Abstract
Fluorescent 2′-deoxynucleotides containing a protecting group at the 3′-O-position are reversible terminators enabling array-based DNA sequencing by synthesis (SBS) approaches. Herein, we describe the synthesis of a new family of 3′-OH unprotected cleavable fluorescent 2′-deoxynucleotides and their evaluation as reversible terminators for high-throughput DNA SBS strategies. In this first version, all four modified nucleotides bearing a cleavable disulfide Alexa Fluor® 594 dye were assayed for their ability to act as a reversible stop for the incorporation of the next labeled base. Their use in SBS leaded to a signal–no signal output after successive addition of each labeled nucleotide during the sequencing process (binary read-out). Solid-phase immobilized synthetic DNA target sequences were used to optimize the method that has been applied to DNA polymerized colonies or clusters obtained by in situ solid-phase amplification of fragments of genomic DNA templates.
INTRODUCTION
The development of accurate and high-throughput DNA-sequencing methods appears as essential for very large-scale analysis such as the exploration of entire genomes in an efficient and cost-effective manner. This new generation of massively parallel DNA-sequencing methods are expected to have a strong impact on biomedical research, health care and clinical medicine (1). A series of high-throughput technologies have been explored (2) and in particular Sequencing by Synthesis (SBS) approaches have been investigated and implemented using arrays of single DNA molecules (3) and clusters or polymerized colonies (4–6) generated by solid-phase in situ amplification of DNA. In SBS methods, a primer hybridized to its target sequence is extended after nucleotide incorporation into the growing DNA strand using a DNA polymerase. The detection of the incorporated nucleotide immediately after each incorporation reaction allows the sequence assignment along the DNA synthesis process. Furthermore, the removal of the reporter signal, such as a fluorophore, after each base identification is essential to ensure that the residual fluorescence from the previous nucleotide incorporation does not affect the identification of the next incorporated fluorescent nucleotide.
In the design of fluorescently labeled reversible chain terminators for SBS, the linker used as a chemically cleavable moiety to attach the fluorophore to 2′-deoxynucleotides, must satisfy several requirements: (1) stability during the polymerase-mediated extension step, (2) its structure (geometry and size) and its location within the 2′-deoxynucleotide moiety must not prevent the recognition of the resulting labeled nucleotide by standard DNA polymerases, (3) cleavage under mild conditions compatible with the stability of DNA biopolymers (single and double strands) and the functionalized surface of DNA biochips, (4) easy synthetic access and high bioconjugation efficiency towards modified 2′-deoxynucleotides and/or fluorophores. Thus, in the context of DNA sequencing, the chemistry of enzyme-labile, Pd-cleavable and photocleavable linkers has been already extensively studied by some academic groups (7–18) and genomic Industry (19–21). A viable sequencing scheme involving the use of 3′-O-allyl Pd-cleavable fluorescent nucleotide analogs as reversible terminators, was recently reported by Ju et al. (22). With the goal in mind to adapt our DNA colonies technology (4,6) to high-throughput DNA sequencing, we have explored a new SBS strategy evaluating the use of cleavable disulfide fluorescent nucleotides as chemically cleavable reversible terminators. Indeed, disulfide bridge containing linkers exhibits some attractive features: inertness towards most of standard DNA polymerases, cleavage under mild reducing conditions (i.e. short treatment with a thiol or a water-soluble trialkylphosphine) compatible with DNA biomolecules and surface immobilization chemistries, and they can be easily introduced into modified nucleotides by using commercially available heterobifunctional cross-linking reagents or by applying the promising chemistry of some thiol-labile protecting groups recently published by us (23) and other groups (24). Application of such cleavable linkers to the preparation of bioconjugates (25) and as an efficient reversible covalent chemistry in drug delivery (26) and solid-phase synthesis (27,28) have been already reported.
Herein we report a new family of 3′-OH unprotected cleavable fluorescent 2′-deoxynucleotides and preliminary experimental optimization for their use as reversible terminators for DNA sequencing by synthesis. Thus, immobilized DNA moieties containing an hairpin structure or hybridized with a primer and solid-phase amplified template DNA molecules (DNA polymerized colonies or clusters) in a microfluidics device, were sequenced with sufficient accuracy up to about 10 nucleotides using this novel class of fluorescent nucleotide analogs, commercial polymerases and an automated inverted fluorescence microscope equipped with a CCD imaging system.
MATERIALS AND METHODS
5-(3-Amino-1-propynyl)-2′-deoxycytidine 5′-triphosphate [dCTP(AP3)] and 5-(3-amino-1-propynyl)-2′-deoxyuridine 5′-triphosphate [dUTP(AP3)] were prepared in two steps [i.e. Pd(0)-catalyzed Sonogashira cross-coupling with N-propargyltrifluoroacetamide and triphosphorylation reaction] from the commercially available 5-iodo-2′-deoxynucleosides (respectively, 5-iodo-2′-deoxycytidine from Berry & Associates, Inc. and 5-iodo-2′-deoxyuridine from Fluka) according to literature procedures (29,30). 7-(3-Amino-1-propynyl)-7-deaza-2′-deoxyadenosine 5′-triphosphate [dATP(AP3)] was prepared in the same manner by using 7-deaza-7-iodo-2′-deoxyadenosine which was readily synthesized from 7-deaza-2′-deoxyadenosine (i.e. 2′-deoxytubercidin from Berry & Associates, Inc.) according to a known method (31). 7-(3-Amino-1-propynyl)-7-deaza-2′-deoxyguanosine 5′-triphosphate [dGTP(AP3)] was prepared from 2-amino-4-methoxy-7-(β-D-2-deoxyribofuranosyl)pyrrolo-[2,3-d]pyrimidine (Berry & Associates, Inc.) by using the multi-step synthetic procedure initially developed by Hobbs Jr et al. and slightly modified by Ramzaeva and Seela (31,32). Texas Red-X succinimidyl ester (TR-X-SE) was prepared in four steps from sulforhodamine 101 according to literature procedures (33). Alexa Fluor® 594 carboxylic acid was synthesized by using a multi-step synthetic procedure described in a WO Patent Application from Invitrogen (formerly Molecular Probes, Inc.) and entitled ‘Sulfonated Xanthene Derivatives’ (34). The two isomers of this water-soluble rhodamine derivative were resolved by flash-chromatography on a RP-C18 silica gel column. The first eluted isomer (i.e. the most polar) was named isomer I and the second one isomer II. Structural assignment of each isomer was achieved by 1H NMR spectroscopy: isomer I corresponds to the 1,4-dicarboxylic acid derivative, whereas isomer II is the 1,3-dicarboxylic acid derivative. N-Succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) cross-linking reagents was purchased from Pierce. Synthetic oligonucleotides were from Eurogentec and were subjected to RP-HPLC purification and ESI-MS characterization before use (see below for name and sequence). Chemical modification of their 5′-end with the cleavable disulfide linker or fluorescent labels were performed ‘in house’ as recently described by us (6). Klenow (exo-) pol (MO212L, 5 U/μl) was purchased from New England Biolabs, Inc. The HPLC-gradient grade CH3CN was obtained from Panreac. 2.0 M Triethylammonium acetate (TEAA) and 1.0 M triethylammonium bicarbonate (TEAB) buffers were purchased from Glen Research Corp. and Fluka, respectively. Buffers and aq. mobile-phases for HPLC were prepared using water purified with a Milli-Q system (purified to 18.2 MΩ cm).
DNA biochips
DNA biochips resulting from glass channels functionalization (BTA chemistry), primers and/or templates grafting and formation of DNA colonies by solid-phase amplification, were prepared by using a microfluidic platform and conditions recently reported by us (35,36).
Synthetic oligonucleotides
Oligonucleotides sequences used in model sequencing experiments were purchased from Eurogentec (Belgium) (Table 1).
Table 1.
Oligonucleotides sequences
| Oligo # | Sequence | Model |
|---|---|---|
| 1 | 5′-amino (C6)-CACTCCATCTGGGACCCTGGCTTGGGTG GTGCGAAGCACCACCCA | Hairpin sequence 1 |
| 2A | 5′-amino (C6)-CACTCCATCTGGGACCCTGGCTCTCGACTCTGGGAA AACACTGGGTTTTCCCAGAGTCGAG | Hairpin sequence 2 For A first base incorporation |
| 2C | 5′-amino (C6)-CACTCCATCTGGCTGACCTATGCTCGACTCTGGGAAA ACACTGGGTTTTCCCAGAGTCGAG | Hairpin sequence 2 For C first base incorporation |
| 2G | 5′-amino (C6)-CACTCCATCTAGCCGTTATTACCTCGACTCTGGGAAA ACACTGGGTTTTCCCAGAGTCGAG | Hairpin sequence 2 For G first base incorporation |
| 2U | 5′-amino (C6)-CACTCCATCTCTTTGGTCAGGACTCGACTCTGGGAAA ACACTGGGTTTTCCCAGAGTCGAG | Hairpin sequence 2 For U first base incorporation |
| 3 | 5′-amino (C6)-GAGGAAAGGGAAGGGAAAGGAAGGGAATTCTACATGCCG ATGACCTGCAGAAGGATCCTTCCTTTCCCTTCCCTTTCCTC | Linear sequence |
| 4 | GAGGAAAGGGAAGGGAAAGGAAGG | Sequencing primer of oligo 3 |
| 5 | 5′-amino (C6)-CAAGCAGAAGACGGCATACGATCCGACAGCTTCAGTTGAA GATATTACACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG | Synthetic Template, Lambda phage fragment F |
| 6 | 5′-amino (C6)-CAAGCAGAAGACGGCATACGATCCGACAGCTTAAGGTTAA ATTAGAACAC TGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG | Synthetic Template, Lambda phage fragment M |
| 7 | 5′-amino (C6)-CAAGCAGAAGACGGCATACGATCCGACAGCTTAGGTGAGA ACATCCCCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAG | Synthetic Template, Lambda phage fragment G |
| 8 | CTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTG | Primer for sequencing synthetic Lambda templates |
| 9 | TCCCGACTGGAAAGCGGGCAGTGATACTGGGCTTCAGTAAGCTGTCGG ATCGTATGCCGTCTTCTGCTTG | Synthetic Template. 33P-5′ |
| 10 | CAAGCAGAAGACGGCATACGATCCGACAGCTT | Primer for sequencing oligo 9 |
Instruments
Analytical and semi-preparative HPLC were performed on a Waters Breeze instrument equipped with a dual wavelength detector. UV-visible spectra were obtained on a Jasco V550 spectrophotometer. Fluorescence spectroscopic studies were performed with a Jasco FP-750 spectrophotometer. Mass spectra were obtained with a Micromass Quattro Micro QAA118 apparatus equipped with an electrospray source.
For fluorescence measurements in solid phase we used an inverted fluorescence microscope equipped with an arc mercury lamp (Axiovert 200 + HBO 100W/2, Carl Zeiss) and coupled to a CCD camera ORCA ER from Hamamatsu. A main concern to quantitative analysis of fluorescence intensities is the fluorophore bleaching. The focus position was approached under regular top light. This method allowed reaching the final focus within a few seconds, minimizing the fluorophore bleaching. Moreover, data analysis was always processed from at least 5 images per sample (channel). Fluorescent signals were measured by integration of the signal in images using in house developed software.
HPLC separations
Several chromatographic systems were used for the analytical experiments and the purification steps. Each one of these systems was optimized in order to improve separation conditions.
System A: RP-HPLC (Waters Xterra MS C18 column, 5 μm, 7.8 × 100 mm) with CH3CN and 0.1% aq. trifluoroacetic acid (aq. TFA, 0.1%, v/v, pH 2.0) as eluents [100% TFA (5 min), linear gradient from 0% to 8% (5 min) and 8% to 50% (40 min) of CH3CN] at a flow rate of 2.5 ml/min. Dual UV-visible detection was achieved at 260 and 595 nm.
System B: RP-HPLC (Waters Xterra MS C18 column, 5 μm, 7.8 × 100 mm) with CH3CN and TEAA buffer (100 mM, pH 7.0) as eluents [100% TEAA (5 min), linear gradient from 0% to 10% (5 min) and 10% to 50% (50 min) of CH3CN] at a flow rate of 3.0 ml/min. Dual UV detection was achieved at 260 nm and 272, 280, 293 nm for dGTP(AP3), dATP(AP3) and dCTP(AP3) [or dUTP(AP3)], respectively.
System C: System B with the following gradient [95% TEAA (5 min), linear gradient from 5% to 35% (60 min) of CH3CN] at a flow rate of 3.0 ml/min. Dual UV-visible detection was achieved at 595 nm and 272, 280, 293 nm for dGTP(AP3), dATP(AP3) and dCTP(AP3) [or dUTP(AP3)], respectively.
System D: RP-HPLC (Waters Xterra MS C18 column, 2.5 μm, 2.1 × 50 mm) with CH3CN and TEAA buffer (50 mM + 2 mM EDTA, pH 7.5) as eluents [linear gradient from 0 to% 20% (8 min) and 20% to 50% (12 min) of CH3CN] at 30°C and with a flow rate of 0.5 ml/min. Dual UV-visible detection was achieved at 595 nm and 272, 280, 293 nm for dGTP(AP3), dATP(AP3) and dCTP(AP3) [or dUTP(AP3)], respectively.
Synthesis of Alexa Fluor® 594-thiol 6 (see Supplementary Figure1)
Preparation of N-hydroxysuccinimidyl ester (single isomer): An amount of 1.38 mg of Alexa Fluor® 594 carboxylic acid (1.90 μmol, weighed in a 0.5 ml Eppendorf tube) was dissolved in 10 μl of dry DMF containing 0.69 mg of TSTU (2.28 μmol). After complete solubilization by vortexing, 1.32 μl of dry TEA (9.5 μmol) was added and the resulting reaction mixture was protected from light and periodically vortexed for 30 min. The conversion of Alexa Fluor® 594 carboxylic acid into its N-hydroxysuccinimidyl ester was checked for completion by ESI-MS. This NHS ester was used without further purification (yield was assumed to be quantitative). MS (ESI, negative mode): m/z 408.26 [M-2H]2−, calcd for C39H37N3O13S2 819.87.
Coupling reaction: A fresh solution of cysteamine [7.32 mg weighed in a 2.0 ml Eppendorf tube and dissolved in 320 μl of 0.1 M sodium bicarbonate buffer (pH 8.0)] was added to the crude N-hydroxysuccinimidyl ester. The resulting reaction mixture was protected from light and periodically vortexed for 30 min. Thereafter, a 1.0 M aq. solution of DTT (133 μl) was added and the resulting solution was again periodically vortexed for 30 min. Finally, the reaction mixture was quenched by dilution with aq. TFA 0.1% (1 ml) and purified by RP-HPLC (system A). The product-containing fractions were lyophilized to give the fluorophore-thiol 6 as a dark purple solid. Quantification was achieved by UV-visible measurements at λmax = 588 nm of Alexa Fluor® 594 dye by using the ɛ value 80 400 M−1 cm−1 [yield after RP-HPLC purification: 40% (isomer I), 70% (isomer II)]. The stock solution of Alexa Fluor® 594-thiol was partitioned into several sealed vials (∼250 nmol/vial), lyophilized and stored at 4°C under an argon atmosphere. MS (ESI, negative mode): m/z 389.48 [M-2H]2−, calcd for C37H39N3O10S3 781.93.
Synthesis of cleavable disulfide Alexa Fluor® 594 nucleotides [dNTP(AP3)-SS-AF594]
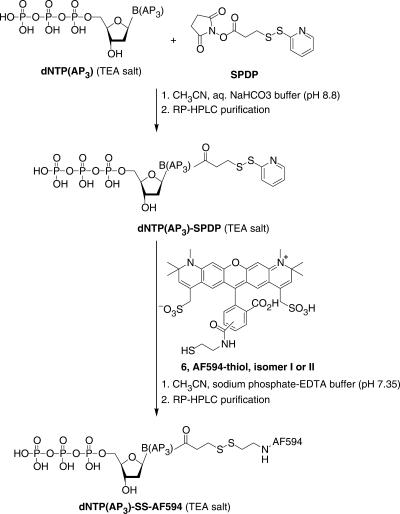
Preparation of dNTP(AP3)-SPDP conjugates: dNTP(AP3) (300 nmol, lyophilized in a 2.0 ml Eppendorf tube immediately after sampling from the stock solution) was dissolved in 0.1 M sodium bicarbonate buffer (pH 8.8, 150 μl) and the resulting solution was stored at room temperature. A solution of SPDP cross-linking reagent (1.40 mg in 80 μl, 4.48 μmol) in dry CH3CN was added and the resulting reaction mixture was periodically vortexed for 1 h at room temperature (after several min, it is essential to add 1 μl of TEA to reach again a pH ∼9.0). After lyophilization, the resulting residue was dissolved in 700 μl of TEAA buffer (100 mM, pH 7.0) and purified by RP-HPLC (system B). The product-containing fractions were lyophilized to give the corresponding dNTP(AP3)-SPDP conjugate as a colorless oily residue. As illustrated in Supplementary Figure 2, the RP-HPLC elution profile of the crude mixture of such reaction exhibits numerous peaks assigned to degradation products of SPDP reagent. The following retention times were found for the dNTP(AP3)-SPDP conjugates: 17.0 min [dCTP(AP3)-SPDP], 17.8 min [dUTP(AP3)-SPDP], 19.5 min [dGTP(AP3)-SPDP] and 20.5 min [dATP(AP3)-SPDP].
Disulfide exchange reaction between dNTP(AP3)-SPDP and fluorophore-thiol 6: dNTP(AP3)-SPDP (see earlier) was dissolved in a mixture of oxygen-free buffer [50 mM sodium phosphate, 10 mM EDTA (pH 7.4, 75 μl)] and CH3CN (25 μl). The resulting solution was added into a sample of lyophilized fluorophore-thiol (isomer I for dCTP and dGTP, isomer II for dATP and dUTP, 720 nmol, 2.5 equiv.). Disulfide exchange reaction was achieved at room temperature in the absence of light with periodic vortexing for 5 h (or overnight). After dilution with TEAA buffer (100 mM, pH 7.0), the reaction product was purified by RP-HPLC (system C). The following retention times were found for the dNTP(AP3)-SS-AF594 derivatives: ∼32.0 min [dCTP(AP3)- and dGTP(AP3)-SS-AF594, isomer I] and ∼38.0 min [dATP(AP3)- and dUTP(AP3)-SS-AF594, isomer II]. The product-containing fractions were twice lyophilized to remove significant amounts of TEAA salts. Stock solutions of dNTP(AP3)-SS-AF594 were prepared in HPLC grade water and UV-visible quantification was achieved at λmax of the Alexa Fluor® 594 dye by using the ɛ value 80 400 M−1 cm−1 (overall yield ∼50% for the two steps). The structure of each dNTP(AP3)-SS-AF594 was confirmed by ESI mass spectrometry and the purity (>95%) was checked by RP-HPLC (system D). MS (ESI, negative mode. Mass found after deconvolution analysis): m/z 1410.49 [dATP(AP3)-SS-AF594, isomer I], 1409.19 [dATP(AP3)-SS-AF594, isomer II] calcd for C54H61N8O23P3S4 1411.31; 1424.50 and 1408.32 [K+ and Na+ adducts of dCTP(AP3)-SS-AF594, isomer I], 1425.10 and 1408.67 [K+ and Na+ adducts of dCTP(AP3)-SS-AF594, isomer II] calcd for C52H60N7O24P3S4 1388.27; 1447.50 [Na+ adduct of dGTP(AP3)-SS-AF594, isomer I], 1446.86 [Na+ adduct of dGTP(AP3)-SS-AF594, isomer II] calcd for C54H61N8O24P3S4 1427.30; 1412.12 [Na+ adduct of dUTP(AP3)-SS-AF594, isomer I], 1430.84 [K+ adduct of dUTP(AP3)-SS-AF594, isomer II] calcd for C52H59N6O25P3S4 1389.25.
The preparation of dNTPs(AP3)-SS-TR was accomplished in the same manner; Texas Red-X-thiol was used instead of Alexa Fluor® 594-thiol in the disulfide exchange reaction.
DNA attachment
The methods for attaching 5′-aminated DNA species on glass activated surfaces of microfluidic channels and the procedures for surface amplification of templates have been previously described in detail by us (6). Briefly, the main steps of the process for grafting DNA using glass microfabricated devices are:
(1) Glass slide surface activation and aminosilanization, (2) carboxylation of ATS glass slide, (3) Attachment of 5′-aminated oligonucleotides to carboxylated surfaces.
DNA extension using dNTP(AP3)-SS-AF594
The procedure optimized using Klenow (exo-) is as follows:
Selected isomer for dNTP(AP3)-SS-AF594: isomer II of dATP-SS-AF594, isomer I of dCTP-SS-AF594, isomer I of dGTP-SS-AF594 and isomer II of dUTP-SS-Alexa594.
Sequencing buffer: buffer for dCTP-SS-AF594, dGTP-SS-AF594 and dUTP(AP3)-SS-AF594 contains 250 mM NaCl, whereas buffer for dATP(AP3)-SS-AF594 contains 500 mM NaCl. Preparation was done as follows: use 5× Klenow buffer (250 or 500 mM NaCl), add BSA (10 mg/ml) to a final concentration of 0.1 mg/ml BSA, add DMSO to a final concentration of 20% (v/v) and finally add NaCl to a final concentration of 250 or 500 mM.
Enzyme solution: add Klenow (exo-) pol to the corresponding sequencing buffer (vide supra) to a final concentration of 0.066 U/μl.
The polymerase extension reactions were performed within microchannels of DNA biochip as follows:
Step 1: Dilute the dNTP(AP3)-SS-AF594 in their respective buffers at a final concentration of 600 nM.
Step 2: Wash the microchannels of biochip using sequencing buffer according to the base being incorporated [buffer 250 mM NaCl for dCTP-SS-AF594, dGTP-SS-AF594 and dUTP(AP3)-SS-AF594 or 500 mM NaCl for dATP(AP3)-SS-AF594] for 5 min.
Step 3: Fill the microchannel with the enzyme solution. Run pulses of 2.5 s followed by 15 s stop for a total time of 4.5 min.
Step 4: Wash for 3 min using 0.1% SDS into 0.1 M Tris–HCl buffer pH 7.5.
Step 5: Wash for 2 min with 4× SSC, 20% DMSO.
Step 6: Wash for 4 min with 4× SSC, 20% DMSO containing 50 mM sodium ascorbate.
Step 7: Fluorescence microscopy measurement using the instrumental setup described earlier equipped with a 20× objective (NA 0.4) and an optical filter appropriate for Alexa Fluor® 594 dye (XF3081, Omega). The irradiation times were variable depending on the experiment, but usually in the range of 1 s for most of the experiments reported here.
RESULTS AND DISCUSSION
3′-O-blocking group
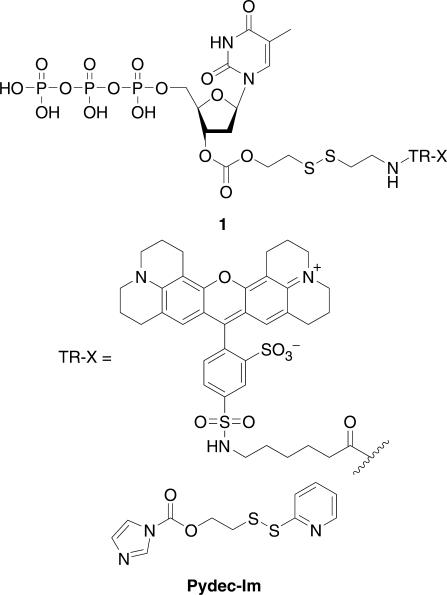
The considerations outlined earlier led us to explore firstly the design and synthesis of nucleotides with a 3′-O-blocking group that is both thiol-labile and fluorescent. Thus, the use of 2-pyridyldithioethyloxycarbonyl imidazole (Pydec-Im) has enabled to label efficiently thymidine with Texas Red® fluorophore to give the reversible terminator 1 (Figure 1). However, preliminary experiments on the incorporation of 1 by DNA polymerases [i.e. T7 Sequenase v2.0 and Klenow (exo-)] have shown that this thymidine carbonate is not incorporated by these DNA replicating enzymes. The unsuccessful DNA polymerase-mediated incorporation of 3′-O-modified nucleotides has also been reported by the Burgess group (37) and was explained as follows: the 3′ position on the deoxyribose is very close to the amino acid residues in the active site of the polymerase, and the polymerase is therefore sensitive to modification in this area of the ribose ring, especially with a bulky fluorophore such as rhodamine derivatives.
Figure 1.
Structures of cleavable fluorescent dNTPs 1 and heterobifunctional cross-linking reagent Pydec-Im.
Unprotected 3′-OH dNTP(AP3)-SS-AF594 nucleotides as reversible terminators
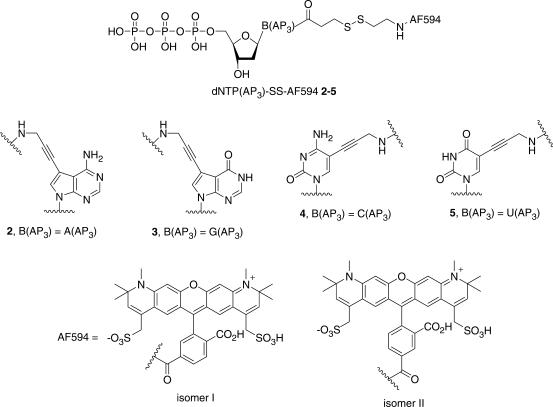
We thus focused on the design and synthesis of nucleotide analogs, in which the fluorophore is linked to the pyrimidine or purine bases through a cleavable disulfide linker. Indeed, it is known that most of DNA polymerases are highly tolerable to extensive modifications with large groups such as fluorescent dyes at the 5 position of pyrimidines (C and T) and 7 position of purines (A and G) (38,39). Furthermore, we postulated that the steric hindrance of the cleavable fluorophore may confer some terminating properties to these free 3′-OH-modified nucleotides. Recently, the use of 3′-OH-free fluorescent deoxynucleotides as terminators of DNA polymerization has been reported for single bases extensions in microarrays (40) supporting the fact that 3′-OH-modified nucleotides can act as terminators for DNA polymerase-mediated elongation under certain experimental conditions. It is clear that for sequencing applications the removal of the label is needed and in this context, our goal was to prepare the four chemically cleavable fluorescent nucleotide analogs 2–5, with an Alexa Fluor® 594 dye attached to the 5 (or 7) position of pyrimidine (or purine) base via a cleavable disulfide linker (Figure 2). Alexa Fluor® 594 (AF594) is a water-soluble analogue of Texas Red® (TR) fluorophore; it was chosen to avoid fluorescent background causing by the non-specific adsorption of fluorescent species (e.g. fluorescent nucleotides and/or free dye) over DNA biochip surface after the incorporation and cleavage steps of binary sequencing cycle. Such phenomenon was observed with Texas Red® despite of adding further washing steps to the SBS scheme. In addition to their excellent water-solubility, (1) the wide variety of Alexa Fluor® dyes spanning the visible and near-infrared spectrum would allow us to develop a series of different fluorescent nucleotides for the design of a four-color DNA-sequencing approach, and (2) they exhibit excellent emission intensity and photostability when conjugated to a biomolecule such as DNA (41,42).
Figure 2.
Structures of dNTP(AP3)-SS-AF594.
The synthesis of dNTP(AP3)-SS-AF594 2–5 bearing an original cleavable disulfide linker between the base and water-soluble rhodamine AF594 (Scheme 1), started with the chemical derivatization of the aminopropargyl (AP3) arm of the nucleotide analogs dNTP(AP3) using the commercially available N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) cross-linking reagent. Thereafter, the fluorophore labeling was readily achieved via a disulfide exchange reaction between dNTP(AP3)-SPDP and Alexa Fluor® 594-thiol 6. As this red dye exists as a mixture of two isomers, two distinct labeling reactions of each dNTP(AP3)-SPDP conjugate were performed with the single labels namely isomer I (i.e. the most polar) and isomer II (i.e. the second eluted on RP-HPLC). All dNTP(AP3)-SS-AF594 were isolated in a pure form (purity >95%, overall yield ∼50%) by semi-preparative RP-HPLC and characterized by ESI-MS and UV-visible spectroscopy.
Scheme 1.
Synthesis of cleavable disulfide Alexa Fluor® 594 nucleotides 2–5.
To use the dNTP(AP3)-SS-AF594 for DNA sequencing, it is critical that these nucleotide analogs be incorporated faithfully and efficiently into a growing DNA strand during a polymerase reaction and that they act as transient terminators. As these cleavable disulfide fluorescent nucleotides are available as two resolved isomers, fluorescence properties (after polymerase-extension reaction) and non-specific binding to biochip surface of isomers I and II of each dNTP(AP3)-SS-AF594 were compared to select the best one. Furthermore, the development of a reliable and simple procedure for the clean removal of the fluorescent label after readout is required.
Sequencing by synthesis optimization using dNTP(AP3)-SS-AF594
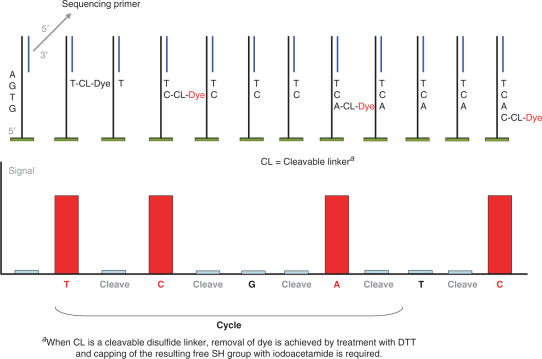
In general, a SBS-based approach would require all the four transient terminators labeled with different dyes that are added simultaneously with the polymerase for decoding the sequence by assigning the dye detected at each cycle of DNA extension. The experiments we performed aimed at evaluating and optimizing the performance of an alternative SBS approach using an unique label and adding one base at a time for subsequent monitoring of the sequencing progress with a signal–no signal read-out according to the base incorporated (see Figure 3 for a diagram exemplifying the theoretical outcome of the method).
Figure 3.
Concept of the binary Sequencing by Synthesis approach: Example for illustrating the base-by-base DNA sequencing method using cleavable fluorescent nucleotide such as those described in Figure 2. The image-based binary (signal/no signal) read-out allows sequence assignment of the target sequence. Prior to sequencing, the ssDNA template sequence attached to a solid support via its 5′-end is hybridized with a sequencing primer.
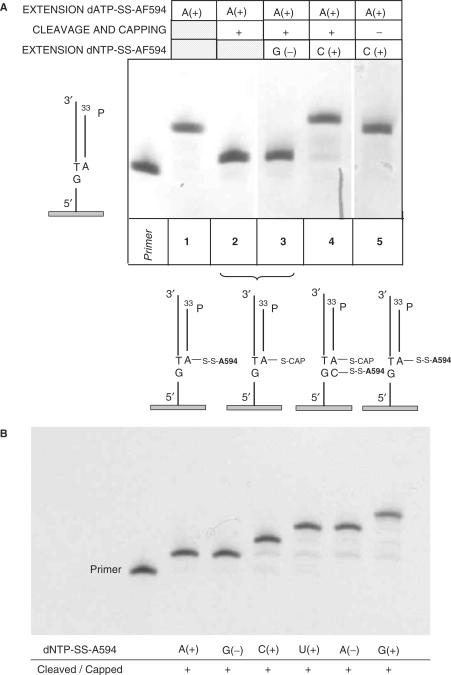
In order to characterize and optimize fluorescent cleavable dNTPs in polymerase-mediated extension reactions for their ability to behave as DNA polymerization transient terminators, numerous experiments using model oligonucleotides grafted on Nucleolink™ (NUNC) or glass surfaces have been carried out. Polyacrylamide gels using radioactive primers were used for verifying and confirming fluorescence data generated. These preliminary experiments were indicative about the performance of enzymes used for incorporating the correct base and gave insights related to the improvements needed for increasing complete filling, reducing mis-incorporation and other undesired events. In the example reported in Figure 4, the autoradiogram illustrates that the sequencing of two bases is possible only if the first labeled incorporated base is cleaved (Figure 4A, lane 5) and there is no band shift if a negative (not expected to incorporate) modified nucleotide is added (Figure 4A, lane 3).
Figure 4.
Evaluation of dNTP(AP3)-SS-AF594 as transient terminators. Autoradiogram of 12% polyacrylamide gels after overnight exposition. 33P-labeled oligo 10 was hybridized to oligo 9 attached to Nucleolink™ well strips (NUNC) The dNTP(AP3)-SS-AF594 modified nucleotides were added as described in the ‘Materials and Methods’ section. (A) Primer: 33P- primer (oligo 10) hybridized to synthetic template, oligo 9. Lane 1: extended primer with dATP(AP3)-SS-AF594. Lane 2: As lane 1 followed by disulfide cleavage and capping. Lane 3: As lane 2 followed by addition of dGTP(AP3)-SS-AF594 (negative base, not expected to elongate the sequencing primer). Lane 4: As lane 2 followed by addition of dCTP(AP3)-SS-AF594 (expected to elongate the sequencing primer). Lane 4: As lane 1 followed by addition of dCTP(AP3)-SS-AF594 without previous cleavage and capping (not expected to elongate the sequencing primer). (B) Sequential addition of fluorescently labeled nucleotides as follows: A, G, C, U, A, G. Cleavage of disulfide bridges and capping of free thiols with iodoacetamide were performed after incubation with each base included negative bases. The sequence assigned is ACTG.
Moreover, a short sequencing run is reported for the same sequence in Figure 4B for an order of sequential nucleotide addition A, G, C and U where the reaction products were loaded onto the gel after each cleavage step. Some minor intermediate products are visible indicating that even in the absence of major undesired effects, the cumulative yield for the expected polymerized product will certainly limit the reading length with the necessary accuracy for sequence assignment. An improved version of the experiment reported in Figure 4 would include an extra lane where the extension reaction is performed with a mix of all the four modified nucleotides for comparison with the outcome reported in Figure 4B. Therefore, for further research on this type of labeled dNTPs, one can design such a conclusive experiment followed by the characterization of extension products by MALDI-TOF mass spectrometry (MS) as recently reported for another type of cleavable fluorescent nucleotides analysis (22).
The sequences containing repeats of identical adjacent bases were useful as models for testing different labels and cleavable linkers. This is exemplified in Supplementary Figure 4 where the termination properties of dGTP(AP3)-SS-AF594 were assayed and compared with dGTP(AP3)-SS-TR under the same experimental conditions using a DNA sequence context where the same base is repeated three times. Texas Red derivatives do not terminate DNA synthesis or have limited termination properties as evidenced by our data. Indeed, a reduced signal for the second and third base in respect to the first inserted one means that the polymerization has not been stopped after the first insertion but rather that more than one of the repeated bases has been introduced in a single reaction. Quenching of the fluorophores for adjacently introduced bases accounts for the ‘leaking’ or not full termination properties of the combination used in the extension reaction. In Supplementary Figure 4, it can be seen that the signal for each consecutive inserted AF594-labeled dGTP, is similar while a constant decrease in signal is observed when using the corresponding Texas Red® derivative.
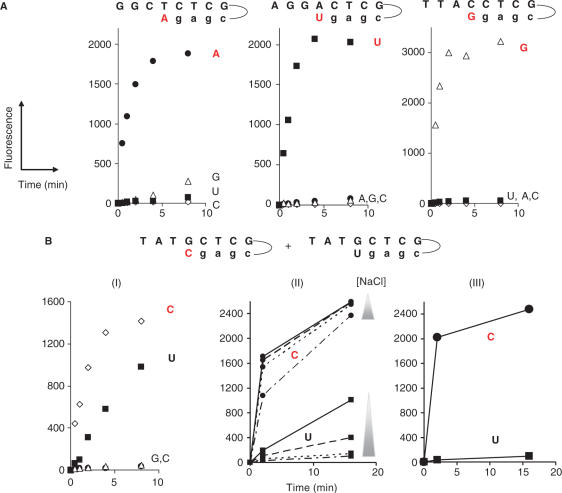
Reaction monitoring over time was also performed for determining the extent of specific signal under different experimental conditions and selecting the optimal reaction time for end-point measurements. As an example, we show in Figure 5 the signal obtained at different reaction times for all the nucleotides in separated reactions using pre-designed hairpin molecules. Using Bst at 50°C, and 250 mM NaCl we observed specific incorporation for A, U and G (Figure 5A), while for C an important mis-incorporation is detected due to the formation of a GU pair (Figure 5B, Picture I). The GT mismatch was not significant when G was assayed using the hairpin specific for A as the first base to be incorporated. In Figure 5B, picture II, we tested increasing NaCl concentrations and the results indicate that this non-specific event can be satisfactorily reduced (500 mM was therefore used for incorporating C with Bst). It has to be noted that in the reaction with Klenow (exo-) at 250 mM salt, no significant mis-incorporation was detected (Figure 5B, picture III).
Figure 5.
Monitoring specificity of the incorporation reaction for dNTP(AP3)-SS-AF594 using hairpin oligonucleotides. (A, B, I) Reactions performed at 50°C using Bst (exo-) and varied NaCl concentration for each nucleotide: A, 650 mM; C, 300 mM; G and U, 250 mM. Hairpin nucleotides are described in the ‘Materials and Methods’ section. All the sequences have a common sequence for hairpin formation and common 5′ portion, the variable region was designed for having a different nucleotide to incorporate for the first extension reaction of self-hybridized oligonucleotides, namely oligos 2A, 2U, 2G and 2C for respectively incorporate A, U, G and C. (B, II) To reduce mis-incorporation of U (UG mis-pair), different concentrations of NaCl (300, 400, 500 and 600 mM) were assayed for the extensions with dUTP(AP3)-SS-AF594 and dCTP(AP3)-SS-AF594. (B, III) Specificity of dCTP(AP3)-SS-AF594 using Klenow (exo-) using conditions described in the ‘Materials and methods’ section (NaCl concentration = 250 mM).
In addition, we tested the influence of the position for attachment of the dye (i.e. position isomers of AF594) as shown in Supplementary Figure 5 for the polymerases Bst and Klenow (exo-). Both isomers I and II of dUTP(AP3)-SS-AF594 5 are efficient transient terminators for Klenow (exo-), but only isomer I is an efficient terminator of Bst-catalyzed polymerization in a run of repeated bases context. Indeed, in the case of isomer II, a second AF594-labeled dUTP is introduced into the extended DNA strand during the incorporation reaction resulting in a quenched signal due to the proximity of both dye labels attached to adjacent positions in the DNA chain (Figure 5B). Both position isomers of AF594 (described in Figure 2) have been tested for all the four dNTPs (A, C, G, U) in different context sequences, in particular those having repeats of the base tested, for assessing the synthesis termination behavior. The preferred combinations of enzyme/isomers for efficient transient termination were as follows: for Klenow (exo-); isomer I for C and G and isomer II for A and U. For Bst (exo-); isomer I for C, G and U and isomer II for A.
Experimental conditions assayed for empirically improving the sequencing performance include salt effects, addition of antioxidants (to avoid the strong quenching of photoexcited AF594 dye through PET from easily oxidized 7-deazaguanine base to this dye), organic co-solvents (DMSO, ethanol) and salt concentration. Some examples of the optimization procedure are described in the Supporting material section in Supplementary Figures 4–7. Optimization of conditions using Klenow (exo-) or Bst (exo-) allowed us to accurately read about 10 bases for most of our model sequences. An example of a model sequencing using a synthetic oligonucleotide (hairpin 1) is reported in Figure 6. For this particular example, about 9–10 bases can be assigned. Among the non-specific signals obtained, some could be explained by the extension of remaining minor incomplete fillings generating some asynchrony or ‘echo’. This limitation in reading length is the consequence of incomplete incorporation or partial termination behavior. Indeed, the fact that the incorporation of some bases is not quantitative generates lag sequences limiting the length of the sequence stretch that can be unambiguously assigned. Nevertheless, these results generated using single and evenly surface distributed synthetic DNA sequences were promising enough for applying our preliminary sequencing method to DNA colonies using optimized standard procedures that include among others the presence of additives such as sodium ascorbate that improved our sequencing approach and other fluorescence-based SBS methods (36).
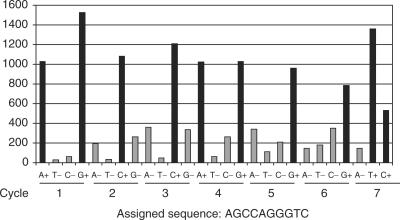
Figure 6.
Application of the binary sequencing approach using hairpin 1 (see oligonucleotide sequences in the ‘Materials and Methods’ section) attached on glass channel using Klenow (exo-) described in the ‘Materials and Methods’ section. Modified nucleotides, dNTP(AP3)-SS-AF594 were sequentially added using the following order A, U, C, G. After recording the signal from each extension, a cleavage and capping reactions were performed followed by imaging (signal after cleavage is in the range of background level and is not reported in this graph). Averaged signals of more than five acquisitions at different x–y positions of the channel are reported. The label T in the graphs stands for the U(AP3) derivative.
As an illustrative example, in Figure 7, we report the application of our binary SBS approach to DNA colonies generated from the amplification of three different DNA templates using the surface chemistry and conditions recently reported by us (6). For illustrating the application and to identify the random distribution of the three DNA populations we applied one cycle of binary sequencing (as described in Figure 3). Furthermore, genomic DNA from Lambda phage was digested and engineered for generating a mixture of 14 solid-phase amplifiable DNA templates. In Figure 8, the DNA colonies generated and two examples of deciphered sequences are reported. The sequences generated allowed to assign the sequences stretches resulting from Lambda phage Hind III digestion using image and data analysis software developed in house. It is important to mention that not all the validated optimizations were implemented in this proof of principle experiment.
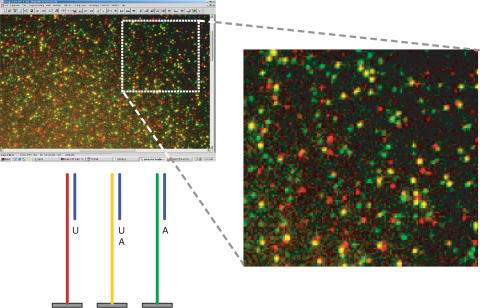
Figure 7.
Binary Sequencing by Synthesis on DNA clusters amplified from three different templates. Fluorescence microscopy image of three populations of DNA clusters generated by in situ amplification of three different DNA template sequences co-attached with amplification primers to the functionalized microfluidic chip surface (6) and detection by incorporation of dNTP(AP3)-SS-AF594 reversible terminators (Figure 3). A first image was acquired with dsDNA clusters using Sybr Green I (diluted 10 000-fold in TE buffer) for detecting all the 2D-addressable amplified products. False color image was generated by superimposition of images generated after addition of dUTP(AP3)-SS-AF594 5 (image 1) followed by reductive cleavage of the dye label and subsequent incorporation of dATP(AP3)-SS-AF594 2 (image 2). Red spots represents DNA colonies that incorporated U. Green spots incorporated A. Yellow spots were detected after both incorporations, U and A. The total number of spots was about 20 000 for a field of view of 0.143 mm2 representing a cluster density of 1.5 × 107 DNA clusters/cm2. Main steps of the process: (1) DNA colonies processing (linearization from dsDNA to ssDNA), (2) Hybridization of a common sequencing primer, (3) dUTP(AP3)-SS-AF594 5 added using Klenow (exo-). Image acquisition revealed those hybridized primers that were extended with U; red and yellow DNA sequences. Reductive cleavage using DTT, capping with iodoacetamide and washes. Dark image no significant detection of spots, (4) dATP(AP3)-SS-AF594 2 added using Klenow (exo-). Image acquisition revealed DNA clusters that incorporated A; yellow and green DNA sequences.
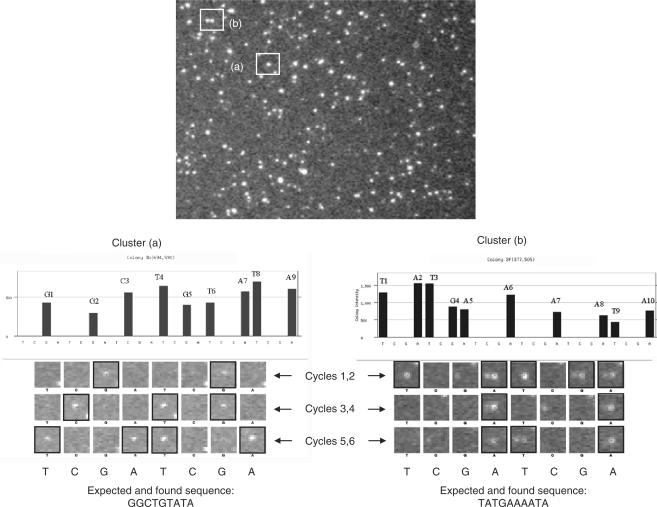
Figure 8.
Binary Sequencing by Synthesis on DNA clusters amplified from 14 different templates prepared from genomic Lambda phage. Sequencing results obtained by fluorescence microscopy with compounds 2–5 and the polymerase Klenow (exo-) for DNA colonies or clusters resulting from amplification of 14 templates obtained from HindIII digestion of Lambda Phage. Panel A is a zoomed view of the in situ amplified ds DNA visualized after incubation with Sybr green I. For illustrating the binary sequencing approach, clusters (1) and (2) are followed over the image-based sequencing process. The graphs in panels B and C are the patterns of the fluorescence intensity evolution measured for each spot after each base addition while at bottom of the figure the spots evolution for the exemplified clusters are shown. After six sequencing cycles with the insertion order UCGA, nine and ten bases can be unambiguously assigned: GGCTGTATA and TATGAAAATA in panels B and C, respectively.
One important remark concerning our experiments is that the sequencing approach tend to perform better in the multi-template DNA colonies context compared to mono-template ones (only one sequence amplified for the whole array) or unique model synthetic DNA sequences. The presence of different sequences in the same reaction mixture increase the probability for the enzyme to incorporate a positive base at certain positions in the array while in mono sequences, the frequency of negative bases is much higher. This suggests to us that some non-specific events such as mis-incorporation are reduced in a real sequencing situation of multiple sequences. The same argument would apply for a four labels approach because the simultaneous addition of all four bases always allows a positive incorporation within each sequence.
CONCLUSIONS
The synthesis of this new class of reversible ‘steric terminators’ is fully described and practical examples reported for illustrating the systematic search for addressing sequence context dependence during the optimization process. Usually, modified nucleotides or synthetic precursors are not commercially available for synthesizing new labeled species with novel properties for their exploration of new sequencing approaches or other applications. In the present article we describe in detail the synthesis, purification and analyses of the reported fluorescent cleavable nucleotides that could be readily adapted for preparing new entities bearing spacers of different length and structure and for attachment of any functionalized fluorophore. Combinatorial approaches for generating a great variety of new species could be generated for further screening of their properties. In particular, this could lead to better combinations of cleavable fluorescent nucleotides/enzymes for sequencing and other genomic applications.
The current termination performance of this new class of cleavable fluorescent nucleotides is suitable for application to simultaneous sequencing of short stretches for more than 10 millions DNA templates per cm2 when used in combination with DNA polymerized colonies, as shown in this manuscript. In particular, our method is presently suitable for sequencing tags illustrated by the utilization of this binary sequencing strategy for decoding sequence tags in a modified version of the MPSS approach (43). Indeed, DNA colonies have been generated, processed and subjected to hybridization with encoded adaptors followed by sequencing of the code using our sequencing by synthesis cleavable fluorescent nucleotides and methods, instead of elucidation of the code by successive hybridization of oligonucleotides probes as originally described in the MPSS method (results to be reported separately).
Our preliminary sequencing results show that the partially optimized method could be used for some applications where reading lengths can be limited to about 10 bases but we believe that further improvements and variants performed by a larger community of scientists could lead to an increase of the reading lengths for targeting genomic applications such as whole-genome re-sequencing (44).
Deeper studies for defining the structural terminator determinants would be beneficial in the future for a rational design of these chemicals in SBS approaches developed for a given polymerase. This also includes the development of a four-color approach that has not only the obvious advantage of being faster than the single fluorophore one, but it also limits the mis-incorporation rate because of the presence of all the four bases simultaneously that would increase the sequencing accuracy. Finally, the development of engineered polymerases customized for specifically recognizing and incorporating these labeled unnatural dNTPs appears as an interesting path to explore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank the whole research team of Manteia for their remarkable multidisciplinary team effort and resulting outstanding scientific contribution that allowed the generation of new generation DNA analytical technologies that recently became commercially available from Illumina. In particular we wish to thank for their contribution in the present article, Mr Didier Bertin for analytical chemistry support and Drs Ilia Leviev and Scott Williams for DNA templates preparation and biochemical characterization of SBS products. Funding to pay the Open Access publication charges for this article was provided by EPFL.
Conflict of interest statement. None declared.
REFERENCES
- 1.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 2.Chan EY. Advances in sequencing technology. Mutat. Res Fundam. Mol. Mech. Mutagen. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc. Natl Acad. Sci. USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adessi C, Matton G, Ayala G, Turcatti G, Mermod J-J, Mayer P, Kawashima E. Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms. Nucleic Acids Res. 2000;28:e87/1–e87/8. doi: 10.1093/nar/28.20.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra RD, Shendure J, Olejnik J, Krzymanska-Olejnik E, Church GM. Fluorescent in situ sequencing on polymerase colonies. Anal. Biochem. 2003;320:55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 6.Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 2006;34:e22/21–e22/13. doi: 10.1093/nar/gnj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzker ML, Raghavachari R, Richards S, Jacutin SE, Civitello A, Burgess K, Gibbs RA. Termination of DNA synthesis by novel 3′-modified-deoxyribonucleosides 5′-triphosphates. Nucleic Acids Res. 1994;22:4259–4267. doi: 10.1093/nar/22.20.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarfati RS, Berthod T, Guerreiro C, Canard B. Synthesis of fluorescent derivatives of 3′-O-(6-aminohexanoyl)pyrimidine nucleosides 5′-triphosphates that act as DNA polymerase substrates reversibly tagged at C-3′. J. Chem. Soc. [Perkin Trans. 1]: Organic Bio-Organic Chem. 1995:1163–1171. [Google Scholar]
- 9.Rasolonjatovo I, Sarfati SR. 6-N-(N-methylanthranylamido)-4-oxo-hexanoic acid: a new fluorescent protecting group applicable to a new DNA sequencing method. Nucleosides Nucleotides. 1998;17:2021–2025. [Google Scholar]
- 10.Welch MB, Burgess K. Synthesis of fluorescent, photolabile 3′-O-protected nucleoside triphosphates for the base addition sequencing scheme. Nucleosides Nucleotides. 1999;18:197–201. doi: 10.1080/15257779908043067. [DOI] [PubMed] [Google Scholar]
- 11.Rasolonjatovo I, Sarfati SR. Development of a new DNA sequencing method: 3′-ester cleavage catalyzed by Taq DNA polymerase. Nucleosides Nucleotides. 1999;18:1021–1022. doi: 10.1080/15257779908041636. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Bai X, Ruparel H, Kim S, Turro NJ, Ju J. A photocleavable fluorescent nucleotide for DNA sequencing and analysis. Proc. Natl Acad. Sci. USA. 2003;100:414–419. doi: 10.1073/pnas.242729199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo TS, Bai X, Ruparel H, Li Z, Turro NJ, Ju J. Photocleavable fluorescent nucleotides for DNA sequencing on a chip constructed by site-specific coupling chemistry. Proc. Natl Acad. Sci. USA. 2004;101:5488–5493. doi: 10.1073/pnas.0401138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi L, Kim DH, Ju J. Design and synthesis of a chemically cleavable fluorescent nucleotide, 3′-O-Allyl-dGTP-allyl-Bodipy-FL-510, as a reversible terminator for DNA sequencing by synthesis. J. Am. Chem. Soc. 2006;128:2542–2543. doi: 10.1021/ja057136n. [DOI] [PubMed] [Google Scholar]
- 15.Ruparel H, Bi L, Li Z, Bai X, Kim DH, Turro NJ, Ju J. Design and synthesis of a 3′-O-allyl photocleavable fluorescent nucleotide as a reversible terminator for DNA sequencing by synthesis. Proc. Natl Acad. Sci. USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo TS, Bai X, Kim DH, Meng Q, Shi S, Ruparel H, Li Z, Turro NJ, Ju J. Four-color DNA sequencing by synthesis on a chip using photocleavable fluorescent nucleotides. Proc. Natl Acad. Sci. USA. 2005;102:5926–5931. doi: 10.1073/pnas.0501965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu G, Burgess K. A diversity oriented synthesis of 3′-O-modified nucleoside triphosphates for DNA sequencing by synthesis. Bioorg. Med. Chem. Lett. 2006;16:3902–3905. doi: 10.1016/j.bmcl.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 18.Meng Q, Kim DH, Bai X, Bi L, Turro NJ, Ju J. Design and synthesis of a photocleavable fluorescent nucleotide 3′-O-allyl-dGTP-PC-Bodipy-FL-510 as a reversible terminator for DNA sequencing by synthesis. J. Org. Chem. 2006;71:3248–3252. doi: 10.1021/jo060300k. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Boyce-Jacino MT, Goelet P. USA: Orchid Biocomputer, Inc; 1999. Nucleotide analogs with 3′-pro-fluorescent fluorophores in nucleic acid sequence analysis. WO patent no. 9964437. [Google Scholar]
- 20.Parce JW, Nikiforov TT, Burd Mehta A, Chow AW, Knapp MR. USA: Caliper Technologies Corp; 2000. DNA sequencing by incorporation of nucleotides with labile chain-terminating groups. WO patent no. 2000050642. [Google Scholar]
- 21.Odedra R, Simmonds A, Pickering L. UK: Amersham Pharmacia Biotech UK Ltd; 2001. Preparation of nucleotide analogs comprising a reporter moiety and a polymerase enzyme blocking moiety. WO patent no. 2001092284. [Google Scholar]
- 22.Ju J, Kim DH, Bi L, Meng Q, Bai X, Li Z, Li X, Marma MS, Shi S, et al. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc. Natl Acad. Sci. USA. 2006;103:19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapeyre M, Leprince J, Massonneau M, Oulyadi H, Renard P-Y, Romieu A, Turcatti G, Vaudry H. Aryldithioethyloxycarbonyl (Ardec): a new family of amine protecting groups removable under mild reducing conditions and their applications to peptide synthesis. Chemistry. 2006;12:3655–3671. doi: 10.1002/chem.200501538. [DOI] [PubMed] [Google Scholar]
- 24.Semenyuk A, Foeldesi A, Johansson T, Estmer-Nilsson C, Blomgren P, Braennvall M, Kirsebom LA, Kwiatkowski M. Synthesis of RNA Using 2′-O-DTM Protection. J. Am. Chem. Soc. 2006;128:12356–12357. doi: 10.1021/ja0636587. [DOI] [PubMed] [Google Scholar]
- 25.Hermanson GT. Bioconjugate Techniques. 1st. Inc., San Diego: Academic Press; 1996. [Google Scholar]
- 26.West KR, Otto S. Reversible covalent chemistry in drug delivery. Curr. Drug Discov Technol. 2005;2:123–160. doi: 10.2174/1570163054866882. [DOI] [PubMed] [Google Scholar]
- 27.Salo H, Guzaev A, Loennberg H. Disulfide-tethered solid supports for synthesis of photoluminescent oligonucleotide conjugates: hydrolytic stability and labeling on the support. Bioconjug. Chem. 1998;9:365–371. doi: 10.1021/bc970194g. [DOI] [PubMed] [Google Scholar]
- 28.Lack O, Zbinden H, Woggon W-D. A useful disulfide linker for single-bead analysis of peptide libraries. Helv. Chim. Acta. 2002;85:495–501. [Google Scholar]
- 29.Hobbs FW., Jr Palladium-catalyzed synthesis of alkynylamino nucleosides. A universal linker for nucleic acids. J. Org. Chem. 1989;54:3420–3422. [Google Scholar]
- 30.Lee SE, Sidorov A, Gourlain H, Mignet N, Thorpe SJ, Brazier JA, Dickman MJ, Hornby DP, Grasby JA, et al. Enhancing the catalytic repertoire of nucleic acids: a systematic study of linker length and rigidity. Nucleic Acids Res. 2001;29:1565–1573. doi: 10.1093/nar/29.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobbs FW, Jr, Cocuzza AJ. 1988. EP patent no. 251786. [Google Scholar]
- 32.Ramzaeva N, Seela F. 7-Substituted 7-deaza-2′-deoxyguanosines: regioselective halogenation of pyrrolo[2,3-d]pyrimidine nucleosides. Helv. Chim. Acta. 1995;78:1083–1090. [Google Scholar]
- 33.Lefevre C, Kang HC, Haugland RP, Malekzadeh N, Arttamangkul S, Haugland RP. Texas Red-X and Rhodamine Red-X, new derivatives of Sulforhodamine 101 and Lissamine Rhodamine B with improved labeling and fluorescence properties. Bioconjug. Chem. 1996;7:482–489. doi: 10.1021/bc960034p. [DOI] [PubMed] [Google Scholar]
- 34.Mao F, Leung W-Y, Haugland RP. USA: Molecular Probes, Inc; 1999. Sulfonated xanthene derivatives synthesis and applications as fluorescent stains. WO patent no. 9915517. [Google Scholar]
- 35.Dodge A, Turcatti G, Lawrence I, de Rooij NF, Verpoorte E. A microfluidic platform using molecular beacon-based temperature calibration for thermal dehybridization of surface-bound DNA. Anal. Chem. 2004;76:1778–1787. doi: 10.1021/ac034377+. [DOI] [PubMed] [Google Scholar]
- 36.Fedurco M, Romieu A, Turcatti G. UK: Solexa Limited; 2006. Improved method of nucleotide detection. WO 2006/064199. [Google Scholar]
- 37.Welch MB, Martinez CI, Zhang AJ, Jin S, Gibbs R, Burgess K. Syntheses of nucleosides designed for combinatorial DNA sequencing. Chemistry. 1999;5:951–960. [Google Scholar]
- 38.Giller G, Tasara T, Angerer B, Muhlegger K, Amacker M, Winter H. Incorporation of reporter molecule-labeled nucleotides by DNA polymerases. I. Chemical synthesis of various reporter group-labeled 2′-deoxyribonucleoside-5′-triphosphates. Nucleic Acids Res. 2003;31:2630–2635. doi: 10.1093/nar/gkg370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tasara T, Angerer B, Damond M, Winter H, Doerhoefer S, Huebscher U, Amacker M. Incorporation of reporter molecule-labeled nucleotides by DNA polymerases. II. High-density labeling of natural DNA. Nucleic Acids Res. 2003;31:2636–2646. doi: 10.1093/nar/gkg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tebbutt SJ, Mercer GD, Do R, Tripp BW, Wong AWM, Ruan J. Deoxynucleotides can replace dideoxynucleotides in minisequencing by arrayed primer extension. BioTechniques. 2006;40:331–338. doi: 10.2144/000112111. [DOI] [PubMed] [Google Scholar]
- 41.Berlier JE, Rothe A, Buller G, Bradford J, Gray DR, Filanoski BJ, Telford WG, Yue S, Liu J, et al. Quantitative comparison of long-wavelength Alexa Fluor dyes to Cy dyes: fluorescence of the dyes and their bioconjugates. J. Histochem. Cytochem. 2003;51:1699–1712. doi: 10.1177/002215540305101214. [DOI] [PubMed] [Google Scholar]
- 42.Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, Leung W-Y, Haugland RP. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J. Histochem. Cytochem. 1999;47:1179–1188. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- 43.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotech. 2000;18:630. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 44.Bentley DR. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 2006;16:545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.