Abstract
Chromatin immunoprecipitation combined with DNA microarrays (ChIP-chip) is a powerful technique to detect in vivo protein–DNA interactions. Due to low yields, ChIP assays of transcription factors generally require amplification of immunoprecipitated genomic DNA. Here, we present an adapted linear amplification method that involves two rounds of T7 RNA polymerase amplification (double-T7). Using this we could successfully amplify as little as 0.4 ng of ChIP DNA to sufficient amounts for microarray analysis. In addition, we compared the double-T7 method to the ligation-mediated polymerase chain reaction (LM-PCR) method in a ChIP-chip of the yeast transcription factor Gsm1p. The double-T7 protocol showed lower noise levels and stronger binding signals compared to LM-PCR. Both LM-PCR and double-T7 identified strongly bound genomic regions, but the double-T7 method increased sensitivity and specificity to allow detection of weaker binding sites.
INTRODUCTION
DNA microarray location analysis of DNA-binding proteins using chromatin immunoprecipitation (ChIP–chip) is making important contributions to the investigation of DNA-bound proteins such as the regulatory factors that control gene expression. ChIP-chip studies have already been successfully used for high-resolution mapping of nucleosome positions (1), for identification of transcription factor binding sites (2) and for elucidating how histone modifications relate to transcription (3). Increased resolution of microarray technology and ongoing optimization of experimental protocols will boost the efficiency of this technique, further increasing its already widespread use.
The basic principle of ChIP is straightforward and typically involves in vivo coupling of proteins to DNA using a crosslinking agent such as formaldehyde (4,5). DNA is then fragmented, by sonication or nuclease digestion, followed by immunoprecipitation with specific antibodies against the protein of interest, thereby enriching for crosslinked genomic fragments. After reversal of the crosslinks, the immunoprecipitated DNA is purified, labeled and hybridized onto a DNA microarray (6). Probes corresponding to regions in the genome bound by the protein of interest will show enrichment for the immunoprecipitated sample compared to input. Alternative methods for microarray detection of protein–DNA interactions exist, such as tethering DNA-binding proteins to DNA adenine methyltransferase, resulting in a localized methylation around genomic binding sites (7). Similar to ChIP, in DamID the location of genomic binding sites can then be inferred following isolation of the methylated DNA fragments and hybridization to whole-genome DNA microarrays.
A crucial step in these techniques is obtaining sufficient material (∼1–5 μg) for DNA microarray hybridization. Whereas immunopreciptiation of abundant proteins such as histones often readily provides the required amounts of bound DNA, the yields for transcription factors with a limited number of genomic binding sites is in the sub-nanogram (ng) range (8). One way to address this problem is to increase the amount of starting material, but for transcription factors with only a few genomic binding sites this is hardly feasible. Therefore, the amount of ChIP material for hybridization is often increased by amplification. PCR-based methods like ligation-mediated PCR (LM-PCR) or whole-genome amplification display an unwanted bias toward certain DNA sequences. This is exaggerated by the exponential nature of the amplification. Previously, it was shown that a linear amplification method based on T7 RNA polymerase transcription prevents this bias (9). Using this protocol, 2.5 ng of endonuclease-digested genomic DNA was increased to 10 μg amplified RNA (aRNA). Several groups have now successfully used T7 amplification for ChIP-chip analysis of nucleosomes and histone modifications (10,11). However, for transcription factors, single round T7 amplification still yields insufficient amounts of aRNA for microarray hybridizations.
Here, we present a two-step T7 amplification protocol that can accurately and reproducibly amplify as little as 0.4 ng of chromatin immunoprecipitated DNA for microarray analysis. Importantly, in our experiments, the double-round T7 amplification method also shows improved signal-to-noise ratios when compared to conventional LM-PCR, resulting in increased sensitivity and specificity to detect genomic regions bound by DNA-sequence-specific transcription factors.
MATERIALS AND METHODS
Construction of strains
GSM1 was TAP-tagged at the carboxy-terminus by integration of a PCR fragment encoding the TAP tag and TRP1 marker into Saccharomyces cerevisiae wild-type YPH499 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1) strain (12). TBP was avitag-tagged at the amino terminus in wild-type W303B (MATα ade2-1 ura3-1 his3-11,15 leu2-3,112 trp1-1 can1-100) as described previously (13). The TBP-avitag and GSM1-TAP strains were verified by immunoblot analysis and by PCR of genomic DNA, using primers corresponding to the tagged allele.
Chromatin immunoprecipitation
Preparation of chromatin extracts and immunoprecipitation steps were performed as described previously (13). In short, yeast cells were grown in synthetic complete (SC) medium containing 2% glucose. 400 ml of cells at an OD600 0.5–0.6 was crosslinked with 1% formaldehyde for 20 min before chromatin was extracted. The TBP-avitag strain was transformed with pRS313-BirA-NLS plasmid (13) and grown in SC medium containing 2% glucose, 250 nM biotin and lacking histidine. After harvesting, cells were disrupted by vortexing in FA lysis buffer (50 mM HEPES–KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS), centrifuged and chromatin pellet was washed in FA lysis buffer followed by sonication (Bioruptor, Diagenode: 7 cycles, 30 s on/off, medium setting) to produce a fragment size of ∼400 bp. For the TAP IPs, 400 μl of chromatin extract and 10 μl BSA (25 mg/ml) was incubated with 40 μl of IgG Sepharose beads (Amersham) for 1 h and 45 min. For histone 3 (H3) and RNA polymerase II (pol II) IPs, 2 μl of anti-H3 (Abcam, ab1791) and 10 μg anti-RNA pol II (8WG16) antibodies were coupled to 30 μl of protein A magnetic beads (Invitrogen). After the IP, the beads were washed and eluted accordingly. TBP IP was performed using 80 μl of streptavidin magnetic beads (Invitrogen), and was processed as described (13). For reversal of formaldehyde crosslinking, input and ChIP samples were incubated at 65°C overnight in 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 1% SDS, 0.5 M NaCl and ribonuclease A (75 μg/ml). Samples (100 μl) were subsequently treated with 100 μg proteinase K (Roche) at 37°C for 2 h. Finally, DNA was obtained by phenol extraction followed by purification on PCR purification kit columns (Qiagen).
First round of T7 amplification
Input and ChIP samples were amplified using an adapted version of the T7 based protocol described by Liu et al. (9), adding a second amplification round to increase the yield for low amounts of ChIP starting material (Figure 1). Prior to the amplification, samples were treated with 1 U shrimp alkaline phosphatase (Roche) at 37°C for 2 h, and purified using the MinElute PCR purification Kit (Qiagen), eluting in 20 μl EB buffer. This step can also be performed directly after reverse crosslinking before proteinase K treatment. Between 0.4 and 5 ng of DNA was typically used in the first round of amplification. A poly(dT) tail was added to the 3′-end of ChIP DNA by a terminal transferase (TdT) (NEB) in a reaction volume of 9 μl reaction mixture [containing 5 μl of sample, 1xTdT buffer (Roche), 75 μM CoCl2, 0.4 μM ddCTP, 4.6 μM dTTP and 0.1 μg linear acrylamide] was denatured at 95°C for 2 min, and then put on ice for 2 min. Samples were incubated at 37°C for 20 min with 20 U TdT enzyme (NEB). The tailed DNA fragments were purified using the MinElute PCR purification Kit by elution in 20 μl EB buffer.
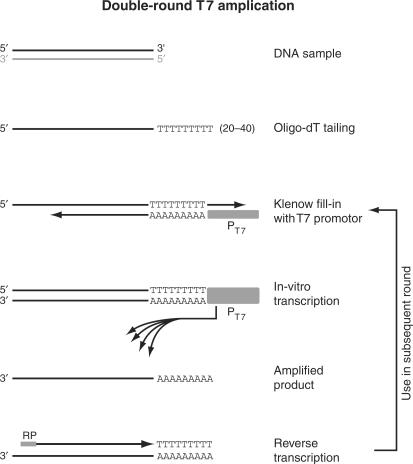
Figure 1.
Overview of the double-T7 RNA polymerase based linear amplification method. The steps in the T7 amplification protocol are illustrated for the top strand (black) of a double-stranded DNA template. The bottom strand (gray) is analogously amplified but omitted from this figure for clarity. After addition of poly(dT), an anchored T7-(dA)18 oligo is annealed and the overhangs are filled in using the Klenow fragment of DNA polymerase I. The resulting double-stranded DNA fragment contains a functional T7 promoter that drives the in vitro transcription reaction, generating multiple RNA copies of the DNA template. To increase the yield for low starting amounts of ChIP material, a second amplification round is performed after reverse transcription using random primers (RP = random primers).
For the second strand synthesis an anchored T7-(dA)18 oligo (5′-GGA GGC CGG AGA ATT GTA ATA CGA CTC ACT ATA GGG AGA CGC GTG A18 B-3′) was used. A volume of 9.5 μl reaction mixture containing, 7.5 μl of sample, 1x NEB buffer 2, 0.25 mM dNTPs and 25 nM T7-(dA)18 oligo was first denatured at 95°C for 2 min, subsequently annealed at 35°C for 2 min (ramp rate, 1°C/s) and incubated at 25°C for 2 min (ramp rate, 0.5°C/s). At this point, 1 U Klenow enzyme (NEB) was added, and the reaction mixture was incubated at 37°C for 90 min. The double-stranded DNA fragments were purified using the MinElute PCR purification Kit.
In vitro transcription was performed using the MEGAscript T7 kit (Ambion). A volume of 20 μl containing, 7 μl of sample, 7.5 mM of each NTP, 1x reaction buffer, 0.1 μg linear acrylamide and 2 μl enzyme mix was incubated at 37°C for 4 h. The reaction was heat-inactivated at 70°C for 10 min. In vitro transcribed RNA was purified using the RNeasy kit (Qiagen) according to the manufacturer's instructions.
Second round of T7 amplification
For the second amplification round typically between 50 and 150 ng of first-round amplified material was used for the reverse transcription reaction. A reaction mixture of 10 μl containing 1 μg random primers (Invitrogen), and 9 μl of sample was denatured at 70°C for 10 min and kept on 48°C. A first-strand mixture of 10 μl consisting of 2x first-strand buffer (Invitrogen), 10 μM DTT, 40 U of RNase inhibitor (Roche), 2 mM of dNTPs, 0.1 μg linear acrylamide and 200 U Superscript III (Invitrogen) was preheated at 48°C before combining with the sample/random primer mixture. The reaction mixture was then incubated at 48°C for 2 h, denatured at 95°C for 2 min and placed on ice for 5 min. Single stranded DNA fragments were purified using the MinElute PCR purification Kit, and eluted in 20 μl EB buffer.
Second-strand synthesis was performed similarly as described for the first round of amplification except that the reaction volume and T7-(dA)18 oligo concentration differed. A reaction mixture of 20 μl containing 15 μl of sample, 1x NEB buffer 2, 0.25 mM dNTP mix and 250 nM T7-(dA)18 oligo was denatured and annealed. Two units of Klenow enzyme were added followed by incubation at 37°C for 90 min. The double-stranded DNA fragments were purified using the MinElute PCR purification Kit.
Samples were amplified using MEGAscript T7 kit as described for the first round except that the 20 μl reaction mixture consisted of 7 μl of sample, 1x reaction buffer, 7.5 mM ATP, 7.5 mM CTP, 7.5 mM GTP, 2.25 mM UTP, 5.25 mM of 5-(3-aminoallyl)-UTP and 2 μl enzyme mix. In vitro transcribed RNA was purified using the RNeasy kit according to the manufacturer's instructions.
LM-PCR
Ligation mediated PCR was performed as described in (14).
RNA quantification and determination of fragment sizes
The amount of DNA used in the first round of amplification and the resulting RNA yield were assayed using PicoGreen (Invitrogen) dsDNA and RiboGreen (Invitrogen) RNA quantification kits, respectively. The amount of fluorescence was measured on a NanoDrop ND3300 (NanoDrop Technologies) fluorospectrometer and compared to a set of DNA and RNA standards. Second-round amplified RNA was quantified on a spectrophotometer (UVmini 1240, Shimadzu). The size distribution of RNA fragments after single or double-T7 amplification round was determined on a Bioanalyzer (Agilent Technologies).
Labeling reactions
A reaction volume of 22 μl consisting of 6 μg aRNA (double-T7) or 5 μg DNA (LM-PCR), 4 μl monofunctional NHS-ester Cy3 or Cy5 dye (GE Healthcare) and 68 mM sodium bicarbonate was incubated at 25°C for 1 h. The reaction was quenched by addition of 12 μl 4M hydroxylamine at 25°C for 15 min. Unbound dyes were removed using Chromaspin-30 (Clontech) columns, and the efficiency of dye incorporation was measured using a spectrophotometer (UVmini 1240, Shimadzu).
Blocking oligo for preventing aspecific hybridization with T7 promoter sequences
During the initial microarray experiments a small subset of microarray probes (∼200; 0.5% of all probes) showed consistently high signal intensity, in both input and ChIP samples amplified with the double-T7 protocol (Supplemental Figure 1A). These microarray probes did not show enrichment after LM-PCR amplification of the same samples. We examined these probes for common sequence characteristics using Gibbs motif analysis (15) (Supplemental Figure 1B). A small, but highly significant CCCTCTG motif was identified and found to correspond to the small section of T7 promoter that is incorporated in all amplified fragments during IVT (15) (Supplemental Figure 1C). Considering that this motif is highly GC-rich and present in great excess in the amplified, labeled material, we surmised that cross-hybridization with this T7 promoter fragment is responsible for the high signal intensity on a small subset of probes.
This cross-hybridization affects both ChIP and input samples equally and is therefore unlikely to result in the selection of false positives. Nevertheless, the resulting increase in signal intensity obscures specific binding sites that may otherwise be detected with these probes. Future microarray designs should thus preferably avoid probes containing CCCTCTG motifs. In case of tiling arrays with oligo's covering the complete genome, this is however not possible. We addressed this by adding an unlabeled blocking oligo with a T(14) CAC GCG TCT CCC sequence in the hybridization step. An 80-fold molar excess of blocking oligo indeed effectively reduced the signals from the affected probes (Supplemental Figure 1D).
Addition of blocking oligo could conceivably inhibit hybridization at another subset of microarray probes that contain the complementary GGGAGAC sequence motif. Alternatively, the blocking oligo could actually improve detection at these probes by preventing the T7 promotor fragment from sequestering the corresponding labeled genomic target sequences, thereby increasing availability for hybridization with the microarray probes. To test which of these effects dominates, we investigated the subset of microarray probes showing significant enrichment (P < 0.05 and fold-enrichment >2) in Gsm1p binding for probe sequences containing one or more GGGAGAC motifs. Two probes were identified, SCAD029882 and SCAD002834, corresponding to the SFC1 and MRH4 promoter regions. A comparison to the fold-enrichment in the original data revealed that the reported binding ratios at these probes improved after addition of blocking oligo (Supplemental Figure 1E). This indicates that the use of a blocking oligo increases availability for hybridization and thereby improves the quality of ChIP-chip data, also on the complementary GGGAGAC-containing probes. The use of blocking oligo is also expected to benefit other applications that make use of T7 amplification, such as microarray gene expression studies.
Microarray hybridization and analysis
For the hybridization mixture, 4 μg aRNA (double-T7) or 4 μg DNA (LM-PCR) of reciprocal labeled input and ChIP samples, with label incorporations between 2 and 4%, were combined together with 750 ng of herring sperm DNA, 40 μg tRNA, 10 μg human Cot1 DNA (Invitrogen), 50 mM Na-MES buffer (pH 6.9), 500 mM NaCl, 6 mM EDTA, 0.5% N-Lauroylsarcosine and 30% formamide in a total volume of 500 μl. For the indicated double-T7 experiments 30-μg blocking oligo (5′-TT TTT TTT TTT TTT CAC GCG TCT CCC-3′) was added to the hybridization mix. Samples were denatured at 95°C for 3 min and hybridized at 42°C for 20 h to high-resolution oligonucleotide arrays (Agilent Technologies) that contain 60 mer oligonucleotide probes covering the complete yeast genome at an average 266-bp resolution. The microarray slides were washed for 5 min each in wash buffer 1 (6xSSPE, 0.005% N-Lauroylsarcosine) and wash buffer 2 (0.06x SSPE), while stirring. Slides were scanned in an Agilent DNA Microarray Scanner (Model G2565BA, Agilent Technologies) and spot quantification was carried out using Imagene 4.0 (Biodiscovery). Genome-wide localization analysis data were generated from four independent hybridizations and two biological samples. For each biological replicate, a dye-swap technical duplicate was performed. The data can be viewed in Supplementary Table 1 that contains binding ratios [log2(ChIP/Input)] for the four hybridizations of T7 and LM-PCR-amplified samples, mapped against the ENSEMBL yeast genome version 37.1 (February 2006).
Following quantification, the microarray data was lowess-normalized over all probes with a span of 0.4. A binding ratio (ChIP enrichment) was calculated for each microarray probe by dividing the signal intensity of the immunoprecipitated DNA sample by that of input, consisting of input DNA. A confidence value was calculated for each microarray probe using the MAANOVA package and corrected for multiple testing using Benjamini–Hochberg false-discovery rate control. A combination of confidence value and binding ratio cutoffs was used to identify significantly bound genomic regions. Signals from neighboring probes were considered to be part of a single binding event if they showed a binding ratio >1.3 and ANOVA q < = 0.1.
Average binding analysis
In order to calculate averaged binding profiles, the genomic region around genes in the yeast genome were divided in two regions consisting of 800-bp promoter and the ORF, each further subdivided in three segments. Within each segment, the probe with the highest binding ratio was used as representative. For segments not covered by microarray probes, a binding ratio was calculated by linear interpolation from directly neighboring probes within a distance of maximally 300 bp. Averaged binding profiles were constructed for a selection of genes by calculating the geometric mean across each of the segments. Random averaged binding profiles were derived from a random selection of an equally sized set of genes from the genome.
Motif analysis
Motif analysis was performed with the GIBBS motif sampler at the Regulatory Sequence Analysis Tools website (http://rsat.ulb.ac.be/rsat/) (16). The resulting profile obtained from the GIBBS sampler was used as input for WebLogo (http://weblogo.berkeley.edu/logo.cgi) to generate a visual representation (15).
Real-time PCR analysis
Real-time PCR was performed as described (13). The mean and standard deviation of each ChIP was calculated from two biological replicates that were assayed in duplicate. The input control normalized signals are presented as fold occupancy over HMR silent mating-type locus region. Oligonucleotide sequences are available upon request.
RESULTS AND DISCUSSION
Double-round T7-based amplification of ChIP samples
The original T7 RNA polymerase-based amplification protocol described by Liu et al. (9) was shown to work efficiently on purified endonuclease-digested genomic DNA. However, the quality of a DNA sample obtained after ChIP is affected by formaldehyde crosslinking and sonication, which reduces the efficiency of the amplification. As a result, the starting amount of ChIP material required to obtain sufficient amounts for hybridization on DNA microarray increases to ∼80 ng (Table 1), which is considerably higher than the yield of material obtained after ChIP of transcription factors with a limited number of genomic targets (8).
Table 1.
Quantification of single and double round T7 amplification of input ChIP DNA
| Input (ng) | Yield after single T7 (ng) | Yield after double T7 (ng) |
|---|---|---|
| 0 | <100a | 112 |
| 0.4 | <100a | 4960 |
| 1 | <100a | 20160 |
| 2.5 | <100a | 35840 |
| 5 | 420 | 50240b |
| 20 | 2480 | NDc |
| 80 | 9880 | NDc |
aexact quantification was not possible because of low yield.
b150 ng from single T7 was used for second round of double T7.
cNot determined.
In order to improve the yield of T7-based amplification, several modifications were made to the original protocol (9) (Figure 1). The main change was the addition of a second round of amplification, using reversed-transcribed cDNA from the first round as a template. Prior to in vitro transcription (IVT), the T7 promoter is incorporated during a Klenow fill-in reaction, using the poly(dT) tail that was added in the first steps as an anchor (Figure 1). An optimized T7 promoter oligo sequence was used to increase the IVT yield (17). The incubation time of the two IVT reactions was reduced to 4 h each, to minimize the chance of amplification-induced artifacts. In the final round of IVT, aminoallyl-UTP was included for indirect labeling with NHS-activated cyanine dyes. Between 4 and −5 μg of labeled aRNA obtained from ChIP and input samples is typically used for DNA microarray hybridization.
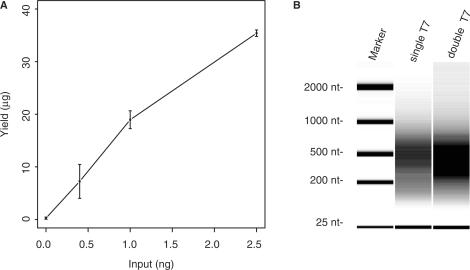
The performance of the double-round T7 amplification (double-T7) protocol was tested on sheared input DNA that was isolated from a chromatin extract of S. cerevisiae. After reversal of the formaldehyde crosslinking, 0.4 to 80 ng of DNA was used in the amplification reactions. Input material ranging from 0.4 to 2.5 ng was amplified to <100 ng after a single round of T7 amplification (single-T7). These amounts of input are in the lower range of what is typically found after ChIP of DNA-binding proteins (8). Double-T7 procedure gave consistent, high yields that increased approximately linearly with these amounts of starting material (Table 1 and Figure 2A). At least 80 ng of input was needed for single-T7 to obtain sufficient material. For the double-T7, 0.4 ng starting material was sufficient for several hybridizations. No difference of the aRNA size was observed when we compared the single-T7 and double-T7 products, suggesting there is no shortening of aRNA during the second round of T7 amplification (Figure 2B). At higher levels of input DNA (>2.5 ng) the amplification efficiency using double-T7 decreased, which is likely related to the depletion of some reaction components. We also observed that the first round of amplification was less efficient (∼50-fold) compared to the second round (∼450-fold) (Table 1). This may be attributed to the lower quality of reverse-crosslinked DNA and low starting amount in the first round. The modified double-T7 protocol therefore significantly decreases the lower threshold for the amount of ChIP material needed for amplification by ∼200-fold compared to the original protocol.
Figure 2.
Yields obtained from amplification of different amounts of input DNA. An input DNA sample was obtained by reverse crosslinking chromatin extracts from a wild-type yeast culture, and used as starting material for a double-round T7 amplification. (A) Averaged RNA yields after two rounds of amplification are plotted as function of the amount of starting material used, which ranged from 0.4 to 2.5 ng. The concentration of T7 oligo added to the Klenow reactions was scaled proportionally with the amount of starting material, and set to 25 nM and 250 nM for the first and second round, respectively. Assays were performed in duplicate and error bars indicate the standard deviation. (B) Analysis of fragment size of amplified material after single round or double round of T7 amplification. One hundred and fifty nanograms of each sample was analyzed on an Agilent 2100 Bioanalyzer together with a RNA size marker. The fragment sizes of the RNA marker are indicated.
Next, we compared the single- and double-round T7 amplification in ChIP assays for transcription regulatory proteins (Table 2). Whereas the histone H3 ChIP yielded sufficient material after single-round T7 amplification, the yields for RNA polymerase II (pol II) and TATA-box binding protein (TBP) were not sufficient. In contrast, the double-T7 method provided sufficient material for hybridization of the pol II and TBP ChIPs. Table 2 also indicates that smaller cultures could be used for the TBP ChIPs. A control ChIP using empty protein A beads also resulted in sufficient material. Together, these results show that our double-T7 amplification protocol can yield sufficient material for DNA microarray hybridization of ChIP assays of transcription regulatory factors.
Table 2.
Quantification of single and double round T7 amplification of several ChIP samples
| ChIP sample | Culture volume (ml) (OD600 = 0.6) | Yield after ChIP (ng) | Yield after single T7 (ng) | Yield after double T7 (ng) |
|---|---|---|---|---|
| H3 | 200 | 140 | 18 000 | NDa |
| pol II | 200 | <20b | 500 | 31 680c |
| Prot A | 200 | <20b | 144 | 32 000 |
| TBP | 400 | <20b | 784 | 46 080c |
| TBP | 200 | <20b | 300 | 21 840c |
| TBP | 100 | <20b | 200 | 22 880c |
| TBP | 25 | <20b | 150 | 37 600 |
aNot determined.
bExact quantification was not possible because of low yield.
c150 ng from single T7 was used for second round of double T7.
Double-T7 amplification exhibits higher signal-to-noise ratios compared to LM-PCR
To test whether the increased amplification yield still allows proper detection of protein–DNA interactions, the performance of the double-T7 method was compared to the commonly used LM-PCR amplification in ChIP. DNA microarrays were used that contain 60 mer oligonucleotide probes covering the complete yeast genome at an average 266 bp resolution. During initial experiments a small subset of microarray probes (∼200; 0.5% of all probes) showed consistently high signal intensity in both input and ChIP samples amplified with the double-T7 protocol (Supplemental Figure 1). As described in the Materials and Methods section, addition of a blocking oligonucleotide corresponding to part of the T7 RNA polymerase promotor sequence completely circumvents this problem and this blocking oligo was included in the subsequent hybridizations.
We compared double-T7 and LM-PCR performance by analyzing binding sites of the putative S. cerevisiae transcription factor GSM1p encoded by YJL103C (Saccharomyces Genome Database; http://www.yeastgenome.org). Gsm1p was recently characterized as a putative regulator of energy metabolism (18) and binding sites for Gsm1p have only been characterized in vitro, thus far. Two biological replicate samples were generated and the ensuing ChIP material from each replicate was subsequently divided equally, to be amplified by either double-T7 or by LM-PCR. Both the double-T7 and LM-PCR methods yielded sufficient material for multiple hybridizations. Amplified samples were labeled according to the same protocols and hybridized in duplicate on DNA microarrays. The label of input and ChIP was swapped between replicates (dye-swap) resulting in a total of four hybridizations for each method.
Because immunoprecipitation enriches for protein-bound DNA fragments, the distribution of binding ratios relative to an input is expected to be skewed toward the ChIP sample. The double-T7 amplified samples indeed shows this expected asymmetry in binding ratio distribution, consistent with ChIP enrichment of specific genomic DNA fragments (Figure 3A). The majority of probes are close to baseline levels, consistent with the expectation that Gsm1p only binds a limited number of targets in the genome. In contrast, LM-PCR-derived binding ratios of the same samples are rather symmetrically distributed (Figure 3B), indicative of a higher degree of technical variation.
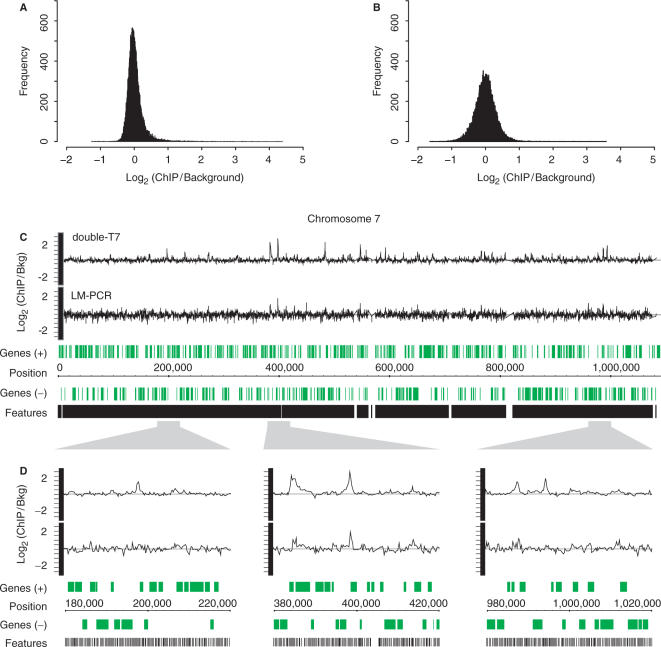
Figure 3.
Double-round T7 amplification results in reduced technical variation compared to LM-PCR. Histograms of averaged binding ratios of four hybridizations with double-T7 (A) or LM-PCR amplified samples (B) across all DNA microarray probes. (C) Overview plot of chromosome 7 showing locations of array probes (black) and genes (green). Graphs of binding ratios of input versus ChIP show the background and binding signals for double-round T7 amplification (double-T7) (top) and LM-PCR (bottom). (D) Enlargement of indicated regions in (C).
An overview plot of chromosome 7 shows the average signals of the four hybridizations for both double-T7 and LM-PCR (Figure 3C and D). The noise around the baseline is considerably reduced for double-T7 amplified samples compared to LM-PCR samples and enriched regions can more readily be identified. Taken together, these data indicate that the double-T7 method results in increased signal-to-noise ratios.
Double-T7 amplification improves detection of Gsm1p-binding sites
The reduction in noise by the double-T7 method is expected to result in an improved detection of Gsm1p-binding sites. To test this we first analyzed the ChIP-chip data using both stringent and less stringent statistical tests. The stringent analysis (ANOVA q < 0.05 and binding ratio ≥ 2) identified 140 genomic-binding events for Gsm1p using double-T7, compared to 66 events for LM-PCR (Figure 4A). There is considerable agreement between the two methods, with 68% of LM-PCR-identified binding events overlapping double-T7 identified regions.
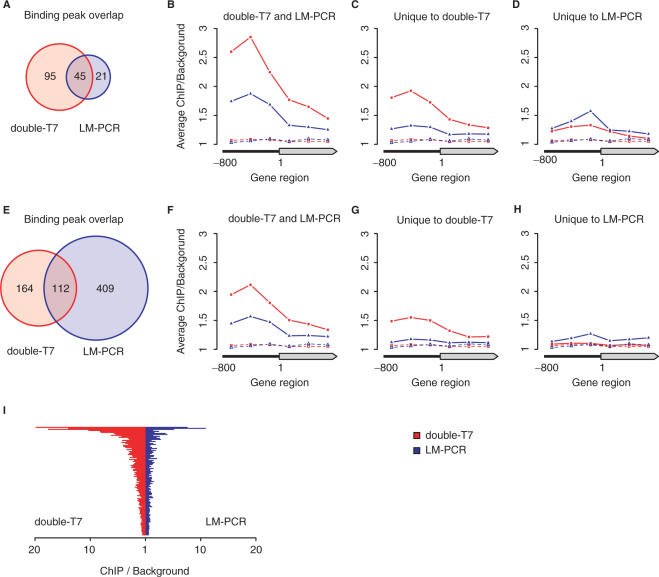
Figure 4.
Comparison of significantly Gsm1p-bound regions after double-round T7 or LM-PCR amplification. The Gsm1p-bound regions identified after double-round T7 amplification (double-T7) and LM-PCR were compared after ANOVA analysis with high- (q < 0.05 and binding ratio>2) (A–D) or low-stringency (q < 0.05 and binding ratio > 1.5) cutoffs (E–H). Venn diagrams (A and E) indicate the degree of overlap between the binding peaks identified after double-T7 or LM-PCR amplification. Average binding profiles were determined for gene regions with significant binding on the ORF or 800 bp of promoter region as indicated (B–D, F–H). Averaged background binding profiles derived from randomly selected sets of genes of similar size are shown as dashed lines. (I) Direct comparison of the maximal peak height after double-T7 (red) or LM-PCR (blue) for all overlapping binding peaks identified in (E).
Gsm1p is a DNA sequence-specific transcription factor and as such it is expected to bind preferentially to promoter regions. Following this assumption, the binding profiles should therefore show enrichment at promoters. To determine which method most accurately reproduced this pattern, we computed and compared the average binding profiles of double-T7 and LM-PCR amplified samples for significant gene regions selected by either methods, or each method individually (Figure 4B–D). As expected, Gsm1p shows clear association with promoters for both the double-T7 and LM-PCR methods (Figure 4B). Overall, the average enrichment with double-T7 was 2-fold greater. Gene regions uniquely identified by each method also showed enrichment at the promoter compared to random selections, albeit with reduced averaged binding ratios (Figures 4C and D). We concluded that many of the binding sites uniquely identified by only a single method are still likely to constitute valid Gsm1p targets, prompting us to relax the selection criteria.
With a less stringent analysis (ANOVA q < 0.05 and binding ratio ≥1.5), the number of significant binding sites identified by LM-PCR amplification increases to 521, compared to 276 for the double-T7 method (Figure 4E). Concomitantly, the overlap between double-T7 and LM-PCR rose to 112, now including many of the gene regions that were identified as unique to one method using the more stringent selection criteria. Again, T7-amplified samples showed higher ChIP enrichment for all overlapping binding events compared to LM-PCR (Figure 4F and I). The relative increase in determined binding sites of almost 8-fold for LM-PCR is striking and suggests the inclusion of many false positives. This is supported by the absence of a clear promoter enrichment in the averaged binding profile for gene regions uniquely identified in LM-PCR samples (Figure 4H). In contrast, the number of binding sites determined by the T7 method increases only modestly (2-fold) and these show a ChIP profile that is consistent with transcription factor binding (Figure 4G). Taken together, this indicates that double-T7 amplification method detects Gsm1p binding with higher specificity and sensitivity.
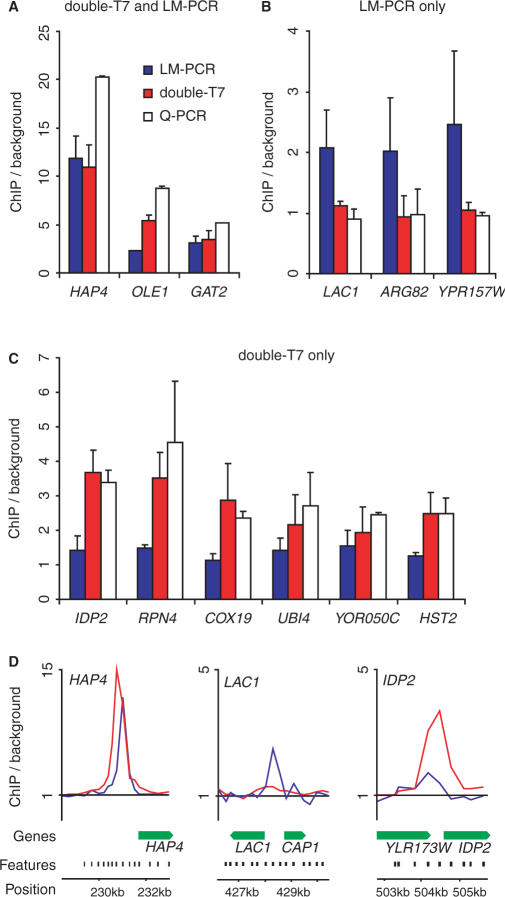
The validity of the binding sites identified with the double-T7 and/or LM-PCR methods was further investigated by quantitative PCR (qPCR) analysis. We randomly selected genes that were significantly enriched according to the stringent analysis from the following groups: double-T7/LM-PCR overlap; LM-PCR only; double-T7 only (Figure 5D). qPCR analysis on the HAP4, OLE1 and GAT2 loci confirmed that double-T7 and LM-PCR had both scored these loci as true positives (Figure 5A). However, genes that were significantly enriched by the LM-PCR method only (LAC1, ARG82 and YDR157W), showed no enrichment with qPCR, in agreement with the double-T7 method (Figure 5B). This indicates that the significantly bound genes uniquely detected by LM-PCR are likely to be false positives. All six loci (IDP2, RPN4, COX19, UBI4, YOR050C and HST2) that were detected with the double-T7 method only, and not with LM-PCR, were confirmed by qPCR (Figure 5C). The qPCR validation therefore shows that the double-T7 method is more accurate and more easily capable of finding weaker binding sites.
Figure 5.
Quantitative PCR validation of Gsm1p-bound regions identified after double-round T7 amplification and LM-PCR amplification. Quantitative PCR validation of Gsm1p-binding peaks identified by both double-round T7 amplification (double-T7) and LM-PCR (A), LM alone (B) or double-T7 alone (C). Binding to tested regions is represented as enrichment over the HMR silent mating-type locus. (D) Enlarged overview of Gsm1p-binding ratios on the indicated loci using either double-T7 (red) or LM-PCR (blue). Locations of array probes (black) and genes (green) are indicated.
CONCLUSIONS
We present an improved T7 amplification method that significantly reduces the amount of starting material required for ChIP-chip, making it feasible to apply T7 amplification for ChIP experiments with low yields. This will benefit studies of DNA-binding proteins with a limited number of genomic binding sites as demonstrated here. Specialized applications such as ChIP-reChIP and ChIP-chip on tissues could also benefit using this method. In our experiments, the double-round T7 amplification method also shows superior performance when compared to LM-PCR. Although there is a good degree of overlap between both methods, many putative sites were uniquely picked up by each method individually. Average binding profiles across promoter regions and validation by qPCR analysis confirmed that the double-T7 method identified binding sites with a much higher degree of accuracy. We also find that the signal-to-noise ratios are higher when using double-T7 method. Taken together, this indicates that the double-T7 method represents an important improvement for obtaining accurate and reproducible genome-wide binding profiles.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Marian Groot Koerkamp and Diane Bouwmeester from the UMC Utrecht/Utrecht University microarray facility for technical assistance. This work is supported by grants (825-06-033, 805-47-080, 016-026-003 and 901-01-238) of the Netherlands Organization for Scientific Research (NWO), the Netherlands Proteomics Centre (NPC), and the European Union (LSHG-CT-2004-502950). Funding to pay the Open Access publication charges for this article was provided by NWO.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 2.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Hecht A, Strahl-Bolsinger S, Grunstein M. Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol. 1999;119:469–479. doi: 10.1385/1-59259-681-9:469. [DOI] [PubMed] [Google Scholar]
- 5.Strutt H, Paro R. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol. Biol. 1999;119:455–467. doi: 10.1385/1-59259-681-9:455. [DOI] [PubMed] [Google Scholar]
- 6.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 7.van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 8.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 9.Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4:19. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 11.Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 13.van Werven FJ, Timmers HT. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 2006;34:e33. doi: 10.1093/nar/gkl003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harismendy O, Gendrel CG, Soularue P, Gidrol X, Sentenac A, Werner M, Lefebvre O. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 2003;22:4738–4747. doi: 10.1093/emboj/cdg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuwald AF, Liu JS, Lawrence CE. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 1995;4:1618–1632. doi: 10.1002/pro.5560040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moll PR, Duschl J, Richter K. Optimized RNA amplification using T7-RNA-polymerase based in vitro transcription. Anal. Biochem. 2004;334:164–174. doi: 10.1016/j.ab.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc. Natl Acad. Sci. USA. 2006;103:9940–9945. doi: 10.1073/pnas.0509185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.