Abstract
Integration is an essential step in the retroviral lifecycle, and the lentiviral integrase binding protein lens epithelium-derived growth factor (LEDGF)/p75 plays a crucial role during human immunodeficiency virus type 1 (HIV-1) cDNA integration. In vitro, LEDGF/p75 stimulates HIV-1 integrase activity into naked target DNAs. Here, we demonstrate that this chromatin-associated protein also stimulates HIV-1 integration into reconstituted polynucleosome templates. Activation of integration depended on the LEDGF/p75-integrase interaction with either type of template. A differential requirement for the dominant DNA and chromatin-binding elements of LEDGF/p75 was however observed when using naked DNA versus polynucleosomes. With naked DNA, the complete removal of these N-terminal elements was required to abate cofactor function. With polynucleosomes, activation mainly depended on the PWWP domain, and to a lesser extent on nearby AT-hook DNA-binding motifs. GST pull-down assays furthermore revealed a role for the PWWP domain in binding to nucleosomes. These results are completely consistent with recent ex vivo studies that characterized the PWWP and integrase-binding domains of LEDGF/p75 as crucial for restoring HIV-1 infection to LEDGF-depleted cells. Our studies therefore establish novel in vitro conditions, highlighting chromatinized DNA as target acceptor templates, for physiologically relevant studies of LEDGF/p75 in lentiviral cDNA integration.
INTRODUCTION
HIV-1 infection proceeds through a series of ordered events. Soon after entering a target cell, the viral enzyme reverse transcriptase converts the viral RNA genome into linear, double-stranded cDNA containing long terminal repeats (LTRs). This cDNA is the substrate for the enzyme integrase, which binds to U3 and U5 sequences at the outer edges of the upstream and downstream LTRs, respectively. Integrase possesses two enzyme activities, known as 3′ processing and DNA strand transfer, that are essential for integration. Integrase hydrolyzes a dinucleotide from each LTR end during the 3′ processing reaction. After entering the cell nucleus and locating an effective target DNA site, integrase uses the recessed 3′-hydroxyl groups to cut the host DNA in a staggered fashion, which at the same time joins the viral cDNA ends to the resultant 5′ overhangs. Repair of the single-stranded gaps in the resulting DNA recombination intermediate, which is likely accomplished by host DNA repair enzymes, completes the integration process. See Ref. (1) for a recent review on HIV-1 integration.
Purified recombinant integrase proteins possess 3′ processing and DNA strand transfer activities (2–5). Simplified forms of in vitro integration assays utilize a synthetic LTR oligonucleotide for the donor DNA substrate that is processed by integrase, and a second LTR oligonucleotide as the target acceptor molecule into which the processed DNA integrates (3,4). These naked DNA substrates contrast sharply to the situation in vivo, where both the viral and host DNAs exist as higher order nucleoprotein complexes: integrase functions as part of a large preintegration complex (PIC) that is derived from the virion core (6), and the target DNA is ensconced within chromatin.
Chromatin structure can significantly influence the frequency and distribution of retroviral integration. For example, HIV-1 and Moloney murine leukemia virus (Mo-MLV) integrases favor the widening of the major groove that occurs as the DNA wraps around the nucleosome core (7–11). By contrast, these viruses target chromatin quite differently on the genomic scale. HIV-1 preferentially integrates into active genes, fairy equally along their lengths (12,13). Mo-MLV in contrast displays much less preference for gene activity, yet targets regions within 5 kb of transcriptional start sites ∼25% of the time (13–15). Analyses of HIV-1/Mo-MLV chimera viruses has revealed integrase as the principle viral determinant responsible for differential gene targeting during integration (16). The mechanism whereby Mo-MLV integrase preferentially targets gene start sites is unknown. In contrast, the propensity for HIV-1 to integrate fairly equally along the lengths of active genes is in large part governed by its interaction with the cellular chromatin-associated protein lens epithelium-derived growth factor (LEDGF)/p75 (15,17).
LEDGF/p75 can be thought of as a molecular tether (18–22) and both its integrase and chromatin-binding functions are critical for HIV-1 infection (15,23). LEDGF/p75 binds to integrase via a conserved, C-terminally located integrase-binding domain (IBD) that spans residues 347–429 of the 530 residue human protein (21,24). LEDGF/p75 mutants lacking the IBD, or containing the substitution of asparigine for Asp-366 that abrogates the interaction with integrase in vitro (25) and in yeast cells (26), fail to rescue the infection defect accrued via LEDGF/p75 depletion (15,23). LEDGF/p75 interacts with chromatin utilizing conserved regions within the N-terminal half of the protein including a PWWP domain, nuclear localization signal (NLS), and a tandem copy of the AT-hook DNA-binding motif (27,28). Deletion mutants lacking the PWWP domain and AT-hooks failed to rescue HIV-1 infection in LEDGF/p75-depleted cells (15,23). As a deletion that lacked just the PWWP domain functioned at ∼17% of the level of wild-type LEDGF/p75 and a mutant lacking only the AT-hooks displayed wild-type function, the PWWP domain would appear to contribute the dominant chromatin-binding function for HIV-1 integration (15). The molecular mechanism of LEDGF/p75 chromatin binding is currently unknown. One study indicated that the PWWP domain directly bound to DNA (29), whereas a separate study ascribed DNA binding to the NLS and AT-hooks, inferring that the PWWP domain likely mediates interaction(s) with protein components(s) of chromatin (27).
Purified LEDGF/p75 protein potently stimulates the 3′ processing and DNA strand transfer activities of purified HIV-1 integrase in vitro (30). Stimulation strictly depends upon an intact IBD (24,25), but the N-terminal chromatin and DNA-binding regions were in large part dispensable for enzymatic stimulation (27). As these properties contrasted with the crucial role of the N-terminal regions during HIV-1 integration in vivo (15,23), we investigated if this discrepancy depended upon the nature of the target DNA template. By using chromatinized integration templates, we have established an in vitro assay that relies on the LEDGF/p75 PWWP domain for maximum stimulation of integrase function. We therefore conclude that future in vitro studies of LEDGF/p75's role in HIV-1 integration will benefit from utilizing nucleosome templates as targets for DNA strand transfer reactions.
MATERIALS AND METHODS
Reconstitution and purification of nucleosomal templates
The 2.56 kb 5S-G5E4 fragment for polynucleosome (PN) assembly was isolated from p2085S-G5E4 following digestion with Asp718I and ClaI (31). The fragment was labeled at the Asp718I end using [α-32P]dATP and the Klenow fragment of DNA polymerase. Products of DNase I digestion were separated on a 1 % agarose native gel and visualized using a phosphorImager and ImageQuant software. Mononucleosomes (MNs) were assembled on a PCR-amplified 157 bp TPT fragment harboring a tandem repeat of the 20 bp GT phasing sequence (32). PN and MN templates were assembled with purified HeLa core histones (33) by gradient salt dialysis utilizing NaCl and KCl, respectively (34,35).
Expression and purification of recombinant proteins
Hexa-histidine (His6)-tagged HIV-1 integrase from the NL4-3 strain was expressed in Escherichia coli and purified by Ni-nitrilotriacetate chromatography in the absence of detergent using 10 μM ZnSO4 in the column elution buffer as described (36).
The following His6-tag LEDGF/p75 fusion proteins were expressed in E. coli, purified, and treated with PreScission protease to remove the tags prior to integration assays as described (27): LEDGF1–530 (wild-type), LEDGFMutL1 (containing six amino acid substitutions in the NLS and six more in the AT-hooks), LEDGFMutL7 (residues 93–530), LEDGFMutL8 (LEDGFMutL7 carrying the 12 MutL1 substitutions), LEDGF1-226 and LEDGF347–530. Untagged LEDGFD366N was expressed and purified as described (25). The following glutathione S-transferase (GST) fusion proteins were expressed in E. coli and purified as described (24): LEDGF1–530, LEDGF1–325, LEDGF326–530, LEDGF1–471, LEDGF326–471 and LEDGF347–471. The region of LEDGF/p75 that lies between the PWWP domain and AT-hooks is poorly conserved among orthologs and highly susceptible to proteolysis (24), precluding the removal of the GST tag via engineered thrombin sites for a subset of proteins. As LEDGF326–530 lacks this region, the tag was removed from this protein by thrombin cleavage prior to integration assays.
Plasmids encoding GST fused to LEDGF93–471, LEDGF226–471 or LEDGF1–100 were constructed in pGEX-4T1 (GE Healthcare) as described (24), and GST-LEDGF226–471 and GST-LEDGF1–100 were purified from E. coli strain BL21 following induction of protein expression with isopropyl-thio-β-galactopyranoside (IPTG) at 37°C essentially as previously described (24). As GST-LEDGF93–471 was unstable under these conditions, it was isolated from 7 L of E. coli strain XL1-Blue grown at 28°C to an optical density at 600 nm of 3.0 in the absence of IPTG induction using a bench top fermentor. GST-LEDGF93–471 was then purified essentially as previously described (24).
In vitro integration assays
Assays were conducted in 50 mM NaCl, 20 mM HEPES–HCl, pH 7.0, 4 mM MgCl2, 4 μM ZnCl2, 100 μg/ml bovine serum albumin (BSA) and 1 mM dithiothreitol (DTT). In condition 1, mini-HIV donor DNA (16 nM) mixed with integrase (1 μM) were pre-incubated for 10 min at 4°C followed by 10 min at 37°C. LEDGF/p75 was added, and mixtures were incubated for 10 min at 37°C prior to adding naked DNA target or PN template to the final concentration of 5 ng/μl (35 nM equivalent of nucleosomes for PN). Other chronologies of addition of reaction components are summarized in Figure 2B. After 90 min at 37°C, reactions were stopped by the addition of EDTA and SDS to 25 mM and 0.5%, respectively, and treated with proteinase K (1 μg/ml) for 45 min at 55°C. DNAs recovered following precipitation with ethanol were used as template for real-time quantitative PCR, using a three primer design adapted from Ref. (27): AE2397; pT-For, 5′-GCTGTGGAAGCGCTGTATGTTGTTC-3′; pT-Rev, 5′-GGCAGCCATAACAGTCAGCCTTACC-3′. Each integration sample was analyzed in duplicate by real-time PCR, and each integration assay was repeated at least three times.
Figure 2.
LEDGF/p75-dependent stimulation of HIV-1 integrase activity using naked DNA and PN acceptor templates. (A) Integration efficiencies into naked DNA versus PNs. Integration products obtained using the 5S-G5E4 fragment (DNA) or PNs in the absence or presence of LEDGF/p75 (150 nM, 500 nM or 1.5 μM) were quantified by real-time PCR. Error bars represent the variation obtained following duplicate PCRs of a minimum of three integration assays. AU, arbitrary units. (B) Order of addition experiments. Integration assays performed using naked DNA or PNs, in the absence of presence of LEDGF/p75 (150 or 500 nM) (top histogram). The various components, integrase (IN), mini-HIV (Donor), LEDGF and integration acceptor template, were added to the reaction mixtures using the noted five different chronologies (detailed in the bottom diagram). (C) Effect of LEDGF/p75 on DNase I accessibility of PN templates. PNs were incubated for 15 min at 37°C with the indicated levels of LEDGF/p75 prior to a 2 min DNAse I digestion, performed with following quantities of nuclease: 0 U, 1, 1′, 1″; 0.01 U, 2, 2′, 2″; 0.03 U, 3, 3′, 3″; 0.1 U, 4, 4′, 4″; 0.3 U, 5, 5′, 5″ and 1 U, 6, 6′, 6″.
GST pull-down assays
Assays were adapted from Ref. (24). Purified GST fusion proteins were absorbed onto Glutathione-Sepharose beads (GE Healthcare) in buffer 1 (200 mM NaCl, 5 mM DTT, 25 mM Tris–HCl, pH 8.0) using 50 μl (settled volume) of beads per 160 pmol of protein. After 4 h at 4°C, beads were washed in excess volume of buffer 1 and stored on ice. GST-LEDGF-containing beads (10 μl settled volume) resuspended in buffer 2 (50–250 mM NaCl as indicated, 5 mM DTT, 25 mM Tris–HCl, pH 8.0, 4 mM MgCl2, 0.1% NP40, 100 μg/ml BSA, 0.2 mM phenylmethysulfonyl fluoride) were incubated with 800 ng of radiolabeled template DNA or assembled nucleosomes. Samples gently rocked at 4°C for 90 min were left to stand without mixing for an additional 30 min. Beads washed twice in excess volume of buffer 2 without BSA were resuspended in a minimal volume of this buffer, and portions were either separated by SDS–PAGE for western blotting with anti-histone H3 antibody (Abcam Ab1791) or digested with proteinase K as described above. DNAs recovered following this treatment were fractionated through native gels and detected by phosphorImager.
RESULTS AND DISCUSSION
Experimental strategy
To study the effects of chromatin structure and LEDGF/p75 protein on HIV-1 integrase activity, we established an in vitro assay utilizing purified proteins, mini-HIV donor DNA, and a linearized target DNA fragment that was either left naked or complexed with nucleosomes. The mini-HIV donor, linearized by digestion with ScaI, harbors blunt ends corresponding to the unprocessed viral cDNA ends (37) (Figure 1A). The 5S-G5E4 acceptor fragment was chosen because it contains an array of sequences that mandate nucleosome positioning, which, upon nucleoprotein complex formation, yields a PN (Figure 1B). This template has been used to study chromatin remodeling, histone modifications and several enzymatic processes occurring on chromatin in vitro (38–40). The PN template was assembled by salt gradient dialysis using native histones purified from Hela cells and different ratios of DNA/histone octamers. Assemblies were monitored by agarose gel electrophoresis following digestion with EcoRI, which cleaves the starting DNA substrate into multiple 196 bp 5S repeat fragments and a central 431 bp Gal 4-containing fragment (Figure 1B and C).
Figure 1.
DNA substrates and nucleosome construction. (A) The mini-HIV plasmid is linearized by digestion with ScaI (recognition sequence in capital letters), which generates blunt ends corresponding to the authentic unprocessed HIV-1 LTRs (shaded boxes). (B) The 5S-G5E4 acceptor template. Gray ovals, 5S rRNA positioned nucleosomes; black ovals, two nucleosomes covering five Gal 4-binding sites and adenovirus 2 E4 minimal promoter, which do not adopt defined positions; vertical arrows, EcoRI sites, with restriction fragment sizes indicated. The relative positions of PCR primers used to quantify integration activity are noted in panels A and B. (C) Electrophoretic analysis of PN assembly. The starting template DNA (lane 2) and PN templates assembled using the indicated mass ratios (μg/μg) of DNA/histone (lanes 3–5) were digested with EcoRI and separated on a 0.8% agarose gel. The migration positions of liberated 5S nucleosomes (Nuc) and unliganded digestion products are indicated to the right of the ethidium-stained gel; the positions of molecular mass standards are indicated to the left. (D) Electrophoretic analysis of the MN. The assembled MN template (lane 3) was visualized on a 5% polyacrylamide gel relative to the starting 157 bp TPT DNA fragment (lane 2). The positions of molecular mass standards are indicated to the left.
Integration reactions were initiated by preincubating integrase (1 μM) and mini-HIV DNA (16 nM) in a magnesium-dependent reaction buffer at 4°C for 10 min, followed by 10 min at 37°C. LEDGF/p75 was added, the mixture was incubated at 37°C for 10 min, followed by addition of PN template or corresponding DNA fragment (35 nM). DNA strand transfer was then allowed to proceed for 90 min at 37°C. The integration products were deproteinized and quantified by real-time PCR, using one primer complementary to the U5 end of mini-HIV (Figure 1A) and two centrally positioned, outward facing primers complementary to the 5S-G5E4 fragment (Figure 1B). This design monitors integration by quantifying the extent of U5 DNA end joining (27) independent of what may occur at the downstream U3 end. Single U3 end integration as well as products resulting from the combined integration of both U3 and U5 ends are expected to occur, but the formation of these products was not monitored here. The extent of U5 end integration provided a perfect quantitative platform to evaluate the influence of PNs on LEDGF/p75-dependent HIV-1 integration in vitro.
LEDGF/p75 activates HIV-1 integrase activity on naked DNA and PN acceptor templates
In the absence of LEDGF/p75, integration into the PN template was 12 times more efficient than when utilizing the corresponding naked DNA fragment (Figure 2A, inset), a result consistent with previous studies utilizing nucleosome cores (9). LEDGF/p75 stimulated integration into both types of acceptor templates (Figure 2A). The efficiency of integration into naked DNA increased with increasing LEDGF/p75 concentration, up to the highest level tested (1.5 μM). Stimulation of integration into PNs in contrast was maximal at 500 nM LEDGF/p75. Accordingly, at this concentration and below, integration was more efficient using PNs than the naked DNA template. f
Various parameters contribute to the mechanism of LEDGF/p75 stimulation of HIV-1 integrase activity in vitro. Foremost is the protein–protein interaction (24,25,41). The integrase-to-LEDGF/p75 ratio (42), as well as the order by which individual components are added into the reaction mixture (41,43), can also influence product formation. In our initial experiment (Figure 2A and B, condition 1), LEDGF/p75 was added to a preformed integrase–mini-HIV donor complex before the addition of the naked DNA or PN template. Due to the propensity of LEDGF/p75 to bind up to three separate components in these reaction mixtures (DNA, nucleosomes, integrase), four additional chronologies (Figure 2B, conditions 2–5) were tested to gain insight into the mechanism of LEDGF/p75 action.
Maximal levels of PN-dependent integration occurred under conditions whereby integrase and LEDGF/p75 interacted with each other early on in the reaction pathway (Figure 2B, conditions 1, 3 and 4); less stimulation was observed when LEDGF/p75 was initially prebound to PNs (condition 2) or to mini-HIV donor DNA (condition 5). In contrast, integration into naked DNA was most efficient under condition 2, when LEDGF/p75 was preincubated with the target DNA prior to mixing with the integrase–mini-HIV complex. To investigate whether the lower level of PN-dependent integration under this condition was due to changes in nucleoprotein structure that may have occurred during LEDGF/p75 binding, the susceptibility of PNs to DNase I digestion in the absence and presence of LEDGF/p75 was examined (Figure 2C). LEDGF/p75 did not detectably alter nucleosome positioning along the PN array, however, more DNase I was required to obtain the same extent of digestion in the presence of LEDGF/p75 than in its absence (compare lanes 4, 4′ and 4″). These results indicate that LEDGF/p75 may preferentially bind to the free linker DNA sequences that lie between the positioned nucleosomes, but that binding did not induce gross structural alterations within the PN template.
LEDGF/p75 regions important for stimulating HIV-1 integration into naked DNA versus PNs
We next investigated LEDGF/p75 domains and amino acid residues important for stimulating HIV-1 integration into naked DNA versus PN acceptor templates. Binding to non-specific DNA in vitro is predominantly mediated by the NLS and AT-hook DNA-binding motifs (27), whereas the PWWP domain in concert with the AT-hooks mediates the binding of the protein to chromatin in live cells (27,28); charged regions (CRs) 1–3 located within residues 93–325 also contribute to chromatin binding (28). The IBD located within the C-terminal part of LEDGF/p75 mediates the interaction with integrase (21,24); Asp-366 within this domain is a key hotspot residue for the interaction (25,26,44). Deletion or missense mutant proteins altered at one or more of these functional determinants (Figure 3A), purified following their expression in E. coli, were tested alongside wild-type LEDGF/p75 to assess their ability to stimulate HIV-1 integrase activity under reaction 1 conditions.
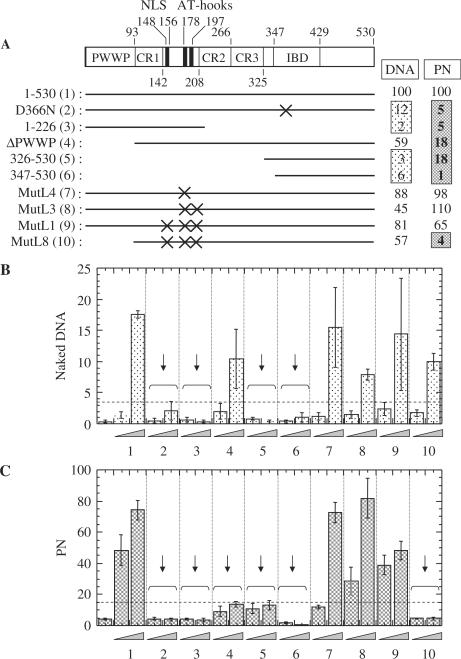
Figure 3.
Characterization of LEDGF/p75 elements important for stimulation of HIV-1 integration into naked DNA versus PN templates. (A) Mutant proteins used in the analysis. The PWWP domain, charged regions (CRs) 1–3, NLS, AT-hooks and IBD are noted in the diagram of full-length LEDGF/p75 (27,28). Residues retained in mutant constructs are indicated by a bold line, with positions of missense mutations denoted by X. Activities of the nine mutant proteins relative to wild-type LEDGF/p75 (500 nM; set to 100%) on naked versus PN templates are summarized at the right; values <20% are boxed. (B) Relative levels of HIV-1 integration in the presence of the noted LEDGF/p75 variant and naked target DNA template. The initial value is the level of integrase activity in the absence of added LEDGF/p75, with the following sets conducted in the presence of 150 or 500 nM protein (indicated by the triangle). Error bars are the variation obtained from duplicate PCRs of integration assays performed in triplicate. The dashed horizontal line denotes 20% of the level of activity obtained in the presence of 500 nM of wild-type LEDGF/p75. The vertical arrows highlight those constructs that failed to attain this level of activity. (C) Same as in panel B, except that PNs were used in place of naked DNA.
Experiments conducted with naked target DNA generated results in agreement with previous studies (27) (Figure 3B). LEDGF1–266 missing the IBD or the full-length LEDGFD366N missense mutant stimulated integration at ∼2% and 12% of the wild-type protein, respectively, highlighting the central role of the LEDGF/p75–integrase interaction in activating enzyme function under these conditions (Figure 3B, panels 2 and 3; results summarized in Figure 3A). Though the IBD is necessary for stimulation, it alone is insufficient (24,27) as evidenced by the fact that the LEDGF326–530 C-terminal fragment containing the intact IBD failed to effectively stimulate integrase activity (Figure 3B, panel 5). Also in agreement with previous findings, DNA binding was in large part dispensable for activation, as full-length proteins altered at the AT-hooks or AT-hooks and NLS (MutL4, MutL3, MutL1) were reduced by 55% at most in their abilities to stimulate integration (Figure 3A and B). The PWWP domain was also not crucial for activation, since its removal only resulted in an ∼40% decrease in function (Figure 3B, panel 4). LEDGFMutL8 lacking the PWWP domain and furthermore mutated in the NLS/AT-hook DNA-binding elements was likewise only about 2-fold defective in its ability to stimulate integrase activity (panel 10). We therefore conclude that LEDGF/p75's ability to stimulate integration into naked DNA depends on its interaction with integrase as well as the PWWP domain, NLS, AT-hooks and CR sequences that encompass the N-terminal 325 residues of the protein.
Results obtained utilizing PNs were compared to those observed using naked DNA (Figure 3A, right and C). As anticipated, the integrase–LEDGF/p75 interaction was central to stimulating integration into PNs. The NLS/AT-hook mutants also behaved similarly, in that each of these stimulated integrase activity similar to the wild-type protein. A major difference was however observed when the PWWP domain deletion mutants were assayed. The LEDGF93–530 mutant lacking the domain functioned at ∼18% of the wild-type level when PNs were the integration target, whereas its activity was reduced only about 2-fold in stimulating integrase to function with naked target DNA (compare panels 4 in Figure 3B and C; P = 1.1 × 10−3 using Student's t-test). More dramatically, knocking out the AT-hooks and NLS reduced the function of LEDGFMutL8 to only ∼4% of wild type under conditions where protein function was again reduced only ∼2-fold from the wild type when naked DNA served as the target (panels 10 in Figure 3B and C: P = 1.1 × 10−10 using Student's t-test). Experiments conducted under higher protein concentrations revealed the same overall trend. LEDGFMutL8 was most active at 1.5 μM (the highest concentration tested; data not shown), but integration into PNs was still ∼5-fold reduced from the suboptimal level of wild-type activity observed under these conditions (Figure 2A). We therefore conclude that LEDGF/p75 stimulation of HIV-1 integration into PNs in vitro depends on its interaction with integrase as well as the PWWP domain, with secondary contributions coming from the NLS and AT-hooks that were for the most part unnoticed unless assayed together with the PWWP domain deletion (Figure 3C, compare panel 10 with panels 7–9). It is noteworthy that these requirements are nearly parallel to those obtained when LEDGF/p75 mutants were tested for their abilities to reconstitute HIV-1 infection to cells severely depleted (23) or knocked out (15) for the host factor. Mutants lacking the IBD (15,23) or carrying the conservative D366N substitution within the otherwise intact domain (15) failed to rescue HIV-1 infection. Mutants lacking both the PWWP domain and AT-hooks likewise failed to rescue infectivity (15,23). The PWWP domain was moreover determined to act as the dominant N-terminal determinant ex vivo: expressing the LEDGF93–530 PWWP domain deletion mutant rescued ∼17% of HIV-1 function, whereas expressing a full-length missense mutant carrying the MulL3 AT-hook mutations fully restored HIV-1 infection (15). We note that other host DNA-binding proteins like HMGA (45,46), HMGB (45) and INI1 (47) have also been shown to stimulate HIV-1 integration in in vitro assays. However unlike LEDGF/p75, ex vivo infections of genetically null HMGA (48) or INI1 (49) cells have failed to reveal a role for either of these factors in integration under physiologically relevant conditions.
Characterization of LEDGF/p75 regions important for DNA and chromatin binding in vitro
The PWWP domain in large part mediates LEDGF/p75 binding to chromatin in live cells (27,28), but the mechanism underlying this interaction is unknown. Unclear is whether the PWWP domain principally interacts with DNA or a protein component(s) of chromatin: one study failed to detect significant binding between the isolated domain and DNA in vitro (27), whereas a separate study indicated that a 58 amino acid residue stretch within the domain contributed to DNA binding (29). The results of the previous experiments revealed a significant role for the PWWP domain in LEDGF/p75-dependent activation of integration into PNs, but not naked DNA. To ascertain if the PWWP domain mediated binding to nucleosomes, an interaction assay was designed to simultaneously monitor LEDGF/p75 binding to free DNA versus nucleosomes. For this, full-length or truncated LEDGF/p75 GST fusion proteins (Figure 4A) pre-bound to glutathione-sepharose beads were incubated with radiolabeled DNA or assembled nucleosomes. Bound DNA was detected following deproteinization, gel electrophoresis and phosporImager analysis. Bound nucleoprotein complexes were analyzed for core histone H3 content by western blotting or for DNA content after deproteinization and agarose gel electrophoresis.
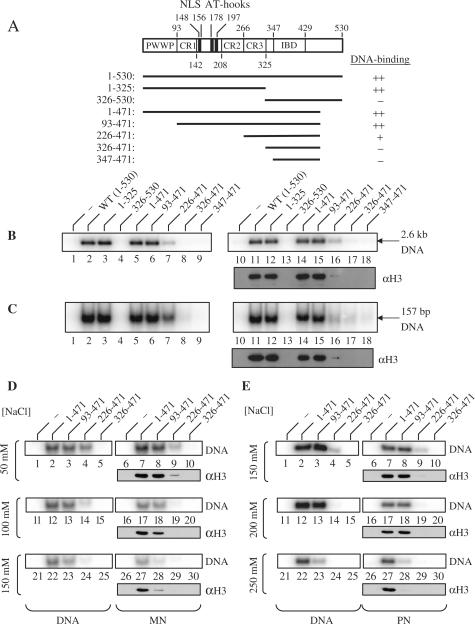
Figure 4.
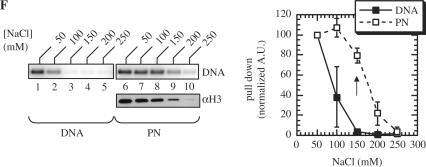
Characterization of the DNA and nucleosome-binding domains of LEDGF/p75. (A) Scheme of the 530 residue human LEDGFp75 protein, highlighting the PWWP domain, charged regions (CRs) 1–3, the NLS, AT-hooks and IBD (27,28). Residues retained in the GST fusion proteins are indicated by bold lines. Relative levels of DNA binding at 50 mM NaCl are summarized at the right. (B) Wild-type and mutant LEDGF/p75 binding to 5S-G5E4 DNA (lanes 1–9) and PN templates (lanes 10–18). Purified GST was substituted for LEDGF/p75 in the reactions (50 mM NaCl) in lanes 1 and 10. Lanes 2 and 11 contained wild-type LEDGF1–530; lanes 3 and 12, LEDGF1–325; lanes 4 and 13, LEDGF326–530; lanes 5 and 14, LEDGF1–471; lanes 6 and 15, LEDGF93–471; lanes 7 and 16, LEDGF226–471; lanes 8 and 17, LEDGF326–471; lanes 9 and 18, LEDGF347–471. The lower panel on the right was developed using anti-histone H3 antibodies. (C) TPT DNA (lanes 1–9) and MN (lanes 10–18) binding. The reactions in lanes 1–18 were identical to those in panel B except for the identity of the indicated DNA and nucleosome templates. (D) Same as in panel C, except that binding of LEDGF mutants to TPT DNA (lanes 1–5, 11–15 and 21–25) or MNs (lanes 6–10, 16–20 and 26–30) was performed at 50 mM NaCl (lanes 1–10), 100 mM NaCl (lanes 11–20) or 150 mM NaCl (lanes 21–30). GST alone was analyzed in lanes 1, 6, 11, 16, 21 and 26. Lanes 2, 7, 12, 17, 22 and 27 contained LEDGF1–471; lanes 3, 8, 13, 18, 23 and 28 harbored LEDGF93–471; lanes 4, 9, 14, 19, 24 and 29 used LEDGF226–471; lanes 5, 10, 15, 20, 25 and 30 used LEDGF326–471. (E) Same as in panel D, except that binding assays were conducted using 5S-G5E4 DNA (lanes 1–5, 11–15 and 21–25) or PNs (lanes 6–10, 16–20 and 26–30) in the presence of 150, 200 or 250 mM NaCl, as indicated. (F) Capture of free 5S-G5E4 DNA (lanes 1–5) or PN templates (lanes 6–10) by GST-LEDGF1–100 at different salt concentrations. The right panel shows a plot of quantified radiolabeled DNA recovery (naked or assembled in PN) as a function of salt concentration; error bars represent the variation obtained from four independent experiments.
Initial experiments were performed at 50 mM NaCl, which corresponds to the salt concentration present in the enzymatic reactions. Under these conditions, the C-terminal deletion mutant LEDGF1-325 bound the 5S-G5E4 restriction fragment as efficiently as wild-type LEDGF1–530 and the C-terminal domain fragment LEDGF326–530 failed to detectably bind DNA (Figure 4B, lanes 2–4). Consistent with our earlier report (27), the internal LEDGF93–471 fragment also bound DNA as efficiently as full-length LEDGF/p75 (Figure 4B, lane 6). Truncation of this fragment to remove the tripartite NLS/AT-hook element revealed residual activity for LEDGF226–471 that was lost upon further N-terminal truncation to residue 326 (Figure 4B, lanes 7 and 8). These results are fully consistent with the notion that the NLS/AT-hook elements confer the majority of DNA-binding activity under these conditions, with minor contributions from sequences that lie within flanking CR3 (Figure 4A) and the PWWP domain (see below).
Strikingly similar results were obtained using PNs instead of free 5S-G5E4 DNA, as the regions of the protein that mediated PN capture were coincident to those that mediated DNA binding (Figure 4B, compare lanes 10–18 with lanes 1–9). Because cell-based assays have revealed a role for the PWWP in chromatin binding (27,28), we considered that LEDGF/p75 might indirectly capture nucleosomal complexes under these conditions via binding to exposed linker DNA sequences present within the PN template. Thus, to constrict the analysis to nucleosome-specific binding, the pull-down experiment was repeated using MNs, which lack linker DNA sequences. The MN template was assembled on a 157 bp TPT fragment containing two GT phasing sequences (50) that uniquely position the DNA around the histone octamer (32) (Figure 1D). As expected from the results of the previous experiments, LEDGF93–471 bound to the 157 bp naked DNA template as efficiently as wild-type LEDGF/p75 (Figure 4C, compare lane 6 with lane 2). LEDGF226–471 again revealed residual binding to naked DNA (Figure 4C, lane 7). As LEDGF93–471 bound MNs as efficiently as LEDGF1–471 and the full-length wild-type protein (Figure 4C, lanes 11, 14 and 15), the level of salt in the binding assays was altered to ascertain effects of increasing ionic concentration on the protein–DNA and protein–protein interactions. The residual binding of LEDGF226–471 to naked DNA (Figure 4D and E, lanes 4, 14 and 24) and both chromatin templates (MN and PN; lanes 9, 19 and 29) was very sensitive to increasing salt, as expected since the CR3 sequences within LEDGF226–471 are likely to mediate electrostatic interactions with DNA and nucleosomes (28). Elevated levels of salt importantly unveiled a preferential role for the PWWP domain in binding to nucleosomes as compared to naked DNA. At 150 mM NaCl, LEDGF93–471 lacking the domain bound-free TPT DNA ∼70% as efficiently as wild-type LEDGF1–530 (data not shown) and LEDGF1–471 (Figure 4D, lanes 22 and 23) whereas MN capture was reduced more than 3-fold as compared to the control proteins (Figure 4D, lanes 27, 28, and data not shown). This level of salt however failed to reveal a role for the PWWP domain in PN binding (Figure 4E, compare lanes 7 and 8 with lanes 2 and 3). A higher concentration of salt (250 mM NaCl) did reveal a role for the PWWP domain in PN capture (Figure 4E, lanes 27 and 28) although this effect seemed primarily attributable to a loss in DNA binding (lanes 22 and 23). We therefore conclude that the absence of DNA linkers in the MN templates emphasized the requirement of the PWWP domain in the interaction between LEDGF/p75 and nucleosomes.
These data suggested that the PWWP domain likely provides a direct link between LEDGF/p75 and nucleosomes during PN-dependent activation of HIV-1 integration. Isotonic (150 mM) NaCl nonetheless failed to reveal a difference in LEDGF1–471 versus LEDGF93–471 binding to free DNA versus PN templates. To directly test a role for the PWWP domain in PN binding under physiological salt conditions, a GST-PWWP domain fusion protein containing the N-terminal 100 residues of LEDGF/p75 was purified and tested in the pull-down assay. GST-PWWP displayed some affinity for naked DNA, however, unlike the case for the GST-LEDGF93–471 deletion mutant (Figure 4E, lane 13), this interaction was very sensitive to salt concentration (Figure 4F, lanes 1–5). In stark contrast, PN binding by GST-PWWP was nearly as efficient at 150 mM and 50 mM NaCl (Figure 4F, compare lane 8 with lane 6; quantification indicated by vertical arrow in rightward panel). We therefore conclude that the PWWP domain possesses affinity for binding to PNs at physiologic NaCl concentration under these conditions. We have however not addressed whether the PWWP domain might interact directly with a core histone protein or other chromatin factor that becomes part of our reconstituted nucleosomal templates.
In conclusion, we have determined that LEDGF/p75-dependent stimulation of HIV-1 integrase activity in vitro is in most cases favored by using chromatinized as compared to naked target DNA acceptor templates (Figure 2A). Stimulation of integration into either template strictly depended on the LEDGF/p75–integrase interaction. Three lines of experimentation however support the notion that the mechanism of stimulation differs when using naked DNA versus PNs. Among these were differential responses to the concentration of LEDGF/p75 (Figure 2A) and to the order by which different components were added into the reaction mixtures (Figure 2B). Chromatinized templates were furthermore critical to unveil the stimulatory requirements of the PWWP domain and AT-hooks of LEDGF/p75, which up until now were only detected in the context of HIV-1-infected cells. Pull-down assays moreover indicated the PWWP domain might preferentially interact with core components of reconstituted human nucleosomes. We therefore conclude that chromatinized templates afford an in vitro system that best recapitulates the role of LEDGF/p75 in HIV-1 integration as it occurs in vivo.
ACKNOWLEDGEMENTS
This work was supported by grant AI39394 from the US National Institutes of Health (to A.E.) and by grants 2005/004 and 2006/124 from the Agence Nationale de Recherche sur le SIDA (to M.L.). M.L. and Y.B. thank Prof. Moshe Yaniv, Dr Félix Rey and Dr Jean François Mouscadet for support, advice and valuable discussion. Y.B. thanks Dr Eric Batsché for help with real-time PCR. Funding to pay the Open Access publication charges for this article was provided by the Pasteur Institute of Paris, France.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vandegraaff N, Engelman A. Molecular mechanisms of HIV integration and therapeutic intervention. Expert Rev. Mol. Med. 2007;9:1–19. doi: 10.1017/S1462399407000257. [DOI] [PubMed] [Google Scholar]
- 2.Bushman FD, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 3.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 4.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 5.Sherman PA, Fyfe JA. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl Acad. Sci. USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 7.Pryciak PM, Sil A, Varmus HE. Retroviral integration into minichromosomes in vitro. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pryciak PM, Varmus HE. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 9.Pruss D, Bushman FD, Wolffe AP. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl Acad. Sci. USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruss D, Reeves R, Bushman FD, Wolffe AP. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 11.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 15.Shun M-C, Ragahvendra NK, Vandergraaf N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specifc HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, et al. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 18.Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- 19.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, et al. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 2005;280:25517–25523. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- 21.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 22.Maertens GN, Cherepanov P, Engelman A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006;119:2563–2571. doi: 10.1242/jcs.02995. [DOI] [PubMed] [Google Scholar]
- 23.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 24.Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- 25.Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 26.Rahman S, Lu R, Vandegraaff N, Cherepanov P, Engelman A. Structure-based mutagenesis of the integrase-LEDGF/p75 interface uncouples a strict correlation between in vitro protein binding and HIV-1 fitness. Virology. 2007;357:79–90. doi: 10.1016/j.virol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Turlure F, Maertens G, Rahman S, Cherepanov P, Engelman A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 2006;34:1663–1675. doi: 10.1093/nar/gkl052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 29.Singh DP, Kubo E, Takamura Y, Shinohara T, Kumar A, Chylack L.T., Jr, Fatma N. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J. Mol. Biol. 2006;355:379–394. doi: 10.1016/j.jmb.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 30.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 31.Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 32.Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 33.Workman JL, Taylor IC, Kingston RE, Roeder RG. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 1991;35:419–447. doi: 10.1016/s0091-679x(08)60582-8. [DOI] [PubMed] [Google Scholar]
- 34.Aalfs JD, Narlikar GJ, Kingston RE. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 2001;276:34270–34278. doi: 10.1074/jbc.M104163200. [DOI] [PubMed] [Google Scholar]
- 35.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leh H, Brodin P, Bischerour J, Deprez E, Tauc P, Brochon JC, LeCam E, Coulaud D, Auclair C, et al. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry. 2000;39:9285–9294. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]
- 37.Cherepanov P, Surratt D, Toelen J, Pluymers W, Griffith J, De Clercq E, Debyser Z. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda K, Steger DJ, Eberharter A, Workman JL. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hassan AH, Neely KE, Workman JL. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell. 2001;104:817–827. doi: 10.1016/s0092-8674(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 40.Lavigne M, Francis NJ, King IF, Kingston RE. Propagation of silencing; recruitment and repression of naive chromatin in trans by polycomb repressed chromatin. Mol. Cell. 2004;13:415–425. doi: 10.1016/s1097-2765(04)00006-1. [DOI] [PubMed] [Google Scholar]
- 41.Raghavendra NK, Engelman A. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology. 2007;360:1–5. doi: 10.1016/j.virol.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey KK, Sinha S, Grandgenett DP. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J. Virol. 2007;81:3969–3979. doi: 10.1128/JVI.02322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu F, Jones GS, Hung M, Wagner AH, MacArthur HL, Liu X, Leavitt S, McDermott MJ, Tsiang M. HIV-1 integrase preassembled on donor DNA is refractory to activity stimulation by LEDGF/p75. Biochemistry. 2007;46:2899–2908. doi: 10.1021/bi602387u. [DOI] [PubMed] [Google Scholar]
- 44.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl Acad. Sci. USA. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J. Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao K, Gorelick RJ, Johnson DG, Bushman F. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 2003;77:1598–1603. doi: 10.1128/JVI.77.2.1598-1603.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 48.Beitzel B, Bushman F. Construction and analysis of cells lacking the HMGA gene family. Nucleic Acids Res. 2003;31:5025–5032. doi: 10.1093/nar/gkg684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boese A, Sommer P, Gaussin A, Reimann A, Nehrbass U. Ini1/hSNF5 is dispensable for retrovirus-induced cytoplasmic accumulation of PML and does not interfere with integration. FEBS Lett. 2004;578:291–296. doi: 10.1016/j.febslet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 50.Shrader TE, Crothers DM. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]