Abstract
MicroRNA-122 (miR-122) is an abundant liver-specific miRNA, implicated in fatty acid and cholesterol metabolism as well as hepatitis C viral replication. Here, we report that a systemically administered 16-nt, unconjugated LNA (locked nucleic acid)-antimiR oligonucleotide complementary to the 5′ end of miR-122 leads to specific, dose-dependent silencing of miR-122 and shows no hepatotoxicity in mice. Antagonism of miR-122 is due to formation of stable heteroduplexes between the LNA-antimiR and miR-122 as detected by northern analysis. Fluorescence in situ hybridization demonstrated uptake of the LNA-antimiR in mouse liver cells, which was accompanied by markedly reduced hybridization signals for mature miR-122 in treated mice. Functional antagonism of miR-122 was inferred from a low cholesterol phenotype and de-repression within 24 h of 199 liver mRNAs showing significant enrichment for miR-122 seed matches in their 3′ UTRs. Expression profiling extended to 3 weeks after the last LNA-antimiR dose revealed that most of the changes in liver gene expression were normalized to saline control levels coinciding with normalized miR-122 and plasma cholesterol levels. Combined, these data suggest that miRNA antagonists comprised of LNA are valuable tools for identifying miRNA targets in vivo and for studying the biological role of miRNAs and miRNA-associated gene-regulatory networks in a physiological context.
INTRODUCTION
MicroRNAs (miRNAs) are an abundant class of short endogenous non-coding RNAs that act as important post-transcriptional regulators of gene expression by base-pairing to their target mRNAs, thereby mediating mRNA cleavage or translational repression (1). An increasing body of research shows that animal miRNAs play fundamental roles in cell growth, development and differentiation (1,2). Recent data suggest that miRNAs are aberrantly expressed in many human cancers and that they may play significant roles as oncogenes or tumour suppressors (3–6). Apart from cancer, miRNAs have also been linked to several other diseases. For example, a mutation in the target site of miR-189 in the human SLITRK1 gene was shown to be associated with Tourette's syndrome (7), while other recent studies have implicated miRNAs in controlling HIV replication (8) and in coronary artery disease (9). Hence, disease-associated human miRNAs could represent a novel group of viable targets for therapeutic intervention. One such example is miR-122, an abundant liver-specific miRNA, with suggested roles in cholesterol, fatty acid and lipid metabolism (10,11). It has also been shown that miR-122 interacts with the hepatitis C virus genome facilitating viral replication in the host cell (12).
A major challenge in understanding the biological functions of miRNAs in animal development and human disease is to identify their target mRNAs. Although computational analyses suggest that miRNAs may be responsible for regulating up to 30% of the human protein-coding genes (13–15), only a few target genes have been experimentally confirmed (16). Microarray expression profiling has been used to detect genes down-regulated in response to exogenous miRNAs (17). However, introduction of an exogenous miRNA into cells that do not normally express it may lead to identification of non-physiological targets. In contrast, specific inhibition of endogenous miRNAs in vivo using chemically modified antisense oligonucleotides has the potential to pinpoint the physiological targets and their sequence determinants. Furthermore, development of miRNA-targeting oligonucleotides with enhanced pharmacological activity and optimized pharmacokinetic properties holds promise as therapeutic agents against disease-associated miRNAs. LNAs (locked nucleic acids) comprise a class of bicyclic conformational analogues of RNA, which exhibit high binding affinity to complementary RNA target molecules and high stability in blood and tissues in vivo (18,19). The unprecedented thermal stability of short LNA-modified oligonucleotide probes together with their improved mismatch discrimination has enabled sensitive and specific miRNA detection by northern blot analysis and by in situ hybridization (ISH) in developing animal embryos and tissue sections (20–24). LNA oligonucleotides have also been shown to mediate potent and specific inhibition of miRNA function in vitro (25–27). In the present study, we set out to assess the utility of LNA-modified oligonucleotides in silencing of miRNAs in vivo by antagonizing miR-122 in the murine liver. We report here that a systemically administered, 16 nt, unconjugated LNA-antimiR oligonucleotide, complementary to the 5′ end of miR-122 leads to specific and dose-dependent miRNA-122 antagonism in mice. Our data suggest that miR-122 regulates the expression of a large number of target mRNAs in adult mouse liver. Most of the identified miR-122 targets showed only slight to moderate de-repression implying that miR-122 might work by fine-tuning several liver gene-regulatory networks.
MATERIALS AND METHODS
Design and synthesis of LNA oligonucleotides
The LNA oligonucleotides were synthesized as unconjugated and fully phosphorothiolated oligonucleotides. The perfectly matching LNA-antimiR oligonucleotide: 5′-ccAttGtcAcamCtcmCa-3′ (uppercase: LNA; lowercase: DNA; mC denotes methyl cytosine) was complementary to nucleotides 1–16 in the mature miR-122 sequence. The mismatch control LNA oligonucleotide was synthesized with the following sequence: 5′-ccAttGtcTcaAtcmCa-3′.
In vivo experiments
NMRI female mice (Taconic M&B Laboratory Animals, Ejby, Denmark) with 27 g average body weight at first dosing were used in all experiments and received regular chow diet (Altromin no 1324, Brogaarden, Gentofte, Denmark). All substances were formulated in physiological saline (0.9% NaCl) to final concentration allowing the mice to receive a tail vein injection volume of 10 ml/kg. The animals were dosed for three consecutive days with LNA-antimiR, LNA mismatch control or saline (0.9% NaCl), receiving daily doses from 2.5 to 25 mg/kg. In the dose–response study, the mice were sacrificed 24 h after last dose, whereas the mice in the duration of action study were sacrificed 1, 2 or 3 weeks after last dose, respectively.
Real-time quantitative RT-PCR
The dissected mice livers were immediately stored in RNA later (Ambion). Total RNA was extracted with Trizol reagent according to the manufacturer's instructions (Invitrogen), except that the precipitated RNA pellet was washed in 80% ethanol and not vortexed.
The miR-122 and let-7a levels were quantified with mirVana real-time RT-PCR detection kit (Ambion) following the manufacturer's instructions, except that 200-ng total RNA was used in the reverse transcription (RT) reaction. The RT reaction was diluted 10 times in water and 10 µl of aliquots were subsequently used for RT-PCR amplification according to the manufacturer's instructions (Ambion). A 2-fold total RNA dilution series from untreated mouse liver RNA served as standard to ensure a linear range (Ct versus relative copy number) of the amplification.
mRNA quantification of selected genes was done using standard TaqMan assays (Applied Biosystems). The reverse transcription reaction was carried out with random decamers, 0.5 μg total RNA and the M-MLV RT enzyme from Ambion according to protocol. First-strand cDNA was subsequently diluted 10 times in nuclease-free water before addition to the RT-PCR reaction mixture. The Applied Biosystems 7500 real-time PCR instrument was used for amplification. A 2-fold total RNA dilution series from a saline-treated animal served as standard to ensure a linear range (Ct versus relative copy number) of the amplification.
Determination of duplex melting temperature and stability in mouse plasma
Twenty-three microlitres of 15 µM pre-annealed LNA-antimiR:miR-122 duplex or 30 µM LNA-antimiR solution were mixed with 100 µl mouse plasma (Lithium heparin plasma from adult BomTac:NMRI female mice, Taconic, Ry, Denmark) and incubated at 37°C. Twenty microlitre aliquots were taken at the different time points and frozen. Samples were diluted in equal volume of (20 µl) 0.1 M Tris, pH 7.6 and phenol extracted with 40 µl phenol/chloroform/isoamyl alcohol (25:24:1, saturated w. 10 mM Tris, pH 8, 1 mM EDTA). The samples were separated on a 20% non-denaturing TBE gel (Novex, Invitrogen). The nucleic acids were visualized by staining the gel in SYBR Gold (Molecular Probes) and analysed with a Bio-Rad Molecular Imager FX using QuantityOne software. The oligonucleotide:miR-122 duplexes were diluted to 3 µM in 500 µl RNase-free H20 and mixed with 500 µl 2× Tm-buffer (200 mM NaCl, 0.2 mM EDTA, 20 mM Na-phosphate, pH 7.0). The solution was heated to 95°C for 3 min and then allowed to anneal in room temperature for 30 min. The duplex melting temperatures (Tm) were measured on a Lambda 40 UV/VIS Spectrophotometer equipped with a Peltier temperature programmer PTP6 using PE Templab software (Perkin Elmer). The temperature was ramped up from 20°C to 95°C and then down to 25°C, recording absorption at 260 nm. First derivative and the local maximums of both the melting and annealing were used to assess the duplex Tm.
Northern blot analysis
Mouse liver total RNAs (10 µg per sample), were electrophoresed in formamide loading buffer (47.5% formamide, 9 mM EDTA, 0.0125% Bromophenol Blue, 0.0125% Xylene Cyanol, 0.0125% SDS) in 15% denaturing Novex TBE-Urea polyacrylamide gels (Invitrogen) without pre-heating the RNA. The total RNAs were transferred to GeneScreen Plus Hybridization Transfer Membrane (PerkinElmer) at 200 mA for 35 min and probed with 32P-labelled LNA-modified oligonucleotide probes for miR-122 and let-7a (Exiqon, Denmark). LNA probes were labelled and hybridized to the membrane as described (23) with the following modifications: pre-hybridization and hybridization solutions contained 50% formamide, 0.5% SDS, 5× SSC, 5× Denhardt's solution and 20 µg/ml sheared denatured herring sperm DNA. Hybridizations were performed at 45°C. The blots were visualized by scanning in a Storm 860 scanner (Molecular Dynamics). The Decade Marker System (Ambion) was used as size marker.
Construction of 3′untranslated region (UTR) reporter plasmids and luciferase assays
HeLa cells were cultivated in Eagles MEM with 10% FBS, 2 mM Glutamax I, non-essential amino acids and 25 µg/ml Gentamicin (Invitrogen). The 3′UTRs of four predicted miR-122 targets were cloned downstream of the Renilla luciferase gene (XhoI/NotI sites) in the psiCheck-2 plasmid (Promega). PCR primers used for amplification of the 3′ UTRs of the four predicted miR-122 targets were (5′ to 3′): AldoA: forward ccctcgagccagagctgaactaaggctgc (incl. a XhoI site), reverse gggcggccgcggcagtgggctggaggg (incl. a NotI site), Bckdk: forward ccctcgagtccgcatcaacggacat, reverse aatgcggccgcaaactttaataagcagcagg, Cd320: forward aatctcgagggacacatggttaccacctg, reverse aatgcggccgccaatattttcttgttttctg, Ndrg3: forward aatctcgagagatctcctcccctggacca, reverse aatgcggccgctaaaaagtcacactctggcagagtg.
HeLa cells were co-transfected with a miR-122 mimic (miRIDIAN mimic, Dharmacon) and target reporter plasmid using Lipofectamine 2000 (Invitrogen). The transfections and luciferase activity measurements were carried out according to the manufacturer's instructions (Invitrogen Lipofectamine 2000/Promega Dual-luciferase kit). Relative protein levels were expressed as Renilla/Firefly luciferase ratios.
Western blot analysis
Liver proteins extracted from treated mice were separated on NuPAGE Bis Tris 4–12% (Invitrogen) using 100 µg protein per sample. The proteins were transferred to a nitrocellulose membrane using iBlot (Invitrogen) according to manufacturer's instructions. ECL advanced western kit (GE Healthcare Life Sciences) was used for blocking, antibody dilution and detection according to the manufacturer. A primary goat-anti-AldoA antibody (SC-12061, Santa Cruz Biotechnology) and an HRP-conjugated secondary rabbit anti-goat (PD449, DAKO) were used according to manufacturer's instructions.
Plasma cholesterol and transaminase measurements
Immediately before sacrifice, retro-orbital sinus blood was collected in EDTA-coated tubes followed by isolation of the plasma fraction. Total plasma cholesterol was analysed using ABX Pentra Cholesterol CP (Horiba Group, Horiba ABX Diagnostics) according to the manufacturer's instructions. Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in the mouse serum were measured using an enzymatic ALT/AST CP assay (Horiba ABX Diagnostics, France) according to the manufacturer's instructions. The measurements were carried out in duplicates and were correlated to a 2-fold diluted standard curve generated from an ABX Pentra MultiCal solution (Horiba ABX Diagnostics, France).
ISH and histological analysis
In situ detection of miR-122 was performed on 8 μm frozen liver sections of LNA-antimiR treated and saline control mice. Slides were thawed, fixed in 4% paraformaldehyde for 10 min at room temperature and treated in acetic anhydride/triethanolamine followed by rinsing in PBS after each treatment. Slides were pre-hybridized in hybridization solution (50% formamide, 5× SSC, 500 μg/ml yeast tRNA, 1× Denhardt's solution) at 48°C for 30 min. Three picomoles of LNA-modified oligonucleotide probe (Exiqon) complementary to miR-122 was labelled (DIG–oligonucleotide 3′ Tailing Kit, Roche Applied Sciences, USA) and hybridized to the liver sections for 1 h at 48°C. After post-hybridization washes in 0.1× SSC at 55°C, the ISH signals were detected using the tyramide signal amplification system (Perkin Elmer USA) according to the manufacturer's instructions.
Detection of the LNA-antimiR in the liver sections involved ISH employing a 5′ FAM-labelled complementary LNA probe: 5′-tGgaGtgTgamCaaTgg-3′. After a 15-min pre-hybridization step, the LNA probe was hybridized to the liver sections for 5 min at the same temperature followed by a 3 × 10-min post-hybridization wash in 0.1× SSC at 55°C. The slides were mounted in Prolong Gold containing DAPI (Invitrogen) and analysed on a Leica epifluorescence microscope equipped with a CCD camera (Leica Microsystems) and Leica CW4000 CytoFISH software.
Histology of the mouse liver samples was carried out by fixation of the cryosections in 4% formaldehyde for 10 min, followed by three washes in distilled water and staining in Mayer's haematoxylin for 1 min. The sections were rinsed in tap water for 10 min and then quickly washed in distilled water followed by staining in 0.2% eosin for 30 s. The slides were washed in distilled water, dehydrated, mounted with coverslips and analysed in a light microscope.
Gene expression profiling
Total liver RNAs were extracted from mice treated with 25 mg/kg/day of LNA-antimiR or saline for three consecutive days and sacrificed 24 h, 1, 2 or 3 weeks after last dose as well as from mice treated with LNA mismatch control sacrificed 24 h after last dose. RNA quality and concentration was measured using an Agilent 2100 Bioanalyzer and Nanodrop ND-1000, respectively. Total RNA was processed following the GeneChip Expression 3′-Amplification Reagents One-cycle cDNA synthesis kit instructions (Affymetrix Inc, Santa Clara, CA, USA) to produce double-stranded cDNA. This was used as a template to generate biotin-labelled cRNA following manufacturer's specifications. Fifteen micrograms of biotin-labelled cRNA was fragmented and 10 µg were hybridized onto Affymetrix Mouse Genome 430 2.0 arrays overnight in the GeneChip Hybridisation oven 6400 using standard procedures. The arrays were washed and stained in a GeneChip Fluidics Station 450. Scanning was carried out using the GeneChip Scanner 3000 and image analysis was performed using GeneChip Operating Software. Normalization and statistical analysis were done using the LIMMA software package for the R programming environment (28,29). Probes reported as absent by GCOS software in all hybridizations were removed from the dataset. Additionally, an intensity filter was applied to the dataset to remove probes displaying background-corrected intensities below 16. Data were normalized using quantile normalization (30). Differential expression was assessed using a linear model method. P-values were adjusted for multiple testing using the Benjamini and Hochberg method. Tests were considered to be significant if the adjusted P-values were P < 0.05. Clustering and visualization of Affymetrix array data were done using the MultiExperiment Viewer software (31). The differentially regulated genes found in the expression studies were analysed for enriched GO-terms using the DAVID online tool (32).
miRNA target site analysis
Affymetrix probe set identifiers were mapped to Ensembl transcripts using annotation files provided by Affymetrix and the Ensembl Biomart system. Transcript sequences were extracted from Ensembl using the ensembl–Perl–API (33). Target prediction algorithms were applied on full-length transcripts taking note whether the first base of a site occurred in 5′UTR, coding sequence or 3′UTR. The following prediction methods were used: Our own implementation of TargetScanS (15) finding seed matches (perfect Watson–Crick matches between the 6mer from base 2 to 7 of the miRNA from the 5′ end) with none or one of three kinds of extensions: (i) an A across from nucleotide 1 in the miRNA (seedM+t1A), (ii) an additional match between the site and nucleotide 8 in the miRNA (seedM+m8M) and (iii), the combination of (i) and (ii) (seedM+m8M+t1A). miRanda (34) was downloaded from www.microrna.org and run with default parameter setting. RNAhybrid (35) was downloaded from http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/and run with P-value cutoff of 0.05 using parameters calculated from human 3′UTRs.
RESULTS AND DISCUSSION
Silencing of miRNA-122 in vitro and in vivo by short, unconjugated LNA-antimiR
Previously, 2′-O-methyl antisense oligonucleotides have been used as potent and irreversible inhibitors of siRNA and miRNA function in vitro and in Drosophila and Caenorhabditis elegans, thereby inducing a loss-of-function phenotype (36,37). This method was recently applied to mice, where cholesterol-conjugated 2′-O-methyl antisense oligonucleotides (antagomirs) were used to silence miRNAs, including miR-122 (11,38). In a similar study, unconjugated 2′ MOE antisense oligonucleotides were used to inhibit miR-122 in both normal and diet-induced obese mice (10). While complementary LNA oligonucleotides have been shown to mediate specific miRNA inhibition in vitro (25–27), their utility in antagonizing miRNAs in vivo has not been addressed to date. A potential advantage of LNA is its high affinity to miRNA as described by Kloosterman et al. (20) in which highly sensitive and specific detection of miR-206 and miR-124a in zebrafish embryos by whole-mount ISH could be achieved using shortened LNA versions (16- to 14-mers) complementary to the 5′-end of the miRNA. In this study, we designed a 16-nt mixed LNA/DNA oligonucleotide (LNA-antimiR) complementary to the 5′ region of miR-122 similar to the LNA probes reported by Kloosterman et al. (20), by substituting every third nucleotide position with an LNA monomer. Since phosphorothioate modifications have been shown to provide good pharmacokinetic and tissue uptake properties in antisense oligonucleotides along with protection against nucleases (39,40), we synthesized the LNA-antimiR as an unconjugated oligonucleotide with a complete phosphorothioate backbone.
We first evaluated miR-122 inhibition by LNA-antimiR in Huh-7 cells, which express miR-122. Treatment of Huh-7 cells with 1, 10 and 100 nM LNA-antimiR revealed dose-dependent reduction of miR-122 as measured by miR-122-specific RT-PCR compared to unaltered miR-122 levels observed with a double mismatch LNA control oligonucleotide (Supplementary Figure 1). Previous studies have shown that endogenous miRNAs are effective in cleaving reporter mRNAs harbouring miRNA recognition sequences with perfect complementarity to the miRNA (41,42). Hence, we cloned a perfectly complementary miR-122 target site into the 3′ UTR of the Renilla luciferase reporter in the psiCHECK 2 vector, which contains both the Renilla and firefly luciferase genes in the same plasmid. As expected, introduction of the miR-122 sensor into Huh-7 cells resulted in repression of the Renilla luciferase compared to the control reporter without a miR-122 target site. By contrast, transfection of the miR-122 sensor plasmid along with the LNA-antimiR led to dose-dependent de-repression of the Renilla luciferase activity in Huh-7 cells, which was not detected with the LNA mismatch control (Supplementary Figure 2). Considered together, these results confirm effective and specific miR-122 inhibition in vitro by the LNA-antimiR oligonucleotide.
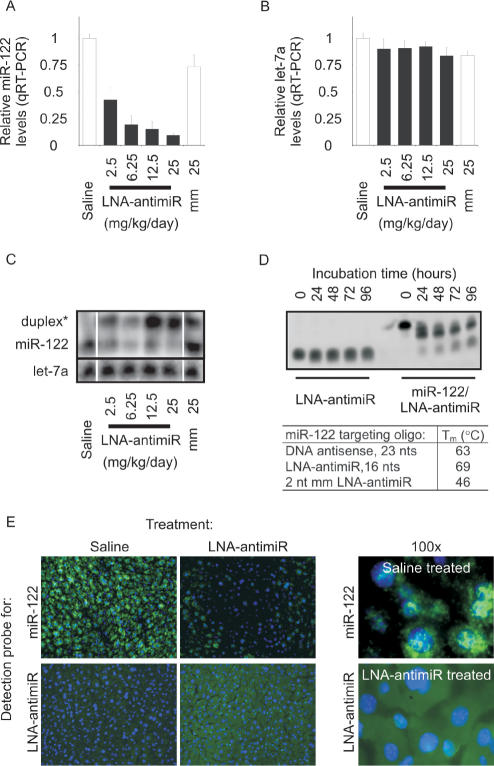
Next, we asked whether the LNA-antimiR could be used to antagonize miR-122 in mice. The LNA-antimiR compound was systemically administered to mice by single intravenous injections on three consecutive days using doses ranging from 2.5 to 25 mg/kg/day. Subsequent real-time RT-PCR analysis of miR-122 levels in total RNA samples extracted from LNA-antimiR-treated mice livers 24 h after last dose revealed a dose-dependent reduction of mature miR-122 (Figure 1A). In contrast, the similarly administered mismatch control LNA dosed at 25 mg/kg/day did not show significant miR-122 reduction, indicating that miR-122 inhibition in vivo by the LNA-antimiR is specific, which is also consistent with our findings in vitro. By comparison, control RT-PCR assays for let-7a, which is also expressed in the liver showed largely unchanged levels across all animal groups (Figure 1B).
Figure 1.
Silencing of miR-122 in mouse liver by LNA-antimiR. (A, B) Real-time RT-PCR assessment of miR-122 and let-7a levels in LNA-antimiR-treated mice livers. The mice were treated with indicated doses of LNA-antimiR along with a mismatch control (mm) and saline for three consecutive days and sacrificed 24 h after last dose. miR-122 and let-7a levels were normalized to the saline-treated group. Shown are mean and SEM, n = 10. (C) Northern blots were probed with a miR-122 specific probe (upper panel) and re-probed with a let-7a specific probe (lower panel). Two miR-122 bands were detected, a lower band corresponding to mature miR-122 and an upper band to a duplex between the LNA-antimiR and miR-122. (D) The stability of the LNA-antimiR and LNA-antimiR:miR-122 duplex was assessed in mouse plasma at 37°C over 96 h. Shown is a SYBR-Gold stained PAGE. The melting temperature (Tm) of the LNA-antimiR:miR-122 duplex was measured along with mismatch control and an unmodified DNA oligonucleotide complementary to miR-122. (E) In situ detection of miR-122 and the LNA-antimiR oligonucleotide in the mouse liver sections. Positive in situ hybridization signals for miR-122 and LNA-antimiR, respectively, are visualized in green, while blue depicts DAPI nuclear stain. 100× magnifications reveal subcellular distribution of miR-122 and LNA-antimiR.
miRNA-122 antagonism by LNA-antimiR leads to stable heteroduplex formation
To better understand the mechanism of miR-122 inhibition by LNA-antimiR, mouse liver RNA samples were subjected to northern blot analysis. The northern results showed significantly reduced levels of mature miR-122 in LNA-antimiR-treated liver RNA, which concurs with our real-time RT-PCR results, whereas the levels of the let-7a control were not altered (Figure 1C). Silencing of miR-122 in mice as reported previously by Krutzfeldt et al. (11,38) using a cholesterol-conjugated 2′-O-Me antagomir and by Esau et al. (10) using 2′-MOE ASO inferred degradation of the targeted miRNA. By contrast, we observed dose-dependent accumulation of a shifted miR-122:LNA-antimiR heteroduplex band, implying that the LNA-antimiR binds stably to the miRNA, thereby antagonizing its function. The fact that a preformed miR-122:LNA-antimiR duplex was stable in mouse plasma over 96 h (Figure 1D) together with the high thermal stability of the LNA-antimiR:miR-122 duplex supports our notion that inhibition of miR-122 by the LNA-antimiR is due to high affinity duplex formation between the two molecules, which is also consistent with recent observations in cell culture (43).
We assessed these findings further by in situ detection of both miR-122 and the LNA-antimiR molecule in frozen liver sections from LNA-antimiR-treated (25 mg/kg) and saline control mice. ISH in frozen liver sections from control animals showed high miR-122 accumulation over the entire liver section, whereas those from the LNA-antimiR-treated mice showed significantly reduced staining for miR-122 (Figure 1E). On the other hand, the LNA-antimiR was readily detected in the liver sections of treated mice, but not in untreated controls. At higher magnification, the mature miR-122 could be localized in distinct cytoplasmic compartments in the control liver sections. By comparison, the LNA-antimiR was evenly distributed in the entire cytoplasm (Figure 1E, right panels), with occasional staining observed also in some, but not all nuclei (Supplementary Figure 3). Combined, these results demonstrate uptake of the systemically administered LNA-antimiR compound by murine liver. Furthermore, our data support the conclusion that antagonism of miR-122 by LNA-antimiR in mouse liver cells is due to formation of stable heteroduplexes between the LNA-antimiR and mature miR-122 in the cytoplasm accompanied by markedly reduced mature miR-122 levels as detected by real-time RT-PCR, northern blots and ISH.
Analysis of miR-122 target mRNAs in the mouse liver
Since most animal target sites are located in the 3′ UTRs and show only limited complementarity to the miRNA (1), precise and reliable experimental identification of targets for miRNAs remains a major bottleneck. Recent studies have reported significant enrichment of predicted target sites in messages that are up-regulated upon miRNA silencing in vivo (10,11,38). Hence, we set out to identify genes with altered transcript levels in response to miR-122 antagonism by LNA-antimiR in murine liver. To this end, we carried out genome-wide expression profiling of total RNA samples from saline, LNA-antimiR and LNA-mismatch-treated mice livers 24 h after the last dose using Affymetrix Mouse Genome arrays. Analysis of the array data revealed 455 up-regulated Affymetrix probe sets corresponding to 395 distinct ENSEMBL genes in the LNA-antimiR-treated mice livers compared to saline and LNA mismatch controls (Figure 2A and Supplementary Table 1). Next, we examined the 3′ UTRs for the presence of the 6-nt sequence CACTCC, which is the reverse complement of the nucleotide 2–7 seed in the mature miR-122 sequence. The number of mRNAs having at least one miR-122 recognition sequence was 199 (50%), while the seed match frequency in all annotated mouse 3′ UTRs was only 21%, implying that a significant pool of the up-regulated mRNAs correspond to de-repressed, direct miR-122 targets in the liver. It is noteworthy that 74% of the mRNAs identified as up-regulated in both this study and in that of Krutzfeldt et al. (11) 24 h after LNA-antimiR or 2′ O-Me antagomir-122 treatment, respectively, have at least one 6-nt seed match in their 3′ UTRs (Supplementary Table 2). In our study, we find a 2.4-fold enrichment for 6-nt seed matches in the up-regulated 3′ UTRs (odds ratio: 2.3, P-value: 1.4E-11, Supplementary Table 3).
Figure 2.
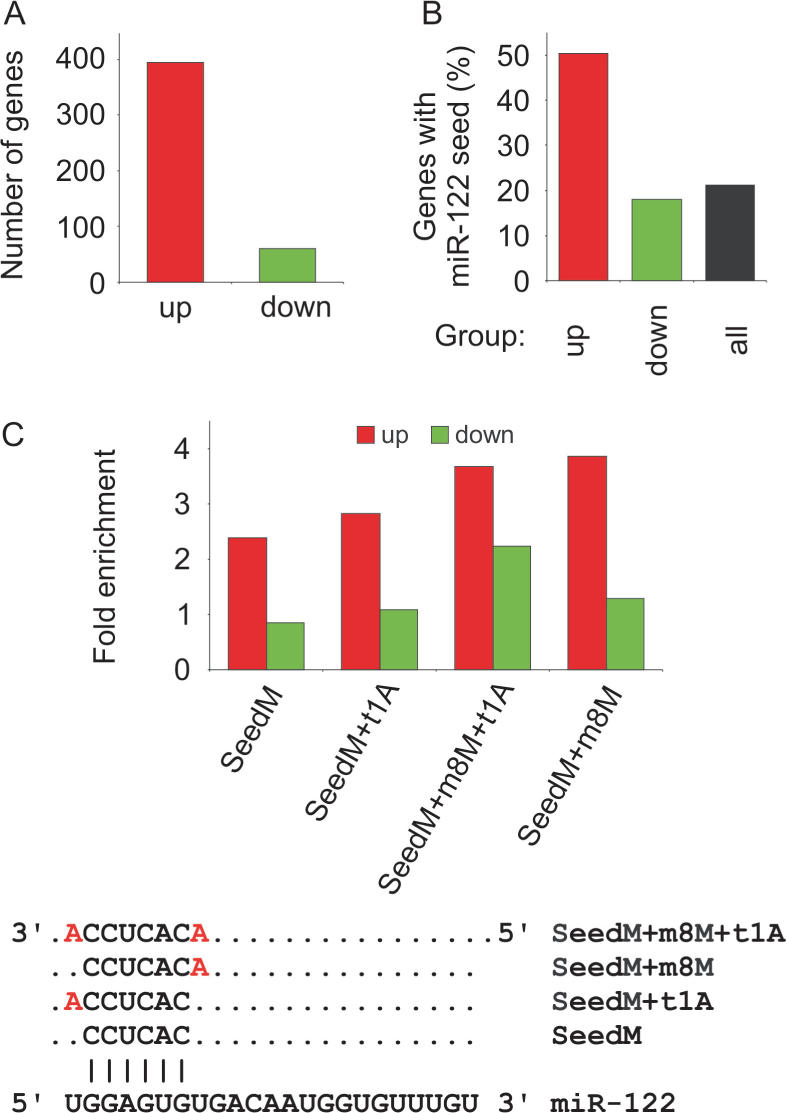
Enrichment of miR-122 target sites in genes up-regulated after LNA-antimiR treatment. (A) The number of significantly (P < 0.05) up- or down-regulated genes (red and green, respectively) in LNA-antimiR-treated mice livers compared to saline and LNA-mismatch-treated mice. (B) The occurrence of miR-122 6-nt seed sequence matches in differentially expressed genes and in all mouse ENSEMBL annotated genes. (C) Enrichment of different seed types (as shown in lower panel) in up- and down-regulated genes relative to all genes.
Detailed in vitro studies using over-expression of miRNAs indicate that the presence of extended seed matches increases the likelihood that a given message is regulated by a miRNA (15,44). We therefore examined the identified seed matches for the presence of an A anchor corresponding to the 5′ most nucleotide of miR-122, as well as for an extended match to base 8 of miR-122 as described by Lewis et al. (15). Both types of 7-nt seed matches were significantly enriched in the de-repressed 3′ UTRs, whereas combined requirement of the A anchor position and M8 match did not show further enrichment in our mouse liver data set (Figure 2C). As a control, we also investigated the frequency of sites for the unrelated let-7a miRNA among the up-regulated mRNAs. Compared to miR-122 seed sites whose enrichment were statistically highly significant, we observed no enrichment of let-7a seed matches in our dataset, which is in good agreement with specific antagonism of miR-122 by LNA-antimiR leading to de-repression of direct miR-122 targets in the murine liver (Supplementary Table 3). Furthermore, we find that simple string matching to miR-122 seed sites yields higher enrichment within the up-regulated mRNAs compared to more complex algorithms, such as RNAhybrid and miRanda (run locally with default settings, data not shown).
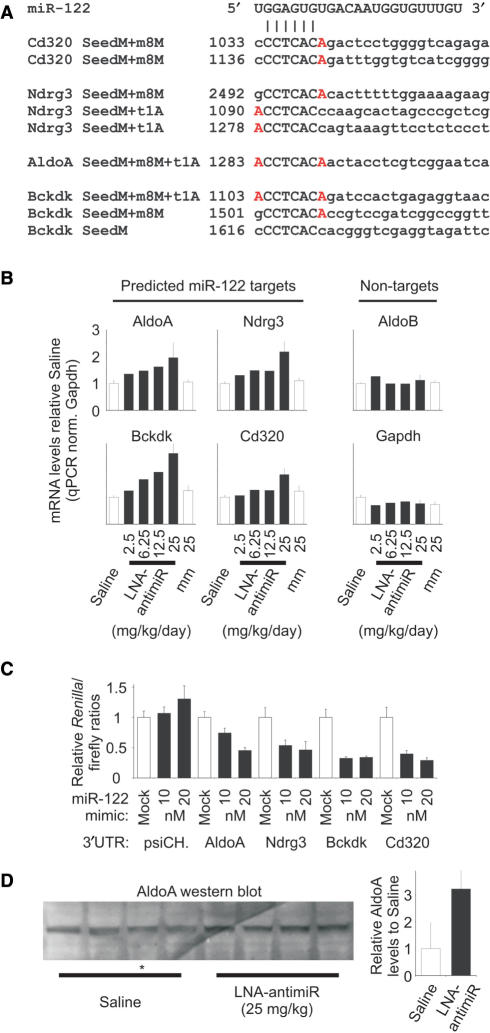
Among the identified miR-122 targets, we chose four mRNAs for further expression analyses by real-time RT-PCR: AldoA (Aldolase A. one SeedM+m8M+t1A site), Bckdk (Branched-chain α-ketoacid dehydrogenase kinase, one SeedM+m8M+t1A, one SeedM+m8M and SeedM site), Cd320 (Cd320 antigen, putative VLDL receptor, two SeedM+m8M sites) and Ndrg3 (N-myc downstream regulated gene 3, one SeedM+m8M and two SeedM+t1A sites) (Figure 3A). At 24 h after the last dose, all four mRNAs were de-repressed in a dose-dependent manner in response to increasing LNA-antimiR dose, as would be expected if miR-122 was functionally inhibited by LNA-antimiR (Figure 3B). In contrast, the expression levels of two non-target mRNAs, aldolase B and GAPDH, were unaltered in the treated mice compared to saline and LNA-mismatch control (Figure 3B). The results obtained here by in vivo antagonism of miR-122 imply that the Ndrg3, AldoA, Bckdk and Cd320 mRNAs are direct targets of miR-122 in the mouse liver. To confirm that the identified target sites in these messages could mediate translational repression by miR-122, we cloned their full-length 3′UTRs downstream of the Renilla luciferase gene and transfected the resulting 3′UTR reporters into HeLa cells alongside a miR-122 mimic. As shown in Figure 3C, all four 3′UTR reporters showed a significant decrease in luciferase activity, whereas the miR-122 mimic had no effect on the luciferase control plasmid without a miR-122 target site. Moreover, AldoA protein levels were 3-fold increased in LNA-antimiR-treated mice compared to saline control mice (Figure 3D), concurring with the observed 2-fold de-repression of AldoA message levels in LNA-treated mice (Figure 3B). Taken together, our data suggest that miR-122 negatively regulates the expression of a large number of target mRNAs in adult mouse liver, which is in good agreement with reports from Drosophila and vertebrates (14,45). On the other hand, the finding that most of these targets showed only slight to moderate de-repression in miR-122-silenced mice livers (Supplementary Tables 1, 2 and 4) is consistent with the proposal that many miRNAs might work by fine-tuning gene-regulatory networks (46,47). Our results are also consistent with the idea that perfect seed pairing is a useful means of predicting animal miRNA targets and provide important in vivo evidence that the presence of extended 7-nt seed matches can be used to improve the specificity of miRNA target site prediction.
Figure 3.
Expression of four miR-122 targets. (A) Alignment showing the seed matches in Cd320, Ndrg3, AldoA and Bckdk. The 6-nt seed is shown in capital letters, with the m8M or t1A extensions highlighted in red. (B) Real-time RT-PCR analysis of AldoA, Nrdg3, Bckdk and Cd320 was carried out to investigate their expression as response to increasing LNA-antimiR doses. Shown are mean and SD (where n = 3) of mRNA levels relative to the saline control group. Shown is also the mismatch control (mm). (C) miR-122-mediated repression of 3′UTR reporters for AldoA, Ndrg3, Bckdk and Cd320 in HeLa cells. The 3′UTRs of the four predicted miR-122 targets were cloned downstream of the Renilla luciferase gene and the resulting reporters were transfected into HeLa cells along with a miR-122 mimic. Shown are mean values and SD, n = 4. (D) Western blot analysis of AldoA in liver protein extracts of LNA-antimiR-treated and saline control mice. Shown is one representative western blot and corresponding quantification. Values are related to the mean of the saline values (mean and SD, n = 4, asterisk: 75 µg protein instead of 100 µg, compensated for in quantification).
Antagonism of miR-122 by LNA-antimiR is reversible and shows no hepatotoxicity in mice
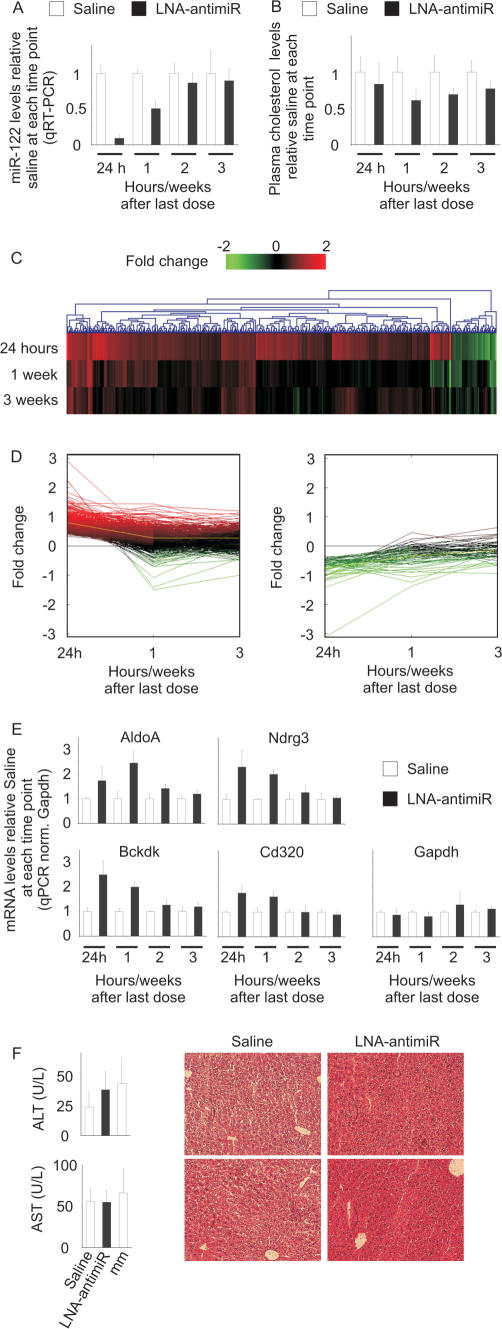
To gain further insight into the long-term effects of LNA-antimiR-mediated antagonism of miR-122 in vivo, we treated mice with single intravenous doses of LNA-antimiR for three consecutive days at 25 mg/kg/day, and then extended monitoring of the study animals to 3 weeks after last dose. The LNA-antimiR treatment resulted in maximal reduction of miR-122 levels at 24 h, followed by an increase of mature miR-122 to ∼50% relative to saline control levels at 1 week leading to completely normalized levels at 3 weeks after last dose (Figure 4A). This coincided with transient de-repression within 24 h of 199 predicted miR-122 targets, which gradually returned to saline control levels by 3 weeks post-treatment, which along with temporal normalization of most changes in liver gene expression (Figure 3 and Supplementary Tables 1, 2 and 4) implies that antagonism of miR-122 by LNA-antimiR in mice is reversible. Contrary to a recent report (48), we did not observe any hepatotoxicity in the LNA-antimiR or LNA-mismatch control-treated mice as shown by unaltered levels of the serum transaminases ALT and AST 24 h after treatment with total LNA doses of 75 mg/kg (3 × 25 mg/kg/day) compared to saline controls (Figure 4F). This is consistent with the absence of morphological changes in liver sections from the LNA-treated animals (Figure 4F, Supplementary Figure 4). It is also noteworthy that we did not observe any perturbations in the treated livers for genes associated with the miRNA pathway (Supplementary Table 6).
Figure 4.
Duration of functional miR-122 inhibition in LNA-antimiR-treated mice. (A) Real-time RT-PCR analysis of miR-122 levels in LNA-antimiR-treated mice livers over time. The mice were treated with 25 mg/kg/day LNA-antimiR or saline for three consecutive days followed by sacrificing the animals after 24 h, 1, 2 or 3 weeks. miR-122 levels were normalized to the mean of the saline control group at each individual time point. Shown are mean and SD, n = 7 (24 h n = 10). (B) Total plasma cholesterol in LNA-antimiR-treated mice over time. The data were normalized to the saline group at each time point. Shown are mean and SD (n = 7, 24 h n = 10). (C) Hierarchical clustering was performed on the expression profiles of genes identified as differentially expressed between LNA-antimiR and saline-treated mice 24 h, 1 week or 3 weeks post-treatment. The LNA-antimiR/saline log2-transformed expression ratios are indicated by colour where red indicates higher expression level in LNA-antimiR-treated mice compared to saline-treated mice, whereas green indicates lower expression level and black indicates equal expression level. (D) Expression profiles of genes identified as differentially expressed between LNA-antimiR- and saline-treated mice 24 h post-treatment was monitored over time. (E) Temporal expression of four miR-122 target genes (AldoA, Nrdg3, Bckdk and Cd320) and GAPDH in LNA-antimiR-treated mice livers. Values were normalized to saline control at each time point. Shown are mean and SD (n = 3). (F) Toxicity assessed by measuring liver enzymes and investigating liver morphology 24 h after last dose, shown are ALT and AST levels (mean and SD, n = 5) and histology on liver sections.
Previous studies in mice have reported effective miR-122 knockdown using total intravenous doses of 120–240 mg/kg for cholesterol-conjugated 2′-O-Me antagomirs and weekly intraperitoneal doses of 25–150 mg/kg for 2′ MOE oligonucleotides leading to a low plasma cholesterol loss-of-function phenotype (10,11,38). One week after dosing with LNA-antimiR, plasma cholesterol levels were reduced by ∼40% in treated mice, thereby confirming the low cholesterol phenotype, with levels still being 20% below controls at 3 weeks (Figure 4B). By comparison, the miR-122 targets AldoA, Bckdk, Ndrg3 and Cd320, were all substantially de-repressed 24 h after LNA-antimiR treatment and then reverted towards control levels over the next 2 weeks (Figure 4E). Thus, the observed changes in gene expression appear to precede reduction in cholesterol, which in turn, appears longer lasting than changes in miR-122 target gene expression. It is therefore tempting to speculate that the coordinated changes in miR-122-associated gene networks are responsible for the control of cholesterol and lipid metabolism in the liver and that such pathways take several weeks to revert to normal levels. Interestingly, we find that the up-regulated genes in treated mice 24 h post-treatment are enriched in the Gene Ontology categories lipid metabolism (25 genes, P =3.3E−5) and lipid biosynthesis (12 genes, P = 9.1E−5).
In conclusion, our temporal liver gene expression profile extended to 3 weeks after the last LNA-antimiR dose combined with the low-cholesterol phenotype demonstrate functional antagonism of miR-122 in mice by LNA-antimiR. Our findings suggest that miRNA antagonists comprised of LNA may be valuable tools for identifying miRNA targets in vivo and for studying the biological role of miRNAs and miRNA-associated gene-regulatory networks in a physiological context. In addition, the high metabolic stability of LNA-antimiRs, due in part to increased nuclease resistance, their small size and lack of acute toxicity imply that LNA-antimiRs may be well suited as a novel class of potential therapeutics for disease-associated miRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors wish to thank Anja Konge, Bettina Nordbo, Christina Udesen, Heidi W. Høvring, Janni Juul Jørgensen, Katrine Rishøj Nielsen, Lisbeth Bang, Otto Olsen and Rikke Sølberg and Ulla Steinmeier for excellent technical assistance. This study was supported by grants from the Danish National Advanced Technology Foundation, Danish Medical Research Council and the Lundbeck Foundation to S.K. Wilhelm Johannsen Centre for Functional Genome Research is established by the Danish National Research Foundation, Copenhagen, Denmark (www.dg.dk). Funding to pay the Open Access publication charges for this article was provided by Santaris Pharma, Hørsholm, Denmark.
Conflict of interest statement. J.E., M.L., M.H., J.B.H., H.F.H., E.M.S., K.M., P.K. and S.K. are employed at Santaris Pharma. Santaris Pharma is a biopharmaceutical company engaged in the development of RNA based medicine.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl Acad. Sci. USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 8.Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 9.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 10.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 12.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 13.John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol. Biol. 2006;342:101–113. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- 14.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 15.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 16.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 18.Fluiter K, Ten Asbroek AL, de Wissel MB, Jakobs ME, Wissenbach M, Olsson H, Olsen O, Oerum H, Baas F. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 2003;31:953–962. doi: 10.1093/nar/gkg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 20.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 21.Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 23.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 25.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 26.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 27.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 28.Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. [Google Scholar]
- 29.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 31.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 32.Dennis GJr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 33.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SM, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Krutzfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crooke ST. Antisense Drug Technology. New York: Marcel Dekker; 2001. [Google Scholar]
- 40.Crooke ST. Progress in antisense technology: the end of the beginning. Methods Enzymol. 2000;313:3–45. doi: 10.1016/s0076-6879(00)13003-4. [DOI] [PubMed] [Google Scholar]
- 41.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naguibneva I, Ameyar-Zazoua M, Nonne N, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Pritchard LL, Harel-Bellan A. An LNA-based loss-of-function assay for micro-RNAs. Biomed. Pharmacother. 2006;60:633–638. doi: 10.1016/j.biopha.2006.07.078. [DOI] [PubMed] [Google Scholar]
- 44.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 47.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 48.Swayze EE, Siwkowski AM, Wancewicz EV, Migawa MT, Wyrzykiewicz TK, Hung G, Monia BP, Bennett CF. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.