived from C1C2W2BT3 is indicated. The sequence in italics is from TAV RNA 3. The sequences underlined represent the imperfect 19-nt repeats. The single nucleotide difference in the repeats is indicated by an asterisk.
The TAV RNA 3 sequence in the recombinant RNA 3 retained the first repeat [in order of 5′ to 3′ on the (+) sense strand] of two 163-nt tandem repeats (39), but lacked the second one, which originally was present in the TAV RNA 3 inoculum. The difference between the two 163-nt repeats is an A at position 1966 in the first repeat, with a G at the corresponding position (2129) in the second 163-nt repeat (Figure 3A left) (39). The repeated sequences showed no difference in viral pathogenicity when only one or the other was present (39). The 3′ terminal 310 nt of the recombinant RNA 3 was not only the same as the 310 terminal nucleotides of Q-CMV RNA 1, but was also the same length as Q-CMV RNA 5. Q-CMV RNA 5 is a mixture of subgenomic RNAs derived from the 3′ terminal NTR of RNAs 1, 2 and 3 [(48); our unpublished data]. The level of RNA 5 has a strong effect on the severity of symptoms; the presence of more RNA 5 in the plants results in less severe symptoms (our unpublished data).
Repeated multiple passages of virus generated from the plasmids comprising C1C2T2BT3 in 10 N. glutinosa plants or 10 N. clevelandii plants resulted in the appearance of the same recombinant RNA 3s with the same border sequences among the sequenced clones. These recombinant RNAs were not detected by northern blot hybridization in the inoculated parental plants, even if the plasmid-inoculum was increased 5-fold (50 μg/10 μl) or decreased 20-fold (0.5 μg/10 μl), although the same recombinant RNAs were detected again in the multiple passage plants, when the initial inoculum concentration was changed (data not shown).
The 2b gene determines the source of the recombinant 3′ terminus and crossover site
The observation that recombinant RNAs were detected when the 2b gene of Q-CMV was substituted by that of TAV (Figure 2A, lane 9), and not when the 2b gene of Q-CMV was present in the interspecific hybrid virus C1C2T3 (Figure 2A, lane 11) suggested that the 2b gene has some role in the generation and/or selection of the recombinant viruses. However, as the replacement of the 2b sequence also changed the sequences encoding the C-terminal 41 amino acids of the 2a protein, it cannot be ruled out that the C-terminal 41 amino acids of the 2a protein might play a role in the recombination and/or selection. Therefore, to determine whether the C-terminal 41 amino acids of the 2a protein encoded by the overlapping TAV 2a gene sequence had such a role, we introduced a stop codon upstream of the TAV 2b gene in C2T2B to prevent the expression of the C-terminal 41 amino acids of the 2a protein (see C2T2BC2a in Figure 1). Bioassay of C1C2T2BC2aT3 onto N. glutinosa showed that the virus induced symptoms similar to those induced by C1C2T2BT3 and northern blot hybridization and RT-PCR analyses showed that recombinant RNA 3 still accumulated in the multiple passage plants, but not in the inoculated plants (data not shown). Moreover, the recombinant RNA 3 generated was the same as that derived from C1C2T2BT3 (data not shown). Therefore, the C-terminal 41 amino acids of the 2a protein encoded by the TAV 2b sequence had no role in the recombination and/or selection, indicating that the TAV 2b gene was required for the recombination and/or selection.
That the 2b protein rather than the 2b RNA sequence itself affected the generation and/or selection of recombinant RNA 3s could not be established conclusively, since expression of the 2b gene was shown to be necessary for the cell-to-cell movement, but not the replication of the pseudorecombinant virus C1C2T3 (21). However, if the 2b protein was actually involved in the recombination/selection process, then at a minimum the 2b protein should be capable of binding viral RNA. To determine whether there was some interaction between TAV RNA 3 and the 2b protein, we performed an RNA–protein-binding assay.
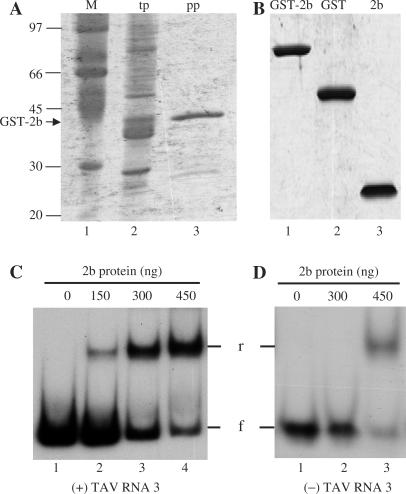
The TAV 2b protein (Figure 4B, lane 3) purified from E. coli (Figure 4A) was able to bind to the T7-transcribed (+) sense TAV RNA 3 (Figure 4C, lanes 2–4) as well as to the T3-transcribed (−) sense TAV RNA 3 (Figure 4D, lane 3). The binding ability increased with an increasing amount of the 2b protein (Figure 4C, compare lanes 1–4). By contrast, comparable amounts of GST alone, which was expressed and purified under the same conditions as the TAV 2b protein (Figure 4B, lane 2), did not bind to TAV RNA 3 (data not shown). This demonstrated that RNA-binding activity was not due to a co-purified, contaminating protein from E. coli. The significance of the more efficient binding of the 2b protein to (+) versus (−) sense RNA is unknown. The different binding abilities probably are due to different sequences and/or structures between RNA 3(+) and RNA 3(−). Since the 2b protein was designated a movement protein (19) and was shown to be required for the movement of pseudorecombinant viruses containing C1C2T3 (21), the 2b protein probably also binds to TAV RNA 3 in vivo.
Figure 4.
TAV 2b protein expression, purification and RNA 3 binding. (A) The TAV 2b protein fused with GST and transformed into the bacteria strain BL21 (DE3) was analysed by SDS–PAGE. Total bacterial lysate after the induction of protein expression (lane 2; tp) was passaged through glutathione beads to purify the GST-2b fusion protein (lane 3; pp). Molecular mass markers (M) are shown in lane 1. (B) Purified GST-2b fusion protein (lane 1) was cleaved using thrombin and the resulting products, GST (lane 2) and 2b proteins (lane 3), were gel-purified. GST protein was treated with thrombin (lane 2). The gels in A and B were stained with Coomassie Blue. (C and D) Autoradiograms of gel retardation electrophoresis assays of the 2b protein binding to RNA 3. Increasing amounts of the 2b protein were incubated in the binding buffer as with 4 ng of either 32P-labelled TAV RNA 3 (+) transcript (C) or 32P-labelled TAV RNA 3(−) transcript (D). The samples were electrophoresed in 4% non-denaturing polyacrylamide gels. The amounts of the 2b protein, the position of free RNA 3 (f) and the position of retarded RNA 3 (r) are indicated.
Another approach to determine the role of the 2b protein in the generation and/or selection of the recombinant RNAs, was to use a different 2b gene in the heterologous interspecific hybrid virus. To this end, we replaced the whole 2b gene of (the subgroup II strain) Q-CMV with that from a subgroup IA strain of CMV, WAII-CMV, generating the hybrid virus C1C2W2BT3 (see C2W2B in Figure 1). The WAII-CMV 2b gene and the Q-CMV 2b gene are only 53.4% identical at nucleotide level and the WAII-2b protein is 10 amino acids longer than the Q-2b protein.
Co-inoculation of C2W2B, C1 and T3 onto N. glutinosa plants followed by multiple passages at 1–2 weekly intervals showed that C1C2W2BT3 induced symptoms as severe as those induced by C1C2T2BT3, which were much more severe than those induced by either of the parental viruses (21). Northern blot hybridization showed that RNA 3 derived from C1C2W2BT3 also had recombined with a CMV sequence in the passage plants, but not in the inoculated plants (Figure 2C and D, compare lanes 27 and 34, and lanes 29 and 36). This was supported by RT-PCR analysis (data not shown). These data strongly indicate that the 2b protein was required for recombination and/or selection or the recombinant RNA 3s, and that this process was not TAV 2b protein-specific.
To determine the nature of the recombinant RNAs derived from C1C2W2BT3, the recombinant RNA 3 was subjected to RT-PCR, cloning and sequence analysis. Surprisingly, the RNA 3 recombinants derived from C1C2W2BT3 differed from those derived from C1C2T2BT3 or C1C2T2BC2aT3 in the following respects: First, the recombinant RNA 3 derived from C1C2W2BT3 was 22 nt longer than that derived from C1C2T2BT3 or C1C2T2BC2aT3 (2392 versus 2370). Second, the 3′ terminal NTR of the recombinant RNA 3 derived from C1C2W2BT3 contained a Q-CMV RNA 2 NTR sequence, rather than the Q-CMV RNA 1 NTR sequence (compare Figure 3A right and B right). Third, the crossover sites on RNA 2 and on RNA 3 were different in the recombinant RNA 3 derived from C1C2W2BT3 versus either C1C2T2BT3 or C1C2T2BC2aT3 (compare Figure 3A and B). And fourth, the recombinant RNA 3 derived from C1C2W2BT3 contained an imperfect repeat of 19 nt near the crossover site preceded by an A between the TAV RNA 3 sequence and the 3′ CMV RNA sequence (Figure 3B). Moreover, the recombinant RNA 3 derived from C1C2W2BT3 also was missing the second 163-nt repeat. These similarities on one hand and differences on the other suggested that the recombinant RNA 3s derived from C1C2T2BT3 and C1C2T2BC2aT3 versus C1C2W2BT3 might be generated using different templates, but occurred via a similar mechanism. Taken together, the above results indicate that the 2b protein determined either the RNA species participating in the recombination and the crossover site, or selected for particular recombinant RNAs generated at random that have the above characteristics.
The 2b protein and a 163-nt tandem repeat on RNA 3 both affect the rate of recombinant selection while a single nucleotide within the repeat determines the crossover site on RNA 3
Both the TAV 2b protein and WAII-CMV 2b protein were able to promote the recombination and/or selection of RNA 3 in the passage plants, but not in the inoculated plants. To determine when the recombinant RNA 3s could first be detected, a time-course experiment was done involving three viruses: C1C2T2BT3, C1C2T2BC2aT3 and C1C2W2BT3. The appropriate plasmid combinations were inoculated onto 10 N. glutinosa plants and sap was then passaged onto healthy N. glutinosa plants for 10 passages, at 6-day intervals. The inoculated leaves of the passage plants were detached 3 days after inoculation, total RNAs were extracted and the RNAs were analysed by northern blot hybridization. These analyses showed that the recombinant RNAs were only detected from the fourth passage and onwards in plants infected with C1C2T2BT3 or C1C2T2BC2aT3, or from the sixth passage and onwards in plants infected with C1C2W2BT3 (Supplementary Figure 1A, data not shown). These results indicated that selection of the recombinant RNA 3s is a slow process and therefore it seems more likely that the 2b proteins were selecting for specific recombinants, rather than being involved in the generation of the recombinant RNA 3s. This is also consistent with the different subcellular localization of the replication proteins and the 2b protein (49–51) and the lack of detectable interaction of the replicase proteins with the 2b protein (52). In addition, virus containing the TAV 2b protein showed selection of the recombinant RNA 3s two passages earlier than did virus containing the WAII-CMV 2b protein. Therefore, it seems likely that the source of the 2b protein also affected the rate of selection.
To examine the role of the two 163-nt repeats in the recombination and/or selection, we deleted each repeat independently in C1C2T2BT3. The resultant deletion mutants were designated C1C2T2BT3Δ163(A) and C1C2T2BT3Δ163(G) (Figure 1), where the subscripts Δ163(A) and Δ163(G) represent the first 163-nt repeat deletion and the second 163-nt repeat deletion, respectively. Bioassay of C1C2T2BT3Δ163(A) and C1C2T2BT3Δ163(G) onto N. glutinosa plants showed that both deletion mutants induced similar symptoms to those induced by C1C2T2BT3 (data not shown). Northern blot hybridization analysis showed that neither deletion affected the presence of recombinant RNA 3s in the multiple passage plants (Figure 2A, lanes 8 and 19, data not shown). However, both deletion mutants yielded recombinant RNA 3s in both the inoculated plants and the passage plants (Figure 2A and B, compare lanes 4 and 15, and lanes 8 and 19, data not shown). Repeated inoculation and northern blot hybridization analysis gave the same results as earlier. This suggests that the two 163-nt repeats affected the rate of the recombination and/or selection of recombinants.
To confirm the role of the two 163-nt repeats in affecting the rate of recombination and/or selection, we generated two other deletion mutants, C1C2W2BT3Δ163(A) and C1C2W2BT3Δ163(G). Bioassay of C1C2W2BT3Δ163(A) and C1C2W2BT3Δ163(G) showed that these two deletion mutants also induced symptoms similar to those induced by C1C2W2BT3 (data not shown). Northern blot hybridization analysis showed that the deletions also resulted in the generation of the recombinant RNA 3s in both the inoculated and passage plants (Figure 2C and D, compare lanes 26 and 33, and lanes 28 and 35, data not shown). These results showed that the two 163-nt repeats indeed affected the rate of the recombination and/or selection.
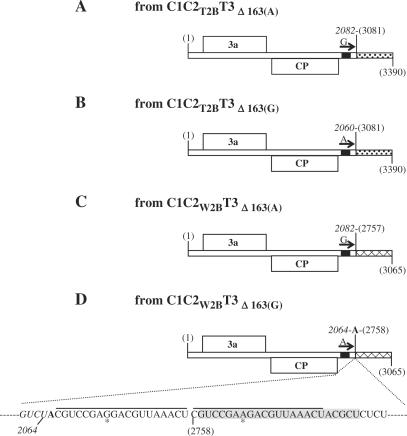
To determine the nature of the recombinant RNA 3s derived from the 163-nt repeat-deletion mutants, we again purified the recombinant RNA 3s of C1C2T2BT3Δ163(A), C1C2W2BT3Δ163(A), C1C2T2BT3Δ163(G) and C1C2W2BT3Δ163(G), used RT-PCR to amplify the recombinant sequences, cloned the RT-PCR products and sequenced them. The sequences of the recombinant RNA 3 derived from C1C2T2BT3Δ163(A) contained the 5′-proximal 2082 nt from TAV RNA 3 and the 3′ proximal 310-nt 3′ NTR of CMV RNA 1 (Figure 5A). The CMV RNA 1 NTR sequence was identical to that present in the recombinant RNA 3 derived from C1C2T2BT3. However, the TAV RNA 3 sequence was 22 nt longer than that of the recombinant RNA 3 derived from C1C2T2BT3 (compare Figures 3A and 5A), suggesting that the first 163-nt repeat might have a role in determining the crossover site on one of the participating RNAs, despite not affecting the crossover site on the other participating RNA. By contrast, the sequence of the recombinant RNA 3 derived from C1C2T2BT3Δ163(G) was exactly the same as that derived from C1C2T2BT3 (compare Figures 3A and 5B), indicating that the second 163-nt repeat had no role in determining the crossover sites on either RNA.
Figure 5.
Genome organization of C1C2T2BT3Δ163(A), C1C2T2BT3Δ163(G), C1C2W2BT3Δ163(A), C1C2W2BT3Δ163(G) and their derived recombinant viruses. The origin of the 3′ NTR of each RNA 3 is differentiated by fill patterns (stippled for RNA 1 and crosshatched for RNA 2). The short black bars on the RNAs represent the 23 nt of sequence identity. The arrows on RNA 3 represent one of the tandem repeats (A or G). The junction site of each RNA 3 recombinant is indicated. The nucleotide sequence in the junction site of the RNA 3 recombinant derived from C1C2W2BT3Δ163(G) is indicated. The sequence in italics is from TAV RNA 3. The sequences underlined represent the imperfect 19-nt repeats. The single nucleotide difference in the repeats is indicated by an asterisk.
The recombinant RNA 3 derived from C1C2W2BT3Δ163(A) consisted of the 5′ proximal 2082 nt from TAV RNA 3 and the 3′ proximal 309-nt 3′ NTR of CMV RNA 2 (Figure 5C). Although the 2082 nt sequence from TAV RNA 3 was exactly the same as that in the recombinant RNA 3 derived from C1C2T2BT3Δ163(A), it was 18 nt longer than the one seen in the recombinant RNA 3 derived from C1C2W2BT3 (compare Figures 3B and 5C). This result supports the conclusion that the first 163-nt repeat might indeed affect the left border of the crossover site. The CMV RNA 2 sequence present in the recombinant RNA 3 derived from C1C2W2BT3Δ163(A) was 19 nt shorter than the CMV RNA 2 present in the recombinant RNA 3 derived from C1C2W2BT3 (due to the absence of the additional A and the first 19-nt repeat shown in Figure 3B, but with an additional U residue, nucleotide 2757), suggesting that the first 163-nt repeat might also affect the right border of the crossover site. In the recombinant RNA 3 derived from C1C2W2BT3Δ163(G), the sequence was exactly the same as that of the recombinant RNA 3 derived from C1C2W2BT3 (compare Figures 3B and 5D), confirming that the second 163-nt repeat had no role in determining the position of the crossover sites on either RNA. Since only one nucleotide differed in the two 163-nt repeats (A versus G), the above results indicate that the position of the crossover sites is actually affected by this single nucleotide difference within each repeat.
Recombinant RNA 3s involve no detectable intermediates and are precisely generated
To determine whether the recombinant RNA 3s derived from C1C2W2BT3 or C1C2W2BT3Δ163(G), both of which contained a 19-nt repeat sequence, might represent an intermediate, buffered extracts from plants infected by viruses containing these recombinant RNA 3s were passaged onto five healthy plants for more than 20 passages done at 1-week intervals. RT-PCR and sequence analyses of the recombinant RNA 3s from the passage plants showed no sequence difference from the original recombinant RNA 3s described above, indicating that the 19-nt repeat was still present in the same position as before (data not shown). These results indicate that recombinant RNA 3 derived from C1C2W2BT3 or C1C2W2BT3Δ163(G) containing the 19-nt repeat was not an intermediate, but the stable recombination end-product.
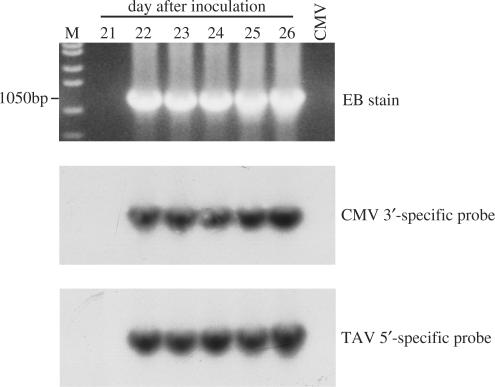
To examine how soon after infection this 19-nt repeat was generated and whether there was an intermediate generated before this 19-nt repeat appeared, a time-course experiment was done, in which two viruses, C1C2T2BT3Δ163(G) and C1C2W2BT3Δ163(G), were inoculated onto N. clevelandii. Nicotiana clevelandii was chosen as the host because the symptoms are more readily seen on this host than on N. glutinosa (unpublished data) and C1C2T2BT3Δ163(G) and C1C2W2BT3Δ163(G) were chosen because both viruses showed more rapid appearance of recombinant RNA 3 (see above). Moreover, the recombinant progeny virus of C1C2W2BT3Δ163(G) contained the 19-nt repeat, which could be used as a marker to determine whether there was an intermediate before this final product. After inoculation of each virus to 200 plants, leaves were detached from groups of five inoculated plants at 1-day intervals for up to 40 days. Northern blot hybridization of the total RNAs extracted from the detached leaves showed that recombinant RNA 3s derived from any of the above viruses did not accumulate to a detectable level before day 35; the earliest time when a recombinant RNA 3 could be detected by northern blot hybridization was at day 35 in the C1C2T2BT3Δ163(G)-infected plants (Supplementary Figure 1B). The recombinant RNA 3 derived from C1C2W2BT3Δ163(G) was not detected during the whole time-course of the experiment (data not shown). However, by using RT-PCR with the primer pair C3′/T3-1187, the recombinant RNA 3 derived from C1C2T2BT3Δ163(G) was detected as early as day 22 (Figure 6 upper panel), while the RNA 3 recombinant derived from C1C2W2BT3Δ163(G) could be detected at day 35 (data not shown). These results also confirmed that the TAV 2b protein promoted the recombination/selection faster than the WAII-CMV 2b protein.
Figure 6.
Agarose gel and Southern hybridization of RT-PCR products from C1C2T2BT3Δ163(G). RT-PCR was done using primer pair C3′/T3-1187 and total RNAs extracted from the plants at day 22–26 after inoculation with C1C2T2BT3Δ163(G). Top row: The RT-PCR products were analysed on a 1% agarose gel and stained with ethidium bromide (EB). The sizes of the products are indicated. M in the left lane indicates the DNA ladder, while CMV in the right lane is used as a negative control showing no RT-PCR product is obtained from the plants infected with Q-CMV alone. Middle row: The RT-PCR products on the gel were blotted to membranes and hybridized with the Q-CMV-specific probe. Bottom row: The same RT-PCR products were blotted and hybridized with the TAV-specific probe.
To characterize the RT-PCR products further, Southern blot hybridization was done using CMV-specific and TAV-specific probes. All the RT-PCR products on the gel were the same size (Figure 6 middle and lower panels) and the size was also the same as for the RT-PCR products obtained from the passage plants (data not shown). In addition, all the RT-PCR products were able to hybridize with both the Q-CMV-specific probe and the TAV-specific probe (Figure 6 middle and lower panels). Sequencing of these RT-PCR products showed that the RT-PCR products from the C1C2T2BT3Δ163(G)-inoculated plants contained 874 nt derived from TAV RNA 3 (up to nucleotide 2060 of TAV RNA 3) and 310 nt from CMV RNA 1. This sequence was exactly the same as that derived from the same virus in the passage plants and also the same sequence as that derived from C1C2T2BT3, either in the inoculated plants or in the passage plants (Figures 3A and 5B; data not shown). All of the RT-PCR products derived from C1C2W2BT3Δ163(G) also were exactly the same in sequence, having 878 nt from TAV RNA 3 and 326 nt from CMV RNA 2 including the 19-nt repeat (Figure 5D; data not shown). The sequence of the RT-PCR products also were the same as those from the passage plants, or those derived from C1C2W2BT3 in both the inoculated plants and passage plants (Figure 3B; data not shown). These results indicate that the recombinant RNA 3s accumulating in the infected plants did not contain a detectable intermediate, and that the crossover sites on both donor and acceptor RNAs occurred at the same position.
The conserved 23-nt sequence and the 3′ NTR of RNA 3 are critical for infection
The conserved 23-nt sequence at the beginning of the 3′ NTR has been implicated in the recombination and nucleotides 1–20 of the 23-nt sequence also have been suggested to be an internal subgenomic promoter site (33,38). This sequence also was always near the crossover sites in the recombinant RNA 3s observed here, and the 19-nt imperfect repeat also comes from this sequence. Therefore, we examined the requirement for this sequence in infection. Three deletion mutants were constructed: C1Δ23, C2T2BΔ23 and T3Δ163(A)Δ23, where the conserved 23-nt sequence was deleted from each of C1, C2T2B and T3Δ163(A), respectively. Co-inoculation of each deletion mutant in the combinations C1Δ23 with C2T2BΔ23 and T3Δ163(A)Δ23, C1Δ23 with C2T2B and T3Δ163(A), C1 with C2T2BΔ23 and T3Δ163(A), or C1 with C2T2B and T3Δ163(A)Δ23, onto N. glutinosa and N. clevelandii plants, showed that none of these combinations was able to infect the plants (Table 1). However, when either C1Δ23 was combined with C2 and C3, or T3Δ163(A)Δ23 was combined with T1 and T2, the mixtures were infectious (Table 1), indicating that the presence of this 23-nt sequence is essential for infection of the pseudorecombinant viruses, but not the parental viruses. Since the pseudorecombinant viruses containing the 23-nt sequence were viable without detectable recombination for some time (Figure 2), these results indicate that the 23-nt sequence is not essential for replication of the pseudorecombinant virus, but may be essential for maintaining infection of the pseudorecombinant viruses through the plants, just as the 2b protein was required for this task, but not for the maintenance of infection of the parental viruses (19,21).
Table 1.
Infectivity of plasmid inocula
| Inoculum | N. clevelandii | N. glutinosa |
|---|---|---|
| C1Δ23C2T2BT3Δ163(A) | − | − |
| C1Δ23C2C3 | + | + |
| C1C2T2BΔ23T3Δ163(A) | − | − |
| C1C2T2BΔ23C3 | − | − |
| C1C2T2BT3Δ163(A)Δ23 | − | − |
| T1T2T3Δ163(A)Δ23 | + | + |
| C1C2T2BT3Δ163(A)Δ141 | − | − |
| C1C2W2BT3Δ163(A)Δ141 | − | − |
| T1T2T3Δ163(A)Δ141 | − | − |
| C1C2T2BT31–2060 | − | − |
| T1T2T31–2060 | − | − |
| C1C2T2BT3Δ163(A)Δ141C13081–3390 | − | − |
| C1C2T2BT3Δ163(A)Δ141C2W2B2757–3065 | − | − |
| C1C2T2BT3Δ163(A)Δ141T31902–2386 | − | − |
| C1C2T2BT3Δ163(A)Δ141T32065–2386 | − | − |
| C1C2T2BT31–2060C13081–3390 | − | − |
| C1C2T2BT31–2060C2W2B2757–3065 | − | − |
| C1C2T2BT31–2060T31902–2386 | − | − |
| C1C2T2BT31–2060T32065–2386 | − | − |
‘+’ indicates infection, while ‘−’ indicates no infection. The nature of the plasmid constructs is described in Figure 1.
The above results showed that the CMV sequence in the recombinant RNA 3 was similar or identical to the CMV RNA 5 sequence. Correspondingly, the TAV RNA 3 sequence in the same recombinant RNA 3s lacked the second 163-nt repeat and the rest of the 3′ NTR (which together corresponds to the TAV RNA 5 sequence). To determine whether CMV RNA 5 itself or the 3′ NTR of CMV RNAs 1 and/or 2 could be used as a template for the synthesis of the recombinant RNA 3, we prepared the appropriate templates that would be expressed transiently in inoculated plants and assessed these for the presence of recombinant virus. In the templates, we deleted 326 nt from the 3′ NTR of T3 to make T31–2060, and deleted the first 163-nt repeat plus 141 nt from the 3′ NTR of T3 to make T3Δ163(A)Δ141 (Figure 1). These two deletions had the same length of the TAV RNA 3 sequence as in the recombinant RNAs. In addition, we also made four RNA 5-like cDNA clones, C13081–3390, C2W2B2757–3065, T31902–2386 and T32065–2386. The former two were based on the CMV RNA 1 and C2W2B sequences in the recombinant viruses, respectively, while the latter two were based on the sequence of TAV RNAs 3B and 5, respectively (39). All of these clones were placed into the pCass1 vector, under transcriptional control of the CaMV 35S RNA promoter and terminator sequences. The infectivity results of the various plasmid mixtures tested on N. glutinosa and N. clevelandii plants are shown in Table 1. The combinations C1, C2T2B, T31–2060, versus C1, C2T2B, T3Δ163(A)Δ141, versus C1, C2W2B, T3Δ163(A)Δ141 all showed no infectivity. The addition of the plasmids expressing the 3′-terminal NTRs C13081–3390, C2W2B2757–3065, T31902–2386 or T32065–2386 into each mixture did not result in infectivity (Table 1). These results indicate that the sequences containing the 3′ NTR are essential for the viral infection and could not be separated functionally from the rest of the TAV RNA 3 sequence. In addition, the results also indicate that the recombination observed in other experiments did not occur during the synthesis of (−) sense RNA from (+) sense templates, since there was no repair of the 3′ NTR from synthesis initiated on either genomic RNAs 1 or 2, or subgenomic RNAs 3B or 5 (Table 1) and template switching to TAV RNA 3 lacking the 3′ NTR. Such repair recombination is common among tombusviruses (8,53) and occurs with the related brome mosaic virus (54–56). Therefore, an alternative explanation is that the recombination occurred during (+) sense RNA synthesis on the (−) sense RNA templates, with recombination occurring as a result of template switching of the viral replicase from the donor (TAV RNA 3) strand to the acceptor (CMV RNA) strand.
A model for the generation of the various recombination products
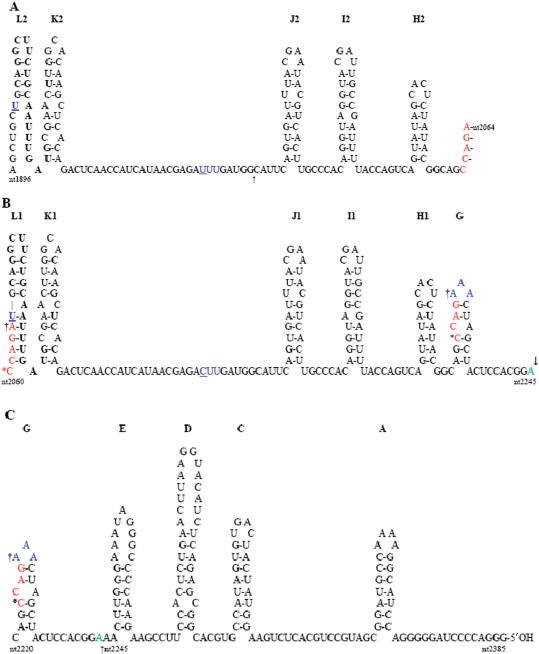
Based on putative secondary structures for the 3′ NTRs of CMV and TAV, models have been proposed to account for the generation of some of the other recombination products previously observed to occur between CMV and TAV (33,38). These models are based on template switching of the viral replicase on a stem-loop structure present in the (−) RNA 3 (L1/L2 in Figure 7) that may be the promoter for the synthesis of subgenomic RNAs 3B and 5 (33,38). Similar structures can be used to explain the various recombinant products obtained here and the effect of the 163-nt repeats and the single G to A substitution on the recombination event.
Figure 7.
Proposed secondary structure of RNA 3 sequences complementary to the 3′ NTR of TAV RNA 3. The residues are numbered according to the corresponding (+) RNA 3, with nucleotide 1902 (underlined) as the position complementary to the 5′ first nucleotide of TAV RNA 3B. Stem-loops are labelled starting from the 5′ end of the (−) TAV RNA 3, as complementary to those stem-loops of the 3′ end of (+) TAV RNA 3 (A–H) described previously (64). Those stem-loops present in the first 163-nt repeat (L2-H2) are shown in (A), while those present in the second 163-nt repeat (L1-H1) and loop G are shown in (B), with the remaining 5′ proximal stem-loops and non-base-paired sequences of (−) TAV RNA 3 shown in (C). The sites of recombination observed here are indicated by an asterisk (nucleotide 2060), a dagger (nucleotide 2064) or an arrow (nucleotide 2245) in (B and C). The site of recombination described in Ref. (30) is at position 1973 and is indicated by an arrow in (A). The single nucleotide difference (nucleotides 1966 and 2129) between the first and second 163-nt repeat is shown in blue and is underlined. A possible pseudoknot can be formed between the three As at the top of stem-loop G and the three Us (shown in blue) in the non-base-paired region between stem-loops K2 and J2 in the first 163-nt repeat, but not with the corresponding CUU sequence shown in the second 163-nt repeat. [Note: Neither stem-loops B nor F are indicated, since loop B did not contain a stem in (+) TAV RNA 3 (64) and small stem-loop F was composed of several non-canonical G–U base pairs in (+) TAV RNA 3 (54) and is not present in (−) TAV RNA 3]. The ΔG values (kcal/mol) for the various stem-loops are A = −11.5, C = −7.8, D = −8.4, E = −6.8, G = −1.3, H = −2.9, I = −4.8, J = −3.8, K = −2.7, L1 = −6.9 and L2 = −5.7, as calculated from Ref. (65).
This model assumes that intramolecular recombination is a more frequent event than intermolecular recombination. Therefore, generation of TAV RNA 3 progeny with deletion of one of the 163-nt repeats is more likely to occur than recombination between TAV RNA 3 and another viral genomic RNA. That is, if one repeat is missing, then only intermolecular recombination can occur, which may explain why the recombinants were seen to accumulate more rapidly in the plants directly inoculated with virus containing only one repeat (Figure 2). On the other hand, loss of a repeat was not a prerequisite for intermolecular recombination, since intramolecular recombination would involve loss of the first repeat [163(A)], while the recombinants formed from the full-length T3 all retained the first repeat. Intramolecular recombination would be expected to occur during (+) TAV RNA 3 synthesis, if the polymerase jumped from the beginning of the first repeat in stem-loop L2 (Figure 7A) to the beginning of the second repeat in stem-loop L1 (Figure 7B). These sequences also contain the putative RNA 5 subgenomic promoter sequence (33,38) that would facilitate this recombination and those described previously that involved this ‘hot spot’ for intermolecular recombination between TAV and CMV (33,38). The presence of another stem-loop structure (K1/K2 in Figure 7) immediately adjacent to the putative subgenomic promoter may slow the polymerase down sufficiently to stutter and in some cases switch templates. In the case of the first TAV-CMV RNA 3 recombinant described (30), this occurred just before another stem-loop structure in the first repeat (J2 in Figure 7A; see arrow), but in that instance, the polymerase jumped to the putative RNA 5 subgenomic promoter of (−) CMV RNA 2, leading to an initial recombinant that had a duplication of the two, putative subgenomic RNA promoters, which then led to a second recombination event, with the intervening TAV RNA 3 sequences deleted, giving the final, stable recombinant RNA 3 described (30).
The full-length TAV RNA 3, as well as the T3Δ163(G) variant gave rise to similar recombination products, with the exact switchover position on the donor RNA of the recombinant determined by the 2b gene present (T2B versus W2B), while the switchover position on the receptor RNA is the beginning of the RNA 5 sequence in either CMV RNAs 1 or 2, again determined by the specific 2b gene present. In the case of C1C2T2BT3 and C1C2T2BT3Δ163(G), the recombination occurred immediately after nucleotide 2060. For C1C2T2BT3, switchover would have had to occur at the base of stem-loop L1 (Figure 7B), while for C1C2T2BT3Δ163(G), switchover would have had to occur within stem-loop G (Figure 7B and C), since the sequences comprising stem-loop L1 (plus stem-loops K1 to H1) would not exist in C1C2T2BT3Δ163(G). Since stem-loops L1 and G are quite different in most of their sequence and structure, as well as stability (ΔG = –1.3 kcal/mol for stem-loop G and −6.9 kcal/mol for stem-loop L1), it seems likely that the common structures preceding these stem-loops (J2-I2-H2 versus J1-I1-H1; ΔG = −3.8, −4.8 and −2.9 kcal/mol, for each stem-loop structure) may have slowed the polymerase sufficiently to allow it to pause at nucleotide 2060, in either stem-loop L1 or G (Figure 7B).
In the case of C1C2W2BT3Δ163(G), switchover occurred following nucleotide 2064 in stem-loop G, but did not immediately proceed to the (−) CMV RNA 2 as the receptor RNA. Rather, the polymerase jumped back to the subgenomic promoter stem-loop (L2) and copied this through the A residue in the gap between stem-loops L2 and K2 (Figure 7A), before pausing and jumping to the same subgenomic promoter sequence on (−) CMV RNA 2. The small difference in ΔG between the 6 bp stem-loop K2 in (−) TAV RNA 3 (ΔG = −2.7 kcal/mol) and a weaker 3 bp stem-loop (K′) structure adjacent to the subgenomic promoter in (−) CMV RNA 2 (ΔG = −2.1 kcal/mol) may be sufficient to prevent the polymerase from pausing at the base of stem-loop K′ in (−) CMV RNA 2. This explains the duplication of the 19 nt seen in Figure 5D, following the A residue (from copying the first nucleotide, U-1902 of the RNA 5/3B promoter). In the case of C1C2W2BT3, the same recombinant progeny RNA 3 is seen, but it must have been generated from a different starting point, since the second repeat is present in the template but missing in the recombinant progeny RNA 3. Here, template switching occurred either immediately after nucleotide 2084 (the A residue in the gap between stem-loops L1 and K1), again leading to a partially duplicated subgenomic promoter with the 19 nt repeat, or it occurred at nucleotide 2064, in the lower region of stem-loop L1 (Figure 7B), and then proceeded as described above for C1C2W2BT3Δ163(G).
In contrast to C1C2T2BT3Δ163(G), C1C2T2BT3Δ163(A) and C1C2W2BT3Δ163(A) underwent recombination 22 nt further along the TAV RNA 3 molecule after nucleotide 2245, adjacent to stem-loop E (in Figure 7C), regardless of the nature of the 2b protein. Therefore, while the nature of the receptor RNA and the apparently slow selection of the recombinants (Figures 2 and 6) were determined by the 2b gene, the generation of recombinants at position 2245 was determined by the absence of the first repeat. Since the first and second repeats differ by only one nucleotide (underlined blue letters in Figure 7A and B, at positions 1966 and 2129), we propose that there must be a difference in structure between the virus containing only one repeat, when this is the second repeat versus the first repeat, and that this difference in structure affects pausing by the polymerase, leading to switchover to another template. This difference could be the presence (or absence) of a weak pseudoknot structure formed between the three A residues in the loop of stem-loop G and the three U residues in the non–base-paired region between stem-loops K2 and J2 (see blue letters in Figures 7). This pseudoknot would not be stable in T3Δ163(A), since it would only contain two A:U base pairs. This weak pseudoknot structure may be sufficient to affect pausing by the polymerase in the T3Δ163(G) variant, if it can re-form after the polymerase has gone through it the first time and then transcribed through the various, following stem-loop structures (stem-loops J, I and H in Figure 7), leading to template switching in stem-loop G. However, as this pseudoknot is absent in T3Δ163(A), the polymerase would continue through stem-loop G until it pauses at the base of stem-loop E (Figure 7C; ΔG = −6.8 kcal/mol), before switching to the receptor RNA.
It would seem likely that the polymerase is as equally able to use RNA 1 as RNA 2 for the receptor RNA, since the polymerase is essentially the same in all cases, and the presence or absence of the 2a protein C-terminal sequences (where the recombinant polymerases showed some differences in sequence) did not affect the nature of the recombinant RNAs. However, since only one type of recombinant progeny was observed in each case, this demonstrates that there must have been selection for a particular progeny RNA, which depended on the source of the 2b gene and its encoded protein.
How could the nature of the 2b protein affect the selection of recombinant RNAs? It is conceivable that the selection of recombinant RNAs is a manifestation of the RNA silencing suppression-related functions of the 2b proteins, since some of these also vary between strains of CMV (57,58). However, based on the spatial separation of the 2b protein from the replication machinery within the cell (49–51), it is difficult to envision how different 2b proteins provide the selection observed through their RNA silencing suppressor functions. In the case of the tombusvirus cucumber necrosis virus, inhibition of expression of the 20-kDa protein now known to be the RNA silencing suppressor of this virus, led to the rapid appearance of defective-interfering RNAs (59). Thus, in that, the absence of the silencing suppressor led to enhanced generation or selection of recombinant RNAs, while in the case of CMV, the source of the 2b protein determined the nature of the recombinant RNAs selected and pseudorecombinant viruses could not be selected in its absence (21). Moreover, in the case of influenza virus, where the NS1 gene encodes an RNA silencing suppressor (60), mutation in a different gene (NS2) led to the rapid appearance of defective interfering particles (61). At the time it was thought that the influenza NS2 protein might be involved in replication. In fact, the NS2 protein is involved in selecting ribonucleoprotein complexes for transport from the nucleus to the cytoplasm and is now called the nuclear export protein (62).
Based on previous observations that the 2b protein is also a movement protein (19,63), that the presence of the 2b protein was necessary for the cell-to-cell movement of pseudorecombinants viruses (21), and that the 2b protein is a viral RNA-binding protein (Figure 4), it seems likely that the 2b binds to and selects specific recombinant viral RNAs for cell-to-cell movement. The different forms of recombinant RNAs generated may have different affinities for the various 2b proteins, leading to selection of one recombinant form over another by a particular 2b protein, which leads eventually to selection of the recombinant RNA over the parental RNA. Since selection to reach detectable levels was only observed after several weeks, the recombinant viral RNAs do not appear to be under strong selection pressure, although there is clearly a gradual selection for the recombinant RNAs over the parental RNA 3. This was seen in the slow appearance of the recombinant RNAs in both sets of time course experiments (Figure 6 and Supplementary Figure 1). The rate of selection could be tested further by direct competition experiments. The Q-CMV 2b protein did not select for any recombinants, while the TAV and WAII-CMV 2b proteins selected for different recombinants, possibly reflecting differences in their RNA-binding properties. It would also be interesting to determine whether various 2b mutants would show differences in their selection of different recombinants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Xiang-Ming Li for her valuable technical assistance. We also thank Peter Langridge and Geoff Fincher for their critical reading of this manuscript. This article is dedicated to our colleague, co-author and former mentor, the late Robert H. Symons. Funding for this study was provided by Australian Research Council Special Research Centre for Basic and Applied Plant Molecular Biology and Scottish Government, Rural and Environment Research and Analysis Directorate (to SCRI). The Open Access publication charges for this article have been partially waived by Oxford University Press and the Australian Centre for Plant Functional Genomics will pay the balance.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hirst GK. Genetic recombination with Newcastle disease virus, polioviruses, and influenza. Cold Spring Harb. Symp. Quant. Biol. 1962;27:303–309. doi: 10.1101/sqb.1962.027.001.028. [DOI] [PubMed] [Google Scholar]
- 2.Ledinko N. Genetic recombination with poliovirus type 1. Studies of crosses between a normal horse serum-resistant mutant and several guanidine-resistant mutants of the same strain. Virology. 1963;20:107–119. doi: 10.1016/0042-6822(63)90145-4. [DOI] [PubMed] [Google Scholar]
- 3.King AM, McCahon D, Saunders K, Newman JW, Slade WR. Multiple sites of recombination within the RNA genome of foot-and-mouth disease virus. Virus Res. 1985;3:373–384. doi: 10.1016/0168-1702(85)90437-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCahon D. The genetics of aphthovirus. Brief review. Arch. Virol. 1981;69:1–23. doi: 10.1007/BF01315261. [DOI] [PubMed] [Google Scholar]
- 5.Luytjes W, Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khatchikian D, Orlich M, Rott R. Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature. 1989;340:156–157. doi: 10.1038/340156a0. [DOI] [PubMed] [Google Scholar]
- 7.Mayo MA, Jolly CA. The 5′-terminal sequence of potato leafroll virus RNA: evidence of recombination between virus and host RNA. J. Gen. Virol. 1991;72:2591–2595. doi: 10.1099/0022-1317-72-10-2591. [DOI] [PubMed] [Google Scholar]
- 8.Nagy PD, Simon AE. New insight into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 9.Figlerowicz M, Nagy PD, Bujarski JJ. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl Acad. Sci. USA. 1997;94:2073–2078. doi: 10.1073/pnas.94.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim MJ, Kao C. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc Natl Acad. Sci. USA. 2001;98:4972–4977. doi: 10.1073/pnas.081077198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panaviene Z, Nagy PD. Mutations in the RNA-binding domains of tombusvirus replicase proteins affect RNA recombination in vivo. Virology. 2003;317:359–372. doi: 10.1016/j.virol.2003.08.039. [DOI] [PubMed] [Google Scholar]
- 12.Gmyl AP, Korshenko SA, Belousov EV, Khitrina EV, Agol VI. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA. 2003;9:1221–1231. doi: 10.1261/rna.5111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallei A, Pankraz A, Thiel H-J, Becher P. RNA recombination in vivo in the absence of replication. J. Virol. 2004;78:6271–6281. doi: 10.1128/JVI.78.12.6271-6281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RIB. Cucumber mosaic virus. Adv. Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 15.Palukaitis P, García-Arenal F. Cucumoviruses. Adv. Virus Res. 2003;62:241–323. doi: 10.1016/s0065-3527(03)62005-1. [DOI] [PubMed] [Google Scholar]
- 16.Nitta N, Takanami Y, Kuwata S, Kubo S. Comparative studies on the nucleotide sequence of cucumber mosaic virus RNA 3 between Y strain and Q strain. Ann. Phytopathol. Soc. Japan. 1988;54:516–522. [Google Scholar]
- 17.Hayes RJ, Buck KW. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- 18.Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Ding SW, Li WX, Symons RH. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 1995;14:5762–5772. doi: 10.1002/j.1460-2075.1995.tb00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li HW, Lucy AP, Guo HS, Li WX, Ji LH, Wong SM, Ding SW. Strong host resistance targeted against a viral suppressor of the plant gene silencing defense mechanism. EMBO J. 1999;18:2683–2691. doi: 10.1093/emboj/18.10.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi BJ, Miller J, Symons RH, Palukaitis P. The 2b protein of cucumoviruses has a role in promoting the cell-to-cell movement of pseudorecombinant viruses. Mol. Plant Microbe Interact. 2003;16:261–267. doi: 10.1094/MPMI.2003.16.3.261. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki M, Kuwata S, Kataoka J, Masuta C, Nitta N, Takanami T. Functional analysis of deletion mutants of cucumber mosaic virus RNA 3 using an in vitro transcription system. Virology. 1991;183:106–113. doi: 10.1016/0042-6822(91)90123-s. [DOI] [PubMed] [Google Scholar]
- 23.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA 3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan IB, Shintaku MH, Li QB, Zhang L, Marsh LE, Palukaitis P. Complementation of virus movement in transgenic tobacco expressing the cucumber mosaic virus 3a gene. Virology. 1995;209:188–199. doi: 10.1006/viro.1995.1242. [DOI] [PubMed] [Google Scholar]
- 25.Ding SW, Anderson BJ, Haase HR, Symons RH. New overlapping gene encoded by the cucumber mosaic virus genome. Virology. 1994;198:593–601. doi: 10.1006/viro.1994.1071. [DOI] [PubMed] [Google Scholar]
- 26.Shi BJ, Ding SW, Symons RH. In vivo expression of an overlapping gene encoded by the cucumoviruses. J. Gen. Virol. 1997;78:237–241. doi: 10.1099/0022-1317-78-1-237. [DOI] [PubMed] [Google Scholar]
- 27.Schwinghamer MW, Symons RH. Translation of the four major RNA species of cucumber mosaic virus in plant and animal cell-free system and toad oocytes. Virology. 1977;79:88–108. doi: 10.1016/0042-6822(77)90337-3. [DOI] [PubMed] [Google Scholar]
- 28.Rao ALN, Grantham GL. Amplification in vivo of brome mosaic virus RNAs bearing 3′ noncoding region from cucumber mosaic virus. Virology. 1994;204:478–481. doi: 10.1006/viro.1994.1559. [DOI] [PubMed] [Google Scholar]
- 29.Sivakumaran K, Bao Y, Roossinck MJ, Kao CC. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicase of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 2000;74:10323–10331. doi: 10.1128/jvi.74.22.10323-10331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez-Cuartero B, Burgyan J, Aranda MA, Salanki K, Moriones E, Garcia-Arenal F. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology. 1994;203:373–377. doi: 10.1006/viro.1994.1496. [DOI] [PubMed] [Google Scholar]
- 31.Masuta C, Ueda S, Suzuki M, Uyeda I. Evolution of a quadripartite hybrid virus by interspecific exchange and recombination between replicase components of two related tripartite RNA viruses. Proc. Natl Acad. Sci. USA. 1998;95:10487–10492. doi: 10.1073/pnas.95.18.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aaziz R, Tepfer M. Recombination between genomic RNAs of two cucumoviruses under conditions of minimal selection pressure. Virology. 1999;263:282–289. doi: 10.1006/viro.1999.9973. [DOI] [PubMed] [Google Scholar]
- 33.de Wispelaere M, Gaubert S, Trouilloud S, Belin C, Tepfer M. A map of the diversity of RNA3 recombinants appearing in plants infected with Cucumber mosaic virus and Tomato aspermy virus. Virology. 2005;331:117–127. doi: 10.1016/j.virol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 34.Chen YK, Goldbach R, Prins M. Inter- and intramolecular recombinations in the cucumber mosaic virus genome related to adaptation to alstroemeria. J. Virol. 2002;76:4119–4124. doi: 10.1128/JVI.76.8.4119-4124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin HX, Rubio L, Smythe AB, Falk BW. Molecular population genetics of Cucumber mosaic virus in California: evidence for founder effects and reassortment. J. Virol. 2004;78:6666–6675. doi: 10.1128/JVI.78.12.6666-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnet J, Fraile A, Sacristan S, Malpica JM, Garcia-Arenal F. Role of recombination in the evolution of natural populations of Cucumber mosaic virus, a tripartite RNA plant virus. Virology. 2005;332:359–368. doi: 10.1016/j.virol.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 37.Canto T, Choi SK, Palukaitis P. A subpopulation of RNA 1 of Cucumber mosaic virus contains 3′ termini originating from RNAs 2 or 3. J. Gen. Virol. 2001;82:941–945. doi: 10.1099/0022-1317-82-4-941. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki M, Hibi T, Masuta C. RNA recombination between cucumoviruses: possible role of predicted stem-loop structures and an internal subgenomic promoter-like motif. Virology. 2003;306:77–86. doi: 10.1016/s0042-6822(02)00050-8. [DOI] [PubMed] [Google Scholar]
- 39.Shi BJ, Ding SW, Symons RH. Two novel subgenomic RNAs derived from RNA 3 of tomato aspermy cucumovirus. J. Gen. Virol. 1997;78:505–510. doi: 10.1099/0022-1317-78-3-505. [DOI] [PubMed] [Google Scholar]
- 40.Shi BJ, Palukaitis P, Symons RH. Stable and unstable mutations in the 5′ non-coding region of tomato aspermy virus RNAs 1 and 2 generated de novo from infectious cDNA clones containing a cauliflower mosaic virus 35S promoter. Virus Genes. 2004;28:277–283. doi: 10.1023/b:viru.0000025775.20862.50. [DOI] [PubMed] [Google Scholar]
- 41.Ding SW, Rathjen JP, Li WX, Swanson R, Healy H, Symons RH. Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J. Gen. Virol. 1995;76:459–464. doi: 10.1099/0022-1317-76-2-459. [DOI] [PubMed] [Google Scholar]
- 42.Ding SW, Shi BJ, Li WX, Symons RH. An interspecies hybrid RNA virus is significantly more virulent than either parental virus. Proc. Natl Acad. Sci. USA. 1996;93:7470–7474. doi: 10.1073/pnas.93.15.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi BJ, Ding SW, Symons RH. Plasmid vector for cloning infectious cDNAs from plant RNA viruses: high infectivity of cDNA clones of tomato aspermy cucumovirus. J. Gen. Virol. 1997;78:1181–1185. doi: 10.1099/0022-1317-78-5-1181. [DOI] [PubMed] [Google Scholar]
- 44.Peden KWC, Symons RH. Cucumber mosaic virus contains a functionally divided genome. Virology. 1973;53:487–492. doi: 10.1016/0042-6822(73)90232-8. [DOI] [PubMed] [Google Scholar]
- 45.Shi BJ. University of Adelaide; 1997. Expression and function of cucumoviral genome. pp. 81–82. Ph.D. Thesis. [Google Scholar]
- 46.Rajendran KS, Pogany J, Nagy PD. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 2002;76:1707–1717. doi: 10.1128/JVI.76.4.1707-1717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi BJ, Palukaitis P, Symons RH. Differential virulence by strains of Cucumber mosaic virus is mediated by the 2b gene. Mol. Plant-Microbe Interact. 2002;15:947–955. doi: 10.1094/MPMI.2002.15.9.947. [DOI] [PubMed] [Google Scholar]
- 48.Blanchard CL, Boyce PM, Anderson BJ. Cucumber mosaic virus RNA 5 is a mixed population derived from the conserved 3′-terminal regions of genomic RNAs 2 and 3. Virology. 1996;217:598–601. doi: 10.1006/viro.1996.0155. [DOI] [PubMed] [Google Scholar]
- 49.Lucy AP, Guo HS, Li WX, Ding SW. Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 2000;19:1672–1680. doi: 10.1093/emboj/19.7.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayers CN, Palukaitis P, Carr JP. Subcellular distribution analysis of the cucumber mosaic virus 2b protein. J. Gen. Virol. 2000;81:219–226. doi: 10.1099/0022-1317-81-1-219. [DOI] [PubMed] [Google Scholar]
- 51.Cillo F, Roberts IM, Palukaitis P. In situ localization and tissue distribution of the replication-associated proteins of Cucumber mosaic virus in tobacco and cucumber. J. Virol. 2002;76:10654–10664. doi: 10.1128/JVI.76.21.10654-10664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang MS, Kim SH, Lee JH, Bae JM, Paek KH, Park YI. Evidence for interaction between the 2a polymerase protein and the 3a movement protein of Cucumber mosaic virus. J. Gen. Virol. 2005;86:3171–3177. doi: 10.1099/vir.0.81139-0. [DOI] [PubMed] [Google Scholar]
- 53.White KA, Morris TJ. Recombination between defective tombusvirus RNA generates functional hybrid genomes. Proc. Natl Acad. Sci USA. 1994;91:3642–3644. doi: 10.1073/pnas.91.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rao ALN, Hall TC. Requirements for a viral trans-acting factor encoded by brome mosaic virus RNA-2 provides strong selection in vivo for functional recombinants. J. Virol. 1990;64:2437–2441. doi: 10.1128/jvi.64.5.2437-2441.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bujarski JJ, Dzianott AM. Generation and analysis of nonhomologous RNA-RNA recombinants in brome mosaic virus: sequence complementarities at crossover sites. J. Virol. 1991;65:4153–4159. doi: 10.1128/jvi.65.8.4153-4159.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Greene AE, Allison RF. Recombination between viral transcripts and transgenic plant transcripts. Science. 1994;263:1423–1425. doi: 10.1126/science.8128222. [DOI] [PubMed] [Google Scholar]
- 57.Lewsey M, Robertson FC, Canto T, Palukaitis P, Carr JP. Selective targeting of miRNA-regulated plant development by a viral counter-silencing protein. Plant J. 2007;50:240–252. doi: 10.1111/j.1365-313X.2007.03042.x. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Yuan YR, Pei Y, Lin S-S, Tuschl T, Patel DJ, Chua N-H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rochon DM. Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20-kDa nonstructural protein. Proc. Natl Acad. Sci. USA. 1991;88:11152–11157. doi: 10.1073/pnas.88.24.11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W-X, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EWA, Johnson KL, García-Sastre A, et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl Acad. Sci. USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Odagiri T, Tobita K. Mutations in NS2, a nonstructural protein of influenza A virus, extragenically causes aberrant replication and expression of the PA gene and leads to generation of defective interfering particles. Proc. Natl Acad. Sci USA. 1990;87:5988–5992. doi: 10.1073/pnas.87.15.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boulo S, Akarsu H, Ruigrok RHW, Baudin F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 2007;124:12–21. doi: 10.1016/j.virusres.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 63.Soards AJ, Murphy AM, Palukaitis P, Carr JP. Virulence and differential local and systemic spread of Cucumber mosaic virus in tobacco are affected by the CMV 2b protein. Mol. Plant-Microbe Interact. 2002;15:647–653. doi: 10.1094/MPMI.2002.15.7.647. [DOI] [PubMed] [Google Scholar]
- 64.Wilson PA, Symons RH. The RNAs of Cucumoviruses: 3′-terminal sequence analysis of two strains of tomato aspermy virus. Virology. 1981;112:342–345. doi: 10.1016/0042-6822(81)90639-5. [DOI] [PubMed] [Google Scholar]
- 65.Mathews DH, Sabina J, Zucker M, Turner H. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.