Abstract
Human chromosome ends are protected with kilobases repeats of TTAGGG. Telomere DNA shortens at replication. This shortening in most tumor cells is compensated by telomerase that adds telomere repeats to the 3′ end of the G-rich telomere strand. Four TTAGGG repeats can fold into G-quadruplex that is a poor substrate for telomerase. This property has been suggested to regulate telomerase activity in vivo and telomerase inhibition via G-quadruplex stabilization is considered a therapeutic strategy against cancer. Theoretically G-quadruplex can form anywhere along the long G-rich strand. Where G-quadruplex forms determines whether the 3′ telomere end is accessible to telomerase and may have implications in other functions telomere plays. We investigated G-quadruplex formation at different positions by DMS footprinting and exonuclease hydrolysis. We show that G-quadruplex preferentially forms at the very 3′ end than at internal positions. This property provides a molecular basis for telomerase inhibition by G-quadruplex formation. Moreover, it may also regulate those processes that depend on the structure of the very 3′ telomere end, for instance, the alternative lengthening of telomere mechanism, telomere T-loop formation, telomere end protection and the replication of bulky telomere DNA. Therefore, targeting telomere G-quadruplex may influence more telomere functions than simply inhibiting telomerase.
INTRODUCTION
Chromosomes in human cells are capped at both ends with telomere DNA of tandem (TTAGGG)n repeats. Due to the end-replication problem, telomere cannot fully replicate its 3′ end and, as a result, shortens during each round of DNA replication (1). Telomere length homeostasis is essential for the generation and growth of cancer cells. More than 85% tumors use telomerase for telomere maintenance (2), others use the alternative lengthening of telomere (ALT) mechanism (3). Four TTAGGG repeats can fold into a four-stranded structure called G-quadruplex and inhibit telomere addition by telomerase (4). Therefore, G-quadruplex has been suggested to act as a negative regulator of telomere elongation by telomerase in vivo and is currently considered a potential target for cancer therapy (5–7). Intense interest has arisen recently in searching for small molecules that stabilize G-quadruplex to inhibit telomere elongation by telomerase.
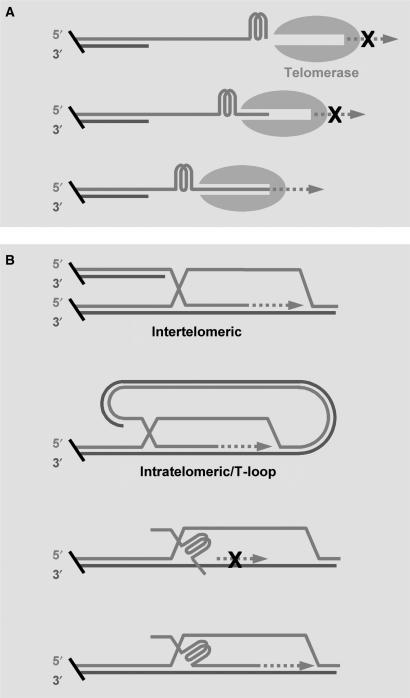
It has been assumed that telomere G-quadruplex can form anywhere at an increment of 6 nucleotides along the G-rich strand that stretches for several kilobases long (8–11). Telomere DNA terminates beyond the double stranded region with a G-rich single-stranded 3′ overhang that can be as long as several hundred bases (12,13). Several G-quadruplex-stabilizing compounds have been shown to displace telomerase (14,15) and other single telomere strand-binding proteins (16–18), suggesting that G-quadruplex can efficiently form at the telomere overhang. Apparently, where G-quadruplex forms may have implications in the biological functions and pharmaceutical applications of telomere G-quadruplex. For instance, a telomere G-rich strand can possibly have at least four forms of terminal structures with respect to the number of single stranded TTAGGG repeats at the 3′ end (i.e. 0–3 repeats). Among those structures, the one with a G-quadruplex formed at the extreme 3′ end is expected to inhibit telomerase while the others may be less or no inhibitory (Figure 1A). Similar requirement of terminal structure may also apply to the ALT mechanism in which a single stranded G-rich 3′ end has to invade and basepair with a C-rich strand for its extension (Figure 1B). Formation of G-quadruplex has also been employed in constructing molecular devices (19–21) and the position where G-quadruplex forms may also have implication in such applications.
Figure 1.
Expected effect of G-quadruplex formation on telomere DNA elongation by (A) telomerase or (B) through the ALT (alternative lengthening of telomeres) mechanism involving strand exchange. A free single-stranded G-rich 3′ end of sufficient size is required for the elongation in both pathways. A terminal G-quadruplex may carry zero to three TTAGGG repeats before the 3′ end. The G-quadruplex formed at the very 3′ end is expected to inhibit telomere elongation (dashed arrow) while the others may be less or not inhibitory. Two examples of ALT involving strand exchange are given, other examples based on the same local interaction can be found in a review (3).
At present, however, no information is available regarding G-quadruplex formation at different positions along the long telomere G-rich strand. We addressed the question with a quantitative DNA footprint method specifically designed for this purpose and a 3′ exonuclease hydrolysis stop assay using (TTAGGG)7 in which one G-quadruplex can potentially form at four different positions. Our work revealed that G-quadruplex preferentially forms at both the 5′ and 3′ ends than at internal positions. The probability at the 3′ end is higher than at the 5′ end. When an open 5′ end is not present, which mimics the telomere G-rich strand in vivo, G-quadruplex tends to form at the very 3′ end and the probability of G-quadruplex formation rapidly decreases as the position moves inwards. Experiments with longer strands revealed that the very first G-quadruplex from the 3′ end preferentially forms at the 3′ terminus and the probability rapidly decreases as it moves inwards. The polarized G-quadruplex formation may be important for the function of telomere DNA. It not only provides a molecular basis for the regulation of telomere extension by both telomerase and the ALT mechanism, but may also have implications in other processes, such as telomere end protection, T-loop formation and replication of bulky telomere DNA. Therefore, drugs targeting telomere G-quadruplex may have effects on more telomere-related processes than simply inhibiting telomerase activity.
MATERIALS AND METHODS
Principle of identifying G-quadruplex formation at different positions by footprinting
Dimethyl sulfate (DMS) footprinting that specifically cleaves at guanines (22) was used in this work. This technique uses oligonucleotide labeled at the 5′ end with 32P. DMS methylates guanine at the N7 position and subsequent treatment with piperidine breaks the DNA backbone at the methylated sites. The DNA sample is then resolved by gel electrophoresis and the 32P-containing fragments are visualized by autoradiography. The size of a band identifies the first nucleotide from the 5′ end that has been attacked.
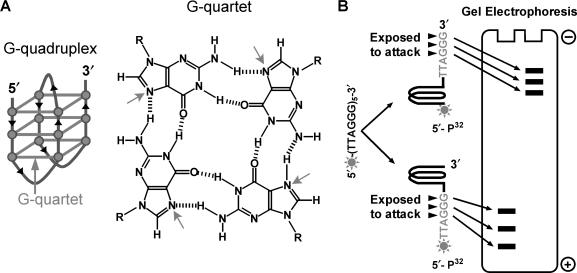
A G-quadruplex requires four TTAGGG repeats. The N7 of those guanines in the G-quartets of a quadruplex is involved in hydrogen bonding (Figure 2A) and thus protected from the attacks by DMS (23, 24). This property has been used to probe the formation of quadruplex (25, 26). To simplify the analysis, we used oligonucleotides of 4–7 TTAGGG repeats in which only one quadruplex can form. We initially assumed that a quadruplex involves only four consecutive repeats. The guanines not involved in a quadruplex will be exposed to attacks, thus the position where a quadruplex forms determines whether a G triplet (GGG) is protected from or exposed to the attacks (Figure 2B). As a result, the sizes of cleaved fragments reflect where the quadruplex forms. The mathematical equations describing the production of DNA fragments used for analyzing quadruplex formation at different positions were given in the Supplementary data. Qudraplex formation with longer loops was also considered and details are available in the Supplementary data.
Figure 2.
Schematic illustration of quadruplex and the principle of quantifying quadruplex formation at different positions by DMS footprint. (A) (TTAGGG)4 can fold back to form G-quadruplex structure hold together by three stacked G-quartets. The Hoogsteen hydrogen bonding between adjacent guanines prevents the N7 position (arrowed) from methylation by DMS. (B) The position at which quadruplex forms determines whether a G triplet (GGG) is exposed to or protected from attack by DMS and the size of cleavage product. In a five-repeat sequence, for example, quadruplex at the very 5′ end will produce 32P-labeled fragment four repeats larger than that produced by quadruplex at the very 3′ end.
Oligonucleotides
The oligonucleotides were purchased from TaKaRa Biotech (Dalian, China). When used in gel electrophoresis, oligonucleotides were labeled at the 5′ end with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas, Lithuania).
Quantitative footprint of G-quadruplex
DNA treatment and gel electrophoresis was carried out essentially as previously described (25) using 32P-labeled oligonucleotide (∼20 000 cpm, 5 ng). The oligonucleotides were treated with DMS for 30, 60, 90 s, respectively, in the presence or absence of 150 mM K+. DNA cleavage products were resolved by gel electrophoresis, autoradiographed on a Typhoon phosphor imager (Amersham Biosciences, Sweden) and the intensity of the bands corresponding to attacks at each G triplet quantified with the software ImageQuant 5.2.
The equations describing the production of DNA fragments were solved by the standard Steepest Descent Method (27, 28). The algorithm was implemented with Visual C++ in a stand-alone application. In this program, the sum of the intensity of the bands produced by cleavage at each G triplet can be input through a friendly graphical interface and the parameters, R, Qi and p, can be obtained simultaneously with an optional initial value calculation.
Exonuclease hydrolysis analysis
32P-labeled oligonucleotide was dissolved in 14 μl of 1× T4 polymerase buffer (33 mM Tris–acetate, pH 7.8, 66 mM CH3COOK, 10 mM (CH3COO)2Mg, 0.5 mM DTT) with the concentration of K+ made to 150 mM by adding KCl, heated at 95°C for 5 min and then cooled down to room temperature and kept for 30 min. Hydrolysis was carried out by adding 1 μl of T4 polymerase (5 U/μl, Takara Biotechnology, Dalian, China) and incubating at 37°C for 3 min. The reaction was stopped by adding equal volume of stop buffer (10 mM NaOH, 10 mM EDTA, 96% deionized formamide and 100 ng/μl salmon sperm DNA) and heated at 90°C for 5 min. Hydrolysis products were run on 12% sequencing gel for 2 h at 40 V/cm. The gel was autoradiographed on a Typhoon phosphor imager (Amersham Biosciences, Sweden) and band intensity was quantified with the software ImageQuant 5.2.
Incomplete hydrolysis was observed (see the first three reference lanes in Figure 5A). Calibration was made for this in each of the structural isoforms in T24(TTAGGG)7, which was calculated as (intensity ratio of the undigested/digested band in the relevant reference lane) × (intensity of the digest product of a corresponding structural isoform of T24(TTAGGG)7).
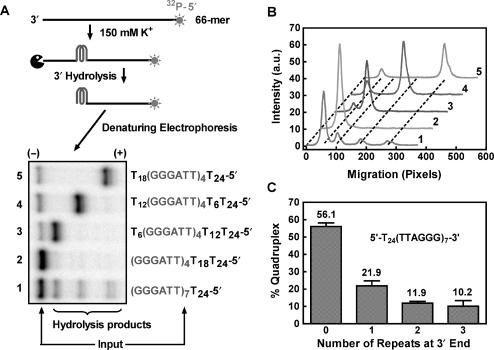
Figure 5.
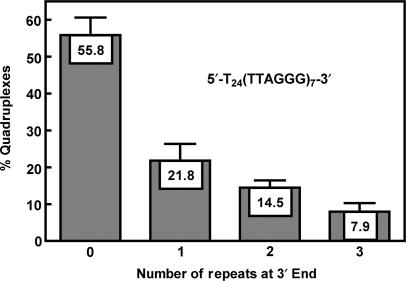
Distribution of G-quadruplex formation at different positions in T24(TTAGGG)7 detected by the 3′ exonuclease activity of T4 polymerase. (A) Gel electrophoresis of hydrolysis products of T24(TTAGGG)7 and the four reference oligonucleotides designated to form G-quadruplex at desired positions leaving a tail of 3, 2, 1, 0 repeats of T6 at the 3′ end, respectively. (B) Densitometry scans of the gel shown in A). (C) Quantified distribution of G-quadruplex formation at different positions in T24(TTAGGG)7 with incomplete hydrolysis calibrated. Data are the mean of three independent experiments with standard deviation.
UV thermal melting analysis
Thermal denaturation was carried out as described previously (29,30) on a DU-640 UV–VIS spectrophotometer (Beckman, USA) equipped with a digital circulating water bath. The absorbance of oligonucleotide in 10 mM phosphate buffer (pH 7.4), 150 mM KCl was monitored at 295 nm and the temperature was simultaneously measured using a thermal probe immersed in sample cell when a sample was heated at a rate of about 1° C/min. A solution containing no oligonucleotide was used as reference.
RESULTS
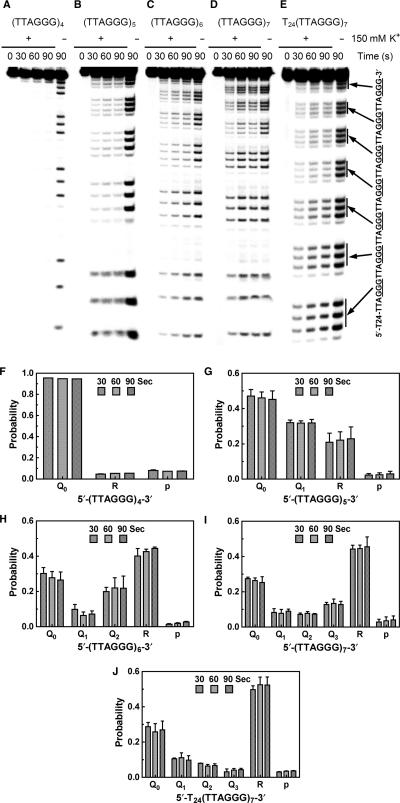
DMS footprinting of 5′-32P-labeled (TTAGGG)4, (TTAGGG)5, (TTAGGG)6, (TTAGGG)7 and T24(TTAGGG)7 was carried out (25) in physiological concentration (150 mM) of K+ that is well known to stabilize G-quadruplex (31). The number of TTAGGG repeats was limited to be no more than seven to allow at most one G-quadruplex to form so that the analysis could be simplified. All these oligonucleotides formed intra-molecular G-quadruplexes as examined by gel electrophoresis (Figure S4, Supplementary data), which is consistent with previous results (32). DMS methylates guanine, when it is not involved in a quartet, at the N7 position and subsequent treatment breaks the DNA backbone at the methylated sites. We derived a set of mathematical equations for each of the oligonucleotides to describe the DNA cleavage using an assumption that G-quadruplex involves only four consecutive TTAGGG repeats. The equations allowed us to calculate the probability of G-quadruplex formation at different positions based on the autoradiographs of footprint.
Representative autoradiographs are given in Figure 3A–E and calculated probability of G-quadruplex formation at different positions in Figure 3F–J. The chemical cleavage selectively occurred at the guanines as expected and weak cleavage was observed at adenines (Figure 3A–E). The extremely faint footprint signals of (TTAGGG)4 in K+ solution indicate protection of the guanines in G-quadruplex, which is in sharp contrast to that of the same sample without added salt (Figure 3A). For (TTAGGG)5, G-quadruplex either forms at the 3′ or 5′ end, leaving one repeat exposed at the 5′ or 3′ end, respectively. The footprint of (TTAGGG)5 shows that the intensity of the bands produced by attacks at the first G triplet (GGG) from the 5′ end was higher than that of the rest bands (Figure 3B). This suggests that G-quadruplex preferred to form at the 3′ than at the 5′ end resulting in protection of the four G triplets at the 3′ end and exposure of one GGG triplet at the 5′ end. Indeed, calculation shows that among the G-quadruplexes, more were formed at the 3′ end (Figure 3G). For (TTAGGG)6 and (TTAGGG)7, the preference could not be intuitively judged from the gel (Figure 3C–D). However, calculations revealed that G-quadruplex formation in the middle of the sequences had a lower probability than those at both the 3′ and 5′ ends. Similarly, the probability at the 3′ end was also higher than at the 5′ end (Figure 3H–I).
Figure 3.
(A–E) Representative gels showing DMS footprint of 5′-32P-labeled (TTAGGG)4, (TTAGGG)5, (TTAGGG)6, (TTAGGG)7 and T24(TTAGGG)7. DMS treatments were carried out for 0, 30, 60 and 90 s, respectively in the presence (+) or absence (–) of 150 mM K+. (F–J) Quantification of the footprints showing the probability of G-quadruplex formation at different positions (Qi, where the subscript indicates the number of TTAGGG repeats at the 3′ side of the G-quadruplex), the probability of being in the relaxed form (R) of the oligonucleotides and the probability of a GGG triplet being attacked (p). Results represent the mean of two experiments for (TTAGGG)4 and three experiments for the rest oligonucleotides with standard derivation.
Since the G-rich strand is much longer than seven repeats and does not have an open 5′ end, we attached a 24mer poly-T to the 5′ end of (TTAGGG)7 to better mimic this situation. Calculation showed that the probability at the 5′ end became the lowest and the G-quadruplex at the very 3′ end became the dominant species (Figure 3J) among the four possible positions. Similar to (TTAGGG)5, the suppression at the 5′ and dominance at the 3′ end of G-quadruplex formation in the telomeric section can be intuitively judged from the much stronger bands for smaller fragments than for larger ones (Figure 3E). In Figure 4, the G-quadruplex at each specific position as percentage of the total G-quadruplexes in T24(TTAGGG)7 was presented. It shows that more than half of the G-quadruplexes formed at the immediate 3′ end and the probability of G-quadruplex formation rapidly decreased as it moved inwards from the 3′ end.
Figure 4.
Distribution of G-quadruplexes at different positions in T24(TTAGGG)7 calculated from the data in Figure 3J.
The above calculations were based on the assumption that a G-quadruplex only involves four consecutive TTAGGG repeats. Although G-quadruplexes with longer loops consisting of a TTA and one or more GGGTTA repeats (Figure S1, Supplementary data) is possible, they are relatively unstable (Table S1, Supplementary data) and constitute only a small fraction of the total G-quadruplexes (Table S2, Supplementary data). Our calculations show that omitting these structures had little effect on our conclusion (Figures S2 and S3, Supplementary data).
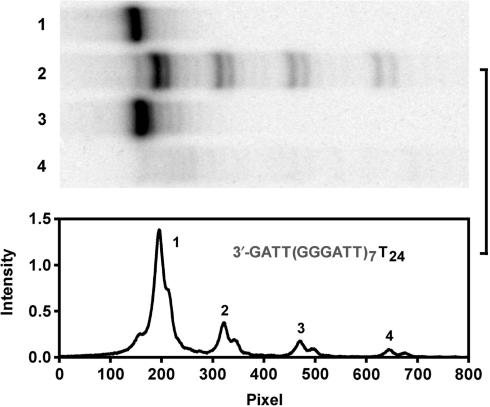
The preferential formation of G-quadruplex at the very 3′ end was further confirmed by a hydrolysis assay of T24(TTAGGG)7 using the 3′ exonuclease activity of T4 polymerase (33). Four reference oligonucleotides were employed, each forms G-quadruplex at a position corresponding to a specific G-quadruplex isomer in T24(TTAGGG)7 (Figure 5A). G-quadruplex has been shown to be resistant to nuclease hydrolysis (34). T4 polymerase trimmed off nucleotides from the 3′ end until the G-quadruplex was met. The hydrolysis of T24(TTAGGG)7 produced four distinct bands corresponding to the blockage of hydrolysis by G-quadruplexes formed at the four different positions (Figure 5A). The preferential formation of G-quadruplex at the very 3′ end was indicated by the fact that the majority of the T24(TTAGGG)7 was protected from digestion. The scanned intensity distribution of these bands (Figure 5B) and further calculations (Figure 5C) resulted in a distribution similar to that revealed by the footprint (Figure 4). In human cells, the ending nucleotides at the 3′ end of human telomeric DNA varies from GGGT to GGGTTAG with a preference for the later (35). We also examined G-quadruplex formation in T24(TTAGGG)7TTAG by the T4 polymerase hydrolysis. The result (Figure 6) shows that the introduction of TTAG to the 3′ end of T24(TTAGGG)7 did not affect the distribution of G-quadruplex formation.
Figure 6.
Distribution of G-quadruplex formation at different positions in T24(TTAGGG)7TTAG detected by T4 polymerase digestion. Top panel: gel electrophoresis of hydrolysis products. Lane 1: T24(TTAGGG)7TTAG without digestion; 2: hydrolysis products of T24(TTAGGG)7TTAG; 3: T24(TTAGGG)7TTAG with the (TTAGGG)7 randomized without digestion; 4: hydrolysis products of T24(TTAGGG)7TTAG with the (TTAGGG)7 randomized. Bottom panel: Densitometry scans of lane 2 in the top panel.
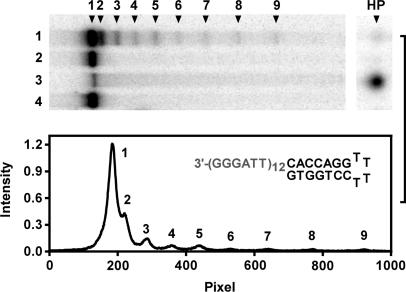
We also analyzed an oligonucleotide with 12 repeats of TTAGGG by the T4 polymerase hydrolysis and the results are given in Figure 7. This oligonucleotide has nine potential positions for G-quadruplex to form and can accommodate a maximum of three G-quadruplexes. The bands of hydrolysis arrest indicate the positions of the very first G-quadruplex from the 3′ end. Similar to the oligonucleotides of seven repeats, the hydrolysis arrests in this longer oligonucleotide also indicate that such G-quadruplex preferentially formed at the 3′ end and the probability of leaving an unstructured tail at the 3′ side rapidly decreased with tail length. Hydrolysis of oligonucleotide with nine repeats also produced similar results (data not shown). These assays did not reveal the distribution of G-quadruplexes along the strand. However, the information on the first G-quadruplex from the 3′ end should reflect the probabilities of G-quadruplex formation along the DNA strand.
Figure 7.
Distribution of the first G-quadruplex from the 3′ end in long G-rich strand with 12 TTAGGG repeats detected by T4 polymerase digestion. Top panel: representative gel electrophoresis of hydrolysis products. Lane 1: hydrolysis products of HP-(TTAGGG)12; 2: HP-(TTAGGG)12 without digestion; 3: hydrolysis products of HP-(T6)12; 4: HP-(T6)12 without digestion. Bottom panel: Densitometry scans of lane 1 in the top panel. HP indicates a hairpin structure shown in the lower panel to prevent full digestion. Numbers 1–9 at the top of gel indicate hydrolysis arrests by G-quadruplexes at nine positions and HP that at the hairpin.
G-quadruplex formed in sequences with more than four repeats is flanked by repeats at one or both the 3′ and 5′ side. The presence of flanking sequence has been shown to destabilize G-quadruplex formed by G3(T2AG3)n (n = 3–16) (36) and TnG4 (n =1–8) (37). To examine the stabilities of G-quadruplexes at different positions, we performed thermal melting analysis with the four reference oligonucleotides used in the exonuclease hydrolysis assay. The result revealed that G-quadruplex at the very 3′ end is thermally more stable with a higher melting temperature (Tm) than those at internal positions (Table 1). This may partially explain the preferential formation of G-quadruplex at the very 3′ end. Formation of G-quadruplex in these sequences had to overcome the physical restraint and static electric repulsion experienced when moving along the flanking repeats. The G-quadruplex at the very 3′ end was restrained only at the 5′ end rather than at both the 3′ and 5′ ends for an internal G-quadruplex.
Table 1.
Melting temperature (Tm) of quadruplexes with 0, 1, 2 and 3 repeats of T6 at the 3′ end
| Sequence | Tm (°C) |
|---|---|
| T24T18(TTAGGG)4 | 57.1 |
| T24T12(TTAGGG)4T6 | 50.8 |
| T24T6(TTAGGG)4T12 | 50.5 |
| T24(TTAGGG)4T18 | 49.1 |
DISCUSSION
We revealed the preferential formation of G-quadruplex at the very 3′ end of the G-rich strand of telomeric DNA by two independent methods. Although the precise analysis was made with DNA in which only one G-quadruplex can form but at several potential positions, such polarity also holds for the very first G-quadruplex from the 3′ end in longer DNA that can accommodate multiple G-quadruplexes. This property should have implications in the function of telomere DNA and the pharmaceutical applications targeting the telomere G-quadruplex.
Recent reports suggest G-quadruplex exists in telomeres of human (38–41) and other species (42–46). The position where the first G-quadruplex from the 3′ end forms determines the size of the single stranded G-rich tail before the 3′ end. In a work where double-stranded telomere DNAs carrying various lengths of single-stranded G-rich overhangs were examined, a minimum tail of approximately six nucleotides was found to be required for extension by telomerase (47). Since G-quadruplex is more bulky than duplex, it might impose a more stringent length requirement than duplex does for telomerase. The polarity of G-quadruplex formation at the 3′ end keeps the size of the 3′ tail minimal to warrant the inhibition on telomerase (Figure 1A). While more than 85% tumors use telomerase (2), a subset adopts the ALT mechanism for telomere maintenance that can be achieved by telomere strand exchange (3) or telomere sister chromatid exchange (t-SCE) (48). When using telomerase inhibitors as anticancer treatment, there is a concern that inhibition of telomerase will have no effect on cancer cells using the ALT mechanism or, to be even worse, select for cells that activate the ALT mechanism for telomere maintenance (49). We expect that the preferential G-quadruplex formation at the very 3′ end should also be inhibitory to the ALT involving telomere strand exchange because in this mechanism the single stranded G-rich 3′ end has to invade and anneal with a C-rich strand to initiate the templated extension (3). The limited tail size and the presence of G-quadruplex nearby should discourage both the invasion and annealing (Figure 1B). In agreement with this, a few studies have reported antiproliferative effect in cells using the ALT mechanism for telomere maintenance (50–53).
The polarity of G-quadruplex formation may also possibly play roles in other processes or events that involve the very 3′ end of the G-rich strand. Telomere DNA terminates with a single stranded G-rich overhang beyond the double stranded region (12,13). Its 3′ end can invade the double-stranded telomere region and anneal with the C-rich strand to form circle-shaped T-loop structure (Figure 1B, second drawing without extension) (54) that is assumed to provide protection to the telomere end (55). G-quadruplex formation at the very 3′ end may potentially interfere with the formation of T-loop. G-quadruplex is resistant to nuclease hydrolysis (34). G-quadruplex formation at the 3′ end may provide protection against such hydrolysis by, for instance, exonuclease. G-quadruplex formed at the 3′ end should be less prone to the unwinding by helicases that require single-stranded DNA of sufficient size to load at the 3′ side of the structure (56–58). On the other hand, formation of G-quadruplex interferes with DNA replication in vitro (59). The lower probability of G-quadruplex formation at internal positions reduces such potential problem in the replication of the bulky telomere DNA. For the above reasons, small molecules targeting at telomere G-quadruplex may potentially affect more processes than telomerase catalyzed telomere lengthening.
Several proteins in human cells have been found capable of unfolding telomere G-quadruplex (60–62). The hPOT1 protein in human cells that binds the single-stranded telomere overhangs has been reported to disrupt G-quadruplexes allowing telomerase extension in vitro (61). Deletion of the pot1+ gene in fission yeast resulted in the loss of telomere DNA (63). These facts imply that the lack of activity to unfold G-quadruplex impaired telomere maintenance suggesting that the formation of G-quadruplex plays a role in such processes. By coordinating with such proteins, the opening and formation of G-quadruplex at the very 3′ end seems to provide a reasonable mechanism for regulating those processes and events where the 3′ end of the G-rich telomere strand is involved.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grant numbers 2007CB507402 from MSTC, 20572082, 30670451 and the Science Fund for Creative Research Groups from NSFC. Funding to pay the Open Access publication charges for this article was provided by grant from MSTC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 2.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 3.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 4.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 5.Han H, Hurley LH. G-quadruplex DNA: a potential target for anti-cancer drug design. Trends. Pharmacol. Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 6.Neidle S, Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nat. Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 7.Jing N, Sha W, Li Y, Xiong W, Tweardy DJ. Rational drug design of G-quartet DNA as anti-cancer agents. Curr. Pharm. Des. 2005;11:2841–2854. doi: 10.2174/1381612054546761. [DOI] [PubMed] [Google Scholar]
- 8.Phan AT, Mergny JL. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Perry PJ, Jenkins TC. DNA tetraplex-binding drugs: structure-selective targeting is critical for antitumour telomerase inhibition. Mini Rev Med Chem. 2001;1:31–41. doi: 10.2174/1389557013407304. [DOI] [PubMed] [Google Scholar]
- 11.Lansdorp PM. Major cutbacks at chromosome ends. Trends. Biochem. Sci. 2005;30:388–395. doi: 10.1016/j.tibs.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 13.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65:1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 15.Phatak P, Cookson JC, Dai F, Smith V, Gartenhaus RB, Stevens MF, Burger AM. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer. 2007;96:1223–1233. doi: 10.1038/sj.bjc.6603691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 17.Gomez D, O’Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 18.Gomez D, Wenner T, Brassart B, Douarre C, O’Donohue MF, El Khoury V, Shin-Ya K, Morjani H, Trentesaux C, et al. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J. Biol. Chem. 2006;281:38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 19.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu D, Balasubramanian S. A proton-fuelled DNA nanomachine. Angew. Chem. Int. Ed. Engl. 2003;42:5734–5736. doi: 10.1002/anie.200352402. [DOI] [PubMed] [Google Scholar]
- 21.Alberti P, Bourdoncle A, Sacca B, Lacroix L, Mergny JL. DNA nanomachines and nanostructures involving quadruplexes. Org. Biomol. Chem. 2006;4:3383–3391. doi: 10.1039/b605739j. [DOI] [PubMed] [Google Scholar]
- 22.Maxam AM, Gilbert W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 24.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 25.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc. 1999;121:3561–3570. [Google Scholar]
- 26.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 Promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zarantonello EH. The closure of the numerical range contains the spectrum. Bull. Am. Math. Soc. 1964;70:781–787. [Google Scholar]
- 28.Vainberg MM. On the convergence of the process of steepest descent for nonlinear equations. Sibirsk Math. Z. 1961;2:201–220. [Google Scholar]
- 29.Zhao Y, Kan ZY, Zeng ZX, Hao YH, Chen H, Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J. Am. Chem. Soc. 2004;126:13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 30.Minhas GS, Pilch DS, Kerrigan JE, Lavoie EJ, Rice JE. Synthesis and G-quadruplex stabilizing properties of a series of oxazole-containing macrocycles. Bioorg. Med. Chem. Lett. 2006;16:3891–3895. doi: 10.1016/j.bmcl.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 31.Simonsson T. G-quadruplex DNA structures – variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 32.Yu HQ, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. J. Am. Chem. Soc. 2006;128:15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 33.Huang WM, Lehman IR. On the exonuclease activity of phage T4 deoxyribonucleic acid polymerase. J. Biol. Chem. 1972;247:3139–3146. [PubMed] [Google Scholar]
- 34.Cao Z, Huang CC, Tan W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal. Chem. 2006;78:1478–1484. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 35.Sfeir AJ, Chai W, Shay JW, Wright WE. Telomere-end processing: the terminal nucleotides of human chromosomes. Mol. Cell. 2005;18:131–138. doi: 10.1016/j.molcel.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 36.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Q, Lu M, Kallenbach NR. Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry. 1993;32:3596–3603. doi: 10.1021/bi00065a010. [DOI] [PubMed] [Google Scholar]
- 38.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- 40.Granotier C, Pennarun G, Riou L, Hoffschir F, Gauthier LR, De Cian A, Gomez D, Mandine E, Riou JF, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Res. 2005;33:4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC, Chu JF, Kao FJ, Chiu YC, Lou PJ, Chen HC, Chang TC. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 42.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown BA, 2nd, Li Y, Brown JC, Hardin CC, Roberts JF, Pelsue SC, Shultz LD. Isolation and characterization of a monoclonal anti-quadruplex DNA antibody from autoimmune "viable motheaten" mice. Biochemistry. 1998;37:16325–16337. doi: 10.1021/bi981354u. [DOI] [PubMed] [Google Scholar]
- 44.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 46.Brown BA, 2nd, Lin Y, Roberts JF, Hardin CC. Antibodies specific for the DNA quadruplex [d(CGC G4 GCG)4] isolated from autoimmune mice. Nucleic Acids Symp. Ser. 1995;33:134–136. [PubMed] [Google Scholar]
- 47.Rivera MA, Blackburn EH. Processive utilization of the human telomerase template: lack of a requirement for template switching. J. Biol. Chem. 2004;279:53770–53781. doi: 10.1074/jbc.M407768200. [DOI] [PubMed] [Google Scholar]
- 48.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 2005;14:R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 49.Mokbel K. The evolving role of telomerase inhibitors in the treatment of cancer. Curr. Med. Res. Opin. 2003;19:470–472. doi: 10.1185/030079903125002081. [DOI] [PubMed] [Google Scholar]
- 50.Kim MY, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The Different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003;63:3247–3256. [PubMed] [Google Scholar]
- 51.Riou JF, Guittat L, Mailliet P, Laoui A, Renou E, Petitgenet O, Megnin-Chanet F, Helene C, Mergny JL. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl Acad. Sci. USA. 2002;99:2672–2677. doi: 10.1073/pnas.052698099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 53.Pennarun G, Granotier C, Gauthier LR, Gomez D, Hoffschir F, Mandine E, Riou JF, Mergny JL, Mailliet P, et al. Apoptosis related to telomere instability and cell cycle alterations in human glioma cells treated by new highly selective G-quadruplex ligands. Oncogene. 2005;24:2917–2928. doi: 10.1038/sj.onc.1208468. [DOI] [PubMed] [Google Scholar]
- 54.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 55.de Lange T. T-loops and the origin of telomeres. Nat. Rev. Mol. Cell. Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- 56.Baran N, Pucshansky L, Marco Y, Benjamin S, Manor H. The SV40 large T-antigen helicase can unwind four stranded DNA structures linked by G-quartets. Nucleic Acids Res. 1997;25:297–303. doi: 10.1093/nar/25.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 58.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 59.Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J. Biol. Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. [DOI] [PubMed] [Google Scholar]
- 61.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.