Abstract
Human tryptophanyl-tRNA synthetase (hTrpRS) differs from its bacterial counterpart at several key positions of the catalytic active site and has an extra N-terminal domain, implying possibly a different catalytic mechanism. We report here the crystal structures of hTrpRS in complexes with Trp, tryptophanamide and ATP and tryptophanyl-AMP, respectively, which represent three different enzymatic states of the Trp activation reaction. Analyses of these structures reveal the molecular basis of the mechanisms of the substrate recognition and the activation reaction. The dimeric hTrpRS is structurally and functionally asymmetric with half-of-the-sites reactivity. Recognition of Trp is by an induced-fit mechanism involving conformational change of the AIDQ motif that creates a perfect pocket for the binding and activation of Trp and causes coupled movements of the N-terminal and C-terminal domains. The KMSAS loop appears to have an inherent flexibility and the binding of ATP stabilizes it in a closed conformation that secures the position of ATP for catalysis. Our structural data indicate that the catalytic mechanism of the Trp activation reaction by hTrpRS involves more moderate conformational changes of the structural elements at the active site to recognize and bind the substrates, which is more complex and fine-tuned than that of bacterial TrpRS.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) are a family of ancient enzymes that are responsible for catalysis of the attachment of amino acids to their cognate tRNAs and thus play a critical role in maintaining the fidelity of protein synthesis. For all aaRSs, the aminoacylation reaction takes place in two steps: the amino acid is first activated with ATP to form aminoacyl-AMP (amino acid activation step), which is then transferred to the 3′ end of its cognate tRNA (acyl transfer step). The two processes are usually separable and occur sequentially. Based on sequence homology and structure similarity, aaRSs have been classified into two classes I and II, each comprising aaRSs for 10 of the 20 standard amino acids (1,2). Class I aaRSs consist of a Rossmann fold (RF) catalytic domain containing two highly conserved sequence motifs HIGH and KMSKS at the catalytic active site and a C-terminal α-helical domain containing the anticodon-binding site (also called the anticodon-binding domain) (3). These enzymes can be further divided into three subclasses, Ia, Ib and Ic, each containing some specific signature motifs which vary from one to the other in both sequence and structure (4). Tryptophanyl-tRNA synthetase (TrpRS) and tyrosyl-tRNA synthetase (TyrRS) belong to the Ic subclass which share considerable structural homology and contain a conserved GXDQ motif in addition to the KMSKS and HIGH motifs in the RF catalytic domain that are involved in the substrate binding.
Baccillus stearothermophilus TrpRS (bsTrpRS) is one of the aaRSs that have been extensively characterized with both structural and biochemical methods. Carter and coworkers (5–11) have reported the structural snapshots of bsTrpRS representing different enzymatic states in the random, sequential kinetic scheme of the Trp activation reaction (9). The structural results together with biochemical data have revealed that the dimeric bsTrpRS uses an induced-fit mechanism in the substrate binding and the Trp activation reaction. Specifically, ligand binding induces conformational change of the KMSKS loop and movement of the C-terminal domain in a coordinated manner during the catalytic reaction, forming three allosteric enzymatic states: an open state in the absence of substrate or in the presence of Trp or ATP, a closed pretransition state in the presence of both Trp and ATP, and a closed product state in the presence of tryptophanyl-AMP (TrpAMP). Sequence comparison indicated that substantial sequence divergence exists between eukaryotic TrpRSs and prokaryotic TrpRSs (12). In particular, human TrpRS (hTrpRS) differs from bsTrpRS at several key positions of the catalytic active site, including the characteristic HIGH, GXDQ and KMSKS motifs. In addition, hTrpRS has acquired an extra N-terminal domain of about 150 amino acids during evolution that not only confers the enzyme an angiostatic activity, but also plays an important role in the aminoacylation reaction (13,14). We speculate that hTrpRS might develop a different catalytic mechanism of the aminoacylation reaction to adapt these changes. Recently, several crystal structures of hTrpRS have been reported, including the apo form, in complex with TrpAMP, and in complex with tRNATrp and Trp (15–20), which have provided some insights into the catalytic mechanism. However, due to the lack of the structures at several intermediate states of the catalytic reaction, especially the pretransition state with both Trp and ATP bound, the catalytic mechanism of the Trp activation reaction by hTrpRS is still unclear.
In this study, we characterized three different enzymatic states of hTrpRS during the Trp activation reaction: the complex with Trp, the complex with tryptophanamide (TrpNH2O, a Trp analog) and ATP, and the complex with TrpAMP, respectively. Structural analyses of these complexes enable us to define the structural elements and key residues that are involved in the recognition and binding of the substrates in different enzymatic states and their functional roles in the catalysis. Comparisons among these structures and with those reported previously reveal the conformational changes occurring when the substrates bind and the activated intermediate is formed, and their functional correlations with the catalysis. These results reveal more insights into the catalytic mechanism of the Trp activation reaction by hTrpRS and provide the structural basis for the mechanistic differences between hTrpRS and bsTrpRS.
MATERIALS AND METHODS
Crystallization and diffraction data collection
Expression and purification of hTrpRS were carried out as described (17). In our previous studies, the N-terminal part of the enzyme (about residues 1–96) was hydrolyzed during crystallization, yielding the structure of the apo T2-hTrpRS (17). Later, biochemical studies showed that addition of protease inhibitor PMSF in protein purification and crystallization could prohibit hydrolysis of the protein. Thus, in this study, 1 mM PMSF was added in the protein solution to prevent proteolysis of the full-length enzyme (Supplementary Figure S1).
Sparse-matrix crystallization screening with the Crystal Screen I and II kits (Hampton Research) was performed using the hanging-drop vapor diffusion method at 4°C. The crystallization drops were prepared by mixing equal volumes (2 μl) of the protein solution (∼15 mg/ml of hTrpRS, 20 mM K2HPO4, pH 6.8, 10 mM MgCl2, 5 mM 2-mercaptoethanol and 1 mM PMSF) and the crystallization solution. Crystals of hTrpRS in complex with Trp were grown from drops containing the protein solution and the crystallization solution (1.0 M Na citrate and 100 mM HEPES, pH 6.8) supplemented with 0.5 μl of 10 mM Trp. Crystals of hTrpRS in complex with TrpAMP were grown from drops containing the protein solution and the crystallization solution (1.0 M Na citrate and 100 mM HEPES, pH 7.2) supplemented with 0.5 μl of 10 mM Trp and 0.5 μl of 100 mM ATP. TrpAMP was formed enzymatically from Trp and ATP during the crystallization solution. Crystals of hTrpRS in complex with ATP and TrpNH2O were grown from drops containing the protein solution and the crystallization solution (1.15 M Na citrate and 100 mM HEPES, pH 7.0) supplemented with 0.5 μl of 10 mM TrpNH2O and 0.5 μl of 100 mM ATP. Crystals of these three complexes (hTrpRS–Trp, hTrpRS–TrpAMP and hTrpRS–TrpNH2O–ATP) have the same tetragonal bipyramid shape belonging to space group P43212 with similar unit cell parameters (Table 1). Attempts to grow crystals of hTrpRS in complex with ATP yielded two types of hTrpRS crystals from drops containing the protein solution and the crystallization solution (0.2 M Na citrate, pH 8.2, 4% PEG3350 and 16% PEG1500) supplemented with 0.5 μl of 100 mM ATP. One type of crystals has the same hexagonal bipyramid shape as that of the apo hTrpRS crystals described previously (17) with space group P6522 and comparable unit cell parameters. The other type of crystals has long hexagonal prism shape belonging to space group P622 with different unit cell parameters. Structure determination indicated that both crystal forms are the apo hTrpRS (data not shown). Diffraction data were collected from flash-cooled crystals at 100 K using synchrotron beamline BL5A at Photon Factory, Japan. The diffraction data were recorded on an ADSC Quantum315 detector and processed and scaled using the HKL2000 suite (21). The statistics of the diffraction data are summarized in Table 1.
Table 1.
X-ray diffraction data and structure refinement statistics
| E-Trp | E-TrpNH2O–ATP | E-TrpAMP | |
|---|---|---|---|
| Statistics of diffraction data | |||
| Space group | P43212 | P43212 | P43212 |
| Cell parameters a = b/c (Å) | 79.9/382.3 | 79.7/383.2 | 80.0/383.0 |
| Resolution range (Å) | 50.0–2.40 (2.49–2.40)a | 50.0–2.40 (2.49–2.40) | 50.0–2.40 (2.51–2.40) |
| Number of observed reflections | 440 440 | 334 480 | 267 613 |
| Number of unique reflections (I/σ > 0) | 43 430 | 47 608 | 43 231 |
| Average redundancy | 10.1 (4.0) | 7.0 (4.8) | 6.2 (3.3) |
| I/σ (I) | 12.1 (1.0) | 16.9 (1.8) | 25.0 (2.9) |
| Completeness (%) | 86.7 (46.2) | 95.1 (69.9) | 88.1 (43.0) |
| Rmerge (%)b | 16.3 (75.6) | 11.4 (57.5) | 6.8 (28.9) |
| Mosaicity | 0.34 | 0.37 | 0.15 |
| Statistics of refinement and model | |||
| Resolution range (Å) | 50.0–2.40 (2.55–5.40) | 50.0–2.40 (2.55–2.40) | 50.0–2.40 (2.55–2.40) |
| Number of reflections (Fo ≥ 0σ (Fo)) | 43 267 | 47 455 | 43 113 |
| Working set | 41 099 | 45 092 | 40 958 |
| Test set | 2168 | 2363 | 2155 |
| Completeness | 86.5 (48.9) | 94.9 (76.8) | 87.9 (39.8) |
| R factor (%)c | 20.7 (35.0) | 21.1 (32.3) | 21.4 (29.3) |
| Free R factor (%) | 23.3 (35.8) | 23.8 (36.2) | 23.8 (30.2) |
| Number of protein residues | 760 | 769 | 766 |
| Number of water molecules | 278 | 226 | 198 |
| Average B factor of all atoms (Å2) | 46.1 | 51.9 | 47.9 |
| Protein main-chain atoms | 45.5 | 51.0 | 47.0 |
| Protein side-chain atoms | 46.5 | 52.2 | 48.0 |
| Ligand atoms | 40.5 | 85.2 | 64.7 |
| RMS bond lengths (Å) | 0.007 | 0.007 | 0.007 |
| RMS bond angles (°) | 1.3 | 1.3 | 1.3 |
| Luzzati atomic positional error (Å) | 0.33 | 0.32 | 0.32 |
| Ramachandran plot (%) | |||
| Most favored regions | 91.8 | 91.3 | 92.1 |
| Allowed regions | 7.8 | 8.3 | 7.6 |
| Generously allowed | 0.4 | 0.4 | 0.3 |
aNumbers in parentheses refer to the highest resolution shell.
bRmerge = ||I0 | − 〈 I〉 |/〈 I〉.
cRfactor = ∣∣Fo∣ − ∣Fc∣∣/∣Fo.
Structure determination and refinement
The structure of the hTrpRS–Trp complex was determined by the molecular replacement (MR) method implemented in the program CNS (22) using the apo hTrpRS structure (17) as the search model. The structures of the hTrpRS–TrpAMP and hTrpRS–TrpNH2O–ATP complexes were solved with MR using the hTrpRS–Trp complex as the starting model. Structure refinement was carried out with CNS using standard protocols (energy minimization, simulated annealing and B factor refinement). Model building was performed manually with the program O (23). Five percent of randomly chosen diffraction data were set aside for free R factor cross-validation throughout the refinement. For all three complexes, there are two hTrpRS molecules in the asymmetric unit that were refined independently due to conformational differences. In the hTrpRS–Trp complex, there was strong electron density at the active site in both monomers in the initial difference Fourier map that matches Trp very well (Figure S2). In the hTrpRS–TrpAMP complex, there was very good electron density for TrpAMP at the active site in monomer A but only Trp in monomer B. In the hTrpRS–TrpNH2O–ATP complex, there was good electron density for both TrpNH2O and ATP at the active site in monomer B but only TrpNH2O in monomer A. Water molecules were picked automatically at a height of above 2.5 σ followed by manual check of proper hydrogen-bonding geometry and B factor of <60 Å2. The statistics of structure refinement and structure models are listed in Table 1.
RESULTS
hTrpRS is structurally and functionally asymmetric with half-of-the-sites reactivity
The structures of hTrpRS in complexes with Trp, TrpNH2O and ATP and TrpAMP, respectively, were all determined at 2.4-Å resolution (Table 1). SDS–PAGE analyses showed that the enzyme exists as the full-length form in both the crystals and the crystallization solution (Figure S1). Nevertheless, the majority of the N-terminal region of hTrpRS (about residues 1–94) is invisible in all three structures and only a small portion corresponding to the N-terminal β-hairpin (residues 83–93) could be seen in one monomer of the hTrpRS–TrpNH2O–ATP complex and the hTrpRS–TrpAMP complex (Table 2). Similarly, the N-terminal region is also disordered in the previously reported crystal structures of hTrpRS, except the structure of the hTrpRS–TrpAMP complex reported by Yang et al. (15) in which one monomer contains a partially ordered N-terminal region with residues 7–60 forming a helix–turn–helix (HTH) motif and residues 82–92 forming a β-hairpin and the two structural motifs being connected by a disordered loop of about 20 residues. These results suggest that the N-terminal region of hTrpRS has an intrinsic conformational flexibility and appears to be flexibly linked to the enzyme's core. It is common to observe a particular region or domain disordered in the crystal structures of aminoacyl-tRNA synthetases. For example, in the crystal structure of human glycyl-tRNA synthetase (hGlyRS), the N-terminal domain (residues 1–62), which is also a WHEP-TRS domain corresponding to the N-terminal HTH motif of hTrpRS, is completely disordered (24). Analysis of the crystal packing in the three hTrpRS structures described here indicates that there is sufficient space between the existing portion of the N-terminal domain (residues 95–150) and the C-terminal α-helical domain to accommodate the disordered N-terminal region in the lattice. The disordered region could have contacts with structural elements of these two domains.
Table 2.
Structural comparisons of hTrpRS in different enzymatic states
| Apo form | Trp | TrpNH2O–ATP | TrpAMP | |||||
|---|---|---|---|---|---|---|---|---|
| Mono A | Mono B | Mono A | Mono B | Mono A | Mono B | |||
| Residues | 94–471 | 97–475 | 92–472 | 97–475 | 82–471 | 88–471 | 94–471 | |
| N-terminal β-hairpin | None | None | None | None | Yes | Partial | None | |
| Bound ligand | Trp | Trp | TrpNH2O | TrpNH2O + ATP | TrpAMP | Trp | ||
| Conformations of the characteristic motifs, the substrate-binding pocket, and the N-terminal and C-terminal domains | ||||||||
| RF domain | AIDQ motif | Open | Closed | Closed | Closed | Closed | Closed | Closed |
| Linker region | Open | Closed | Closed | Closed | Closed | Closed | Closed | |
| KMSAS motif | Open | Semi-closed | Closeda | Semi-closed | Closeda | Closed | Closeda | |
| Substrate-binding pocket | Open | Semi-closed | Closed | Semi-closed | Closed | Closed | Closed | |
| N-terminal domain | Open | Closed | Closed | Closed | Closed | Closed | Closed | |
| C-terminal domain | Open | Closed | Closed | Closed | Closed | Closed | Closed | |
| Averaged displacements of the specific structural elements relative to the apo form based on superposition of the RF core regionb (Å) | ||||||||
| AIDQ motif | 3.14 | 3.39 | 3.21 | 3.21 | 3.22 | 3.25 | ||
| KMSAS motif | 3.13 | 2.70 | 2.94 | 2.57 | 2.72 | 2.73 | ||
| N-terminal domain (helix α3)c | 2.88 | 2.87 | 2.87 | 2.90 | 2.89 | 3.03 | ||
| C-terminal domain (anticodon-binding site)c | 3.86 | 3.63 | 3.93 | 3.38 | 3.96 | 3.36 | ||
| KMSAS loop (residues 344–357) | 3.09 | 2.72 | 2.99 | 2.56 | 2.77 | 2.71 | ||
| Linker region (residues 336–343) | 2.35 | 2.17 | 2.34 | 2.16 | 2.28 | 2.14 | ||
| Conformational differences of the substrate-binding pocketd | ||||||||
| AIDQ-KMSAS (Å) (Gln313–Lys349) | 14.6 | 15.9 | 13.4 | 15.4 | 13.8 | 13.5 | 13.8 | |
| AIDQ-helix α8 (Å) (Gln313–Glu199) | 16.9 | 13.6 | 13.6 | 13.5 | 13.6 | 13.6 | 13.6 | |
| KMSAS-helix α8 (Å) (Lys349–Glu199) | 22.3 | 20.8 | 20.6 | 20.7 | 21.0 | 20.3 | 21.0 | |
| Solvent-accessible surface area (Å2) | 1481 | 1089 | 976 | 1107 | 930 | 990 | 995 | |
| Volume (Å3) | 3347 | 2248 | 1608 | 2143 | 1600 | 1436 | 1454 | |
aThe KMSAS loop in these monomers is involved in crystal packing contacts with a symmetry-related molecule.
bThe core region of the RF catalytic domain comprises residues 154–308 and 318–335, excluding the variable regions containing the AIDQ motif and the KMSAS loop.
cThe representative structural element of the N-terminal domain is defined as residues 99–110 of helix α3 and that of the C-terminal domain is defined as residues 379–395 of the anticodon-binding site.
dThe distance between the AIDQ motif and the KMSAS loop is defined as the Cα–Cα distance between Gln313 (AIDQ) and Lys349 (KMSAS). The distance between the AIDQ motif and the bottom of the substrate-binding pocket is defined as the Cα–Cα distance between Gln313 and Glu199 of helix α8. The distance between the KMSAS loop and the bottom of the substrate-binding pocket is defined as the Cα–Cα distance between Lys349 and Glu199. The solvent-accessible surface area and the volume of the substrate-binding pocket are calculated using the CASTp server (46,47).
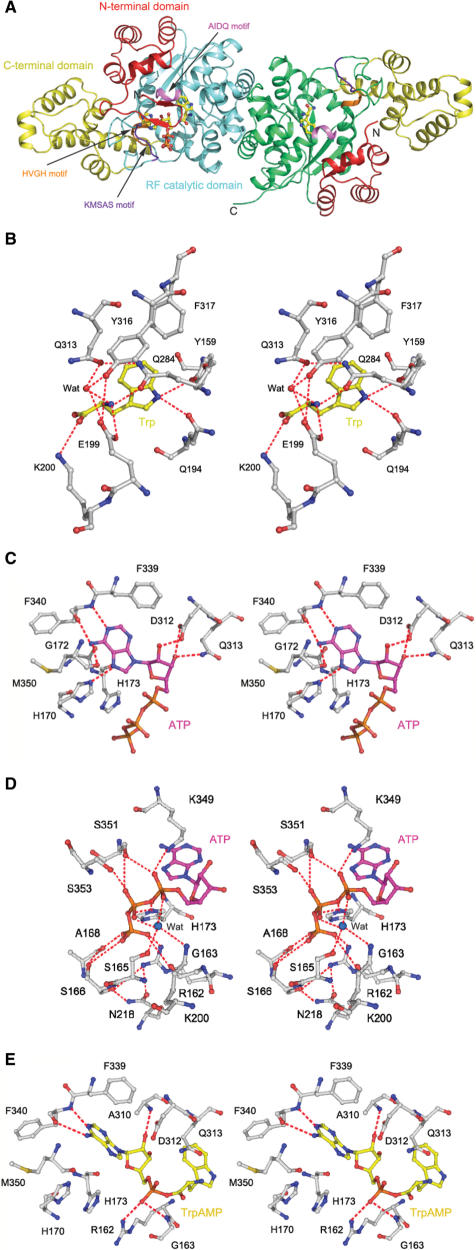
The overall structure of hTrpRS in these complexes is very similar to that of hTrpRS described previously (15–17). It is consisted of three domains: an N-terminal domain (residues 1–150), an RF catalytic domain (residues 151–362 and 453–471) and a C-terminal domain (residues 363–452) [hereafter the nomenclature of the secondary structure of hTrpRS is after Yang et al. (15)] (Figure 1A). The catalytic active site resides in a deep pocket of the RF domain and is surrounded by several conserved structural elements, including the KMSAS loop (the connecting loop between strand β9 and helix α16), the HVGH motif (helix α7) and the AIDQ motif (helix α15). The substrate-binding pocket consists of the Trp-binding site and the ATP-binding site. The anticodon-binding site is located at the tip of the C-terminal domain (residues 479–495).
Figure 1.
Structures of hTrpRS in complexes with different ligands. (A) A ribbon diagram showing the overall structure of the hTrpRS–TrpNH2O–ATP complex. The N-terminal domain, the RF catalytic domain and the C-terminal domain are colored in red, cyan (or green) and yellow, respectively. The characteristic KMSAS, HVGH and AIDQ motifs are colored in purple, orange and violet, respectively. The bound TrpNH2O and ATP are shown in ball-and-stick models. (B) A stereoview showing the interactions of Trp with the surrounding residues in the hTrpRS–Trp complex. The hydrogen bonds are indicated by dashed lines. (C) A stereoview showing the interactions between the adenine and ribose moieties of ATP and the surrounding residues in the hTrpRS–TrpNH2O–ATP complex. (D) A stereoview showing the interactions between the triphosphate group of ATP and the surrounding residues in the hTrpRS–TrpNH2O–ATP complex. (E) A stereoview showing the interactions of TrpAMP with the surrounding residues in the hTrpRS–TrpAMP complex.
Unlike the apo hTrpRS structure, the ligand-bound hTrpRS structures contain two monomers in the asymmetric unit related by a 2-fold noncrystallographic symmetry which form the biologically functional homodimer. However, the two monomers are bound with different ligands at the active site (except the Trp-bound complex) and adopt different conformations of the KMSAS loop. In the structure of the hTrpRS–Trp complex, although both monomers are bound with a Trp, the KMSAS loop assumes a semi-closed conformation in monomer A, but a closed conformation in monomer B (see Discussion section). In the structure of the hTrpRS–TrpNH2O–ATP complex, monomer A is bound with a TrpNH2O with the KMSAS loop in the semi-closed conformation, while monomer B is bound with both TrpNH2O and ATP with the KMSAS loop in the closed conformation. In the structure of the hTrpRS–TrpAMP complex, although the KMSAS loop adopts the closed conformation in both monomers, only monomer A is bound with a TrpAMP, while monomer B with a Trp. This structural asymmetric property of hTrpRS was also observed in the structure of the hTrpRS–TrpAMP complex reported by Yang et al. (15) in which one monomer is bound with a TrpAMP with the KMSAS loop in the closed conformation, while the other has no substrate bound with a disordered KMSAS loop. All three ligand-bound hTrpRS structures belong to the same space group and have the same crystal packing. Analysis of the crystal lattice shows that the KMSAS loop in monomer B has some contacts with a symmetry-related molecule, which might play some role in the formation of the closed conformation. However, in monomer A it is not involved in contact with any symmetry-related molecule and thus its conformation is not constrained by crystal lattice. Since the KMSAS loop in monomer A can assume either semi-closed or closed conformation in these complexes, therefore, both conformations should be biologically relevant rather than artifact of the crystal packing.
These results clearly show that the dimeric hTrpRS can get activation in one monomer and binding of Trp to the other, but not recruitment of the second ATP, suggesting that hTrpRS has ‘half-of-the-sites’ reactivity and the two monomers work in an anticooperative manner. This is consistent with the kinetic data showing that only one monomer of the dimeric bovine TrpRS operates at a time in the aminoacylation reaction and the two monomers function randomly with half-of-the-sites reactivity (25–27). This structural and functional asymmetric property of hTrpRS is reminiscent of bsTrpRS and TyrRS. Nevertheless, in hTrpRS the half-of-the-sites reactivity involves the binding of ATP that is similar to bsTrpRS (9,10), while in TyrRS it is the binding of the amino acid displaying the half-of-the-sites reactivity (28,29).
Recognition of Trp is by an induced-fit mechanism
Previous analyses of the hTrpRS structures in the apo form and in complex with TrpAMP have suggested the structural elements involved in the binding of Trp (15,17). However, the exact recognition mechanism of Trp by hTrpRS and the conformational changes associated with the binding of Trp are still obscure. Thus, we determined the structure of the hTrpRS–Trp complex. Structural analysis of this complex shows that the recognition of Trp is by an induced-fit mechanism involving movement of the AIDQ motif which creates a perfect binding pocket for Trp with complementarity in both hydrogen-bonding property and hydrophobicity (Figures 1B and S3A). The indole nitrogen of Trp is specifically recognized via hydrogen-bonding interactions by the hydroxyl group of Tyr159 (3.0 Å) and the side-chain Οɛ1 of Gln194 (3.0 Å). In addition, Tyr159 also forms π–π stacking interaction with the Trp indole ring. Both Tyr159 and Gln194 are highly conserved in eukaryotic TrpRSs and their importance in the aminoacylation reaction has been confirmed by our biochemical data (18). These interactions provide the structural basis for amino acid discrimination between TrpRSs and TyrRSs (15). The main-chain α-amino group of Trp is positioned by hydrogen-bonding interactions directly with the side chains of Glu199 (3.1 Å) and Gln284 (2.7 Å) and indirectly with the side chains of Gln313 and Tyr316 through a water molecule. The carboxyl group of Trp interacts via a salt bridge with the side chain of Lys200 (3.5 Å). These interactions are responsible for the precise positioning of Trp and play important roles in activating the carboxyl group.
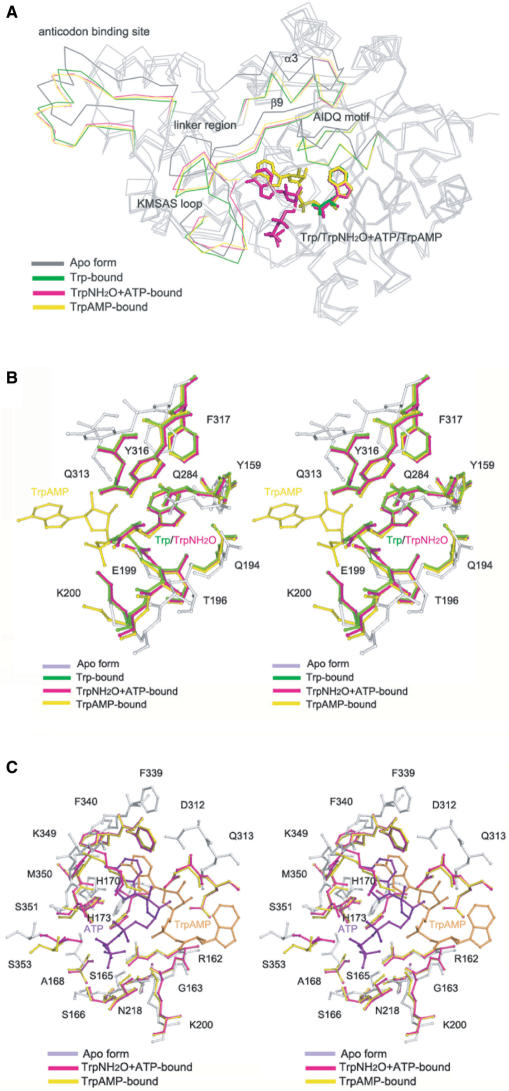
Compared to the apo hTrpRS, the binding of Trp results in the movement of the AIDQ motif toward the active site by ∼3.1 Å to adopt a closed conformation, and thus it can make interactions with the substrate, ensuring the specific recognition and activation of Trp (Figure 2A and Table 2). Coupled with the closure of the AIDQ motif, the N-terminal domain which flanks strand β9 of the RF domain also moves closer toward the RF domain to adopt a closed conformation (residues 99–110 of helix α3 is displaced by ∼2.9 Å). The C-terminal domain also rotates moderately toward the RF domain (the anticodon-binding site is displaced by ∼3.9 Å). This rotation can be viewed as the C-terminal domain being pulled toward the RF domain by the linker region of residues 336–343 (which links the KMSAS loop to strand β9 of the RF domain) with helix α20 (which connects the C-terminal domain back to the RF domain) as the hinge. These domain movements lead to the formation of a compact or closed overall conformation of the enzyme. In addition, the KMSAS loop also has conformational change toward the active site: it adopts a semi-closed conformation in monomer A and a closed conformation in monomer B. Compared to the open conformation in the apo hTrpRS, the KMSAS loop of the semi-closed conformation undergoes a twisted movement (downwards and inwards) toward the active site by ∼3.0 Å but is not in a proper position to interact with ATP modeled based on the TrpNH2O–ATP complex. The KMSAS loop of the closed conformation is shifted inwards toward the active site by ∼2.7 Å, which is similar to that in the TrpNH2O–ATP complex and thus can interact with ATP. As mentioned earlier, the KMSAS loop in monomer B is involved in inter-molecular contacts and thus the closed conformation might be constrained by the crystal packing, but the semi-closed conformation in monomer A is not and thus is biologically relevant. The biological implication of the different conformations of the KMSAS loop will be discussed later.
Figure 2.
Structural comparison of hTrpRS in different enzymatic states. (A) Structural comparison of hTrpRS in different enzymatic states showing the conformational changes of the structural elements at the active site and the overall structure induced by the binding of substrates. The comparison is based on superposition of the core region of the RF catalytic domain relative to the apo hTrpRS. The structural elements displaying the significant conformational changes are color-coded as follows: the apo hTrpRS in gray, the hTrpRS–Trp complex in green, the hTrpRS–TrpNH2O–ATP complex in magenta and the hTrpRS–TrpAMP complex in yellow, respectively. The bound ligands are shown as ball-and-stick models and colored according to the respective complex. (B) Structural comparison of the Trp-binding site. The color-coding of different hTrpRS structures is same as in (A). The Trp substrate/moiety occupies the same position and has similar interactions with the surrounding residues in all three complexes. (C) Structural comparison of the ATP-binding site. ATP in the pretransition state and TrpAMP in the product state (shown in ball-and-stick models) have moderate positional difference with varied interactions with the surrounding residues.
In both the hTrpRS–TrpNH2O–ATP complex and the hTrpRS–TrpAMP complex, the AIDQ motif also adopts the closed conformation and the Trp-binding site has no notable conformational change compared to the hTrpRS–Trp complex (Figure 2A and B). The TrpNH2O analog and the Trp moiety of TrpAMP occupy almost the same position and maintain similar interactions with the surrounding residues as the Trp substrate in the hTrpRS–Trp complex. These results suggest that the structural elements and key residues involved in the recognition and binding of Trp are strongly conserved during the Trp activation reaction.
Recognition and binding of ATP
The structure of the hTrpRS–TrpNH2O–ATP complex is the first structure of hTrpRS bound with ATP and represents the pretransition state of the enzyme. Thus, analysis of this complex allows us to identify the structural elements and residues that are involved in the recognition and binding of ATP and/or play important roles in the aminoacylation reaction. In this complex, both the AIDQ motif and the KMSAS loop assume the closed conformations, forming a closed substrate-binding pocket (Figure 3 and Table 2). Meanwhile, both the N-terminal and C-terminal domains also adopt the closed conformations, assuming the closed overall conformation. The closed conformation of the KMSAS loop is necessary to assemble a proper ATP-binding site to accommodate the substrate and carry out the Trp activation reaction.
Figure 3.
Conformational differences of the substrate-binding pocket. (A) Electrostatic surfaces of the substrate-binding pocket in different enzymatic states, showing the changes of the size and shape of the pocket. (B) A graph diagram showing the changes of the solvent-accessible surface area and the volume of the pocket. For parallel comparison, the N-terminal β-hairpin in the pretransition and product states is omitted in the surface presentation and the calculation of the solvent-accessible surface area and the volume of the pocket.
The bound ATP assumes a bent conformation and has extensive interactions with residues of the conserved HVGH, AIDQ and KMSAS motifs (Figures 1C and D, and S3B). Gly172 of the HVGH motif is strictly conserved in all TrpRSs and the lack of a side chain at this position provides the room to accommodate the adenine moiety of ATP. The N1 atom of the adenine moiety forms a hydrogen bond with the main-chain amide of Phe340 (2.8 Å); the N6 atom forms two hydrogen bonds with the main-chain carbonyls of Phe340 and Met350 of the KMSAS loop (2.9 and 3.1 Å, respectively); and the N7 atom forms a hydrogen bond with the side-chain Nɛ2 of His170 of the HVGH motif (2.8 Å) (Figures 1C and S3B). In addition, the adenine ring has π–π stacking interaction with the side chain of Phe339. The KMSAS and HVGH motifs are coupled together via a hydrogen bond between the main-chain carbonyl of Met350 and the side-chain Nɛ2 of His170 (3.6 Å). The specific recognition and binding of the adenine moiety by these conserved residues form the molecular basis for the enzyme to use only ATP but not GTP as the substrate.
The ribose moiety of ATP has interactions primarily with residues of the AIDQ motif (Figures 1C and S3B). Particularly, the ribose 2′-OH group forms a hydrogen bond with the side-chain Oδ2 of Asp312 (2.3 Å), which appears to be the determinant for distinguishing the ribose moiety of ATP from the deoxyribose moiety of dATP. The ribose 3′-OH group forms two hydrogen bonds with the side-chain Oδ1 of Asp312 (3.1 Å) and the side-chain Nɛ2 of Gln313 (2.7 Å). All of these interactions are present only in the hTrpRS–TrpNH2O–ATP complex, but not in the hTrpRS–TrpAMP complex. Since the AIDQ motif is also involved in the Trp binding, it is very likely that this motif plays a crucial role in bringing ATP and Trp together to form the transition state.
The triphosphate moiety of ATP is stabilized by extensive interactions with residues of the KMSAS loop, the HVGH motif, and the β5 of the RF domain (Figures 1D and S3B). The α-phosphate is stabilized by two hydrogen bonds with the side-chain Nζ of Lys349 (2.9 Å) and the side-chain Oγ of Ser351 (3.0 Å). The β-phosphate forms two hydrogen bonds with the side-chain Oγ atoms of Ser351 and Ser353 (both 3.1 Å). The γ-phosphate has hydrogen-bonding interactions with the main-chain carbonyls of Gly163 (3.5 Å) and Ala168 (2.7 Å), and the side-chain Nη1 of Arg162 (2.5 Å). In addition, the side-chain Nɛ2 of His173 forms two weak hydrogen bonds with the oxygen atoms bridging the α-, β-phosphates and the β-, γ-phosphates (3.9 and 3.5 Å, respectively).
Recently, a structural model of the hTrpRS–ATP complex was constructed based on the bsTrpRS–ATP complex and the hTrpRS–TrpAMP complex by overlapping the adenosine moiety of ATP with that of TrpAMP (20). Based on this model, a distal Arg162 of hTrpRS (which is absolutely conserved in eukaryotic TrpRSs) is proposed to serve as a surrogate for the missing second landmark Lys of the KMSKS motif (which is replaced by Ala in eukaryotic TrpRSs) for ATP binding. However, structural comparison between the hTrpRS–TrpNH2O–ATP complex and the hTrpRS–TrpAMP complex indicates that both adenine and ribose moieties of ATP cannot overlap onto those of TrpAMP even though the Trp substrate and the Trp moiety of TrpAMP are very well superimposed (see Discussion section). Moreover, sequence alignment also shows that the key residues involved in ATP binding are not well conserved between hTrpRS and bsTrpRS, suggesting that the two enzymes might use different residues to recognize and bind ATP. In fact, although most of the residues that were suggested to make interactions with ATP based on the model are involved in interactions with ATP in the crystal structure, the interaction profiles are substantially different. Particularly, Ser351 of the KMSAS motif plays a crucial role in recognizing the α- and β-phosphates through two hydrogen bonds in the crystal structure. However, these interactions are not present in the hTrpRS–ATP model (and the bsTrpRS–ATP complex). In bsTrpRS, these phosphates are mainly recognized by the second Lys (Lys195) of the KMSKS motif. This observation leads us to propose that hTrpRS might use Ser351 from the same sequence motif to compensate the functional role of the missing second Lys in a way that is more applicable and efficient than to employ a spatially adjacent but sequentially distal Arg162. Interestingly, Arg162 is indeed involved in ATP binding by forming a salt bridge with the γ-phosphate in the pretransition state complex which appears to take over the functional role of the conserved residue Lys111 of bsTrpRS, a residue located in the bacterial TrpRS-specific insertion between α-helices α12 and α13 of hTrpRS. In addition, Arg162 also helps to stabilize TrpAMP by forming a salt bridge with the α-phosphate in the product state complex. In the structure context, this functional role cannot be replaced by either Lys195 or Lys111 in bsTrpRS. This explains the biochemical data that the double mutant R162A/A352K hTrpRS can efficiently rescue ATP binding, but not the aminoacylation activity (20). These results suggest that the adaptive changes of hTrpRS at the key positions of the active site have multiple effects on the aminoacylation reaction, including the substrate binding and the catalytic reaction.
Comparison of the pretransition and product states
In this work, we also determined the structure of the hTrpRS–TrpAMP complex that was crystallized at the similar condition as the other complexes with the same space group and comparable unit cell parameters (Table 1). Thus, we have the advantage to compare these complexes in the context of the same crystal-packing effect. Since the overall structure of the hTrpRS–TrpAMP complex reported here accords well with that described previously (15,20) and the bound TrpAMP has almost identical interactions with the surrounding residues of the enzyme, we will focus specifically on the structural differences between the pretransition state and the product state. Structural comparison between the TrpNH2O–ATP-bound complex and the TrpAMP-bound complex reveals no notable conformational change of the enzyme at the active site or in the overall structure (Figure 2A). However, the positions of both adenine and ribose moieties of ATP and TrpAMP differ moderately even though the positions of the TrpNH2O analog and the Trp moiety of TrpAMP are superimposed very well (Figure 2B and C). In the TrpNH2O–ATP-bound complex, the adenine and ribose moieties of ATP are positioned closer to the KMSAS loop and make extensive contacts with residues of the loop. In addition, the AIDQ and HVGH motifs also make some contacts with the ribose and triphosphate moieties of ATP. The α-phosphate of ATP is positioned ∼6 Å away from the carboxyl group of Trp, but is in a nearly ideal position for inline nucleophilic attack similar to that seen in the bsTrpRS–TrpNH2O–ATP complex (9). In the TrpAMP-bound complex, however, the adenine and ribose moieties of TrpAMP are positioned much deeper into the active site with an averaged displacement of ∼3 and 4 Å, respectively, compared to those of ATP in the pretransition state. As a result, although the adenine moiety retains some interactions with the KMSAS loop, the ribose and α-phosphate moieties lost most of the interactions with the AIDQ motif and the KMSAS loop (Figure S3C). These results suggest that the adenine and ribose moieties of ATP in the pretransition state have to move closer to the amino acid in the transition state to carry out the aminoacyl-AMP formation reaction, and the changes of interactions between the substrate and the enzyme might play an important role in the formation and/or stabilization of TrpAMP and the release of the pyrophosphate group of ATP.
DISCUSSION
Human TrpRS differs substantially from prokaryotic TrpRSs in primary sequence, including residues at several key positions of the active site. In addition, hTrpRS has an extra N-terminal domain that plays an important role in the acquisition of an angiostatic activity. Previous structural and biochemical studies have also shown that hTrpRS has substantial differences from bsTrpRS in the structure of the active site and in the overall conformation, and the N-terminal domain is involved in the aminoacylation reaction. Thus, we raised the question of whether and how hTrpRS adapts these changes in the recognition and binding of the substrates and the catalytic mechanism of the aminoacylation reaction. To address this question, we determined the crystal structures of hTrpRS in complexes with Trp, TrpNH2O and ATP and TrpAMP, respectively. Analyses of these structures, together with those reported previously (15–19), enable us to define the structural elements and the key residues involved in the recognition and binding of the substrates, reveal the conformational changes of the enzyme associated with the substrate binding, and provide new insights into the catalytic mechanism of the Trp activation reaction by hTrpRS.
Conformational changes among different enzymatic states
Structural comparisons of hTrpRS in different enzymatic states clearly show that upon the binding of Trp and ATP, several structural elements at the active site, particularly the AIDQ motif and KMSAS loop, undergo substantial conformational changes which are further coupled with movements of the N-terminal and C-terminal domains, forming different conformations of the substrate-binding pocket and the overall structure (Figure 3 and Table 2). In the apo hTrpRS, both the AIDQ motif and the KMSAS loop are positioned away from the active site and assume the open conformations. Thus, the substrate-binding pocket is widely open with a solvent-accessible surface area of 1481 Å2 and a volume of 3347 Å3, respectively. In addition, both the N-terminal and C-terminal domains are apart from the RF domain, forming an open overall conformation. The binding of Trp induces the AIDQ motif to move closer toward the active site and adopt the closed conformation, which further induces both the N-terminal and C-terminal domains moving toward the RF domain to adopt the closed conformations, forming a compact or closed overall conformation. Interestingly, upon the binding of Trp, the KMSAS loop also shifts toward the active site and assumes the semi-closed conformation, which is however not in position to interact with ATP. As a result, the substrate-binding pocket assumes a partially closed conformation (the solvent-accessible surface area and the volume are ∼1100 Å2 and 2200 Å3, respectively). When both TrpNH2O and ATP are bound, the AIDQ motif and the N-terminal and C-terminal domains all maintain the closed conformations. However, the KMSAS loop moves further toward the active site and assumes the closed conformation to interact with ATP, forming a closed substrate-binding pocket (the solvent-accessible surface area and the volume are ∼930 Å2 and 1600 Å3, respectively). In the hTrpRS–TrpAMP complex, the enzyme assumes similar conformations of the overall structure and the substrate-binding pocket as those in the pretransition state. However, it is yet unclear whether any subtle conformational change takes place at the active site in the formation of the transition state.
During the Trp activation reaction, hTrpRS can have two different conformations of the AIDQ motif and the N-terminal and C-terminal domains: an open conformation in the absence of Trp and a closed conformation in the presence of Trp. Formation of the closed Trp-binding site depends solely on the binding of Trp. Amino acid-binding-induced conformational changes have also been observed in other aaRSs, such as MetRS, HisRS and LysRS (30–32). On the other hand, the KMSAS loop can assume three subtly distinct conformations: an open conformation in the absence of the substrates, a semi-closed conformation in the presence of Trp and a closed conformation in the presence of both Trp and ATP. It is intriguing to observe that the KMSAS loop has conformational change when Trp binds because it is not directly involved in the Trp binding. Thus, the cause(s) of the conformational changes of the KMSAS loop is not clear. Nevertheless, it is certain that the binding of both Trp and ATP can lead to and the binding of ATP is indispensable for the formation of the closed ATP-binding site. It is yet unknown whether the binding of ATP alone is sufficient to induce the formation of the closed ATP-binding site and the crystal structure of the hTrpRS–ATP complex should be able to resolve the issue.
Catalytic mechanism of the Trp activation reaction
The Trp activation reaction by hTrpRS appears to follow a typical inline displacement mechanism (33–35). In the pretransition state, the α-amino group of Trp is precisely positioned by three hydrogen bonds with residues Glu199, Gln284 and a conserved water molecule (Figure S3B). The side chain of Gln284 is further stabilized by two hydrogen bonds with the side-chain hydroxyl group of Tyr159 (which plays a crucial role in determining the side-chain specificity of Trp) and the side-chain carbonyl group of Gln313 of the AIDQ motif. The attacking carboxyl group of Trp is stabilized by the side-chain NH3+ group of Lys200 via a salt bridge. Considering that the α-phosphate of ATP is positioned ∼6 Å away from the carboxyl group of Trp in the pretransition state and the AMP moiety of TrpAMP in the product state is positioned much deeper into the active site, it seems necessary that to reach the transition state, ATP has to move closer toward Trp so that the carboxyl group of Trp can carry out the nucleophilic attack on the α-phosphate of ATP to form TrpAMP. In addition to their involvements in the positioning and activation of Trp, residues Asp312 and Gln313 of the AIDQ motif also make hydrogen bonds with the ribose moiety of ATP and Lys200 is also involved in interaction with the triphosphate group of ATP through Ser165 in the pretransition state. Thus, it is very likely that these residues might play important roles in bringing the two substrates together in the transformation from the pretransition state to the transition state. On the other hand, the conserved residues Lys349 and Ser351 of the KMSAS motif interact with the α-phosphate of ATP via salt bridge and hydrogen bonds and thus play a crucial role in positioning the reactive group of ATP. In addition, the positively charged Lys349 might play an important role in stabilizing the negatively charged transition state. The β- and γ-phosphate groups of ATP have extensively interactions with Ser351 and Ser353 (KMSAS motif), His173 (HVGH motif), Arg162, Gly163, Ser166 and Ala168 (the β5–α7 connecting loop), and these residues may be responsible for the stabilization and the release of the pyrophosphate group. In addition, since Arg162 and Gly163 also have interactions with the α-phosphate group of TrpAMP in the product state, they might play an important role in stabilizing TrpAMP (Figure S3C). Taking together, all the HVGH, AIDQ and KMSAS motifs conserved in subclass Ic aaRSs play key roles in the recognition and binding of Trp and ATP and the Trp activation reaction.
For TrpRSs, an Mg2+ ion is required for the amino acid activation reaction (26,36). In the bsTrpRS–TrpNH2O–ATP complex, an Mg2+ ion is bound at the active site that forms hydrogen bonds with all three phosphates of ATP (6). In the hTrpRS–TrpNH2O–ATP complex, a residual electron density peak was found at a similar position that was tentatively assigned as a water molecule. This water molecule has interactions with the α- and β-phosphates of ATP and the side chains of Ser165 and Lys200, but no interaction with the γ-phosphate (Figure 1D). This position could be occupied by an Mg2+ ion in the transition state that may help to stabilize the negative charges of the transition state during catalysis.
Functional role of the N-terminal β-hairpin in Trp activation
Previous biochemical data showed that the full-length, mini- (residues 48–471) and T1-hTrpRSs (residues 71–471) have aminoacylation activity, but T2-hTrpRS (residues 94–471) does not, suggesting that the N-terminal β-hairpin (residues 83–93) is indispensable for the aminoacylation reaction, but the HTH motif is not (13,14). The crystal structure of the hTrpRS–tRNATrp–Trp complex revealed that the N-terminal region is not involved in tRNA binding and particularly the β-hairpin has to be displaced in order for the enzyme to bind the 3′ end CCA of the tRNA, consistent with the kinetic data showing that T2-hTrpRS has no measurable activity in the Trp activation reaction, but weak activity in the acylation reaction (18). Moreover, mutagenesis data showed that two conserved residues Val85 and Val90 of the β-hairpin play an important role in Trp activation (37). All of these data pinpointed that the N-terminal β-hairpin is crucial for the Trp activation reaction, but not for the acylation reaction; however, its exact functional role was still elusive. Our structural data clearly show that the N-terminal β-hairpin is defined in the TrpNH2O–ATP-bound complex and partially defined in the TrpAMP-bound complex, but disordered in the apo form and the Trp-bound complex. In the pretransition and product states, the N-terminal β-hairpin is located on top of the active site and has interactions with both the KMSAS loop and the AIDQ motif, but no direct interaction with TrpNH2O, ATP or TrpAMP. Moreover, the Trp-binding site in the pretransition and product states has no conformational change compared to that in the Trp-bound state, suggesting that the β-hairpin is not essential for Trp binding. Furthermore, isothermal titration calorimetry data have shown that deletion of the β-hairpin would dramatically decrease the binding affinity for ATP, but not for Trp (20). Based on these structural and biochemical data, we conclude that the functional role of the N-terminal β-hairpin in the Trp activation reaction is to facilitate the binding of ATP and the stabilization of TrpAMP through properly positioning the KMSAS loop and the AIDQ motif.
Structural basis for the mechanistic differences between hTrpRS and bsTrpRS
Structural comparison between hTrpRS and bsTrpRS indicates that although both enzymes utilize an induced-fit mechanism in Trp activation, the binding of substrates can cause different conformational changes at the active site and in the overall structure. bsTrpRS assumes an open conformation in the absence of substrate (7). The binding of Trp does not cause any conformational change of the enzyme and the Trp-binding site assumes a closed conformation in all enzymatic states, suggesting that the recognition of Trp by bsTrpRS is via a lock-and-key mechanism (9). Also, no conformational change is observed when ATP binds alone (at physiologically low concentration). When both Trp and ATP bind, the KMSAS loop undergoes a substantial conformational change toward the active site (∼6 Å) which further induces a hinge-like movement of the C-terminal domain toward the RF domain (the anticodon-binding site is displaced by ∼10 Å), forming a compact closed pretransition state. In the transition state, the enzyme assumes a similar conformation as that in the pretransition state; however, it is in a high-energy conformation stabilized by the ligands (10,11). After catalysis, the KMSAS loop shifts slightly outwards (∼2 Å) which causes the C-terminal domain moving moderately away from the RF domain (the anticodon binding site is displaced by ∼5 Å), forming a less compact closed product state (6). In contrast, hTrpRS has only two different overall conformations dependent on the conformations of the AIDQ motif and the N-terminal and C-terminal domains (open and closed), but three subtly distinct conformations of the substrate-binding pocket mainly due to the conformational differences of the KMSAS loop (open, semi-closed and closed). The recognition of Trp by hTrpRS is via an induced-fit mechanism. The conformational changes of the KMSAS loop and the N-terminal and C-terminal domains are not drastic as those of bsTrpRS. These differences can be well explained by the structural differences of the two enzymes.
At the Trp-binding site, because bsTrpRS does not have an N-terminal domain, the AIDQ motif and strand β5 (equivalent to strand β9 of hTrpRS) are exposed to the surface and pack tightly with the core of the RF domain. The AIDQ motif forms extensive interactions with residues 101–107 (equivalent to helix α12 of hTrpRS) and is stabilized in the closed conformation. Thus, the AIDQ motif can interact with Trp in the apo form and no conformational change is necessary when Trp binds. In hTrpRS, however, the AIDQ motif has very few hydrophobic contacts with helix α12 and is sandwiched between the core of the RF domain and the N-terminal domain. In addition, residues of helix α3 of the N-terminal domain have interactions with several residues adjacent to the AIDQ motif. These interactions not only stabilize the conformation of the AIDQ motif, but also couple the N-terminal and C-terminal domains with the RF domain. In the apo hTrpRS, the AIDQ motif assumes the open conformation and cannot interact with Trp. When Trp binds, the AIDQ motif has to move closer toward the active site to interact with Trp. Meanwhile, the N-terminal and C-terminal domains are pulled closer toward the RF domain in a concerted manner.
At the ATP-binding site, bsTrpRS contains a six-residue insertion (residues 108–113) that forms part of an α-helix to cover part of the entrance to the substrate-binding pocket opposite the KMSAS loop and narrows the entrance substantially. Thus, the KMSAS loop has to adopt a widely open conformation in the apo form to allow the substrates to enter into the pocket. When both substrates bind to the enzyme in the pretransition state, the KMSAS loop closes up to form a closed active site and the C-terminal domain moves toward the RF domain in a concerted way to assume a closed overall conformation. Because hTrpRS does not contain the insertion, the entrance to the substrate-binding pocket in the apo form is much wider than that in the apo bsTrpRS. Both Trp and ATP can enter into the pocket without significant conformational change of the KMSAS loop. Therefore, the conformational changes of the KMSAS loop between different enzymatic states are smaller compared to those of bsTrpRS. These results indicate that the catalytic mechanism of the Trp activation reaction by hTrpRS is more complex and fine-tuned than that of bsTrpRS and involves more moderate conformational changes of the structural elements at the active site.
Comparison between TrpRS and TyrRS
As mentioned earlier, TrpRS shares high homology in both sequence and structure with TyrRS, the other member of the Ic subclass, which has also been extensively studied at the biochemical and structural levels (19,38–45). So it is feasible and valuable to make a comparison between the two closely related enzymes. Due to the lack of an N-terminal domain equivalent to that of hTrpRS, the Tyr-binding site in TyrRSs from different species assumes a rigid conformation similar to that of bsTrpRS and the binding of Tyr does not cause substantial conformational change of the Tyr-binding site, suggesting that the recognition and binding of Tyr by TyrRSs are more similar to that by bsTrpRS (19,38,42). On the other hand, moderate conformational changes of the KMSKS loop are observed at the ATP-binding site between different enzymatic states, which are more similar to those of hTrpRS (38,41,43,45). Moreover, the dramatic relative domain movements observed in bsTrpRS are not seen in TyrRSs due to the structural differences of the linker region between the RF domain and the C-terminal domain. In bsTrpRS, the linker is exposed on the surface and tilted against the β-sheet of the RF domain, thus allowing the relative movement of the two domains coupled with the conformational change of the KMSAS loop upon the binding of ATP and Trp (9). In TyrRSs, however, the linker is sandwiched between two β-strands of the RF domain and becomes a part of the β-sheet, and therefore the two domains are strongly coupled together (38,41,43,45).
Protein Data Bank accession codes
The structures of hTrpRS in complexes with Trp, TrpNH2O and ATP and TrpAMP, respectively, have been deposited with the Protein Data Bank under accession codes 2QUH, 2QUI and 2QUJ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the staff members of Photon Factory, Japan for technical support in diffraction data collection and other members of our group for helpful discussion. This work was supported by grants from the Ministry of Science and Technology of China (2004CB720102, 2006CB806501, and 2006AA02Z112), the National Natural Science Foundation of China (30570379, 30770480, and 30730028) and the Science and Technology Commission of Shanghai Municipality grants (07XD14032 and 07ZR14131). Funding to pay the Open Access publication charges for this article was provided by MOST.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cusack S. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 1997;7:881–889. doi: 10.1016/s0959-440x(97)80161-3. [DOI] [PubMed] [Google Scholar]
- 2.Woese C, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hountondji C, Dessen P, Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986;68:1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- 4.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 5.Carter CW, Jr, Doublié S, Coleman DE. Quantitative analysis of crystal growth – tryptophanyl-tRNA synthetase crystal polymorphism and its relationship to catalysis. J. Mol. Biol. 1994;238:346–365. doi: 10.1006/jmbi.1994.1297. [DOI] [PubMed] [Google Scholar]
- 6.Doublie S, Bricogne G, Gilmore C, Carter C.W., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995;3:17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- 7.Ilyin V, Temple B, Hu M, Li G, Yin Y, Vachette P, Carter C.W., Jr 2.9 Å crystal structure of ligand-free tryptophanyl-tRNA synthetase: domain movements fragment the adenine nucleotide binding site. Protein Sci. 2000;9:218–231. doi: 10.1110/ps.9.2.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Retailleau P, Yin Y, Hu M, Roach J, Bricogne G, Vonrhein C, Roversi P, Blanc E, Sweet RM, Carter C.W., Jr High-resolution experimental phases for tryptophanyl-tRNA synthetase (TrpRS) complexed with tryptophanyl-5′AMP. Acta Cryst. 2001;D57:1595–1608. doi: 10.1107/s090744490101215x. [DOI] [PubMed] [Google Scholar]
- 9.Retailleau P, Huang X, Yin Y, Hu M, Weinreb V, Vachette P, Vonrhein C, Bricogne G, Roversi P, Ilyin V, et al. Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations. J. Mol. Biol. 2003;325:39–63. doi: 10.1016/s0022-2836(02)01156-7. [DOI] [PubMed] [Google Scholar]
- 10.Retailleau P, Weinreb V, Hu M, Carter C.W., Jr Crystal structure of tryptophanyl-tRNA synthetase complexed with adenosine-5′ tetraphosphate: evidence for distributed use of catalytic binding energy in amino acid activation by class I aminoacyl-tRNA synthetases. J. Mol. Biol. 2007;369:108–128. doi: 10.1016/j.jmb.2007.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapustina M, Weinreb V, Li L, Kuhlman B, Carter C.W., Jr A conformational transition state accompanies tryptophan activation by B. stearothermophilus tryptophanyl-tRNA synthetase. Structure. 2007;15:1272–1284. doi: 10.1016/j.str.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F, Chen X, Xin L, Chen L, Jin Y, Wang D. Species-specific differences in the operational RNA code for aminoacylation of tRNA(Trp) Nucleic Acids Res. 2001;29:4125–4133. doi: 10.1093/nar/29.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otani A, Slike BM, Dorrell MI, Hood J, Kinder K, Ewalt KL, Cheresh D, Schimmel P, Friedlander M. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl Acad. Sci. USA. 2002;99:178–183. doi: 10.1073/pnas.012601899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl Acad. Sci. USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang XL, Otero FJ, Skene R, McRee DE, Schimmel P, de Pouplana LR. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl Acad. Sci. USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kise Y, Lee SW, Park SG, Fukai S, Sengoku T, Ishii R, Yokoyama S, Kim S, Nureki O. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2004;11:149–156. doi: 10.1038/nsmb722. [DOI] [PubMed] [Google Scholar]
- 17.Yu Y, Liu Y, Shen N, Xu X, Xu F, Jia J, Jin Y, Arnold E, Ding J. Crystal structure of human tryptophanyl-tRNA synthetase catalytic fragment. J. Biol. Chem. 2004;279:8378–8388. doi: 10.1074/jbc.M311284200. [DOI] [PubMed] [Google Scholar]
- 18.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, Schimmel P. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang XL, Guo M, Kapoor M, Ewalt KL, Otero FJ, Skene RJ, McRee DE, Schimmel P. Functional and crystal structure analysis of active site adaptations of a potent anti-angiogenic human tRNA synthetase. Structure. 2007;15:793–805. doi: 10.1016/j.str.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276A:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J.-S, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 24.Xie W, Nangle LA, Zhang W, Schimmel P, Yang XL. Long-range structural effects of a Charcot-Marie-Tooth disease-causing mutation in human glycyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 2007;104:9976–9981. doi: 10.1073/pnas.0703908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazat JP, Merle M, Graves PV, Merault G, Gandar JC, Labouesse B. Kinetic anticooperativity in pre-steady-state formation of tryptophanyl adenylate by tryptophanyl-tRNA synthetase from beef pancreas. A consequence of the tryptophan anticooperative binding. Eur. J. Biochem. 1982;128:389–398. doi: 10.1111/j.1432-1033.1982.tb06977.x. [DOI] [PubMed] [Google Scholar]
- 26.Trezeguet V, Merle M, Gandar JC, Labouesse B. On the mechanism of tRNATrp aminoacylation catalysed by beef tryptophanyl-tRNA synthetase using presteady-state kinetics. FEBS Lett. 1983;157:210–214. doi: 10.1016/0014-5793(83)81147-8. [DOI] [PubMed] [Google Scholar]
- 27.Trezeguet V, Merle M, Gandar JC, Labouesse B. Kinetic evidence for half-of-the-sites reactivity in tRNATrp aminoacylation by tryptophanyl-tRNA synthetase from beef pancreas. Biochemistry. 1986;25:7125–7136. doi: 10.1021/bi00370a055. [DOI] [PubMed] [Google Scholar]
- 28.Ward WH, Fersht AR. Asymmetry of tyrosyl-tRNA synthetase in solution. Biochemistry. 1988;27:1041–1049. doi: 10.1021/bi00403a029. [DOI] [PubMed] [Google Scholar]
- 29.Ward WH, Fersht AR. Tyrosyl-tRNA synthetase acts as an asymmetric dimer in charging tRNA: a rationale for half-of-the-sites activity. Biochemistry. 1988;27:5525–5530. doi: 10.1021/bi00415a021. [DOI] [PubMed] [Google Scholar]
- 30.Serre L, Verdon G, Choinowski T, Hervouet N, Risler JL, Zelwer C. How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding. J. Mol. Biol. 2001;306:863–876. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 31.Yaremchuk A, Tukalo M, Grotli M, Cusack S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: comparison with histidyl-tRNA synthetase. J. Mol. Biol. 2001;309:989–1002. doi: 10.1006/jmbi.2001.4712. [DOI] [PubMed] [Google Scholar]
- 32.Onesti S, Desogus G, Brevet A, Chen J, Plateau P, Blanquet S, Brick P. Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding. Biochemistry. 2000;39:12853–12861. doi: 10.1021/bi001487r. [DOI] [PubMed] [Google Scholar]
- 33.Lowe G, Tansley G. An investigation of the mechanism of activation of tryptophan by tryptophanyl-tRNA synthetase from beef pancreas. Eur. J. Biochem. 1984;138:597–602. doi: 10.1111/j.1432-1033.1984.tb07956.x. [DOI] [PubMed] [Google Scholar]
- 34.Belrhali H, Yaremchuk A, Tukalo M, Berthet-Colominas C, Rasmussen B, Bosecke P, Diat O, Cusack S. The structural basis for seryl-adenylate and Ap4A synthesis by seryl-tRNA synthetase. Structure. 1995;3:341–352. doi: 10.1016/s0969-2126(01)00166-6. [DOI] [PubMed] [Google Scholar]
- 35.Desogus G, Todone F, Brick P, Onesti S. Active site of lysyl-tRNA synthetase: structural studies of the adenylation reaction. Biochemistry. 2000;39:8418–8425. doi: 10.1021/bi0006722. [DOI] [PubMed] [Google Scholar]
- 36.Merle M, Trezeguet V, Graves PV, Andrews D, Muench KH, Labouesse B. Tryptophanyl adenylate formation by tryptophanyl-tRNA synthetase from Escherichia coli. Biochemistry. 1986;25:1115–1123. doi: 10.1021/bi00353a026. [DOI] [PubMed] [Google Scholar]
- 37.Guo LT, Chen XL, Zhao BT, Shi Y, Li W, Xue H, Jin YX. Human tryptophanyl-tRNA synthetase is switched to a tRNA-dependent mode for tryptophan activation by mutations at V85 and I311. Nucleic Acids Res. 2007;35:5934–5943. doi: 10.1093/nar/gkm633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brick P, Blow DM. Crystal structure of a deletion mutant of a tyrosyl-tRNA synthetase complexed with tyrosine. J. Mol. Biol. 1987;194:287–297. doi: 10.1016/0022-2836(87)90376-7. [DOI] [PubMed] [Google Scholar]
- 39.Fersht AR. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987;26:8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- 40.Fersht AR, Knill-Jones JW, Bedouelle H, Winter G. Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: a mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry. 1988;27:1581–1587. doi: 10.1021/bi00405a028. [DOI] [PubMed] [Google Scholar]
- 41.Brick P, Bhat TN, Blow DM. Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution: interaction of the enzyme with the tyrosyl adenylate intermediate. J. Mol. Biol. 1989;208:83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- 42.Yang XL, Skene RJ, McRee DE, Schimmel P. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc. Natl Acad. Sci. USA. 2002;99:15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kobayashi T, Nureki O, Ishitani R, Yaremchuk A, Tukalo M, Cusack S, Sakamoto K, Yokoyama S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Mol. Biol. 2003;10:425–432. doi: 10.1038/nsb934. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi T, Takimura T, Sekine R, Kelly VP, Kamata K, Sakamoto K, Nishimura S, Yokoyama S. Structural snapshots of the KMSKS loop rearrangement for amino acid activation by bacterial tyrosyl-tRNA synthetase. J. Mol. Biol. 2005;346:105–117. doi: 10.1016/j.jmb.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 46.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Binkowski TA, Naghibzadeh S, Liang J. CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.