Abstract
Kluyveromyces lactis γ-toxin is a tRNA endonuclease that cleaves Saccharomyces cerevisiae , and between position 34 and position 35. All three substrate tRNAs carry a 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) residue at position 34 (wobble position) of which the mcm5 group is required for efficient cleavage. However, the different cleavage efficiencies of mcm5s2U34-containing tRNAs suggest that additional features of these tRNAs affect cleavage. In the present study, we show that a stable anticodon stem and the anticodon loop are the minimal requirements for cleavage by γ-toxin. A synthetic minihelix RNA corresponding to the anticodon stem loop (ASL) of the natural substrate is cleaved at the same position as the natural substrate. In , the nucleotides U34U35C36A37C38 are required for optimal γ-toxin cleavage, whereas a purine at position 32 or a G in position 33 dramatically reduces the cleavage of the ASL. Comparing modified and partially modified forms of E. coli and yeast reinforced the strong stimulatory effects of the mcm5 group, revealed a weak positive effect of the s2 group and a negative effect of the bacterial 5-methylaminomethyl (mnm5) group. The data underscore the high specificity of this yeast tRNA toxin.
INTRODUCTION
Killer strains of the dairy yeast Kluyveromyces lactis secrete a heterotrimeric toxin (zymocin), which causes an irreversible arrest of sensitive yeast cells, such as Saccharomyces cerevisiae in the G1 phase of the cell cycle (1–4). Zymocin consists of three subunits, α, β and γ, that are encoded by a linear plasmid (1,5). Upon secretion, the α- and β-subunits dock the zymocin to the cell wall of susceptible yeasts and facilitate transfer of the γ-subunit into the cells (4). Cytotoxicity of zymocin resides within the γ-subunit (γ-toxin), since its intracellular expression in sensitive cells mimics the action of exogenous zymocin (6). Recently we showed that γ-toxin is a tRNA endonuclease that cleaves 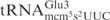 ,
, 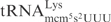 and
and 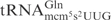 between positions 34 and 35 (7). All substrate tRNAs have the modified nucleoside 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) at position 34 (wobble position). The mcm5 group present on the wobble uridine is important for tRNA cleavage, since tRNAs missing part of, or the entire mcm5 side chain are less efficiently cleaved (7).
between positions 34 and 35 (7). All substrate tRNAs have the modified nucleoside 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) at position 34 (wobble position). The mcm5 group present on the wobble uridine is important for tRNA cleavage, since tRNAs missing part of, or the entire mcm5 side chain are less efficiently cleaved (7).
The γ-toxin and certain Escherichia coli proteins comprise a group of tRNA anticodon nucleases (ACNs) that cleave tRNA in the anticodon region. The PrrC protein is a nuclease found in prr+
E. coli strains in a latent form. When E. coli is infected with phage T4, the nuclease is activated and cleaves 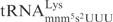 between positions 33 and 34 (8,9). Similar to γ-toxin, the efficiency of tRNA cleavage by PrrC is remarkably affected by the modified nucleosides present in the anticodon region (10,11). Colicin E5 and colicin D are produced by bacteria carrying the corresponding Col plasmids. When secreted, these colicins kill sensitive E. coli strains by inhibiting translation. Colicin E5 cleaves the queuosine (Q) containing tRNAs,
between positions 33 and 34 (8,9). Similar to γ-toxin, the efficiency of tRNA cleavage by PrrC is remarkably affected by the modified nucleosides present in the anticodon region (10,11). Colicin E5 and colicin D are produced by bacteria carrying the corresponding Col plasmids. When secreted, these colicins kill sensitive E. coli strains by inhibiting translation. Colicin E5 cleaves the queuosine (Q) containing tRNAs, 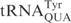 ,
, 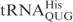 ,
, 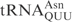 and
and 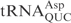 (12) and colicin D targets all tRNAArg isoacceptors (13). Cleavage by all the ACNs generates 5′-OH and 2′,3′-cyclic phosphate termini. The ACNs show no amino acid homology, however, the catalytic domains of colicin E5 and colicin D show similarities in their crystal structures (14,15). These similarities shared by colicin E5 and colicin D are also seen in other ribonucleases, such as ribonuclease T1 (16) and archaeal toxin aRelE (17).
(12) and colicin D targets all tRNAArg isoacceptors (13). Cleavage by all the ACNs generates 5′-OH and 2′,3′-cyclic phosphate termini. The ACNs show no amino acid homology, however, the catalytic domains of colicin E5 and colicin D show similarities in their crystal structures (14,15). These similarities shared by colicin E5 and colicin D are also seen in other ribonucleases, such as ribonuclease T1 (16) and archaeal toxin aRelE (17).
Even though all three γ-toxin substrates, 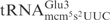 ,
, 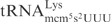 and
and 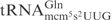 , contain mcm5s2U at the wobble position, the latter two tRNA species are cleaved with much lower efficiency (7). This suggests that in addition to the wobble uridine modification, there are other features in
, contain mcm5s2U at the wobble position, the latter two tRNA species are cleaved with much lower efficiency (7). This suggests that in addition to the wobble uridine modification, there are other features in 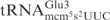 that improve cleavage by γ-toxin. In this study, we have investigated the determinants in tRNA important for efficient cleavage by γ-toxin.
that improve cleavage by γ-toxin. In this study, we have investigated the determinants in tRNA important for efficient cleavage by γ-toxin.
MATERIALS AND METHODS
Plasmid constructions
DNA manipulations, plasmid preparations and bacterial transformations were performed according to standard protocols. The plasmids used in this study are listed in Table 1. Plasmids containing T7 promoter-driven tRNA genes were constructed as previously described (18). Briefly, three pairs of oligonucleotides corresponding to the sequence of the T7 promoter and the various tRNA genes were ligated into the EcoRI/BamHI sites of pUC18 (Roche Applied Science). In the oligonucleotides used to construct genes of 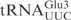 ,
, 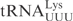 and their derivatives, T1-A72 were changed to G1-C72 to facilitate in vitro transcription. In the plasmid carrying the T7 promoter-driven
and their derivatives, T1-A72 were changed to G1-C72 to facilitate in vitro transcription. In the plasmid carrying the T7 promoter-driven 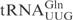 gene, there is an MvaI site in the tRNA gene, which prevents the proper linearization of the plasmid for in vitro transcription. To circumvent this, the C5-G68 was changed to T5-A68 in the oligonucleotides used to construct the
gene, there is an MvaI site in the tRNA gene, which prevents the proper linearization of the plasmid for in vitro transcription. To circumvent this, the C5-G68 was changed to T5-A68 in the oligonucleotides used to construct the 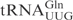 gene. The C34 in the
gene. The C34 in the 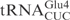 gene in plasmid pABY1539 was mutated to T using QuickChange® Site-Directed Mutagenesis kit (Stratagene), generating pABY1542.
gene in plasmid pABY1539 was mutated to T using QuickChange® Site-Directed Mutagenesis kit (Stratagene), generating pABY1542.
Table 1.
Plasmids used in this study
| Plasmids name | Description | Source |
|---|---|---|
| pABY1537 | pUC18-T7-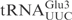 (G1-C72) (G1-C72) |
(22) |
| pABY1539 | pUC18-T7-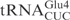 (G1-C72) (G1-C72) |
This study |
| pABY1542 | pUC18-T7-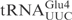 (G1-C72, T34) (G1-C72, T34) |
This study |
| pABY1714 | pUC18-T7-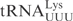 (G1-C72) (G1-C72) |
This study |
| pABY1738 | pUC18-T7-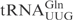 (T5-A68) (T5-A68) |
This study |
| pABY1746 | pUC18-T7-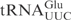 (G1-C72)- (G1-C72)-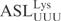
|
This study |
| pABY1747 | pUC18-T7-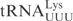 (G1-C72)- (G1-C72)-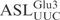
|
This study |
| pABY1650 | pETM-13-GST | (7) |
| pABY1644 | pETM-13-γ-toxin-GST | (7) |
RNA methods
For RNA preparation, S. cerevisiae cells were collected from mid-log phase cultures grown in YEPD medium at 30°C. E. coli cells were collected from mid-log phase cultures grown in LB medium at 37°C. Total RNA was prepared as described (19) followed by LiCl fractionation (20). The T7-transcribed radiolabeled tRNAs were prepared by using MvaI linearized vectors, 5′-[α-32P] UTP (400 Ci/mmol, Amersham Biosciences), and the Riboprobe in vitro transcription system (Promega). The transcripts were purified as described (21). The RNA oligonucleotides used in this study are listed in Table 2. They were synthesized by Invitrogen and 5′-labeled using adenosine [γ32P]-triphosphate (5000 Ci/mmol, Amersham Biosciences) and polynucleotide kinase (Roche Applied Science).
Table 2.
RNA oligonucleotides used in this study
| Oligo name | Sequencea |
|---|---|
| Wt | 5′-UCACGCUUUCACCGUGG-3′ |
| No stem | 5′-UCACGCUUUCACGCACU-3′ |
| GC stem | 5′-CCGCCCUUUCACGGCGG-3′ |
| AU stem | 5′-AUAAACUUUCACUUUAU-3′ |
| C39G | 5′-UCACGCUUUCACGGUGG-3′ |
| C32A | 5′-UCACGAUUUCACCGUGG-3′ |
| C32G | 5′-UCACGGUUUCACCGUGG-3′ |
| C32U | 5′-UCACGUUUUCACCGUGG-3′ |
| U33A | 5′-UCACGCAUUCACCGUGG-3′ |
| U33C | 5′-UCACGCCUUCACCGUGG-3′ |
| U33G | 5′-UCACGCGUUCACCGUGG-3′ |
| U34A | 5′-UCACGCUAUCACCGUGG-3′ |
| U34C | 5′-UCACGCUCUCACCGUGG-3′ |
| U34G | 5′-UCACGCUGUCACCGUGG-3′ |
| U35A | 5′-UCACGCUUACACCGUGG-3′ |
| U35C | 5′-UCACGCUUCCACCGUGG-3′ |
| U35G | 5′-UCACGCUUGCACCGUGG-3′ |
| C36A | 5′-UCACGCUUUAACCGUGG-3′ |
| C36G | 5′-UCACGCUUUGACCGUGG-3′ |
| C36U | 5′-UCACGCUUUUACCGUGG-3′ |
| A37C | 5′-UCACGCUUUCCCCGUGG-3′ |
| A37G | 5′-UCACGCUUUCGCCGUGG-3′ |
| A37U | 5′-UCACGCUUUCUCCGUGG-3′ |
| C38A | 5′-UCACGCUUUCAACGUGG-3′ |
| C38G | 5′-UCACGCUUUCAGCGUGG-3′ |
| C38U | 5′-UCACGCUUUCAUCGUGG-3′ |
aSequences different from the 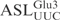 are underlined.
are underlined.
Endonuclease activity assay
Recombinant γ-toxin-GST and GST were purified from an overnight culture of E. coli BL21(DE3) (Novagen) carrying corresponding expression plasmids by using Glutathione–Sepharose 4B (Amersham Biosciences) as described previously (7). Purified proteins were stored in 50% glycerol at −20°C. Under this condition, the proteins are stable for at least 12 months. To compare the cleavage of 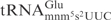 from E. coli and
from E. coli and 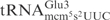 from S. cerevisiae, 5 µg of total tRNA was mixed with purified γ-toxin-GST or GST proteins in buffer M (10 mM Tris–HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, pH 7.5) and incubated at 30°C for 10 min. The tRNAs were separated on 8% polyacrylamide, 8 M urea gels. The cleavage was analyzed by northern blot as previously described (7). The oligonucleotide used to detect E. coli
from S. cerevisiae, 5 µg of total tRNA was mixed with purified γ-toxin-GST or GST proteins in buffer M (10 mM Tris–HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, pH 7.5) and incubated at 30°C for 10 min. The tRNAs were separated on 8% polyacrylamide, 8 M urea gels. The cleavage was analyzed by northern blot as previously described (7). The oligonucleotide used to detect E. coli
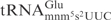 was 5′- CCTCTAGACGAAGGGGAC-3′. For detection of S. cerevisiae
was 5′- CCTCTAGACGAAGGGGAC-3′. For detection of S. cerevisiae
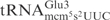 the oligonucleotide 5′-CTCCGCTACGGGGAGTCGAAC-3′ was used as a probe (7). The chemical trimethylamine N-oxide (TMAO) was reported to increase the cleavage of unmodified tRNA by a bacterial anticodon nuclease, PrrC (11). We found that TMAO also enhanced the cleavage of unmodified tRNA and ASLs by γ-toxin-GST (data not shown). Therefore TMAO was included in the endonuclease assay when unmodified substrates were tested. To test cleavage of unmodified tRNAs, 2 nM of T7-transcribed radiolabeled tRNAs were mixed with purified γ-toxin-GST or GST proteins in buffer M containing 1.5 M TMAO and incubated at 30°C for 10 min. The tRNAs were separated on 8% polyacrylamide, 8 M urea gels. The gels were dried and cleavage monitored by autoradiography. The radioactivities of the full-length RNA and cleavage products were quantified. Extent of cleavage was calculated using the formula: extent of cleavage=cleavage products/(full-length tRNA+cleavage products). To compare the cleavage efficiencies of T7-transcribed
the oligonucleotide 5′-CTCCGCTACGGGGAGTCGAAC-3′ was used as a probe (7). The chemical trimethylamine N-oxide (TMAO) was reported to increase the cleavage of unmodified tRNA by a bacterial anticodon nuclease, PrrC (11). We found that TMAO also enhanced the cleavage of unmodified tRNA and ASLs by γ-toxin-GST (data not shown). Therefore TMAO was included in the endonuclease assay when unmodified substrates were tested. To test cleavage of unmodified tRNAs, 2 nM of T7-transcribed radiolabeled tRNAs were mixed with purified γ-toxin-GST or GST proteins in buffer M containing 1.5 M TMAO and incubated at 30°C for 10 min. The tRNAs were separated on 8% polyacrylamide, 8 M urea gels. The gels were dried and cleavage monitored by autoradiography. The radioactivities of the full-length RNA and cleavage products were quantified. Extent of cleavage was calculated using the formula: extent of cleavage=cleavage products/(full-length tRNA+cleavage products). To compare the cleavage efficiencies of T7-transcribed 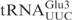 and the RNA oligonucleotide corresponding to the anticodon stem loop of
and the RNA oligonucleotide corresponding to the anticodon stem loop of 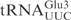 (
(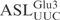 ), 5 µM of labeled tRNA or
), 5 µM of labeled tRNA or 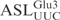 was incubated in buffer M containing 1.5 M TMAO and 2 nM purified γ-toxin-GST at 0°C. At intervals, 10 µl of the reaction was withdrawn, extracted with equal volume of phenol, and kept on ice. Samples were run on 8% (for tRNA transcript) or 10% (for RNA oligonucleotide) polyacrylamide, 8 M urea gels. The gels were dried and cleavage monitored by autoradiography. The extent of cleavage was calculated as described above. To compare
was incubated in buffer M containing 1.5 M TMAO and 2 nM purified γ-toxin-GST at 0°C. At intervals, 10 µl of the reaction was withdrawn, extracted with equal volume of phenol, and kept on ice. Samples were run on 8% (for tRNA transcript) or 10% (for RNA oligonucleotide) polyacrylamide, 8 M urea gels. The gels were dried and cleavage monitored by autoradiography. The extent of cleavage was calculated as described above. To compare 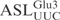 and its derivatives, 5 µM of labeled ASLs were incubated in buffer M containing 1.5 M TMAO and 5 nM purified γ-toxin-GST at 0°C. The extent of cleavage at different time points was monitored as described above.
and its derivatives, 5 µM of labeled ASLs were incubated in buffer M containing 1.5 M TMAO and 5 nM purified γ-toxin-GST at 0°C. The extent of cleavage at different time points was monitored as described above.
Strains
The S. cerevisiae strains are derivatives of W303-1A (MATa ura3-1 leu2-3,112 trp1-1 his3-11,15 can1-100 ade2-1). The elp3 and tuc1 mutant strains have been described previously (22,23). Bacterial strains used in this study, TH168 (F+, asnA, asnB, thi, lacIQ, relA, spoT, fadR3115::Tn10(Km), mnmA107), TH169 (F+, asnA, asnB, thi, lacIQ, relA, spoT, fadR3115::Tn10(Km), val(R), mnmE) and TH170 (F+, asnA, asnB, thi, lacIQ, relA, spoT, fadR3115::Tn10(Km), val(R)) have been described elsewhere (24).
RESULTS
In vitro transcribed 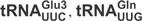 and
and
 are substrates for γ-toxin
are substrates for γ-toxin
Even though in vitro transcribed tRNAs lack modified nucleosides, they provide a useful tool to investigate the RNA sequence elements that are important for the interaction with proteins, e.g. PrrC and colicin E5 (11,12). To investigate whether unmodified tRNAs can be used to identify the primary sequence important for γ-toxin cleavage, in vitro transcribed 32P-labeled 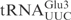 ,
, 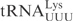 or
or 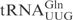 was incubated with different concentrations of purified γ-toxin protein. The extent of cleavage was determined by applying the samples to denaturing polyacrylamide gel followed by autoradiography. Interestingly, the unmodified
was incubated with different concentrations of purified γ-toxin protein. The extent of cleavage was determined by applying the samples to denaturing polyacrylamide gel followed by autoradiography. Interestingly, the unmodified 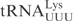 and
and 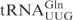 required ∼100-fold higher concentration of γ-toxin to be cleaved to a similar extent as
required ∼100-fold higher concentration of γ-toxin to be cleaved to a similar extent as 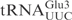 (Figure 1B, data not shown). This difference in cleavage efficiencies was also found between native modified
(Figure 1B, data not shown). This difference in cleavage efficiencies was also found between native modified 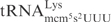 and
and 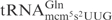 when compared to
when compared to 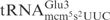 (7), suggesting that it is caused by differences in the primary sequence. Thus, the unmodified tRNAs can be used to study the sequence identity elements for γ-toxin cleavage.
(7), suggesting that it is caused by differences in the primary sequence. Thus, the unmodified tRNAs can be used to study the sequence identity elements for γ-toxin cleavage.
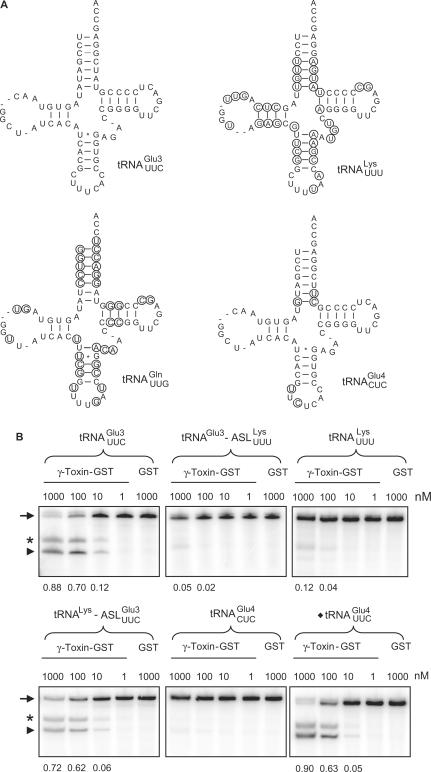
Figure 1.
The determinants for γ-toxin cleavage reside within the anticodon stem loop. (A) Sequence of unmodified 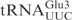 ,
, 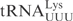 ,
, 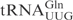 and
and 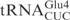 . Nucleotides circled in
. Nucleotides circled in 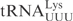 ,
, 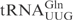 and
and 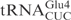 identify differences compared to
identify differences compared to 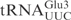 . (B) In vitro cleavage of
. (B) In vitro cleavage of 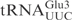 ,
, 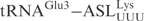 ,
,  ,
, 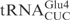 and ♦
and ♦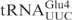 by γ-toxin-GST. The tRNAs were T7 transcribed in the presence of [α-32P] UTP and 2 nM of transcribed tRNAs were incubated with the indicated concentration of γ-toxin-GST or GST protein at 30°C for 10 min. Samples were analyzed on 8% polyacrylamide, 8 M urea gels and the cleavage products were quantified. Complete cleavage of substrate tRNA was set to 1.0 (for details see Materials and Methods section). Full-length, 5′-half and 3′-half of tRNAs are indicated with arrow, arrow head and asterisk, respectively. In ♦
by γ-toxin-GST. The tRNAs were T7 transcribed in the presence of [α-32P] UTP and 2 nM of transcribed tRNAs were incubated with the indicated concentration of γ-toxin-GST or GST protein at 30°C for 10 min. Samples were analyzed on 8% polyacrylamide, 8 M urea gels and the cleavage products were quantified. Complete cleavage of substrate tRNA was set to 1.0 (for details see Materials and Methods section). Full-length, 5′-half and 3′-half of tRNAs are indicated with arrow, arrow head and asterisk, respectively. In ♦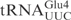 , the anticodon was mutated from CUC to UUC.
, the anticodon was mutated from CUC to UUC.
Anticodon stem loop in  is important for cleavage by γ-toxin
is important for cleavage by γ-toxin
To investigate what domain(s) of 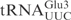 are important for γ-toxin cleavage, chimeric tRNA genes were constructed. Since γ-toxin cleaves the anticodon of substrate tRNAs, the anticodon stem loop (ASL) represents a candidate identity element of γ-toxin cleavage. A recombinant gene was created in which the ASL of
are important for γ-toxin cleavage, chimeric tRNA genes were constructed. Since γ-toxin cleaves the anticodon of substrate tRNAs, the anticodon stem loop (ASL) represents a candidate identity element of γ-toxin cleavage. A recombinant gene was created in which the ASL of 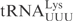 was replaced by the corresponding part of
was replaced by the corresponding part of 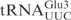 (designated
(designated 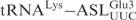 ). In another construct, the ASL in
). In another construct, the ASL in 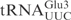 was replaced with the ASL from
was replaced with the ASL from 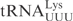 , generating
, generating 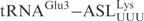 . These chimeric tRNAs were in vitro transcribed and cleavage by γ-toxin was compared. The
. These chimeric tRNAs were in vitro transcribed and cleavage by γ-toxin was compared. The 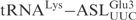 was cleaved with a similar efficiency as
was cleaved with a similar efficiency as 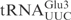 , whereas the cleavage of
, whereas the cleavage of 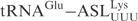 was comparable to
was comparable to 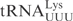 (Figure 1B).
(Figure 1B).
Unmodified 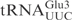 and
and 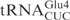 share significant sequence homology: except for 3 nt in the acceptor stem and two in the anticodon loop, all the other nucleotides are identical (Figure 1A). In contrast to
share significant sequence homology: except for 3 nt in the acceptor stem and two in the anticodon loop, all the other nucleotides are identical (Figure 1A). In contrast to 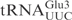 , in vitro transcribed
, in vitro transcribed 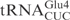 was resistant to γ-toxin cleavage (Figure 1B). Interestingly, a point mutation in the anticodon loop of
was resistant to γ-toxin cleavage (Figure 1B). Interestingly, a point mutation in the anticodon loop of 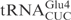 (C34 to U) was enough to make the mutant
(C34 to U) was enough to make the mutant 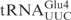 as reactive as
as reactive as 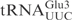 (Figure 1B). Taken together, these data show that the ASL in
(Figure 1B). Taken together, these data show that the ASL in 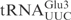 and position 34 are important for the cleavage by γ-toxin.
and position 34 are important for the cleavage by γ-toxin.
Unmodified ASL as γ-toxin substrate
The chimeric tRNAs data presented above showed that the ASL of 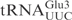 carries the vital information for γ-toxin cleavage. We utilized an in vitro synthesized 17-mer RNA oligonucleotide with identical sequence to the ASL of
carries the vital information for γ-toxin cleavage. We utilized an in vitro synthesized 17-mer RNA oligonucleotide with identical sequence to the ASL of 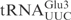 (designated
(designated 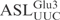 ) to investigate if it acts as a substrate for γ-toxin. Incubation of the 5′-32P-labeled
) to investigate if it acts as a substrate for γ-toxin. Incubation of the 5′-32P-labeled 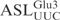 with γ-toxin generated a product of 8 nt in length (Figure 2A), showing that the
with γ-toxin generated a product of 8 nt in length (Figure 2A), showing that the 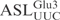 was cleaved between nucleotides 8 and 9, which correspond to positions 34 and 35 in
was cleaved between nucleotides 8 and 9, which correspond to positions 34 and 35 in 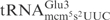 . Thus, the
. Thus, the 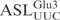 is cleaved at the same site as
is cleaved at the same site as 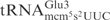 . The cleavage of 5′-labeled
. The cleavage of 5′-labeled 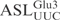 was compared with that of in vitro-transcribed radiolabeled
was compared with that of in vitro-transcribed radiolabeled 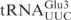 . As shown in Figure 2B,
. As shown in Figure 2B, 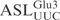 and
and 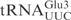 displayed similar cleavage kinetics upon γ-toxin treatment, supporting the notion that the
displayed similar cleavage kinetics upon γ-toxin treatment, supporting the notion that the 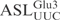 contains all the sequence information required for γ-toxin cleavage.
contains all the sequence information required for γ-toxin cleavage.
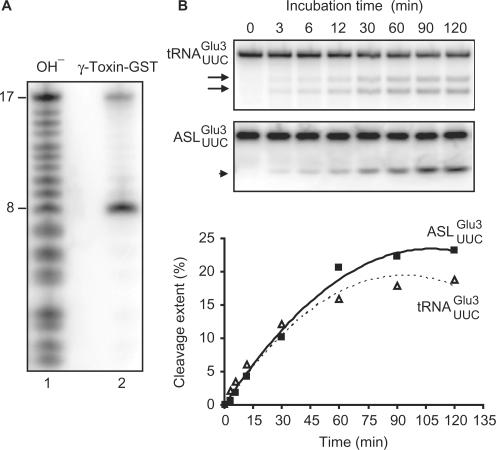
Figure 2.
Cleavage of unmodified 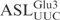 and
and  by γ-toxin. (A) The 5′-32P-labeled
by γ-toxin. (A) The 5′-32P-labeled  was incubated with γ-toxin-GST and the products (lane 2) were separated by 20% polyacrylamide, 8 M urea gel electrophoresis. As marker, a ladder (lane 1) was obtained by partial alkaline hydrolysis of the 5′-32P-labeled
was incubated with γ-toxin-GST and the products (lane 2) were separated by 20% polyacrylamide, 8 M urea gel electrophoresis. As marker, a ladder (lane 1) was obtained by partial alkaline hydrolysis of the 5′-32P-labeled  . Numbers on the left indicate chain length in nucleotides. (B) The [α-32P] UTP-labeled T7-transcribed
. Numbers on the left indicate chain length in nucleotides. (B) The [α-32P] UTP-labeled T7-transcribed  (top panel) and the 5′-32P-labeled anticodon stem loop (
(top panel) and the 5′-32P-labeled anticodon stem loop (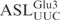 , middle panel), both at a concentration of 5 µM, were incubated with 2 nM γ-toxin-GST at 0°C. Aliquots of the reactions were taken at the indicated time points and analyzed on 8% or 10% polyacrylamide, 8 M urea gels, respectively. The cleavage products of
, middle panel), both at a concentration of 5 µM, were incubated with 2 nM γ-toxin-GST at 0°C. Aliquots of the reactions were taken at the indicated time points and analyzed on 8% or 10% polyacrylamide, 8 M urea gels, respectively. The cleavage products of 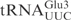 and
and 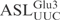 are indicated by arrows and an arrow head, respectively. The extent of cleavage was quantitated for each time point and plotted (bottom panel).
are indicated by arrows and an arrow head, respectively. The extent of cleavage was quantitated for each time point and plotted (bottom panel).
Influence of anticodon stem sequence on the cleavage of ASL
The observation that 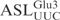 constitutes a substrate equal to the full-length
constitutes a substrate equal to the full-length 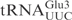 made it possible to use different variants of the ASL to pinpoint the sequence important for γ-toxin cleavage. In order to investigate the importance of the anticodon stem for cleavage by γ-toxin, variants of the
made it possible to use different variants of the ASL to pinpoint the sequence important for γ-toxin cleavage. In order to investigate the importance of the anticodon stem for cleavage by γ-toxin, variants of the 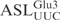 with different stem sequences were synthesized. The 5′-32P-labeled ASLs were incubated with γ-toxin and the cleavage at different time points was analyzed by denaturing PAGE. In one of the ASLs, the sequence of the 3′ strand of the stem was changed to prevent base-pair formation in the stem, thus disrupting the stem structure (designated ‘no stem’). Under the condition used in this study, this linear RNA oligonucleotide is not cleaved by γ-toxin (Figure 3B). A mutation, C39 to G, that disrupts the lowest (31–39) base-pair in the stem expands the anticodon loop size (and at the same time, shortens the stem length) also abolished the cleavage of the ASL (Figure 3B). Changing the stem sequence of
with different stem sequences were synthesized. The 5′-32P-labeled ASLs were incubated with γ-toxin and the cleavage at different time points was analyzed by denaturing PAGE. In one of the ASLs, the sequence of the 3′ strand of the stem was changed to prevent base-pair formation in the stem, thus disrupting the stem structure (designated ‘no stem’). Under the condition used in this study, this linear RNA oligonucleotide is not cleaved by γ-toxin (Figure 3B). A mutation, C39 to G, that disrupts the lowest (31–39) base-pair in the stem expands the anticodon loop size (and at the same time, shortens the stem length) also abolished the cleavage of the ASL (Figure 3B). Changing the stem sequence of 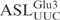 to that of E. coli
to that of E. coli
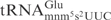 (in total 5 G–C base pairs, Figure 4A) did not alter the cleavage, while a stem with a total of 5 A–U base pairs made the ASL 5-fold less sensitive to γ-toxin (Figure 3B). Taken together, these data suggest that γ-toxin requires the canonical anticodon stem loop structure for efficient cleavage, and that ASL with a stable stem structure seems to be preferred by γ-toxin.
(in total 5 G–C base pairs, Figure 4A) did not alter the cleavage, while a stem with a total of 5 A–U base pairs made the ASL 5-fold less sensitive to γ-toxin (Figure 3B). Taken together, these data suggest that γ-toxin requires the canonical anticodon stem loop structure for efficient cleavage, and that ASL with a stable stem structure seems to be preferred by γ-toxin.
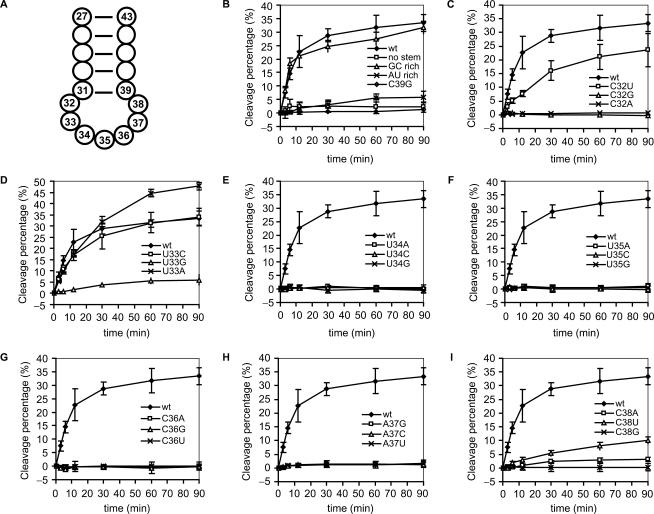
Figure 3.
Cleavage of 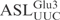 and its derivatives by γ-toxin. (A) Schematic drawing of the anticodon stem loop of tRNA. Positions of nucleotides are numbered according to conventional rules (25). (B–I) The 5′-32P-labeled wild-type and mutant forms of
and its derivatives by γ-toxin. (A) Schematic drawing of the anticodon stem loop of tRNA. Positions of nucleotides are numbered according to conventional rules (25). (B–I) The 5′-32P-labeled wild-type and mutant forms of 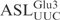 , all at a concentration of 5 µM, were incubated with 5 nM γ-toxin-GST at 0°C. Aliquots of the reactions were taken at the indicated time points and analyzed on 10% polyacrylamide, 8 M urea gels. The extent of cleavage of
, all at a concentration of 5 µM, were incubated with 5 nM γ-toxin-GST at 0°C. Aliquots of the reactions were taken at the indicated time points and analyzed on 10% polyacrylamide, 8 M urea gels. The extent of cleavage of 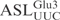 and its mutant derivatives were calculated for each time point and plotted. Each experiment was repeated at least three times, the standard deviations are shown as error bars.
and its mutant derivatives were calculated for each time point and plotted. Each experiment was repeated at least three times, the standard deviations are shown as error bars.
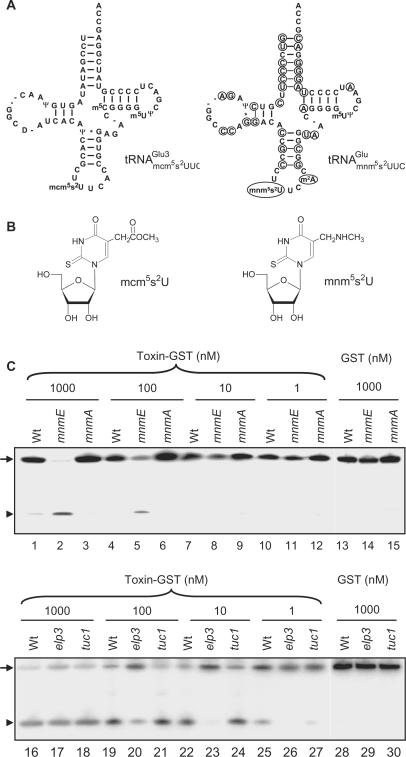
Figure 4.
Effects of mnm5, mcm5 and s2 groups on tRNA cleavage by γ-toxin. (A) Sequences of 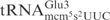 from S. cerevisiae (left) and
from S. cerevisiae (left) and 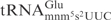 from E. coli (right). The nucleotides deviating from S. cerevisiae
from E. coli (right). The nucleotides deviating from S. cerevisiae
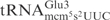 are circled. (B) Structure of mcm5s2U and mnm5s2U. (C) Northern blot analysis of total tRNA isolated from E. coli wild type, mnmE, mnmA and yeast wild type, elp3, or tuc1 strains. In the reactions, 5 µg of total tRNA was incubated with the indicated concentrations of purified γ-toxin-GST or GST protein at 30°C for 10 min. Signals were detected using oligonucleotides specific for E. coli
are circled. (B) Structure of mcm5s2U and mnm5s2U. (C) Northern blot analysis of total tRNA isolated from E. coli wild type, mnmE, mnmA and yeast wild type, elp3, or tuc1 strains. In the reactions, 5 µg of total tRNA was incubated with the indicated concentrations of purified γ-toxin-GST or GST protein at 30°C for 10 min. Signals were detected using oligonucleotides specific for E. coli
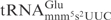 or yeast
or yeast 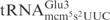 . Full-length and 5′-half of tRNAs are indicated with an arrow and arrow head, respectively.
. Full-length and 5′-half of tRNAs are indicated with an arrow and arrow head, respectively.
Effects of mutations in the anticodon loop on the ASL cleavage by γ-toxin
In the anticodon loop, the three substrate tRNAs, i.e. 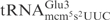 ,
, 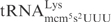 and
and 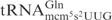 , share identical nucleotides U33U34U35 (25), which might be a sequence important for γ-toxin cleavage. However, ASLs carrying U33U34U35, but with differences in other positions in the anticodon loop, still exhibit different cleavage efficiency (see below). This suggests that other nucleotides are also important for γ-toxin action.
, share identical nucleotides U33U34U35 (25), which might be a sequence important for γ-toxin cleavage. However, ASLs carrying U33U34U35, but with differences in other positions in the anticodon loop, still exhibit different cleavage efficiency (see below). This suggests that other nucleotides are also important for γ-toxin action.
To determine the nucleotides important in the 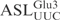 for γ-toxin cleavage, we designed a series of ASLs, each with one of the loop nucleotides changed. The ASLs were 5′-32P-labeled and their cleavage efficiencies examined as described above. Changing C32 to U in
for γ-toxin cleavage, we designed a series of ASLs, each with one of the loop nucleotides changed. The ASLs were 5′-32P-labeled and their cleavage efficiencies examined as described above. Changing C32 to U in 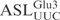 caused a ∼30% drop in cleavage efficiency (Figure 3C), whereas replacement of C32 by A or G abolished cleavage, suggesting that γ-toxin prefers a pyrimidine at position 32. At position 33 almost all tRNAs carry a U, which is required for the formation of U-turn, a signature of the canonical anticodon structure. Replacing U33 by G caused a ∼85% reduction in cleavage, whereas a C33 mutation displayed very similar kinetics as the
caused a ∼30% drop in cleavage efficiency (Figure 3C), whereas replacement of C32 by A or G abolished cleavage, suggesting that γ-toxin prefers a pyrimidine at position 32. At position 33 almost all tRNAs carry a U, which is required for the formation of U-turn, a signature of the canonical anticodon structure. Replacing U33 by G caused a ∼85% reduction in cleavage, whereas a C33 mutation displayed very similar kinetics as the 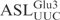 . More surprisingly, ASLGlu3 carrying an A33 mutation was cleaved even more efficiently than the
. More surprisingly, ASLGlu3 carrying an A33 mutation was cleaved even more efficiently than the 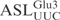 (Figure 3D). Any change of the nucleotides in the anticodon (position 34, 35 and 36) and position 37 abolished ASL cleavage (Figure 3E, F, G and H). Changing C38 to U resulted in a ∼60% reduction in ASL cleavage, while a purine (A or G) at this position led to at least a ∼90% drop in cleavage efficiency (Figure 3I). Taken together, the nucleotides U34U35C36A37C38 in the
(Figure 3D). Any change of the nucleotides in the anticodon (position 34, 35 and 36) and position 37 abolished ASL cleavage (Figure 3E, F, G and H). Changing C38 to U resulted in a ∼60% reduction in ASL cleavage, while a purine (A or G) at this position led to at least a ∼90% drop in cleavage efficiency (Figure 3I). Taken together, the nucleotides U34U35C36A37C38 in the 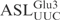 are required for efficient cleavage by γ-toxin. A purine at position 32 or a G at position 33 dramatically reduced the ability of the ASL to act as a substrate.
are required for efficient cleavage by γ-toxin. A purine at position 32 or a G at position 33 dramatically reduced the ability of the ASL to act as a substrate.
The mnm5 group in E. coli
 is a negative element for γ-toxin cleavage
is a negative element for γ-toxin cleavage
In contrast to the mcm5s2U-containing S. cerevisiae
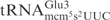 , the E. coli
, the E. coli
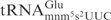 contains a 5-methylaminomethyl-2-thiouridine (mnm5s2U) residue at the wobble position (25). Moreover, the E. coli
contains a 5-methylaminomethyl-2-thiouridine (mnm5s2U) residue at the wobble position (25). Moreover, the E. coli
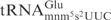 has a 2-methyladenosine (m2A) at position 37, whereas the S. cerevisiae
has a 2-methyladenosine (m2A) at position 37, whereas the S. cerevisiae
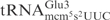 has an unmodified A37 (25). The other nucleotides in the anticodon loop are identical between the two tRNAs (Figure 4A). Total RNA isolated from a wild-type E. coli strain was treated with purified γ-toxin and tRNA cleavage was investigated by northern blot analysis. Compared with the S. cerevisiae
has an unmodified A37 (25). The other nucleotides in the anticodon loop are identical between the two tRNAs (Figure 4A). Total RNA isolated from a wild-type E. coli strain was treated with purified γ-toxin and tRNA cleavage was investigated by northern blot analysis. Compared with the S. cerevisiae
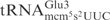 , at least a 1000-fold higher concentration of γ-toxin was required to observe the cleavage of E. coli
, at least a 1000-fold higher concentration of γ-toxin was required to observe the cleavage of E. coli
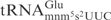 (Figure 4C, compare lane 25 with lane 1). The fact that unmodified E. coli and S. cerevisiae
(Figure 4C, compare lane 25 with lane 1). The fact that unmodified E. coli and S. cerevisiae
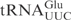 share identical anticodon loop sequence and their corresponding ASLs are cleaved by γ-toxin with the same efficiency (Figure 3B, compare wt and GC rich), makes it likely that mnm5s2U34 and/or m2A37 act negatively on γ-toxin cleavage of E. coli
share identical anticodon loop sequence and their corresponding ASLs are cleaved by γ-toxin with the same efficiency (Figure 3B, compare wt and GC rich), makes it likely that mnm5s2U34 and/or m2A37 act negatively on γ-toxin cleavage of E. coli
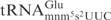 .
.
We individually investigated the effect of the mnm5- or s2-group on cleavage of E. coli
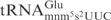 . An E. coli mnmE mutant is defective in the formation of the mnm5 but not the s2 group (26). Similarly, a yeast elp3 mutant lacks the mcm5 side chain but contains the s2 group in tRNA (22). Total tRNA isolated from these mutants and their corresponding wild-type strains were treated with serially diluted γ-toxin and the cleavage of
. An E. coli mnmE mutant is defective in the formation of the mnm5 but not the s2 group (26). Similarly, a yeast elp3 mutant lacks the mcm5 side chain but contains the s2 group in tRNA (22). Total tRNA isolated from these mutants and their corresponding wild-type strains were treated with serially diluted γ-toxin and the cleavage of 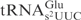 was analyzed by northern blot analysis. Interestingly,
was analyzed by northern blot analysis. Interestingly, 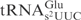 from the mnmE mutant (lacking the mnm5 modification) was more sensitive to γ-toxin than the wild-type
from the mnmE mutant (lacking the mnm5 modification) was more sensitive to γ-toxin than the wild-type 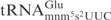 (Figure 4C, compare lane 2 with lane 1, lane 5 with lane 4, respectively), indicating that the mnm5 group has a negative effect on cleavage of
(Figure 4C, compare lane 2 with lane 1, lane 5 with lane 4, respectively), indicating that the mnm5 group has a negative effect on cleavage of 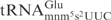 . Estimated by the amount of γ-toxin needed, E. coli
. Estimated by the amount of γ-toxin needed, E. coli
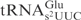 lacking the mnm5 modification served as an equally comparable substrate to yeast
lacking the mnm5 modification served as an equally comparable substrate to yeast 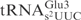 lacking the mcm5 modification (Figure 4C, compare lane 8 and lane 23, lane 5 and lane 20, respectively). This observation has two implications: First, despite sequence variations in other domains, the identical anticodon loop sequence in E. coli and yeast
lacking the mcm5 modification (Figure 4C, compare lane 8 and lane 23, lane 5 and lane 20, respectively). This observation has two implications: First, despite sequence variations in other domains, the identical anticodon loop sequence in E. coli and yeast 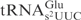 is enough to dictate their similar cleavage efficiencies, reinforcing the importance of previously identified sequence elements important for γ-toxin cleavage. Second, in this scenario, the presence of m2A37 did not make E. coli
is enough to dictate their similar cleavage efficiencies, reinforcing the importance of previously identified sequence elements important for γ-toxin cleavage. Second, in this scenario, the presence of m2A37 did not make E. coli
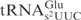 a worse substrate than the yeast
a worse substrate than the yeast 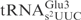 , suggesting that the m2A37 is unlikely to be a negative element in E. coli
, suggesting that the m2A37 is unlikely to be a negative element in E. coli
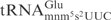 for γ-toxin cleavage.
for γ-toxin cleavage.
The effect of the s2 group was explored using an E. coli mnmA mutant and a yeast tuc1 mutant, both of which lack the s2 modification in tRNA (23,27). When tested with γ-toxin at a concentration of 1000 nM, 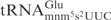 from wild-type E. coli strain was cleaved with low efficiency but
from wild-type E. coli strain was cleaved with low efficiency but 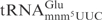 from the mnmA mutant was not detectably cleaved (Figure 4C, compare lane 1 with lane 3). The difference in cleavage between
from the mnmA mutant was not detectably cleaved (Figure 4C, compare lane 1 with lane 3). The difference in cleavage between 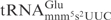 and
and 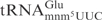 is not dramatic, but was detected repeatedly. A similar tendency was observed using total tRNA isolated from a yeast wild-type strain and a tuc1 mutant, but only at low γ-toxin concentration (Figure 4C, compare lane 25 and lane 27). Taken together, the mcm5 group has a positive effect, while the mnm5 group has a negative effect on cleavage of tRNAGlu. In both yeast and E. coli tRNAs, the s2 group has a weak stimulatory effect on cleavage by γ-toxin.
is not dramatic, but was detected repeatedly. A similar tendency was observed using total tRNA isolated from a yeast wild-type strain and a tuc1 mutant, but only at low γ-toxin concentration (Figure 4C, compare lane 25 and lane 27). Taken together, the mcm5 group has a positive effect, while the mnm5 group has a negative effect on cleavage of tRNAGlu. In both yeast and E. coli tRNAs, the s2 group has a weak stimulatory effect on cleavage by γ-toxin.
DISCUSSION
Sequence determinants for γ-toxin cleavage reside within the anticodon stem loop
The mcm5 group on wobble uridines in tRNA was shown to be important for γ-toxin cleavage (7). However, the three γ-toxin substrate tRNAs are cleaved with different efficiency even though they all contain the same wobble nucleoside (7). This could be attributed to their sequence differences, or other tRNA modification differences, or a combination thereof. For example, 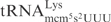 contains t6A37 instead of the unmodified A37 present in
contains t6A37 instead of the unmodified A37 present in 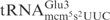 . In addition, there are many sequence differences between these two tRNAs (25). Interestingly, the difference in cleavage was also observed for unmodified in vitro transcribed tRNAs, suggesting that the primary sequence accounts for the different cleavage efficiency of substrate tRNAs (Figure 1B).
. In addition, there are many sequence differences between these two tRNAs (25). Interestingly, the difference in cleavage was also observed for unmodified in vitro transcribed tRNAs, suggesting that the primary sequence accounts for the different cleavage efficiency of substrate tRNAs (Figure 1B).
Using unmodified tRNAs, we have shown that the anticodon stem loop (ASL) contains important sequence information for efficient γ-toxin cleavage (Figure 1B). In addition, an RNA oligonucleotide corresponding to the 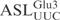 is cleaved in a similar way as the full-length T7-transcribed
is cleaved in a similar way as the full-length T7-transcribed 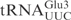 (Figure 2). Assaying sequence variations of
(Figure 2). Assaying sequence variations of 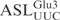 indicated that the canonical stem loop structure is important for γ-toxin cleavage and that γ-toxin may prefer a stable stem structure in the ASL (Figure 3). Of the nucleotides in the
indicated that the canonical stem loop structure is important for γ-toxin cleavage and that γ-toxin may prefer a stable stem structure in the ASL (Figure 3). Of the nucleotides in the 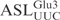 , U34U35C36A37C38 are required for optimal γ-toxin cleavage, whereas a purine at position 32 or a G in position 33 dramatically reduces the cleavage of the ASL (Figure 3). In contrast, a U33 to A mutation stimulated cleavage of the ASL and a U33 to C mutation had no effect. The presence of a U at position 33 is a conserved feature of tRNAs and contributes to a sharp turn of the phosphate backbone, the U-turn, which is crucial for the canonical anticodon loop structure (28). However, lack of U-turn has been reported for unmodified ASLPhe (29) and unmodified ASLLys (30). The modifications normally present in the anticodon region of native tRNAs promote U-turn formation in these ASLs, either by disrupting the intra-loop Watson–Crick base pairs or stabilizing anticodon stacking (29,31,32). Even though the effect of mcm5s2U modification on the structure of ASLGlu is not known, it is conceivable that the unmodified ASLGlu might lack U-turn. Therefore, the different γ-toxin cleavage effects brought about by mutations in position 33 of ASLGlu seem independent of U-turn formation.
, U34U35C36A37C38 are required for optimal γ-toxin cleavage, whereas a purine at position 32 or a G in position 33 dramatically reduces the cleavage of the ASL (Figure 3). In contrast, a U33 to A mutation stimulated cleavage of the ASL and a U33 to C mutation had no effect. The presence of a U at position 33 is a conserved feature of tRNAs and contributes to a sharp turn of the phosphate backbone, the U-turn, which is crucial for the canonical anticodon loop structure (28). However, lack of U-turn has been reported for unmodified ASLPhe (29) and unmodified ASLLys (30). The modifications normally present in the anticodon region of native tRNAs promote U-turn formation in these ASLs, either by disrupting the intra-loop Watson–Crick base pairs or stabilizing anticodon stacking (29,31,32). Even though the effect of mcm5s2U modification on the structure of ASLGlu is not known, it is conceivable that the unmodified ASLGlu might lack U-turn. Therefore, the different γ-toxin cleavage effects brought about by mutations in position 33 of ASLGlu seem independent of U-turn formation.
The ASL also represents the minimal structure required for cleavage by a bacterial anticodon nuclease (ACN), PrrC. However, unlike γ-toxin, PrrC prefers a substrate with partially destabilized stem structure (10). On the other hand, colicin E5, another bacterial ACN, seems to employ a different recognition mechanism. The dinucleotide GpUp corresponding to G34U35, a common moity between the substrate tRNAs, was proposed to be the minimal recognition element for colicin E5 (33). Crystal structure of colicin E5 complexed with the substrate analog dGpdUp revealed a tight binding (34). However, it is noteworthy that the disruption of the stem structure in an RNA oligonucleotide corresponding to the ASL of substrate tRNA reduced the cleavage by colicin E5 (33). It remains to be seen if the dinucleotide GpUp and the ASL are cleaved with similar efficiency.
The effects of wobble uridine modifications on tRNA cleavage by the γ-toxin
Substrate tRNAs lacking part of, or the entire mcm5 side chain are cleaved much less efficiently by γ-toxin, showing the importance of the mcm5 side chain (7). In the present study, we have shown that the s2 group at the wobble position has a positive effect on tRNA cleavage by γ-toxin, even though the effect is less significant than presence of the mcm5 side chain (Figure 4C). On the contrary, the mnm5 side chain that is present on the wobble uridine in E. coli
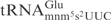 has a negative effect on cleavage. Both mcm5 and mnm5 side chains promote U-turn formation in the anticodon loop of
has a negative effect on cleavage. Both mcm5 and mnm5 side chains promote U-turn formation in the anticodon loop of 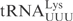 (32), and most likely also in
(32), and most likely also in 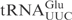 . Therefore, the different effects of these two modifications on the tRNA cleavage by γ-toxin rather suggest that the wobble nucleotide might interact directly with γ-toxin. The positively charged amino group in the mnm5 side chain of
. Therefore, the different effects of these two modifications on the tRNA cleavage by γ-toxin rather suggest that the wobble nucleotide might interact directly with γ-toxin. The positively charged amino group in the mnm5 side chain of 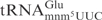 (32) might interfere with binding and/or catalysis by γ-toxin. A direct contact between tRNA endonuclease and wobble nucleotide has also been proposed for the interaction between PrrC and its substrate
(32) might interfere with binding and/or catalysis by γ-toxin. A direct contact between tRNA endonuclease and wobble nucleotide has also been proposed for the interaction between PrrC and its substrate 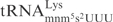 (10,11). Interestingly, although all substrate tRNAs of colicin E5 contain the modified nucleotide queuosine (Q) at the wobble position, it seems that Q is not important for cleavage (12). Consequently, based on the crystal structure of colicin E5 in complex with the substrate analog, it was proposed that Q base does not interact with colicin E5 (34). Obtaining the crystal structure of -toxin complexed with substrate tRNA would provide the detailed picture of the interaction between tRNA and -toxin.
(10,11). Interestingly, although all substrate tRNAs of colicin E5 contain the modified nucleotide queuosine (Q) at the wobble position, it seems that Q is not important for cleavage (12). Consequently, based on the crystal structure of colicin E5 in complex with the substrate analog, it was proposed that Q base does not interact with colicin E5 (34). Obtaining the crystal structure of -toxin complexed with substrate tRNA would provide the detailed picture of the interaction between tRNA and -toxin.
ACKNOWLEDGEMENTS
We thank Dr T. Hagervall for E. coli strains and acknowledge Drs M.J.O. Johansson, G.R. Björk, M.S. Francis and T. Hagervall for critical reading of the manuscript. This work was financially supported by grants from the Swedish Cancer Foundation (Project 3516-B05-12XAB), Swedish Research Council (Project 621-2006-4269) and Margareta Dannbergs Foundation (Project 223-302-06). Funding to pay the Open Access publication charges for this article was provided by Swedish Cancer Foundation, Swedish Research Council, and Margareta Dannbergs Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gunge N, Sakaguchi K. Intergeneric transfer of deoxyribonucleic acid killer plasmids, pGKl1 and pGKl2, from Kluyveromyces lactis into Saccharomyces cerevisiae by cell fusion. J. Bacteriol. 1981;147:155–160. doi: 10.1128/jb.147.1.155-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugisaki Y, Gunge N, Sakaguchi K, Yamasaki M, Tamura G. Kluyveromyces lactis killer toxin inhibits adenylate cyclase of sensitive yeast cells. Nature. 1983;304:464–466. doi: 10.1038/304464a0. [DOI] [PubMed] [Google Scholar]
- 3.White JH, Butler AR, Stark MJR. Kluyveromyces lactis toxin does not inhibit yeast adenylyl cyclase. Nature. 1989;341:666–668. [Google Scholar]
- 4.Schaffrath R, Meinhardt F. Kluveromyces lactis zymocin and other plasmid-encoded yeast killer toxins. Top Curr. Genet. 2005;11:133–155. [Google Scholar]
- 5.Stark MJ, Boyd A, Mileham AJ, Romanos MA. The plasmid-encoded killer system of Kluyveromyces lactis: a review. Yeast. 1990;6:1–29. doi: 10.1002/yea.320060102. [DOI] [PubMed] [Google Scholar]
- 6.Butler AR, Porter M, Stark MJ. Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast. 1991;7:617–625. doi: 10.1002/yea.320070610. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Huang B, Esberg A, Johansson MJ, Byström AS. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Y, Blanga S, Amitsur M, Meidler R, Krivosheyev E, Sundaram M, Bajji AC, Davis DR, Kaufmann G. Structural features of tRNALys favored by anticodon nuclease as inferred from reactivities of anticodon stem and loop substrate analogs. J. Biol. Chem. 2002;277:3836–3841. doi: 10.1074/jbc.M110072200. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, Meidler R, Amitsur M, Kaufmann G. Specific interaction between anticodon nuclease and the tRNALys wobble base. J. Mol. Biol. 2001;305:377–388. doi: 10.1006/jmbi.2000.4282. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- 13.Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl Acad. Sci. USA. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YL, Elias Y, Huang RH. Structural and mutational studies of the catalytic domain of colicin E5: a tRNA-specific ribonuclease. Biochemistry. 2005;44:10494–10500. doi: 10.1021/bi050749s. [DOI] [PubMed] [Google Scholar]
- 15.Graille M, Mora L, Buckingham RH, van Tilbeurgh H, de Zamaroczy M. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 2004;23:1474–1482. doi: 10.1038/sj.emboj.7600162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Oyanedel J, Choe HW, Heinemann U, Saenger W. Ribonuclease T1 with free recognition and catalytic site: crystal structure analysis at 1.5 Å resolution. J. Mol. Biol. 1991;222:335–352. doi: 10.1016/0022-2836(91)90215-r. [DOI] [PubMed] [Google Scholar]
- 17.Takagi H, Kakuta Y, Okada T, Yao M, Tanaka I, Kimura M. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat. Struct. Mol. Biol. 2005;12:327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- 18.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Byström AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avital S, Elson D. A convenient procedure for preparing transfer ribonucleic acid from Escherichia coli. Biochim. Biophys. Acta. 1969;179:297–307. doi: 10.1016/0005-2787(69)90038-0. [DOI] [PubMed] [Google Scholar]
- 21.Johansson MJ, Byström AS. The Saccharomyces cerevisiae TAN1 gene is required for N4-acetylcytidine formation in tRNA. RNA. 2004;10:712–719. doi: 10.1261/rna.5198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B, Johansson MJ, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagervall TG, Pomerantz SC, McCloskey JA. Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypomodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol. 1998;284:33–42. doi: 10.1006/jmbi.1998.2162. [DOI] [PubMed] [Google Scholar]
- 25.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elseviers D, Petrullo LA, Gallagher PJ. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 1984;12:3521–3534. doi: 10.1093/nar/12.8.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan MA, Cannon JF, Webb FH, Bock RM. Antisuppressor mutation in Escherichia coli defective in biosynthesis of 5-methylaminomethyl-2-thiouridine. J. Bacteriol. 1985;161:368–376. doi: 10.1128/jb.161.1.368-376.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 29.Cabello-Villegas J, Winkler ME, Nikonowicz EP. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J. Mol. Biol. 2002;319:1015–1034. doi: 10.1016/S0022-2836(02)00382-0. [DOI] [PubMed] [Google Scholar]
- 30.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J. Mol. Biol. 1999;285:115–131. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 31.Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000;39:13396–13404. doi: 10.1021/bi0013039. [DOI] [PubMed] [Google Scholar]
- 32.Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry. 2005;44:8078–8089. doi: 10.1021/bi050343f. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa T, Inoue S, Yajima S, Hidaka M, Masaki H. Sequence-specific recognition of colicin E5, a tRNA-targeting ribonuclease. Nucleic Acids Res. 2006;34:6065–6073. doi: 10.1093/nar/gkl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res. 2006;34:6074–6082. doi: 10.1093/nar/gkl729. [DOI] [PMC free article] [PubMed] [Google Scholar]