Abstract
DNA repair is an important mechanism by which cells maintain genomic integrity. Decline in DNA repair capacity or defects in repair factors are thought to contribute to premature aging in mammals. The nematode Caenorhabditis elegans is a good model for studying longevity and DNA repair because of key advances in understanding the genetics of aging in this organism. Long-lived C. elegans mutants have been identified and shown to be resistant to oxidizing agents and UV irradiation, suggesting a genetically determined correlation between DNA repair capacity and life span. In this report, gene-specific DNA repair is compared in wild-type C. elegans and stress-resistant C. elegans mutants for the first time. DNA repair capacity is higher in long-lived C. elegans mutants than in wild-type animals. In addition, RNAi knockdown of the nucleotide excision repair gene xpa-1 increased sensitivity to UV and reduced the life span of long-lived C. elegans mutants. These findings support that DNA repair capacity correlates with longevity in C. elegans.
INTRODUCTION
It has been proposed that aging-related disease and dysfunction is caused by cumulative damage in somatic cells and tissues (1,2). DNA is an important target of such age-related cellular damage because DNA encodes all structural and functional cellular components and because somatic cell mutations can have deleterious consequences at the cellular and organismal levels. As the deleterious effects of DNA damage can be prevented by DNA repair, it has been proposed that DNA repair and aging are interrelated.
Defects in some mouse and human DNA repair genes cause a phenotype similar to premature aging, suggesting that lowered capacity for DNA repair might accelerate the process of aging (3–5). For example, mouse mutants with mutations in the Xeroderma Pigmentation Group D (XPDTTD, trichothiodystrophy), XPDTTD/XPA, XPF or ERCC1, which are involved in nucleotide excision repair, show many symptoms of premature aging in mice (6–8). Human patients with the rare hereditary disease Cockayne syndrome (CS), caused by mutations in CSA or CSB, have defects in transcription coupled-nucleotide excision repair (TC-NER) and show features of accelerated aging (5,9). In addition, defects in double-strand break (DSB) repair caused by mutations in ATM, p53, Ku80, DNA-PKcs, BRCA1 or RAD50, also cause progeroid phenotypes in mice and humans (5,10). Thus, it is likely that loss of DNA repair capacity and increased DNA damage are major factors in aging-related dysfunction and disease.
Caenorhabditis elegans is an excellent model system for the study of aging, both because of its relatively short life span (11–13) and because of a large number of identified C. elegans genes that influence life span (14,15). These include age-1 and daf-2, which are involved in the insulin/IGF-1 pathway (16–19), and whose influence on life span is mediated by daf-16, a FOXO family transcription factor (20,21). Long-lived C. elegans mutants with defects in the insulin/IGF-1 pathway are resistant to oxidative stress (12,22), high temperature (23), UV radiation (24) and heavy metal stress (25), suggesting that these mutants live longer because of increased resistance to damage induced by multiple forms of stress and possibly due to higher DNA repair capacity. Recently, it has been reported that NER process slows significantly in aging C. elegans (26). However, direct evidence showing that DNA repair capacity correlates with stress resistance and/or longevity in C. elegans is still lacking.
In this study, gene-specific repair of UV-induced pyrimidine dimers was compared in wild-type C. elegans and long-lived C. elegans mutants. The results show that DNA repair capacity is higher in long-lived mutants than in wild-type animals. Furthermore, RNAi knockdown of C. elegans xpa-1 decreased resistance to UV and oxidative stress and reduced life span in long-lived worms. These findings provide evidence that DNA repair capacity correlates with life span in C. elegans.
MATERIALS AND METHODS
Strains, growth and egg preparation
Caenorhabditis elegans strains were maintained and handled on nematode growth media (NGM) plates with Escherichia coli OP50 lawn at 20°C. Caenorhabditis elegans N2 Bristol was the wild-type hermaphroditic strain and was used as a control. The following mutants from Caenorhabditis Genetics Center (CGC, University of Minnesota, Minneapolis, MN, USA) were used in this study: TJ401 [age-1(hx546) fer-15(b26)], TJ1052 [age-1 (hx546)], CB1370 [daf-2(e1370)], GR1308 [daf-2(e1370);daf-16(mg54)], DR26 [daf-16(m26)] and RB864 [xpa-1(ok698)]. Eggs were prepared by incubating worms in lysis solution (1% NaOCl, 0.5 N NaOH).
Larval development in the presence of paraquat or after UV irradiation
Eggs (20–40) per plate were obtained by incubating two to three egg-laying (gravid) worms. Adult worms were discarded from plates and eggs used for experiments as described subsequently. Eggs were irradiated with UV light and development from eggs to adult worms was monitored on NGM plates. Alternatively, eggs were exposed to various concentrations of paraquat on NGM plates. The developmental stage of each worm was recorded by comparing the length of larva with that of N2 at multiple time points for 2–3 days. Each experiment was repeated 3–5 times.
Life span assays
Life span assays were conducted on NGM agar at 20°C as described previously (23). Briefly, synchronous worms from a large-scale egg-laying procedure were grown on NGM plates with OP50. When larvae reached the L4 stage, 12 worms were transferred onto individual OP50-spotted plates containing 40 μM 5-fluoro-2-deoxyuridine (FUdR). Plates were irradiated with UV. Animals were scored once per day for touch-provoked movement with a platinum wire; animals that failed to respond were considered dead. Each experiment was repeated 3–5 times (N). Mean life span was calculated for each life span assay.
RNA-mediated interference (RNAi) analysis
Worms were treated with RNAi on NGM agar at 20°C as described previously (27). In brief, dsRNA of gene of interest was induced by isopropyl-β-d-thiogalactopyranoside in the RNase-III-deficient E. coli HT115 (DE3) strain. Cells were applied to standard NGM/agar media supplemented with IPTG and antibiotics (tetracycline and ampicillin) and incubated overnight. L4 staged hermaphrodite worms were placed onto these NGM plates. Worms from the F2 generation were tested for resistance to stress.
Irradiation, isolation of C. elegans genomic DNA and restriction digestion
Worms were grown on NGM plates until ≥80% of animals were L4 or higher stage. The plates were irradiated at the indicated dose using a 15-watt germicidal UV lamp and incubated for the indicated time at 20°C. Worms were rinsed in M9 buffer, harvested in a clinical centrifuge, frozen immediately in liquid nitrogen and stored at –80°C. Genomic DNA was purified with a QIAamp DNA mini kit (QIAGEN GmbH, Germany).
Restriction digestion (100 μl) was performed by incubating genomic DNA (50 μg) in EcoR V reaction buffer at 4°C for 5 h before adding EcoR V (New England Biolabs, MA, USA) at a concentration of 2 U/μg. Reactions were then incubated at 37°C for 12–16 h. Products of restriction digestion were analyzed on agarose minigels. Restricted DNA was precipitated, washed and resuspended in TE buffer.
T4 endonuclease V treatment and electrophoresis
DNA (1–2 μg) was treated with (+) or without (−) T4 endonuclease V in 10 μl buffer (Trevigen, Frederick, MD, USA) at 37°C for 2 h. DNA was denatured at 50°C for 1 h in buffer containing 1 M glyoxal, 50% dimethyl sulfoxide, 10 mM sodium phosphate (pH 7.0) and 0.5 mM EDTA (28). DNA was subjected to electrophoresis on a 0.7% agarose gel [10 mM sodium phosphate buffer (pH 7.0) and 0.5 mM EDTA] for 16 h at ∼30 V.
Southern blot
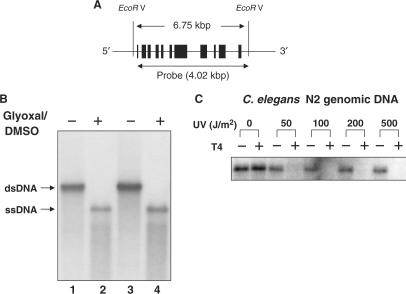
After electrophoresis, gels were soaked in alkaline transfer buffer (0.4 N NaOH/1 M NaCl) at room temperature for 20 min. Hybond XL (Amersham Bioscience, GE life sciences, NJ, USA) membrane was also soaked separately in alkaline transfer buffer for 5 min. DNA was transferred to the membrane using a standard Southern blot protocol. After transfer, the membrane was washed with 2× SSPE, dried and cross-linked using Hoefer UV crosslinker (Model UVC-500, Amersham Bioscience, GE life sciences, NJ, USA). The membrane was incubated in 20 mM Tris–HCl (pH 8.0) at 65°C for 30 min and pre-hybridized in Hybrisol (Chemicon, CA, USA) containing denatured calf thymus DNA (100 μg/ml) at 45°C overnight. Fresh Hybrisol was added and incubation continued for 4 h. Radioactive probe was added to the membrane and hybridization was carried out at 45°C overnight. Membranes were washed twice in 2 × SSPE/0.1% SDS at room temperature for 15 min, twice in 0.1 × SSPE/0.1% SDS at room temperature for 15 min and once in 0.1× SSPE/0.1% SDS at 60°C for 30 min. Membranes were exposed to Kodak BIOMAX Light and/or MR X-ray film (Eastman Kodak, NY, USA) at −70°C for 1–3 days. DNA probes were labeled with [α-32P]dCTP using the Rediprime II Random Prime Labeling kit (Amersham Bioscience, GE life sciences) and were pre-incubated with Hybond XL membrane at 45°C overnight. The probe was a 4 kb PCR product (Figure 3A) of the VPS-45 gene (C. elegans CGC name, human vacuolar protein sorting-associated protein 45, located from 974 786 to 970 617 in chromosome X).
Figure 3.
Detection of UV-induced pyrimidine dimers in VPS-45. C. elegans genomic DNA was digested with EcoR V, electrophoresed, transferred to membrane by Southern blot, and hybridized to radio-labeled DNA probe. (A) Genomic map of the VPS-45 gene. “EcoR V” indicates location of EcoR V restriction sites outside of VPS-45. The black boxes indicate exons. The position of the DNA probe used in this study is shown below the gene. (B) Southern blot in glyoxal-DMSO buffer. Lanes 1 and 2, 1 μg genomic DNA; lanes 3 and 4, 3 μg genomic DNA. DNA was irradiated with UV light, treated with (+) or without (−) T4 endonuclease, electrophoresed in glyoxal/DMSO, and analyzed by Southern blot. Results are shown for EcoR V digested C. elegans genomic DNA (C).
Quantification of pyrimidine dimers
The number of CPDs (cyclobutane pyrimidine dimers) in genomic DNA was determined as described previously (29). Autoradiographic signals were quantified by scanning densitometry using Scion Image software (Scion, MD, USA) or with a Fuji Phosphoimager (Fuji Medical Systems, CT, USA). The percentage of CPD-free fragments (the zero class) was calculated from the ratio of the full-length fragment in T4 endo V-treated and -untreated samples. The average number of CPDs per fragment was calculated from the zero class measurements using a Poisson expression. Results from three times experiments were used to calculate mean values ± SD.
RESULTS
Stress resistance and life span of C. elegans aging mutants
Almost all long-lived C. elegans mutants (also called aging mutants) are resistant to environmental stress, including oxidative stress, heat shock, UV irradiation and exposure to heavy metals (25). This study examines the relationship between resistance to oxidative stress and UV, life span and development in C. elegans aging mutants and daf-16-related mutants.
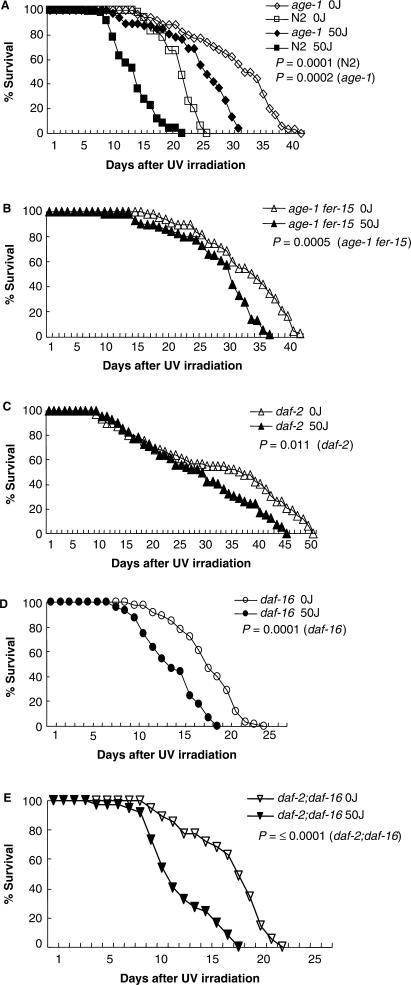
Survival of wild-type C. elegans and aging mutants was measured in the absence or presence of UV irradiation (50 J/m2). In initial studies, worms were irradiated as adults, when the somatic tissues are quiescent and composed of non-dividing cells. We were able to observe an increased life span for C. elegans aging mutants under normal growth conditions (Figure 1A and Table 1) as reported by a previous study (24). The results showed that mean life span of aging mutants (age-1 fer-15, age-1, daf-2) was 26.5–30.5 days after UV irradiation, with an average UV-induced decrease in life span of 16.4%; in contrast, the life span of wild-type C. elegans was 13 days in the presence of UV, an average UV-induced decrease in life span of 37.2% (Table 1). Furthermore, the mean life span and the maximum life span of irradiated aging mutants were longer than for unirradiated wild-type worms (Figures 1A–1C; Table 1). These data indicate that C. elegans aging mutants are somewhat more resistant to UV and may have greater capacity to repair or tolerate UV-induced damage in somatic cells.
Figure 1.
UV sensitivity of adult wild-type C. elegans and aging mutants. Survival of adult wild-type worms and C. elegans hermaphrodite mutants was measured after exposure to 0 or 50 J/m2 UV (see Materials and Methods section). (A) age-1; (B) age-1 fer-15; (C) daf-2; (D) daf-16; (E) daf-2;daf-16. Graph represented the mean value of repeated 3–6 times experiments. The P-values are for the log rank test.
Table 1.
Life span of C. elegans strains after UV irradiation
| 0 J/m2 | 50 J/m2 | |||||
|---|---|---|---|---|---|---|
| Strains | Mean ± SD (days) | Maximum (days) | Repeated experiment (total worms) | Mean ± SD (days) | Maximum (days) | Repeated experiment (total worms) |
| age-1 (hx546) fer-15(b26) | 34.0 ± 1.4 | 43 | 4(48) | 30.5 ± 1.5 | 38 | 4(48) |
| age-1 (hx546) | 33.0 ± 1.6 | 41 | 3(36) | 26.5 ± 1.5 | 31 | 3(36) |
| daf-2(e1370) | 36.0 ± 2.2 | 50 | 4(48) | 29.0 ± 0.9 | 45 | 4(48) |
| daf-2;daf-16 | 17.5 ± 1.1 | 22 | 3(36) | 11.0 ± 0.8 | 18 | 3(36) |
| daf-16(m26) | 17.5 ± 0.8 | 24 | 6(60) | 13.0 ± 1.3 | 19 | 6(60) |
| Wild-type N2 | 20.7 ± 0.8 | 26 | 5(60) | 13.0 ± 0.8 | 21 | 5(60) |
Previous studies report that the phenotype of longer life span and stress resistance conferred by daf-2 and age-1 is suppressed by daf-16 (24,30,31). This result was confirmed here, by measuring the survival of daf-16 and daf-2;daf-16 mutants in the presence and absence of UV irradiation. The results showed that daf-2;daf-16 worms (decrease in life span of 37.1%) were less resistant to UV than daf-2 mutants with a mean life span of 13 days (decrease in life span of 25.7%) after UV treatment (Figures 1D and 1E; Table 1). However, the UV resistance of daf-16 and wild-type worms was similar. The slight UV-resistance difference between daf-16 and daf-2;daf-16 mutant may be due to hypomorphs of daf-16 alleles (mg54 in the double mutant and m26 in the single mutant) employed in two mutants. These data support the observation that UV resistance correlates with adult life span in C. elegans aging mutants (24).
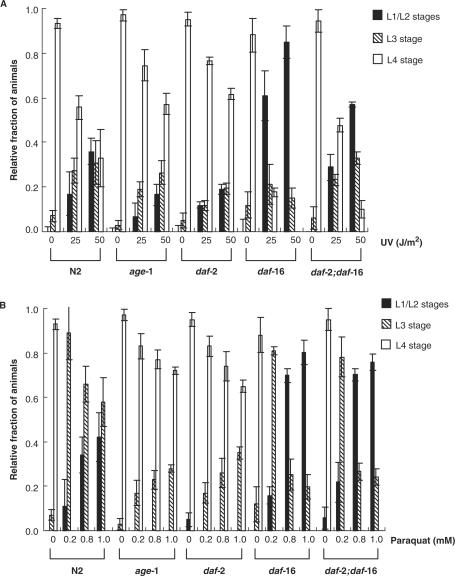
UV sensitivity was also tested in proliferatively dividing cells containing eggs and larvae. Eggs laid from C. elegans mutants were immediately irradiated and the number of larvae at each stage was counted at multiple post-irradiation time points. In the control (no UV irradiation), >90% of all eggs (i.e. age-1 fer-15, age-1, daf-2, daf-16-related and wild-type N2) developed to the L4 stage (Figure 2A and Supplementary data, Table S1). In contrast, ∼50% fewer N2 and daf-16 worms developed to L4 after irradiation at a dose of 25 J/m2 (Figure 2A and Table S1) For daf-2;daf-16 double mutants, 20% of irradiated eggs reached L4 and 60% underwent arrest in the L1/L2 stages. When the UV dose was increased to 50 J/m2, growth arrest increased (Figure 2A and Table S1). However, most age-1 and daf-2 eggs developed to L4 after irradiation with 25 J/m2 and 60% of the eggs reached L4 at a dose of 50 J/m2 (Figure 2A and Table S1) and many fewer age-1 and daf-2 animals arrested in L1-L3 stages than did N2 and daf-16-related mutants. Thus, these results show that long-lived mutants have higher resistance to UV radiation in eggs and larvae during development of the worms. Subsequently, this resistance may affect the resistance of adult worms to UV.
Figure 2.
Stress resistance of wild-type C. elegans and aging mutants during larval development. Twenty to forty eggs per plate from two to three egg-laying worms were collected and exposed to UV or grown on plates containing paraquat. The developmental stage of each worm on each plate was recorded throughout development. (A) Larval development of UV-irradiated eggs. (B) Larval development on paraquat-containing plates. Each experiment was repeated 3–4 times.
Resistance to oxidative stress induced by paraquat was also examined in eggs/larvae of several C. elegans strains. Paraquat is a well-characterized genotoxic agent that generates intracellular superoxide. Increasing doses of paraquat (0.2–1.0 mM) caused many N2 eggs to arrest at L1/L2 or L3 stages (Figure 2B and Table S2) and no L4 larvae were detected when exposed to higher doses of paraquat (Figure 2B and Table S2). In contrast, 70% of age-1 and daf-2 eggs exposed to 1 mM paraquat developed to L4 (Figure 2B and Table S2) with only a small number of eggs undergoing arrest at L3 at high doses of paraquat (Figure 2B and Table S2). The majority of daf-16 and daf-2;daf-16 eggs arrested in L1/L2 stages after exposure to 1 mM paraquat (Figure 2B and Table S2).
While the long-lived mutant strains are slightly more resistant to UV irradiation, their resistance to paraquat is much greater. This undoubtedly reflects the different spectra of damage induced by these two agents.
These results indicate that increased resistance to genotoxic stress during larval stages is likely due to increased genomic maintenance capacity against genotoxic damage and correlates with longer life span in C. elegans.
Detection of repair of UV-induced DNA pyrimidine dimers
Figure 1 indicates that life span correlates with resistance to genotoxic stress in several C. elegans strains. One possible explanation for these data is that C. elegans strains that are more resistant to genotoxic stress have higher DNA repair capacity. This possibility was tested by conducting a gene-specific DNA repair assay that detects repair of UV-induced DNA damage. This assay measures the number of pyrimidine dimers in a target gene (VPS-45) using T4 endonuclease V (T4 endo V), making single-strand cleavages with high specificity for DNA pyrimidine dimers (29). In mammalian cells, gene-specific DNA repair occurs preferentially in expressed genes and is more closely correlated with UV survival than overall DNA repair (32). The VPS-45 gene is a homolog of human vacuolar protein sorting-associated protein 45 and is located from 974 786 to 970 617 on the X chromosome (Figure 3A). VPS-45 expression has been detected in worms by microarray, SAGE and by cDNA cloning (Wormbase database, www.wormbase.org) and a recent report showed that the VPS-45 is an essential gene (33).
The gene-specific pyrimidine dimer repair assay was carried out using EcoR V restricted C. elegans genomic DNA. Control experiments (alkali gel electrophoresis followed by a standard Southern blot protocol) in the absence of T4 endo V showed that restricted genomic DNA was smeared, instead of a distinct band (data not shown). When the restricted genomic DNA was treated with glyoxal/DMSO and subjected to electrophoresis in phosphate buffer (pH 7) (28), the glyoxal/DMSO-treated genomic DNA had a faster electrophoretic mobility than untreated dsDNA (Figure 3B), indicating that the DNA was denatured. Subsequently, pyrimidine dimers were induced in a linear plasmid containing a part of VPS-45 gene and in purified C. elegans genomic DNA using UV doses from 25 to 500 J/m2. The linear plasmid DNA and EcoR V restricted genomic DNA were treated with T4 endo V, incubated with glyoxal/DMSO, subjected to electrophoresis and analyzed by Southern blot. The results were quantified by a scanning densitometer or a Fuji phosphorimager. As expected, the intensities of T4 endo V-treated samples in the linear plasmid (data not shown) and restricted genomic DNA (Figure 3C) were greatly decreased and a dose–response was observed between 0 and 50 J/m2 as percent dimer formation is already close to 100% at >50 J/m2 (Supplementary data, Figure S1). The results indicate that this assay is suitable for detection of UV-induced pyrimidine dimers.
Repair of pyrimidine dimers in wild-type and C. elegans aging mutants
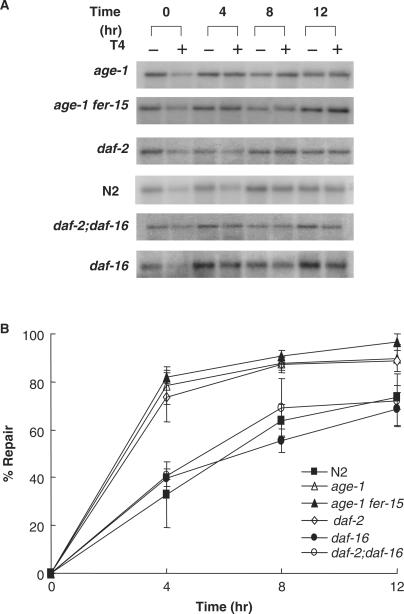
DNA repair of C. elegans was estimated by quantifying pyrimidine dimers of isolated DNA from UV-irradiated C. elegans strains at different time points after UV-irradiation (50 J/m2), providing an estimate of the rate and extent of pyrimidine dimer repair. Percent repair was calculated from the intensity of the full-length restriction fragment at different time points after irradiation as described in Materials and Methods (Figures 4A and 4B). In wild-type N2 worms, percent DNA repair increased from 0 to 8 h after irradiation and then reached a plateau (67% repair) at 12 h. In contrast, the rate and extent of repair was greater for age-1, daf-2 and age-1 far-15 worms than for wild-type N2 worms (Figure 4B). Percent repair reached 85% at 4 h and ∼90% at 12 h after UV irradiation (Figure 4B). These data suggest that C. elegans aging mutants have a higher capacity for repair of UV-induced DNA damage than wild-type worms, resulting in less DNA damage for the rest of their lifetimes. As shown before for UV resistance and life span, the higher stress-resistance of C. elegans aging mutants is daf-16 dependent (24). The extent of repair of UV-induced DNA damage was similar in daf-2;daf-16, wild-type and daf-16 animals. This result suggests that DAF-16 may regulate DNA repair in aging mutants of C. elegans.
Figure 4.
Gene-specific repair in wild-type worms and C. elegans mutants. (A) Worms were irradiated with 50 J/m2 UV and DNA was prepared at various time points (0, 4, 8 and 12 h). Samples were incubated with or without T4 endo V, treated with glyoxal/DMSO, electrophoresed on phosphate agarose gels, transferred to membrane, and hybridized with a probe for VPS-45. (B) Results shown in (A) were quantified using Scion Image software (NIH Image). The average number of CPDs per fragment was calculated from the zero class measurements using a Poisson expression. Data points are mean ± SD from 3–4 independent experiments.
Effect of xpa-1 knockdown (RNAi) on life span of C. elegans aging mutants
The above data demonstrate that higher resistance to UV correlates with higher DNA repair capacity and lower levels of UV-induced DNA damage in C. elegans aging mutants. Thus, we examined the effect of a mutation in the NER pathway on UV resistance and life span in these C. elegans strains. UV-induced pyrimidine dimers are repaired by NER, a DNA repair pathway that is conserved from E. coli to humans (34). Although NER is not well characterized in C. elegans, the C. elegans orthologs of two NER genes, xpa-1 and xpf-1, have been reported and knockout mutants or knockdown worms of these genes are sensitive to UV (27,35,36).
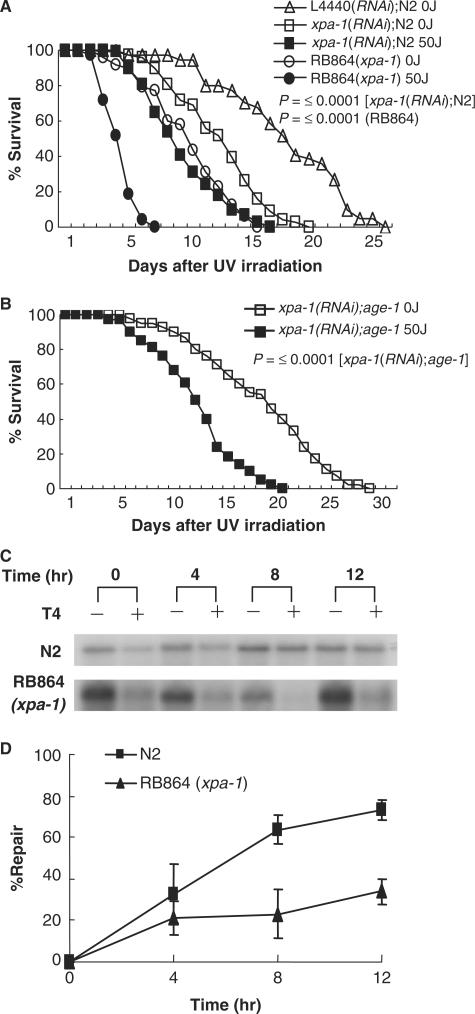
RB864 xpa-1 mutants (C. elegans Gene Knockout Project strain carrying a <1300 bp deletion including xpa-1 gene region) and xpa-1(RNAi) worms were greatly sensitive to UV irradiation (Figure 5A and Table 2). In addition, xpa-1 had lower capacity to repair UV-induced DNA damage than N2 worms (Figures 5C and 5D), indicating that UV sensitivity is related to the extent of DNA repair in these mutants. When xpa-1(RNAi);age-1 knockdown worms were generated by feeding xpa-1 targeted RNAi to age-1 worms, UV resistance and life span decreased significantly. Surprisingly, the mean life span of xpa-1(RNAi);age-1 knockdown worms was similar to that of wild-type N2 worms (Figure 5B and Table 2). These results suggest that levels of DNA damage and/or DNA repair correlate with life span in C. elegans.
Figure 5.
UV sensitivity of xpa-1 knockdown worms. (A) Survival of xpa-1(RNAi) worms and RB864(xpa-1) worms was measured in the presence of FUdR at 20°C after UV irradiation at 0 or 50 J/m2. Each experiment was repeated six times. L4440 is a plasmid vector for RNAi control. (B) Survival of xpa-1(RNAi);age-1 worms was measured after UV irradiation at 0 or 50 J/m2. Each experiment was repeated six times. (C) Gene-specific repair of UV-induced DNA pyrimidine dimers in RB864(xpa-1), performed as described in Figure 4A. (D) Quantification of results shown in (C). The P-values are for the log rank test.
Table 2.
Life span of xpa-1(RNAi) knockdown strains
| xpa-1 (RNAi)N2 | RB864 (xpa-1) | xpa-1 (RNAi); age-1 | L4440 (RNAi)N2 | |
|---|---|---|---|---|
| 0 J/m2 Mean life span ± SD (days) | 12 ± 0.9 | 11.5 ± 0.8 | 18.5 ± 1.8 | 17.5 ± 3.1 |
| Maximum life span (days) | 19 | 15 | 28 | 25 |
| Repeated experiment (total worms) | 6(70) | 6(70) | 6(70) | 3(60) |
| 50 J/m2 Mean life span ± SD (days) | 8 ± 0.8 | 6.5 ± 0.2 | 12 ± 0.8 | |
| Maximum life span (days) | 15 | 7 | 20 | |
| Repeated experiment (total worms) | 6(70) | 6(70) | 6(70) |
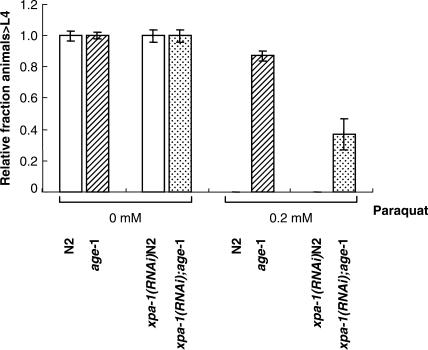
Interestingly, the mean life span of unirradiated xpa-1(RNAi);age-1 or xpa-1(RNAi) worms is shorter than that of unirradiated age-1 mutants or L4440(RNAi)N2 worms, respectively (Figures 5A and 5B, Table 2), suggesting that xpa-1 may influence longevity and/or repair of endogenous DNA damage. This idea was tested by measuring sensitivity of xpa-1(RNAi);age-1 and xpa-1(RNAi)N2 worms to paraquat-induced oxidative damage. The results showed that xpa-1(RNAi);age-1 worms are more sensitive to paraquat than age-1 (Figure 6 and Table S3). Although xpa-1(RNAi)N2 and N2 worms did not develop to L4 at this concentration of paraquat, a majority of N2 worms underwent developmental arrest at L3 stage. Instead, 50% of xpa-1(RNAi)N2 worms were arrested at L1/L2 stages (Table S3). At low concentration of 50 μM paraquat, 60% of N2 worms developed to L4, whereas most of xpa-1(RNAi)N2 worms underwent arrest at L3 (Table S3). Since there are forms of oxidative DNA damage known to be repaired by NER, the oxidative lesions most relevant to aging are those that can be repaired by NER.
Figure 6.
Effect of xpa-1 knockdown on sensitivity to oxidative stress. Eggs were collected from two to three egg-laying worms and grown in the presence or absence of 0.2 mM paraquat. Development of each worm was recorded throughout larval development. Each experiment was repeated four times.
DISCUSSION
This study investigates the relationship between UV resistance, DNA repair capacity and longevity in long-lived mutants of C. elegans. The results suggest that an increase in DNA repair capacity correlates with increased stress resistance in these animals and that this may also lead to longer life span.
Mouse and human NER mutants with symptoms of premature aging also show correlation between longevity, aging-related dysfunction and capacity for DNA repair (5,8,37). To further test this relationship, DNA repair capacity needs to be investigated in long-lived animal mutants. Thus, C. elegans aging mutants with increased UV resistance could be an excellent model to explore the relationship between DNA repair capacity and longevity. A wide range of helix distorting DNA lesions, including UV-induced photoproducts and certain oxidative lesions are repaired by NER (38). NER repair capacity has been quantified by gene-specific repair of UV-induced DNA damage (29). This repair assay is a sensitive assay for evaluating the correlation between UV survival and DNA repair and repair in active, essential genes is likely to be more relevant to survival than global genome DNA repair (32). Results present here show that mean life span is longer and DNA repair capacity is higher in UV-irradiated C. elegans aging mutants than in UV-irradiated wild-type worms, suggesting that C. elegans aging mutants may have higher levels of enzymatic activities involved in repair of UV-induced DNA lesions. Meyer and associates (26) and Hartman et al. (39) have also demonstrated that xpa-1 (formerly known as rad-3) are defective for the repair of UV radiation-induced DNA damage. In addition, the kinetics of repair reasonably approximates those observed by the Meyer and Hartman laboratories. However, future studies will be needed to address how DNA repair capacity varies throughout the C. elegans genome and its relevance for longevity in other long-lived C. elegans mutants such as clk-1, eat-4, mev-1 and gas-1. It is worth noting that gene-specific repair is lower in hepatocytes from old rats than in hepatocytes from young rats (40). Interestingly, caloric restriction does not alter gene-specific DNA repair in young rats.
Previous studies revealed that daf-16 decreases UV resistance and shortens life span of long-lived C. elegans mutants (20,21). This study confirmed that daf-2;daf-16 and daf-16 mutants are less resistant to UV than daf-2 mutants and that DNA repair capacity of them is lower than that of daf-2. However, the precise role of daf-16 in the response to UV and UV-induced DNA damage is unknown. DAF-16 is a forkhead transcription factor involved in insulin-like signaling, stress resistance and longevity in C. elegans (31); thus, it is possible that gene products regulated by daf-16 directly (or indirectly) regulates DNA repair activity. Additional factors that enhance stress resistance in these strains, heat shock proteins, may be involved in increasing DNA repair activity. In supporting this, there is a growing body of evidence linking HSPs to DNA repair activity. For example, inducible heat shock proteins are required for maintenance for of genomic instability in stress conditions (41). In yeast, HSP90 interacts with Ssl2, the yeast homolog of XPB (42).
Two NER orthologs, xpa-1 and xpf-1, have been reported in C. elegans and RNAi knockdown of these genes cause sensitivity to UV (27,36). This study showed that xpa-1(RNAi) worms and xpa-1 knockout (RB864) worms have a reduced life span and that xpa-1 worms have much lower capacity to repair UV-induced DNA damage than wild-type worms (Figure 5A, C and D). In addition, xpa-1(RNAi);age-1 worms are more sensitive to UV than age-1 worms and have a mean life span similar to wild-type worms (Figure 5B). This strongly supports the hypothesis that DNA damage and DNA repair play a major role in determining life span in C. elegans. Recent studies also showed that knockdown of the C. elegans RecQ orthologs causes sensitivity to DNA damage and a shorter life span (43,44).
The mean life span of unirradiated xpa-1(RNAi);age-1 worms was significantly lower than for wild-type worms, suggesting higher susceptibility to endogenous DNA damage in the mutant animals. The nature of these endogenous lesions is unknown, but might include lesions caused by oxidative stress, which is implicated as a critical contributing factor in the process of aging. The observation that xpa-1(RNAi);age-1 worms are more sensitive to paraquat during larval development than age-1 worms (Figure 6) supports this possibility. Previous studies also show that human xpa mutants are sensitive to oxidative damage (45) and that cells from xpd/xpa double mutant mice have a reduced life span and are hypersensitive to paraquat (8). However, as paraquat can induce different types of oxidative stress including protein carbonylation, it is hard to rule out other effect such as protein homeostasis on life span.
The importance of endogenous DNA lesion (oxidative damage) for life span has been also reported in C. elegans. The finding that superoxide anion levels are higher and SOD activity is lower in short-lived C. elegans strains (46,47) also implicates lower oxidative stress and higher antioxidant defense in longevity and aging (48,49). For example, catalase and Cu/Zn superoxide dismutase activity was reported to be higher in age-1 worms than in wild-type worms (22), and expression of stress–response factors such as hsp, mtl-1, gst-1 and sip-1 were reported to be higher in daf-2 worms (50–52). Recently, similar observations have been made for C. elegans mev-1 mutants, in which ROS and nuclear DNA damage are increased (53).
In conclusion, the data presented here demonstrate for the first time that longevity correlates with DNA repair capacity in C. elegans. The results strongly support the hypothesis that DNA damage plays a direct role in the process of aging. In addition, this study demonstrates that C. elegans has an NER-like DNA repair pathway that repairs UV-induced pyrimidine dimers. However, additional studies are needed to determine the exact relationship between specific factors that influence life span, stress resistance and DNA repair capacity in C. elegans.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Mohammad Hedeyati and Dr Nadja de Souza-Pinto for critical reading of the manuscript. Caenorhabditis elegans strains were obtained from C. elegans Genetic Center (University of Minnesota, Minneapolis, MN, USA), which is supported by the National Center for Research Resources. This work was supported in part by the second stage of the Brain Korea 21 Project in 2007 (to B. Ahn) and Research Grant of University of Ulsan (2005-0156 to B. Ahn). This research was supported in part by funds from the National Institute of Aging/National Institutes of Health Intramural Research Program (to V. A. Bohr). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kirkwood TB, Holliday R. Commitment to senescence: a model for the finite and infinite growth of diploid and transformed human fibroblasts in culture. J. Theor. Biol. 1975;53:481–496. doi: 10.1016/s0022-5193(75)80018-x. [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 3.Martin GM. Genetic modulation of senescent phenotypes in Homo sapiens. Cell. 2005;120:523–532. doi: 10.1016/j.cell.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 4.Karanjawala ZE, Lieber MR. DNA damage and aging. Mech. Ageing Dev. 2004;125:405–416. doi: 10.1016/j.mad.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- 6.Dolle ME, Snyder WK, Gossen JA, Lohman PH, Vijg J. Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc. Natl Acad. Sci. USA. 2000;97:8403–8408. doi: 10.1073/pnas.97.15.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van LW, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 8.de Boer J, Andressoo JO, de WJ, Huijmans J, Beems RB, van SH, Weeda G, van der Horst GT, van LW, Themmen AP, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- 9.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 10.Lans H, Hoeijmakers JH. Cell biology: ageing nucleus gets out of shape. Nature. 2006;440:32–34. doi: 10.1038/440032a. [DOI] [PubMed] [Google Scholar]
- 11.Johnson TE, Nelson GA. Caenorhabditis elegans: a model system for space biology studies. Exp. Gerontol. 1991;26:299–309. doi: 10.1016/0531-5565(91)90024-g. [DOI] [PubMed] [Google Scholar]
- 12.Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1993;90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19:1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson TE. Advantages and disadvantages of Caenorhabditis elegans for aging research. Exp. Gerontol. 2003;38:1329–1332. doi: 10.1016/j.exger.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 18.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 19.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 20.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 21.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 22.Vanfleteren JR. Oxidative stress and ageing in Caenorhabditis elegans. Biochem. J. 1993;292(Pt 2):605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl Acad. Sci. USA. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lithgow GJ. Stress response and aging in Caenorhabditis elegans. Results Probl. Cell Differ. 2000;29:131–148. doi: 10.1007/978-3-540-48003-7_7. [DOI] [PubMed] [Google Scholar]
- 26.Meyer JN, Boyd WA, Azzam GA, Haugen AC, Freedman JH, Van HB. Decline of nucleotide excision repair capacity in aging Caenorhabditis elegans. Genome Biol. 2007;8:R70. doi: 10.1186/gb-2007-8-5-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park HK, Suh D, Hyun M, Koo HS, Ahn B. A DNA repair gene of Caenorhabditis elegans: a homolog of human XPF. DNA Repair. 2004;3:1375–1383. doi: 10.1016/j.dnarep.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 28.Tian M, Alt FW. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 29.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 30.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 31.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 32.Bohr VA, Phillips DH, Hanawalt PC. Heterogeneous DNA damage and repair in the mammalian genome. Cancer Res. 1987;47:6426–6436. [PubMed] [Google Scholar]
- 33.Gengyo-Ando K, Kuroyanagi H, Kobayashi T, Murate M, Fujimoto K, Okabe S, Mitani S. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–157. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 35.Boulton SJ, Gartner A, Reboul J, Vaglio P, Dyson N, Hill DE, Vidal M. Combined functional genomic maps of the C. elegans DNA damage response. Science. 2002;295:127–131. doi: 10.1126/science.1065986. [DOI] [PubMed] [Google Scholar]
- 36.Park HK, Yook JS, Koo HS, Choi IS, Ahn B. The Caenorhabditis elegans XPA homolog of human XPA. Mol. Cells. 2002;14:50–55. [PubMed] [Google Scholar]
- 37.Hasty P, Vijg J. Aging. Genomic priorities in aging. Science. 2002;296:1250–1251. doi: 10.1126/science.1071808. [DOI] [PubMed] [Google Scholar]
- 38.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 39.Hartman P, Hevelone J, Dwarakanath V, Mitchell DL. Excision repair of UV radiation-induced DNA damage in Caenorhabditis elegans. Genetics. 1989;122:379–385. doi: 10.1093/genetics/122.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Z, Heydari AR, Wu W, Yang H, Sabia MR, Richardson A. Characterization of gene-specific DNA repair by primary cultures of rat hepatocytes. J. Cell Physiol. 1998;176:314–322. doi: 10.1002/(SICI)1097-4652(199808)176:2<314::AID-JCP9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 41.Hunt CR, Dix DJ, Sharma GG, Pandita RK, Gupta A, Funk M, Pandita TK. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol. Cell. Biol. 2004;24:899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flom G, Weekes J, Johnson JL. Novel interaction of the Hsp90 chaperone machine with Ssl2, an essential DNA helicase in Saccharomyces cerevisiae. Curr. Genet. 2005;47:368–380. doi: 10.1007/s00294-005-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SJ, Yook JS, Han SM, Koo HS. A Werner syndrome protein homolog affects C. elegans development, growth rate, life span and sensitivity to DNA damage by acting at a DNA damage checkpoint. Development. 2004;131:2565–2575. doi: 10.1242/dev.01136. [DOI] [PubMed] [Google Scholar]
- 44.Grabowski MM, Svrzikapa N, Tissenbaum HA. Bloom syndrome ortholog HIM-6 maintains genomic stability in C. elegans. Mech. Ageing Dev. 2005;126:1314–1321. doi: 10.1016/j.mad.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Hayashi M, Araki S, Kohyama J, Shioda K, Fukatsu R. Oxidative nucleotide damage and superoxide dismutase expression in the brains of xeroderma pigmentosum group A and Cockayne syndrome. Brain Dev. 2005;27:34–38. doi: 10.1016/j.braindev.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Ishii N, Takahashi K, Tomita S, Keino T, Honda S, Yoshino K, Suzuki K. A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat. Res. 1990;237:165–171. doi: 10.1016/0921-8734(90)90022-j. [DOI] [PubMed] [Google Scholar]
- 47.Senoo-Matsuda N, Yasuda K, Tsuda M, Ohkubo T, Yoshimura S, Nakazawa H, Hartman PS, Ishii N. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J. Biol. Chem. 2001;276:41553–41558. doi: 10.1074/jbc.M104718200. [DOI] [PubMed] [Google Scholar]
- 48.Honda Y, Honda S. Oxidative stress and life span determination in the nematode Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2002;959:466–474. doi: 10.1111/j.1749-6632.2002.tb02117.x. [DOI] [PubMed] [Google Scholar]
- 49.Wallace DC, Melov S. Radicals r'aging. Nat. Genet. 1998;19:105–106. doi: 10.1038/448. [DOI] [PubMed] [Google Scholar]
- 50.Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, Brooks-Wilson AR, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 52.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 53.Hartman P, Ponder R, Lo HH, Ishii N. Mitochondrial oxidative stress can lead to nuclear hypermutability. Mech. Ageing Dev. 2004;125:417–420. doi: 10.1016/j.mad.2004.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.