Abstract
Telomerase is a ribonucleoprotein enzyme that maintains chromosome ends through de novo addition of telomeric DNA. The ability of telomerase to interact with its DNA substrate at sites outside its catalytic centre (‘anchor sites’) is important for its unique ability to undergo repeat addition processivity. We have developed a direct and quantitative equilibrium primer-binding assay to measure DNA-binding affinities of regions of the catalytic protein subunit of recombinant Tetrahymena telomerase (TERT). There are specific telomeric DNA-binding sites in at least four regions of TERT (the TEN, RBD, RT and C-terminal domains). Together, these sites contribute to specific and high-affinity DNA binding, with a Kd of ∼8 nM. Both the Km and Kd increased in a stepwise manner as the primer length was reduced; thus recombinant Tetrahymena telomerase, like the endogenous enzyme, contains multiple anchor sites. The N-terminal TEN domain, which has previously been implicated in DNA binding, shows only low affinity binding. However, there appears to be cooperativity between the TEN and RNA-binding domains. Our data suggest that different DNA-binding sites are used by the enzyme during different stages of the addition cycle.
INTRODUCTION
Telomeres form a protective cap on the ends of chromosomes and are composed of short G-rich DNA repeats bound by sequence-specific proteins (reviewed in 1). The telomeres of unicellular eukaryotes and of the germ cells of multicellular organisms are maintained by the ribonucleoprotein enzyme telomerase, which was first identified in the ciliated protozoan Tetrahymena thermophila (2). Telomerase activity has been detected in ∼90% of human cancers, providing a telomere maintenance mechanism that is necessary for their unlimited growth (3–5).
The protein catalytic subunit of telomerase, telomerase reverse transcriptase (TERT, ref. 6), contains protein motifs that are conserved with other reverse transcriptases (‘RT motifs’) and motifs in its amino terminal region that are conserved among TERTs from different species. A defined region of the telomerase RNA subunit is used as the template for addition of DNA repeats onto telomeres (7). A complex consisting of TERT and telomerase RNA is sufficient to reconstitute in vitro telomerase activity from Tetrahymena and human enzymes, measured by processive extension of a DNA oligonucleotide with telomere repeats (8,9). The repeated copying of a short segment of RNA, followed by translocation to the beginning of this sequence, leads to the unique property of telomerase known as ‘repeat addition processivity’. While in vitro activity of the recombinant enzyme is robust, recombinant Tetrahymena telomerase is less processive than its endogenous counterpart (9,10).
The interactions of telomerase with its DNA substrate have been studied using purified telomerase from Tetrahymena extracts. High concentrations of short oligonucleotide primers (<10 nt) were required to achieve efficient extension (11,12). For longer oligonucleotides, primer affinity, as measured by activity (Km), varied with different sequences at the 5′ end of the primer (11). Telomerase was able to extend a non-telomeric primer bearing telomeric sequences at its 5′ end with nanomolar affinity (13). Sequences at the 5′ end of the primer also affected the processivity of endogenous human telomerase (14). These observations led to the proposal that telomerase contains a DNA-binding site outside the template region, called the ‘anchor site’ (12,14). It was proposed that the anchor site is necessary for repeat addition processivity, by allowing the enzyme to remain bound to the 5′ end of its DNA substrate during translocation. Thus, in order to fully understand the unique mechanism of telomerase, it is necessary to know more about the nature of the anchor site.
There is evidence for telomerase from several species that there are multiple sites of interaction with different regions of the primer (11,15–17). For example, the affinity of endogenous Tetrahymena telomerase for DNA decreased in a stepwise manner as the primer was reduced in length from 12 to 10 nt then 6 nt (11), and two 5′ regions of a 24 nt primer influenced yeast telomerase activity (15). These different sites have been dubbed the ‘template-proximal’ and ‘template-distal’ anchor sites (17). For Tetrahymena at least, it is possible that there are even more distal sites of interaction between DNA and protein, which may be important for binding to non-telomeric sequences (18). While both the template-proximal and template-distal anchor sites have an effect on repeat addition processivity (14,17,19), the fact that both recombinant and endogenous Tetrahymena telomerase can extend even primers as short as 4 nt with reasonable processivity shows that neither is absolutely necessary for this property (19, our unpublished data).
Although evidence for direct interactions with the TERT protein have been obtained by DNA crosslinking to telomerase from Tetrahymena, the ciliate Euplotes aediculatus and yeast (20–24), it is unclear which anchor sites reside in the TERT protein and which are provided by other members of the enzyme complex. There is some evidence from activity-based assays for interactions of DNA with various regions of TERT. Deletion of the TERT-specific C-terminal extension of yeast TERT led to a modest (∼2-fold) decrease in primer-binding affinity (25); the fact that mutations in this region in human TERT led to a moderate decrease in repeat addition processivity suggests that this function may be conserved among species (26). A mutation in a TERT-specific region of the RT domain reduced repeat addition processivity of yeast telomerase, suggesting that this region may also interact with DNA (27).
Recently, evidence has been presented that a region within the N-terminal 200 amino acids of TERT (known as the GQ or TEN domain) may be involved in anchor site interactions. This region of TERT has the ability to form a soluble, protease-resistant, independently folded structure (24,28,29). Complementation studies with fragments of human TERT indicate that this region (together with an adjacent linker domain) can interact in trans with the rest of the TERT protein (30–32). There may, however, be species differences in the function of this domain, since its removal leads to a loss of processivity of human telomerase (32,33) but a complete loss of activity of Tetrahymena telomerase (29,34).
A mutation in the TEN domain of yeast TERT led to altered utilization of primers with non-telomeric 5′ ends (22). A mutation within this region in human TERT caused a slight decrease in affinity for a 9 nt primer but not an 18 nt primer, indicating that this part of TERT may interact with the 9 nt at the 3′ end of the primer (17). Human TERT proteins containing mutations in this region were also defective in elongation of telomeric primers (35). Direct evidence for the involvement of this region in DNA binding was provided by the demonstration that mutations in this region in Tetrahymena TERT led to decreased crosslinking to a primer containing 5-iodo-deoxyuridine (5-iodo-dU) substitutions at its 5′ end (24), and the site of crosslinking to a short primer was mapped to a particular amino acid within TEN (W187) (23). Finally, a fragment of human TERT encompassing TEN bound to a biotinylated primer, though in a non-sequence-specific manner (36).
While these studies provide an informative start in understanding telomerase–DNA interactions, they do not address relative contributions from the different regions of the TERT protein. To directly and quantitatively measure binding affinities of defined regions of TERT, we developed an activity-independent assay for primer binding to recombinant Tetrahymena telomerase. We used this direct equilibrium binding assay, in combination with UV crosslinking, to assess primer affinities of domains of the TERT protein. Since the assay is independent of activity, we were also able to use it to assess the level of RNA-independent binding of DNA to TERT, bearing in mind that such binding may not be identical to that in the intact enzyme. These quantitative studies provide new insights into the complexity of the telomerase anchor site.
MATERIALS AND METHODS
Plasmids and oligonucleotides
The construction of a synthetic Tetrahymena thermophila TERT gene and its insertion into a pET-28 plasmid encoding an N-terminal FLAG tag and a T7 promoter have been previously described (10,37). A version of this plasmid containing a BamHI site between the FLAG tag and TERT was constructed in order to simplify subsequent cloning steps. This plasmid (FLAG-Bam-TERT) was made with site-directed mutagenesis using the Quick Change II kit (Stratagene) according to the manufacturer's instructions. To construct a plasmid encoding FLAG-tagged amino acids 1–519 of TERT (plasmid FLAG-N519), this fragment was amplified using primers 5′-GTCGCAGGCGTCTGTTTGAATCAGTA CTTTTC TGTC (forward) and 5′-CAGGATCTCGAG TCACAATTTTTCTTCCACTTTCTC (reverse), digested with ScaI and XhoI and cloned into the ScaI and XhoI sites of FLAG-TERT. To construct a plasmid encoding FLAG-tagged amino acids 520-1117 of TERT (plasmid FLAG-C598), this fragment was amplified using primers 5′-GATATACCATGGCTGACTACAAGGACGACGA TGACAAGCATATGGGGGGATCCATCCCAGAAG ATTCATTTCAG (forward) and 5′-CAGTGTCTTAAT GTCCTGTATGAGGTTATTTAAATCTATGAATTC AATAGCCTCCAGTTCTTTTTTGTTCAGCTG (reverse), digested with NcoI and EcoRI and cloned into the NcoI and EcoRI sites of FLAG-TERT. To construct a plasmid encoding FLAG-tagged amino acids 192–519 of TERT (plasmid FLAG-RBD), this fragment was amplified using primers 5′-CATATGGGGGGATCCTTTAA TATGAACGGGAAAGCT (forward) and 5′-CAGGAT CTCGAGTCACAATTTTTCTTCCACTTTCTC (reverse), digested with BamHI and XhoI and cloned into the BamHI and XhoI sites of FLAG-Bam-TERT. To construct a plasmid encoding FLAG-tagged amino acids 1–884 of TERT (plasmid FLAG-ΔC233), this fragment was amplified using primers 5′-GCACTGAATCTG ATTGTACAACTCCAGAACTGTGCAAAC (forward) and 5′-CAGGATCTCGAGTCAGTTCATGTCGATGC TTTTCCC (reverse), digested with BsrGI and XhoI and cloned into the BsrGI and XhoI sites of FLAG-Bam-TERT. pET-28 plasmids containing amino acids 2–191 (TEN or GQ) and 192–1117 (ΔTEN or TERT-ΔGQ) were a kind gift from Drs Thomas Cech and Steven Jacobs at the University of Colorado, Boulder, CO, USA (29). To construct a plasmid encoding FLAG-tagged amino acids 2–191, the TEN/GQ plasmid was digested with NdeI and BlpI and cloned into the NdeI and BlpI sites of FLAG-TERT. For a FLAG-tagged version of ΔTEN, the ΔTEN plasmid was digested with BamHI and XhoI and cloned into the BamHI and XhoI sites of FLAG-Bam-TERT. A plasmid encoding FLAG-tagged amino acids 1–1094 of TERT (plasmid FLAG-ΔC23) was made by site-directed mutagenesis (Quick Change II kit, Stratagene) of FLAG-Bam-TERT using the primers 5′-CTGGAGGCTATTGAATTCATAGATTTAAATTA ACTCATACAGGAC (forward) and 5′-GTCCTGTAT GAGTTAATTTAAATCTATGAATTCAATAGCCTC CAG (reverse).
All oligonucleotides were purchased from Sigma-Genosys. PCR primers were desalted by the manufacturer, while the oligonucleotides listed in Table 1 were gel purified on 20% polyacrylamide/8 M urea gels before use.
Table 1.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| 18GGGG | TTGGGGTTGGGGTTGGGG |
| 18GGT | TGGGGTTGGGGTTGGGGT |
| 18GTT | GGGGTTGGGGTTGGGGTT |
| 18TTG | GGGTTGGGGTTGGGGTTG |
| 18TGG | GGTTGGGGTTGGGGTTGG |
| 18TGGG | GTTGGGGTTGGGGTTGGG |
| 20GTT | TTGGGGTTGGGGTTGGGGTT |
| 12GTT | GGGGTTGGGGTT |
| 6GTT | GGGGTT |
| 37TTG | GGGGTTGGGGTTGGGGTTGGGGTTG GGGTTGGGGTTG |
| PBR | AGCCACTATCGACTACGCGATCAT |
| PBR48 | AGCCACTATCGACTACGCGATCATAGC CACTATCGACTACGCGATCAT |
| (TTAGGG)3 | TTAGGGTTAGGGTTAGGG |
All sequences are listed 5′ to 3′. Some oligonucleotides were modified by addition of a biotin, on either the 5′ (e.g. Bio-18GTT) or the 3′ (e.g. PBR48-Bio) end. 20GTT was modified by incorporation of 5-iodo-deoxyuridine (Figure 5).
In vitro translation and purification of Tetrahymena telomerase
Tetrahymena telomerase RNA was transcribed in vitro from the plasmid pTET-telo (38) as described previously (10). The RNA was gel-purified before inclusion in in vitro translation reactions using the plasmids described above to translate TERT protein or fragments thereof. Translations were carried out using the TnT T7 Quick for PCR rabbit reticulocyte lysate system (Promega) as per the manufacturer's instructions. Translations included 20 ng/μl plasmid DNA, 0.8 μCi/μl [35S]methionine and 20 nM telomerase RNA.
In vitro reconstituted telomerase was purified by immunoprecipitation with anti-FLAG M2 affinity agarose beads (Sigma-Aldrich). Beads (100 μl) were washed four times in 1.3 ml of wash buffer-100 (20 mM Tris acetate, pH 7.5, 10% glycerol, 1 mM EDTA, 5 mM MgCl2, 0.1 mM dithiothreitol [DTT], 100 mM potassium glutamate), centrifuging at 1500 × g for 2 min at 4°C between washes. The beads were incubated twice with 1 ml of blocking buffer [wash buffer-100 plus 0.5 mg/ml lysozyme, 0.5 mg/ml bovine serum albumin (BSA), 0.05 mg/ml glycogen, 0.1 mg/ml yeast RNA] for 15 min at 4°C with agitation. Reticulocyte lysate translation reaction (400 μl) was added to 400 μl of blocking buffer and centrifuged at 16 000 × g for 10 min at 4°C to remove any particulates. The supernatant from that spin was added to the blocked beads and agitated at 4°C for 2 h. The beads were washed four times in 1.3 ml of wash buffer-300 (the same as wash buffer-100 except with 300 mM potassium glutamate), twice in TMG (10 mM Tris acetate pH 8.0, 10% glycerol, 1 mM MgCl2) and once in wash buffer-0 (the same as wash buffer-100 except with no potassium glutamate). For all experiments except those in Figure 1, telomerase was eluted from the beads by competition with a 3× FLAG peptide (Sigma-Aldridge). The beads were resuspended in 200 μl of peptide (0.75 mg/ml) in wash buffer-0 in a Protein LoBind tube (Eppendorf). BSA (New England Biolabs) was added (0.5 mg/ml) to prevent telomerase from sticking to the tube after elution. The bead slurry was rotated at 4°C for 1 h, centrifuged at 1500 × g for 2 min at 4°C and the eluate removed to a fresh LoBind tube. Eluted telomerase had ∼3–4-fold greater affinity (Km) for its DNA primer than did bead-bound telomerase (data not shown), presumably reflecting a faster on-rate.
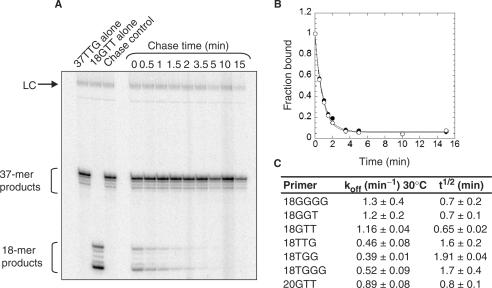
Figure 1.
Rate of dissociation from recombinant Tetrahymena telomerase of DNA primers of different permutations. (A) A bind-and-chase telomerase activity assay (described in the text) was used to measure the dissociation of primer 18GTT (Table 1). The pattern of extension of 18GTT and the chase primer 37TTG alone, when incubated with ddTTP and [α-32P]dGTP, are shown in the first two lanes. As a control to ensure that the excess of 37TTG did not allow the enzyme to rebind the 18GTT, both primers were preincubated with enzyme simultaneously prior to telomerase extension (‘chase control’). LC: 32P-labelled 100-mer DNA oligonucleotide used as a recovery and loading control. (B) The amount of signal from products of 18GTT was normalized to the total telomerase product signal and plotted versus time of chase. Two experiments utilizing the same primer (18GTT) are shown. (C) The curves shown in B were fit with a single exponential equation (see ‘Experimental Procedures’) to give the off-rate (koff) and half-life (t1/2) of a set of 18 or 20 nt oligonucleotides of different permutations (Table 1). The mean ± SD of two to four experiments is shown.
The yield of active enzyme was determined by dot blotting an aliquot of the enzyme along with a dilution series of Tetrahymena telomeric RNA onto a Hybond N+ membrane (Amersham Biosciences/GE Healthcare) and detecting the RNA in the telomerase complex by hybridizing the membrane with a radioactive probe complementary to Tetrahymena telomerase RNA (5′-TATCAGCACTAGATTTTTGGGGTTGAATG-3′). The final concentration of eluted telomerase was 7–9 nM.
In vitro telomerase activity assay
Activity of recombinant telomerase was measured by incubating 5 μl of eluted telomerase in a 10 μl reaction including 1× Telomerase buffer (50 mM Tris–Cl, pH 8.3, 1.25 mM MgCl2, 5 mM DTT), 100 μM dTTP and 10 μM [α-32P]dGTP at 80 Ci/mmol (0.2 μl of non-radioactive dGTP at 487 μM and 0.8 μl of [α-32P]dGTP at 10 mCi/ml, 3000 Ci/mmol; PerkinElmer Life Sciences). Dilutions of the indicated primers were added to the reactions at concentrations of 1 nM to 5 μM (with the concentration range depending on the length of the primer). The reactions were incubated at 30°C for 15 min (which is within the linear phase of the reaction) and then terminated by adding 90 μl of TES (50 mM Tris–HCl pH 8.3, 20 mM EDTA and 0.2% SDS). 5000 cpm of a 32P-labelled 100-mer oligonucleotide was added as a recovery and loading control. The reaction products were phenol/chloroform extracted and ethanol precipitated. The pellet was resuspended in 5 μl of formamide loading dye (90% deionized formamide, 0.1% bromophenol blue, 0.1% xylene cyanol in 1× TBE buffer) and half of each reaction was electrophoresed on a denaturing 10% polyacrylamide/8 M urea sequencing-type gel for 1.25 h at 80 W. The gel was dried for 30 min at 80°C, exposed to a phosphorimager screen and analysed using ImageQuant 5.2 software (Amersham Biosciences/GE Healthcare). Total intensity of extension products at each substrate concentration was normalized against the intensity of the 32P-labelled 100-mer loading control. This value was expressed as percentage of maximum intensity at the highest primer concentration and plotted against substrate concentration. This was fitted to a Michaelis–Menten kinetics equation to yield Km.
Bind-and-chase activity assay to measure primer off-rates
A modified telomerase assay was used to measure the rate of dissociation of primer from recombinant Tetrahymena telomerase, as previously described (10). Telomeric oligonucleotides of 18–20 nt (Table 1) were preincubated at a concentration of 200 nM with 5 μl of immunopurified telomerase (1:1 bead slurry) in 1× Telomerase buffer at 30°C for 15 min. A 37-mer competitor primer (37GTT) was added (final concentration 15 μM) and incubated with the enzyme/primer mixture at 30°C for varying lengths of time (Figure 1). Telomerase activity was initiated by the addition of nucleotides (either [α-32P]dGTP at 10 μM and ddTTP at 100 μM or [α-32P]dTTP at 3.3 μM and ddGTP at 10 μM, depending on the permutation of the primer) and allowed to extend the primers at 30°C for 5 min. The reactions were terminated and electrophoresed as earlier. To determine koff, the intensity of the 18–20-mer products was quantified, normalized to the combined intensities of all major extension products (18- and 37-mer) and plotted against time. The data were fit to the equation y = ae−kt + b, where k = koff and b represents a background in the experiment of about 5%. The half life was calculated by using the equation t½ = −{ln[(0.5 – b)/a]}/k.
DNA-binding assay to measure primer binding to TERT
Oligonucleotides with a 5′ biotin and PBR48 with a biotin at the 3′ end (Table 1) were synthesized and gel-purified by Sigma-Genosys. UltraLink Immobilized NeutrAvidin Protein Plus (Pierce) beads (200 μl) were washed four times in 1 ml of Protein Pulldown (PP) buffer (50 mM Tris chloride, pH 8.3, 10% glycerol, 1.25 mM MgCl2, 5 mM DTT), centrifuging at 1500 × g for 2 min at 4°C between washes. The beads were incubated twice with 0.5 ml of blocking buffer (PP buffer plus 0.75 mg/ml BSA, 0.15 mg/ml glycogen, 0.15 mg/ml yeast RNA) for 15 min at 4°C with agitation and resuspended in 100 μl of blocking buffer. Primer binding of recombinant telomerase was measured by incubating 2.5 μl of eluted protein in a Protein LoBind tube (Eppendorf) in a 20 μl reaction including 1× Telomerase buffer (see earlier), 2500 cpm of 33P-labelled PBR48-Bio and the indicated primers at concentrations of 1 nM to 10 μM (with the concentration range depending on the length of the primer and the fragment of protein). The reactions were incubated at 30°C for 10 min, followed by the addition of 20 μl of blocked NeutrAvidin beads and agitation at 4°C for 15 min. The beads in each reaction tube were washed four times (1500 × g for 2 min at 4°C) in 100 μl of PP-300 buffer (PP buffer plus 300 mM lithium acetate; lithium was used to minimize formation of intermolecular G-quadruplexes) and finally resuspended in 20 μl of PP-300 buffer and 10 μl of NuPage 4× Lithium dodecyl sulfate (LDS) Sample Buffer (Invitrogen) and heated at 95°C × 3 min. Half of each reaction was electrophoresed on 4–12% NuPage gels in MES [2-(N-morpholino) ethane sulfonic acid] buffer (Invitrogen) for 35 min at 200 V. The gels were fixed for 15 min in 25% isopropanol/10% acetic acid, dried at 80°C for 1 h and exposed to a phosphorimager screen. Total intensity of protein captured at each substrate concentration was normalized against the intensity of the 33P-labelled PBR48-Bio loading control. The signal from the sample with no oligonucleotide, representing background binding to the beads, was subtracted from all others. The resulting value was expressed as percentage of maximum intensity at the highest primer concentration and plotted against substrate concentration [S]. This was fitted to the following equation: y = (Bmax × [S]) ÷ (Kd + [S]), where Bmax is the maximal level of binding, to yield Kd.
Crosslinking of 5-iodo-dU substituted primers to TERT
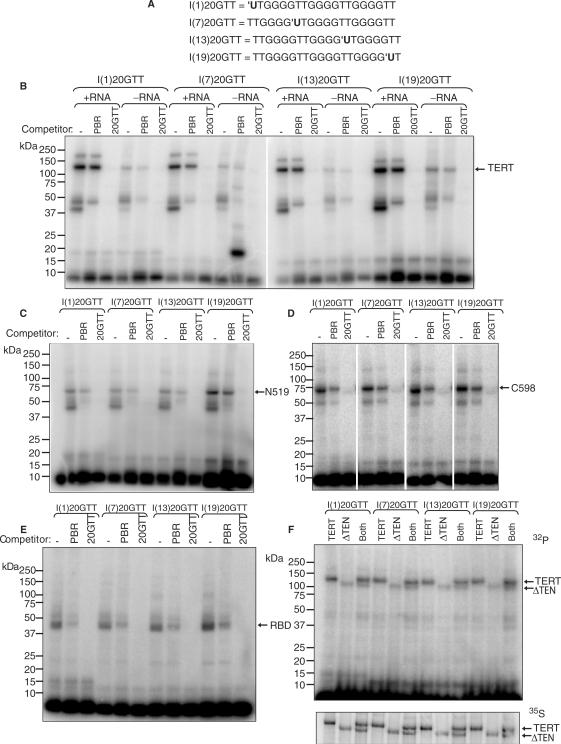
20GTT oligonucleotides with 5-iodo-deoxyuridine in one of four positions (Figure 5A) were synthesized and gel-purified by Sigma-Genosys. The oligonucleotides were 5′-end-labelled with T4 polynucleotide kinase (New England Biolabs) and unincorporated nucleotides were removed by purification on a Mini Quick Spin Oligo column (Roche Applied Science). Labelled oligonucleotides (15–100 nM) were incubated with unlabelled in vitro translated, immunopurified and eluted TERT or fragment of TERT (∼3–4 nM) in 1× Telomerase buffer (see earlier) for 10 min at 30°C. Reactions contained either no competitor or 5 μM unlabelled specific (20GTT) or non-specific (PBR) competitor oligonucleotide, as indicated. Reactions were dispensed onto a parafilm-covered aluminium block sitting on ice, covered with a 4-mm thick 313 ± 3 nm bandpass optical filter (Andover Corporation) and crosslinked in a Stratalinker (Stratagene) with 312 nm bulbs (UviTec) for 45 min. After crosslinking, reactions were electrophoresed on 4–12% NuPage gels in MES buffer (Invitrogen) for 35 min at 200 V. The gels were fixed for 15 min in 25% isopropanol/10% acetic acid, dried at 80°C for 1 h and exposed to a phosphorimager screen. For the experiment in Figure 5F, 35S-labelled proteins were used. A piece of X-ray film was placed between the gel and phosphorimager screen to shield signal from 35S-labelled protein and leave only signal from 32P-labelled DNA (upper panel). A second exposure was obtained with no X-ray film to ensure equal loading of protein (bottom panel).
Figure 5.
Crosslinking of 5-iodo-deoxyuridine substituted 20GTT to TERT and fragments of TERT. (A) Sequence of the four oligonucleotides used for crosslinking in B–F; IU = 5-iodo-deoxyuridine. (B) 32P-labelled DNA (15 nM) was crosslinked to full-length TERT in the absence of competitor or in the presence of a 333-fold excess of cold telomeric (20GTT) or non-telomeric (PBR) competitor primer. Crosslinking was carried out in the presence or absence of telomerase RNA. (C) 32P-labelled DNA (90 nM) was crosslinked to N519 in the absence of competitor or in the presence of a 55-fold excess of cold telomeric (20GTT) or non-telomeric (PBR) competitor primer. (D) 32P-labelled DNA (15 nM) was crosslinked to C598 in the absence of competitor or in the presence of a 333-fold excess of cold telomeric (20GTT) or non-telomeric (PBR) competitor primer. (E) 32P-labelled DNA (100 nM) was crosslinked to RBD in the absence of competitor or in the presence of a 50-fold excess of cold telomeric (20GTT) or non-telomeric (PBR) competitor primer. (F) 32P-labelled DNA (50 nM) was crosslinked to full-length TERT, ΔTEN or a 1:1 mixture of both proteins in the presence of a 100-fold excess of cold non-telomeric (PBR) competitor primer. The upper panel shows the signal from 32P-labelled DNA only (with a piece of X-ray film between the gel and phosphorimager screen to shield 35S signals); the lower panel shows signals from both 32P and 35S-labelled proteins, to ensure the use of an equimolar mixture of proteins.
For the experiment in Figure 6B, in vitro translated TERT was replaced with the TEN domain of TERT that had been bacterially expressed and purified as described (29) and which was a gift from Drs Steven Jacobs and Thomas Cech. These crosslinking reactions were carried out in the presence of 5 μM non-specific competitor (PBR).
Figure 6.
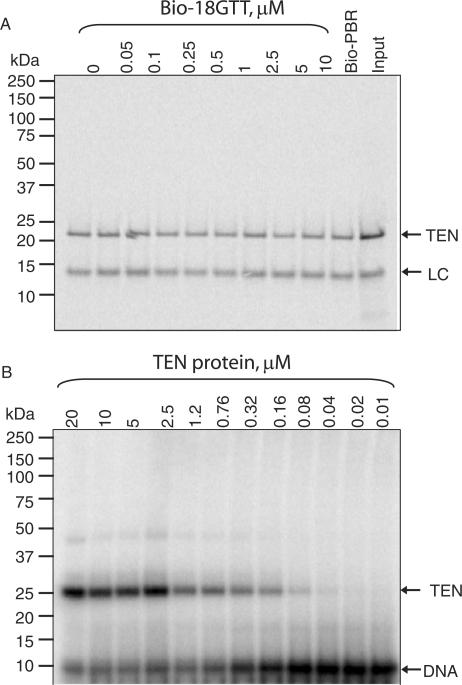
The TEN domain of TERT binds to DNA oligonucleotides with low affinity. (A) DNA immobilization assay using in vitro translated TEN protein. The indicated concentrations of biotinylated 18GTT oligonucleotide were incubated with 35S-labelled TEN protein (+600 nM telomerase RNA) and recovered on NeutrAvidin beads. ‘Input’ represents 40% of the starting material in the bound lanes. LC: 33P-labelled PBR48-Bio oligonucleotide, used as a loading and recovery control. (B) Crosslinking assay using bacterially expressed, purified TEN protein. The indicated concentrations of TEN protein were crosslinked to 5 nM 32P-labelled I(1)20GTT DNA.
Immunoprecipitation to measure RNA binding to TERT
FLAG-tagged fragments of TERT were tested for their ability to immunoprecipitate telomerase RNA as described (39) with the exceptions that anti-FLAG M2 affinity agarose beads (Sigma-Aldrich) were used, and the reactions were electrophoresed on 4–12% NuPage gels (Invitrogen).
RESULTS
Recombinant Tetrahymena telomerase dissociates equally from primers with different 3′ ends
It has been shown that DNA primers dissociate from endogenous human telomerase at different rates, depending on the permutation of the 3′ end of the primer (40). Primers ending in GGG displayed half-lives of greater than 20 h at room temperature. In order to determine the best primer to use for binding experiments, we sought to determine whether the same is true for recombinant Tetrahymena telomerase. We used an activity-based bind-and-chase assay for primer dissociation that we previously developed (10). In this assay, telomerase was allowed to form a complex with a short (18–20 nt) DNA primer of telomeric sequence. A 75-fold excess of a longer primer (37 nt) was added and incubated with the enzyme/primer mixture at 30°C for varying lengths of time. Telomerase activity was then initiated by addition of nucleotides (either ddTTP and [α-32P]dGTP, or [α-32P]dTTP and ddGTP, depending on the permutation of the primer) and allowed to proceed at 30°C for 5 min. The inclusion of ddTTP or ddGTP resulted in termination of extension after incorporation of the first T or G respectively, generating 2 to 5 product bands, depending on the permutation of the primer (Figure 1A and B, illustrated for primer 18GTT). To establish that the excess of longer primer did not allow the enzyme to rebind 18-mer, both primers were preincubated simultaneously with enzyme, which eliminated extension of the 18-mer (Figure 1A, chase control lane). Thus, the amount of addition to 18GTT after varying lengths of chase time is a reflection of the amount of the original primer still bound to enzyme at that time. We observed similar dissociation rates for all six primer permutations; koff ranged from 0.39 to 1.3 min−1, corresponding to half lives of 0.7 to 1.9 min at 30°C (Figure 1C). Thus recombinant Tetrahymena telomerase, like endogenous Euplotes aediculatus telomerase (41), does not show the large (∼100-fold) differences in primer affinities that were observed for endogenous human telomerase. Since 18GTT has been the standard primer in our telomerase assays (10,39), we chose to use this primer for the primer binding assay described later.
Development of a direct equilibrium binding assay for measuring TERT–primer interactions
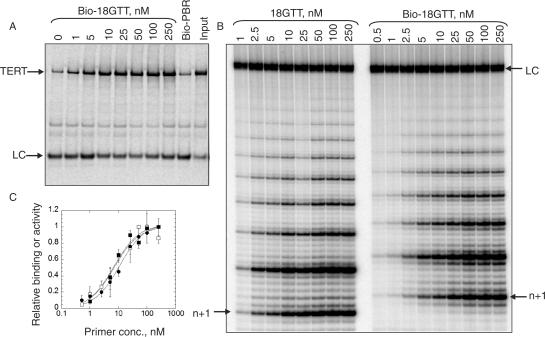
Previous studies examining the interaction of telomerase with DNA have relied on measurement of telomerase activity (10,17,35) or crosslinking of modified oligonucleotides (20–22,24). We sought to develop a quantitative activity-independent equilibrium binding assay for recombinant Tetrahymena telomerase in order to directly measure DNA-binding affinities of TERT protein domains. We developed a simple assay in which purified 35S-labelled in vitro-translated TERT is bound to increasing concentrations of biotinylated 18GTT and the resulting TERT–DNA complex is captured on NeutrAvidin beads. Products are analyzed by PAGE to visualize the amount of TERT that is recovered with the beads. A 33P-labelled biotinylated primer of random sequence (PBR48-Bio; Table 1) is used as an internal recovery and loading control. A similar assay was recently independently reported, with the difference that that assay used unpurified enzyme and a single, presumably saturating concentration of DNA primer (36). Rabbit reticulocyte lysates contain proteins that efficiently bind telomeric oligonucleotides (our unpublished data), so a key requirement to quantitatively measure telomerase binding affinities is to purify the enzyme.
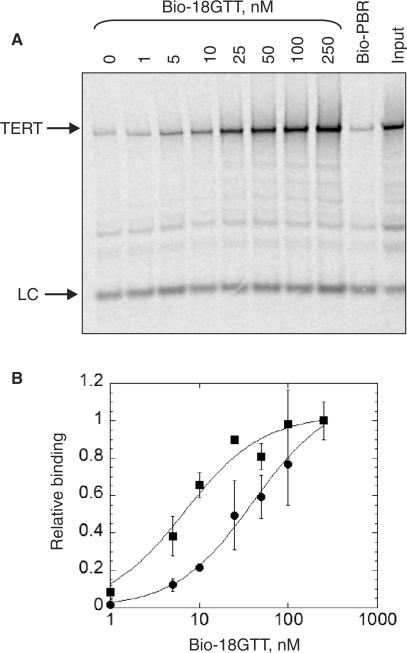
Binding of recombinant TERT to Bio-18GTT resulted in a dissociation constant (Kd) of 8 ± 2 nM (Figure 2A and C), which is comparable to that of telomerase purified from Tetrahymena extracts (20–30 nM for an 18 nt primer, ref. 11 and our unpublished data) despite the higher processivity of the latter.
Figure 2.
A DNA-binding assay to directly measure DNA primer affinity to telomerase. (A) The indicated concentrations of biotinylated telomeric (18GTT) oligonucleotide or 250 nM non-telomeric control (Bio-PBR) were incubated with 35S-labelled full-length recombinant telomerase (TERT + RNA) and recovered on NeutrAvidin beads. ‘Input’ represents 20% of the starting material in the bound lanes. LC: 33P-labelled PBR48-Bio oligonucleotide, used as a loading and recovery control. (B) Primer extension activity of purified recombinant Tetrahymena telomerase, using biotinylated or non-biotinylated 18GTT primer at the indicated concentrations. ‘n + 1’ is the first nucleotide added to each primer; ‘LC’ is a 32P-labelled 100-mer DNA oligonucleotide used as a recovery and loading control. (C) Binding or activity were quantified from gels such as those in A and B, and normalized to the highest value. Filled square, binding of Bio-18GTT to telomerase; filled circle, activity using Bio-18GTT; open square, activity using 18GTT. The mean of three to four independent experiments is plotted; error bars = SD.
As a control for non-specific DNA binding in all experiments, a biotinylated non-telomeric primer was used for the pull-down (Bio-PBR; Table 1); binding to this primer was not apparent above the background levels that are observed in the absence of primer (Figure 2A). Binding of Tetrahymena TERT to the human telomeric sequence (TTAGGG)3 was also not apparent above background levels (Supplementary Figure); thus this assay shows exquisite specificity for the Tetrahymena telomeric sequence, TTGGGG. Tetrahymena telomerase does have the capability to extend other sequences (18), but the affinity of such interactions is low (∼1 μM for (TTAGGG)3, ∼8 μM for Bio-PBR; data not shown) and is presumably beyond the detection limits of our primer binding assay.
To determine whether the binding observed in the DNA-binding assay reflects active telomerase, we also determined the Km of the same purified recombinant enzyme in a direct telomerase primer extension assay (Figure 2B). The biotinylated and non-biotinylated versions of 18GTT displayed similar Kms, which were not significantly different from the Kd as measured by the DNA-binding assay (Table 2 and Figure 2C), indicating that the binding we observe reflects active primer–enzyme complexes.
Table 2.
Km and Kd of oligonucleotides of various lengths
| Primer | Km (nM) | Kd (nM) |
|---|---|---|
| Bio-18GTT | 12 ± 7 | 8 ± 2 |
| 18GTT | 8 ± 3 | n/a |
| Bio-12GTT | 22 ± 7 | 160 ± 140 |
| 12GTT | 60 ± 50 | n/a |
| Bio-6GTT | 900 ± 1200 | n.d. |
| 6GTT | 1900 ± 500 | n/a |
Km was determined using direct telomerase activity assays at different concentrations of primer, and Kd was determined using the DNA immobilization assay at different concentrations of primer. Mean ± SD is shown (n = 3 to 4). n/a, not applicable, since a biotin tag is necessary for the DNA immobilization assay. n.d., not detectable up to 10 μM primer.
Tetrahymena telomerase binds DNA in a length-dependent and RNA-independent manner
It has been demonstrated for endogenous Tetrahymena telomerase that the Km increases in a stepwise manner as the primer gets shorter, reflecting loss of binding sites in different regions of the primer (11). We also observed that both the Km and the Kd increased with a 12 nt primer (Table 2). Within experimental error, the Kd for this 12 nt primer also reflects the Km (it should be noted that the error inherent in these assays is large when dealing with low-affinity interactions with high rates of dissociation).
Decreasing the length of the telomeric primer to 6 nt resulted in a further increase in Km of 1–2 orders of magnitude (Table 2). There was no detectable binding of TERT to this primer using the DNA-binding assay. Primers of 4 or 5 nt, which can be extended by Tetrahymena telomerase (19) with Kms of 1–2 μM (data not shown), also showed no detectable binding in our assay (data not shown). Since this assay involves washing the beads after binding, low-affinity interactions with high off-rates are not detectable. Thus we put an upper limit on Kd measurement for this assay of between 300 (see later) and 900 nM.
It has been reported that yeast and Tetrahymena telomerase are dependent on the telomerase RNA subunit for crosslinking to a DNA primer (22,24). We found that binding of TERT to 18GTT DNA in our binding assay was only partially dependent on the RNA (Figure 3). The Kd for this interaction was 29 ± 5 nM, 3–4-fold greater than that in the presence of RNA. Under these reconstitution conditions, about 50% of TERT protein in the +RNA samples is not bound to telomerase RNA (37), so the actual difference in binding affinities may be somewhat greater than 3–4-fold. Nevertheless, these data clearly show that telomerase RNA contributes to, but is not completely necessary for, binding of Tetrahymena telomerase to a DNA primer.
Figure 3.
DNA interacts with telomerase independently of telomerase RNA. (A) The indicated concentrations of biotinylated 18GTT oligonucleotide were incubated with 35S-labelled full-length recombinant TERT (-telomerase RNA) and recovered on NeutrAvidin beads. ‘Input’ represents 20% of the starting material in the bound lanes. LC: 33P-labelled PBR-48-Bio oligonucleotide, used as a loading and recovery control. (B) Quantification of data in A, together with that in Figure 2A, to show increase in Kd in the absence of telomerase RNA. Filled square, binding of Bio-18GTT to TERT + RNA, filled circle, binding of Bio-18GTT to TERT–RNA. The mean of three to four independent experiments is plotted; error bars = SD.
DNA–protein interactions exist in multiple domains of TERT
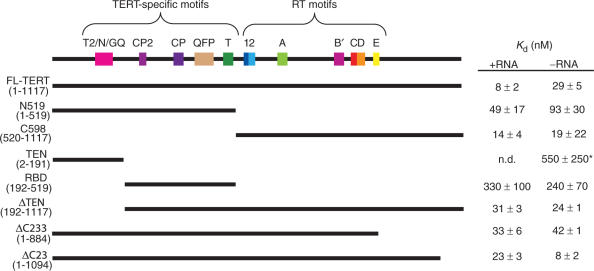
We utilized the DNA-binding assay to explore binding of DNA to domains of the TERT protein. A fragment consisting of the N-terminal half of the protein, which includes several TERT-specific motifs, bound to 18GTT with a ∼6-fold reduction in affinity over full-length protein (N519, Figure 4). In contrast, the C-terminal half of TERT, encompassing the RT motifs, bound to 18GTT with an affinity almost equal to full-length TERT (C598, Figure 4). Binding to each of the fragments in Figure 4 was specific for telomeric DNA, since the oligonucleotide Bio-PBR showed only background levels of binding (data not shown). Thus there are specific high-affinity binding sites in both halves of TERT, especially the C-terminal half. It is not possible to confirm that the same binding sites would be used in the context of an active enzyme, since these fragments of TERT are inactive, but the specificity and high affinity of the interactions point to their biological relevance.
Figure 4.
Summary of binding of oligonucleotide Bio-18GTT to fragments of TERT. At the top is a representation of Tetrahymena thermophila TERT, with conserved protein motifs shown as coloured boxes. The names of the fragments and the amino acids of TERT which they encompass are shown on the left. On the right are the binding constants of DNA and TERT in the presence or absence of telomerase RNA (mean ± SD). *This Kd was measured using crosslinking; n.d. = not determined.
Deletion of the C-terminal 233 amino acids, which are unique to TERT, resulted in a modest increase in Kd of about 4-fold (ΔC233, Figure 4), indicating that the RT motifs likely play a significant role in the high-affinity primer binding of the C-terminal half of the protein. We also made a version of TERT lacking the C-terminal 23 amino acids; this mutant completely lacked telomerase activity (data not shown), but again showed only a modest decrease in primer binding affinity (ΔC23, Figure 4). Thus the extreme C-terminus of Tetrahymena TERT has an essential role in telomerase activity, which may include, but is probably not limited to, interactions with the DNA primer. This result illustrates the importance of a quantitative equilibrium binding assay for assessing the extent of the contribution to DNA binding of regions of TERT.
TERT–DNA binding is direct
To confirm that the binding to the TERT protein and its fragments is direct rather than mediated through a copurifying protein, we utilized UV crosslinking to 5-iodo-dU substituted oligonucleotides. We used a 20 nt telomeric oligonucleotide (20GTT; Table 1) that has a similar off-rate from telomerase to that of 18GTT (Figure 1C). In an attempt to probe for crosslinks between DNA and TERT at different positions along the DNA primer, we used a set of four primers, each of which had one substitution (Figure 5A). When 32P-labelled oligonucleotide was exposed to 312 nm light in the presence of unlabelled immunopurified telomerase and electrophoresed on PAGE gels, a band the size of TERT (125 kDa) was observed (Figure 5B). Specificity was demonstrated by the fact that this band was competed by an excess of telomeric primer (20GTT), but was only minimally competed by a non-telomeric primer (PBR; see Table 1). Some competition from PBR was expected, since Tetrahymena telomerase has a limited ability to extend non-telomeric primers (18); the Km for PBR is ∼8 μM (data not shown). Crosslinking of all four substituted primers was observed. The telomeric primer also crosslinked to proteins of a different size than full-length TERT (Figure 5B); some of these interactions are clearly non-specific, but the specific ones may be due either to crosslinking to a protein that copurifies with TERT, or to incomplete translation products of TERT.
At this concentration of DNA (15 nM), crosslinking was reduced in the absence of telomerase RNA (Figure 5B), which is consistent with the increased Kd of TERT in the absence of telomerase RNA (Figure 4).
We used the same four primers to confirm binding of DNA to the fragments of TERT discussed earlier. Surprisingly, all four primers also crosslinked specifically to N519 and C598, so we were unable to determine which region of the protein interacts with which nucleotide of the primer (Figure 5C and D).
Cooperative binding in the N-terminal half of TERT
A region encompassing the first 191 amino acids of Tetrahymena TERT forms a soluble, independently folded domain that is essential for telomerase activity and crosslinks to a telomeric DNA primer (24,29). In our DNA-binding assay using 35S-labelled in vitro translated protein, there was no detectable binding above background levels of this fragment (the TEN domain) to 18GTT (Figure 6A). Binding of 18GTT to TERT that is missing this domain (ΔTEN) resulted in an increase in Kd of only about 4-fold (Figure 4), despite the fact that ΔTEN completely lacks telomerase activity (29). This suggests that the TEN domain, alone, does not provide significant binding affinity for the DNA primer.
To confirm this finding, we also utilized UV crosslinking of 5-iodo-dU substituted oligonucleotides to this fragment. At the low protein concentrations which we are able to produce from in vitro translation (∼7–9 nM), there was no detectable crosslinking to this fragment (data not shown). However, when we crosslinked the same four primers to high concentrations (80 nM–20 μM) of TEN protein purified from E. coli, we were able to detect crosslinking specific for telomeric DNA, as previously reported (Figure 6B, data not shown and ref. 24). Titration of the TEN protein gave a Kd of ∼550 nM for this interaction (Figure 6B), which is presumably beyond the detection limit of our DNA-binding assay. It has previously been demonstrated that the TEN domain also crosslinks to DNA in the context of the full-length protein, since crosslinking to an equimolar mixture of full-length and ΔTEN proteins resulted in a greater extent of crosslinking to the full-length protein (24). We confirmed this finding, and determined that this was also the case for all four of our substituted oligonucleotides (Figure 5F).
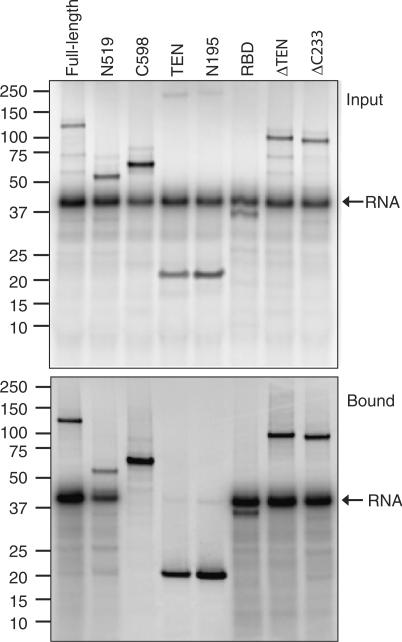
The region encompassing amino acids 192–519 of Tetrahymena TERT is able to independently bind to telomerase RNA (34). This region, the RNA binding domain (RBD), was also able to bind to DNA oligonucleotides in both the DNA binding (Figure 4) and crosslinking (Figure 5E) assays, although the affinity was quite low (∼330 nM). Thus the RBD and TEN domains each show low-affinity interactions with DNA, although in combination (as in fragment N519) they demonstrate relatively high-affinity binding. This suggests that these two fragments act cooperatively to bind DNA in the context of the full-length protein. It is also possible that a high-affinity site at the boundary of these two regions has been disrupted, although addition of an extra four amino acids to the C-terminal end of the TEN domain did not restore binding in our assay (data not shown). There is evidence that these amino acids mark the boundary of a discrete structural domain (29) and beyond this is an unstructured, flexible linker (28,42).
To confirm the structural integrity of the RBD domain and other protein fragments used in our study, we measured their ability to interact with telomerase RNA (Figure 7). RNA interaction was measured by immunoprecipitation of 32P-labelled telomerase RNA with 35S-labelled protein. As previously reported (34,39,43), N519 and RBD showed a robust ability to immunoprecipitate RNA, whereas C598 did not show any detectable binding. The ΔTEN and ΔC233 proteins also bound telomerase RNA, indicating that the RNA binding region of these proteins is correctly folded. The TEN domain showed very weak binding to telomerase RNA, as previously reported (43).
Figure 7.
Test of RNA-binding ability of TERT fragments. The indicated fragments of TERT were in vitro translated in the presence of 35S-methionine and 32P-labelled telomerase RNA and then immunoprecipitated with an antibody recognizing a FLAG peptide on TERT. The upper panel shows input material; the lower panel shows the bound fractions.
DISCUSSION
The ability of telomerase to interact with DNA at sites outside its catalytic centre is important for its unique ability to undergo repeat addition processivity. There have now been a number of studies localizing different DNA-binding regions of the TERT protein. What is unique about this study is our ability to directly and quantitatively measure DNA-binding affinities independent of activity, which has provided insight into the relative contributions of different regions of TERT. It should be borne in mind that the actual affinities of isolated regions may differ to those in the complete enzyme, since there are likely to be complex interactions and cooperativity between TERT, DNA and telomerase RNA in the latter context. Indeed, our data reveal some of this complexity. However, it is clear that together, these sites contribute to very specific and high-affinity DNA binding by recombinant Tetrahymena telomerase, with a Kd of ∼8 nM. Both the Km and Kd increased when the primer length was reduced from 18 to 12 nt, and underwent a further increase of 1–2 orders of magnitude upon a reduction in primer length to 6 nt. This suggests that recombinant Tetrahymena telomerase, like the endogenous enzyme, contains multiple anchor sites. Indeed, the affinity of recombinant Tetrahymena telomerase for an 18 nt primer was similar to that of extract-derived telomerase (11, and our unpublished data), despite the much higher processivity of the latter. There are two possible explanations for this apparent paradox. First, it is possible that recombinant telomerase may have a faster primer on-rate as well as a faster primer off-rate than endogenous telomerase; this may lead to a similar overall Kd. Secondly, it has been noted that there appear to be two populations of extension products of endogenous Tetrahymena telomerase, corresponding to the addition to the primer of a few repeats or many repeats respectively (11,12); the latter result from decreasing probability of product dissociation with increasing product length above 30–50 nt (11). Thus the endogenous Tetrahymena telomerase complex may possess yet another ‘anchor site’, provided by a protein other than TERT that interacts with very long products, that may not affect the affinity of endogenous telomerase for short (e.g. 18 nt) DNA primers.
Previous studies have concluded that a region encompassing the first 200 amino acids of TERT (the TEN or GQ domain) is an important component of the telomerase anchor site (17,22–24,35,36). However, quantitative analysis shows that this region possesses relatively low-binding affinity, two orders of magnitude lower than that of the full-length protein. This domain had the lowest affinity for DNA of all the regions that we tested. This might be expected when analyzing the TEN domain protein in isolation, but the fact that the ΔTEN protein showed only a modest decrease in primer-binding affinity relative to full-length protein indicates that TEN does not provide much of the overall affinity for DNA primer in the context of the full-length protein. While the interactions of TEN with DNA may be crucially important for proper alignment or movement of the primer, they clearly are not necessary for maintaining tight binding to DNA, and hence may only constitute a minor part of the telomerase anchor sites.
Amino acids 1–300 of human TERT, encompassing the TEN domain, were able to be captured by biotinylated telomeric oligonucleotides in an assay similar to that used here (36). Possible reasons for this apparent discrepancy with our results include: (i) species differences in TEN function, (ii) the fact that a longer region of the protein was tested or (iii) the fact that the Wyatt et al. study used a high concentration of DNA, which may have allowed detection of a weak interaction.
In the context of the full-length protein, the Tetrahymena TEN region may contribute to DNA binding cooperatively with the RBD, since a protein encompassing both of these fragments showed much higher DNA binding than either fragment alone (Figure 4). Such cooperativity would be consistent with the observed functional interactions between the TEN domain and the rest of the protein (30–32).
Notwithstanding the above discussion, it is clear that the Tetrahymena TEN domain interacts with the DNA primer (23,24). The question remains, with which part of the primer is it interacting, i.e. if it does in fact constitute a telomerase anchor site, is it the template-proximal or the template-distal anchor site? The study by Romi et al. (23) is unique in that it involved crosslinking of the DNA primer, followed by extension of the primer by telomerase using radiolabelled nucleotides; thus any signal obtained must be due to DNA molecules with their 3′ end in the active site of the enzyme. Crosslinking was observed with DNA primers substituted in a position that is within the template-binding region; this indicates that the Tetrahymena TEN domain is close to a very template-proximal region of the primer. Studies with human and yeast TERT also suggest that the TEN domain interacts with a template-proximal part of the primer (17,22). On the other hand, mutations in Tetrahymena TERT disrupted crosslinking to nucleotides far upstream of the 3′ end of the primer (24) and in fact substitutions at any of four positions throughout a primer support crosslinking of the primer to the TEN domain either in isolation (data not shown), or in the context of the full-length protein (Figure 5F). It is possible that in the latter two studies, crosslinking is reflecting stable non-productive interactions between primer and enzyme such that the 3′ end of the primer is not aligned with the active site. However, the prevalence of these interactions would be inconsistent with the very low Kd that we observe between full-length TERT and the primer, and also with the fact that some of the amino acids involved in mediating the crosslinking reaction are also important for activity (24). An alternative scenario to reconcile these data could be that initial binding of the primer to its specific binding sites within TERT can occur in any of the ‘registers’ of repeat binding, and it is these events that are captured by crosslinking; this is followed by rapid sliding of the DNA to position its 3′ end to be close to the TEN domain and the enzyme active site.
The data in Figure 4 suggest that a minimum of four discrete DNA-interaction sites (in the TEN, RBD, RT and C-terminal domains) exist. The actual number of binding sites is potentially much higher than four. These data directly confirm some interactions that have been inferred based on activity assays, and add a novel function of the RNA-binding region of TERT to DNA binding, which has also recently been suggested based on crosslinking (23). Binding to the RBD was independent of the presence of telomerase RNA (Figure 4), indicating that this domain has two separable functions. Although the TEN domain may contribute to the template-proximal anchor site, the template-distal anchor site remains to be localized, despite ample evidence for its existence (11,13–15,19); this site may reside in any of the RBD, RT or C-terminal domains.
Previous studies have concluded that binding of a DNA primer to telomerase is greatly increased by the presence of telomerase RNA (22,24). On the other hand, RNA-independent binding of human TERT to DNA has recently been reported (36). The present study, due to its quantitative nature, suggests that the RNA component has only a modest impact on binding of DNA to full-length TERT; this result would be expected in the presence of several protein anchor sites. It is possible that the RNA-independent binding we observe does not completely reflect interactions that would occur in the context of the intact enzyme, though the specificity of the interaction (Figure 3A) argues against this. The RNA may positively impact on DNA binding solely through its base-pairing interactions with the primer, or possibly by also inducing a protein conformational change. Only the primer Kd of full-length protein was substantially affected by the RNA; this would be expected since the C598 fragment does not show high-binding affinity for the RNA, and the N519 fragment lacks the active site of the protein, which is where a base-pairing interaction or putative protein conformational change would occur.
An intriguing aspect of our data is that the C598 fragment of TERT binds to DNA with an affinity equal to full-length protein, while the N-terminal half of the protein also binds to DNA with an affinity just 6-fold lower than full-length protein. This suggests that both N- and C-terminal halves of the protein contain multiple binding sites, not all of which are used simultaneously in the context of the full-length protein. Binding contributed by each half of the protein must be independent, since cooperative binding would likely lead to a DNA-binding affinity by the full-length protein even greater than that observed. In the context of binding to telomerase RNA, the two halves of the protein also demonstrate independence in that the N-terminal half is necessary and sufficient for binding (Figure 7 and ref. 39). We propose that different DNA-binding sites are used by the enzyme during different stages of the six nucleotide addition cycle, since the DNA must move relative to the enzyme as each nucleotide is added.
Our data not only confirm that DNA-binding sites are located in multiple regions of the TERT protein, but also suggest that binding to these sites may be more complex than previously envisaged. It is possible that the DNA moves relative to the different sites while preparing for extension; it is also possible that different sites interact with DNA at different stages of the addition cycle. Also, there may be cooperativity in DNA binding between different regions of the TERT protein. Thus there remains much to be learned about the nature of the telomerase anchor sites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Funding was provided by The Wellcome Trust (Senior Research Fellowship GR066727MA). We thank Julie Jurczyluk for technical assistance and Drs Thomas Cech and Steven Jacobs for the kind gift of the purified TEN domain protein and plasmids.
Conflict of interest statement. None declared.
REFERENCES
- 1.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 3.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 4.Hahn WC, Stewart SA, Brooks MW, York SG, Eaton E, Kurachi A, Beijersbergen RL, Knoll JH, Meyerson M, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat. Med. 1999;5:1164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 7.Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- 8.Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 9.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. U.S.A. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryan TM, Goodrich KJ, Cech TR. A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem. 2000;275:24199–24207. doi: 10.1074/jbc.M003246200. [DOI] [PubMed] [Google Scholar]
- 11.Lee MS, Blackburn EH. Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 1993;13:6586–6599. doi: 10.1128/mcb.13.10.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins K, Greider CW. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 13.Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- 14.Morin GB. Recognition of a chromosome truncation site associated with alpha-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- 15.Lue NF, Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. doi: 10.1093/nar/26.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins K. Ciliate telomerase biochemistry. Annu. Rev. Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- 17.Moriarty TJ, Ward RJ, Taboski MA, Autexier C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell. 2005;16:3152–3161. doi: 10.1091/mbc.E05-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Blackburn EH. De nova telomere addition by Tetrahymena telomerase in vitro. EMBO J. 1997;16:866–879. doi: 10.1093/emboj/16.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baran N, Haviv Y, Paul B, Manor H. Studies on the minimal lengths required for DNA primers to be extended by the Tetrahymena telomerase: implications for primer positioning by the enzyme. Nucleic Acids Res. 2002;30:5570–5578. doi: 10.1093/nar/gkf676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond PW, Lively TN, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sperger JM, Cech TR. A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- 22.Lue NF. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 2005;280:26586–26591. doi: 10.1074/jbc.M503028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romi E, Baran N, Gantman M, Shmoish M, Min B, Collins K, Manor H. High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl Acad. Sci. U.S.A. 2007;104:8791–8796. doi: 10.1073/pnas.0703157104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs SA, Podell ER, Cech TR. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 25.Hossain S, Singh SM, Lue NF. Functional analysis of the C-terminal extension of telomerase reverse transcriptase: a putative “thumb” domain. J. Biol. Chem. 2002;277:36174–36180. doi: 10.1074/jbc.M201976200. [DOI] [PubMed] [Google Scholar]
- 26.Huard S, Moriarty TJ, Autexier C. The C terminus of the human telomerase reverse transcriptase is a determinant of enzyme processivity. Nucleic Acids Res. 2003;31:4059–4070. doi: 10.1093/nar/gkg437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lue NF, Lin YC, Mian IS. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol. Cell. Biol. 2003;23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J, Peng Y, Mian IS, Lue NF. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs SA, Podell ER, Wuttke DS, Cech TR. Soluble domains of telomerase reverse transcriptase identified by high-throughput screening. Protein Sci. 2005;14:2051–2058. doi: 10.1110/ps.051532105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beattie TL, Zhou W, Robinson MO, Harrington L. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:6151–6160. doi: 10.1128/MCB.21.18.6151-6160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriarty TJ, Huard S, Dupuis S, Autexier C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 2002;22:1253–1265. doi: 10.1128/MCB.22.4.1253-1265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriarty TJ, Marie-Egyptienne DT, Autexier C. Functional organization of repeat addition processivity and DNA synthesis determinants in the human telomerase multimer. Mol. Cell. Biol. 2004;24:3720–3733. doi: 10.1128/MCB.24.9.3720-3733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beattie TL, Zhou W, Robinson MO, Harrington L. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell. 2000;11:3329–3340. doi: 10.1091/mbc.11.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SR, Wong JM, Collins K. Human telomerase reverse transcriptase motifs required for elongation of a telomeric substrate. J. Biol. Chem. 2003;278:52531–52536. doi: 10.1074/jbc.M311359200. [DOI] [PubMed] [Google Scholar]
- 36.Wyatt HD, Lobb DA, Beattie TL. Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell. Biol. 2007;27:3226–3240. doi: 10.1128/MCB.02368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryan TM, Goodrich KJ, Cech TR. Tetrahymena telomerase is active as a monomer. Mol. Biol. Cell. 2003;14:4794–4804. doi: 10.1091/mbc.E03-07-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]
- 39.Bryan TM, Goodrich KJ, Cech TR. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell. 2000;6:493–499. doi: 10.1016/s1097-2765(00)00048-4. [DOI] [PubMed] [Google Scholar]
- 40.Wallweber G, Gryaznov S, Pongracz K, Pruzan R. Interaction of human telomerase with its primer substrate. Biochemistry. 2003;42:589–600. doi: 10.1021/bi026914a. [DOI] [PubMed] [Google Scholar]
- 41.Hammond PW, Cech TR. Euplotes telomerase: evidence for limited base-pairing during primer elongation and dGTP as an effector of translocation. Biochemistry. 1998;37:5162–5172. doi: 10.1021/bi972988o. [DOI] [PubMed] [Google Scholar]
- 42.Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 2001;21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J. Biol. Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.