Abstract
Expression of genes belonging to the pho regulon in Streptomyces coelicolor is positively regulated (as shown by comparing the wild-type and a ΔphoP mutant) by binding of the response regulator PhoP to 11-nt direct repeats (DRus). These sequences have been found in over 100 genes of Streptomyces coelicolor; 20 of them were cloned and the binding of PhoPDBD to most of their promoters has been shown by electrophoretic mobility shift assays. Deletion experiments showed that at least two DRus are required for proper binding of PhoPDBD. Deletion of 1 nt leaving a 10-nt direct repeat reduced drastically binding of PhoPDBD. Three different types of operators have been identified. Complex operators (class III) contain up to six DRus, some of them with poor conservation of the 11-nt consensus sequence, which however were protected by PhoPDBD in footprinting analyses. A cooperative binding of PhoPDBD molecules initiated at conserved core DRus appears to be the mechanism involved in binding of several PhoPDBD monomers to those complex operators. The information theory-based model that incorporates the positive or negative contribution to the binding of PhoPDBD of adjacent sequences has been used to deduce the structure of PHO boxes and the relevance of each DRu.
INTRODUCTION
Streptomyces species and other actinomycetes are soil-dwelling Gram-positive bacteria that produce an impressive array of secondary metabolites (1), many of which possess important pharmacological activities (2,3). The ability to produce this variety of secondary metabolites is due to a wealth of genes, usually arranged as clusters (4,5) that are being found in the sequenced genomes of Streptomyces species (6,7).
Most of the secondary metabolites are secreted (8) and serve as inter-cellular talk signals in pure culture (e.g. quorum sensing) (9,10) or as inter-generic weapons or communication signals (e.g. anti-bacterial, anti-fungal, plant-growth promoting substances) (11,12). The metabolism of Streptomyces species in soil and also in pure cultures is strictly regulated by the availability of nutrients, particularly inorganic phosphate (13,14).
Defined sets of genes that show specific overexpression patterns are triggered in response to phosphate starvation (15). Expression of genes encoding secondary metabolites and, therefore, production of these compounds takes place only under phosphate-limiting conditions (16,17), whereas high phosphate conditions favour expression of housekeeping genes, particularly those related to nucleic acid and protein synthesis. Silent gene clusters that are not significantly expressed may be triggered if phosphate-controlled mechanisms are disrupted, thus providing an increasing arsenal of rare secondary metabolites with potentially novel biological activities. Not all secondary metabolite genes are equally sensitive to phosphate control. Polyketide and aminoglycoside compounds are more sensitive to phosphate regulation than non-ribosomal peptides (14,18,19) and this is likely to be due to the presence of specific control sequences in their promoter regions and in the genes-encoding enzymes providing precursors for these metabolites (20).
The control of phosphate-regulated genes in Streptomyces coelicolor and Streptomyces lividans is mediated by the two-component system PhoR-PhoP (20–22). Recently, we purified the regulatory fused proteins GST-PhoP and GST-PhoPDBD (a truncated version with the DNA-binding domain) and showed that they bind to PHO boxes, close to the –35 sequence in the pstS and phoU promoters. Each of these PHO boxes is formed by direct repeat units (DRu) with the GG/TTCAYYYRG/CG consensus sequence (23).
The identification of PHO boxes was based on the footprinting analysis of three phosphate-regulated genes. However, a computer search of putative phosphate-controlled genes showed a degree of variability in some of the nucleotide positions of the DRus. The divergence with respect to the consensus sequence in the PHO boxes may affect the binding of phosphorylated PhoP and, therefore, the degree of phosphate control.
Moreover, there are usually two or four DRus in the PHO boxes of phosphate-regulated genes, although the presence of up to eight DRus has been reported in the upstream region of the phoRP cluster of Streptomyces natalensis (19). In early studies in Escherichia coli (24), the presence of an odd number (e.g. seven DRus in the upstream region of the ugp operon) was proposed. It is unclear, however, whether a single DRu will allow formation of stable PhoP–DNA complexes. It is likely that the presence of four or more adjacent DRus in a region may allow binding of phosphorylated PhoP to an otherwise poorly conserved DRu sequence. It was, therefore, of great interest to study the binding of PhoP to a significant number of native putative PHO boxes and also to engineer new promoters with modified DRus in order to establish precisely the optimal nucleotide sequence and the number of DRus required for stringent or ‘relaxed’ phosphate control of gene expression.
In this study, we analyse the importance of the number of DRus for binding of PhoPDBD and the presence of PHO boxes in different phosphate-regulated promoters. We have modified the nucleotide sequence of the PHO boxes or the distance between DRus by directed mutagenesis and this has allowed us to define a model for the binding of PhoP to those boxes.
MATERIALS AND METHODS
DNA manipulations
All the oligonucleotides used in this work are listed in Table S1 (Supplementary Data). Promoters with putative PHO boxes were amplified by PCR and the products were cloned into pBluescript KS+ plasmid (Stratagene). The correct amplification was tested by sequencing using an ABI Prism 3130 Genetic Analyzer (Applied Biosystems). For analysis of pstS promoter, two new restriction targets were created with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) using plasmid pGEM-PpstS (23) as template. The PstS1/PstS2 oligonucleotide pair was used for introducing an Acc65I site, and the PstS3/PstS4 for an SpeI site. The resulting plasmid was digested with these enzymes and the PHO boxes in the pstS promoter region were replaced by new sequences obtained by oligonucleotide pairing. For this, a mixture of 25 nM of oligonucleotides PstS5 and PstS6 or PstS7 and PstS8 was heated to 95°C for 5 min and cooled to 25°C with a cooling slope of 0.02°C/s. The final product was ligated with the vector plasmid (from which the original PHO box had been removed) and transformed into E. coli DH5α. The correct insertion was established by sequencing. Likewise, for modification of SCO1854, SCO1993 and pstS promoters the QuickChange system was used with oligonucleotide pairs PHO21/PHO22, PHO23/PHO24 and PstS9/PstS10, respectively.
Bioinformatic analysis of PhoP operators
The individual information theory programs makebk, delila, encode, rseq, dalvec, makelogo, ri, scan, multiscan and lister (25,26) were used to identify and analyse putative target sequences of the response regulator PhoP. The search for new operator sequences was carried out as described previously (15), using an individual information weight matrix constructed with the alignment of 18 DRus (model 0). These DRus stem from the experimentally validated operators of pstS (two DRus), phoU-phoR (four), phoA (two), phoD (six) and pitH2 (four) [(23,28), Santos-Beneit,F., unpublished data]. Using the newly validated DRus, we constructed model I that comprises 37 DRus that form the core of the binding sites referred in Table 1. Model I was used to evaluate the DRus found in the protected regions, and to generate the logo and the walkers (25,29).
Table 1.
Promoters recognized by PhoP
| Promoter region | Regulated gene and function | Operator structurea | Number of EMSA complexes | Strand | Distance to ATG | Distance to +1 | Profileb | P-valuec | PhoP regulationd |
|---|---|---|---|---|---|---|---|---|---|
| A. Class I: operators with 2 well-conserved DRu | |||||||||
| SCO2286 | phoA, secreted alkaline phosphatase | CC | 1e | Coding | 128 | 103 | AA | >0.25 | Activatione |
| SCO3790-1 | SCO3790, putative phosphatase | CC | 1 | Coding | 79 | N.D. | 0D | 0.165 | Activation |
| SCO4142 | pstS, Pst transporter, phosphate-binding component | (EE)CC | 1f | Coding | 114 | 39 | A0 | <0.000 | Activationf |
| SCO6169-70 | SCO6169, possible regulatory protein | CC | 1 | Coding | 195 | N.D. | 00 | >0.25 | N.D. |
| SCO6169-70 | SCO6170, putative oxidoreductase | CC | 1 | Non-coding | 101 | N.D. | 00 | >0.25 | N.D. |
| B. Class II: operators with 3 conserved DRu | |||||||||
| SCO0033-4 | SCO0034, unknown | CCC | 1g | Non-coding | 127 | N.D. | 0D | 0.034 | Activation |
| SCO1906 | SCO1906, putative phosphatase | CCC | 1 | Coding | 92 | N.D. | 00 | > 0.25 | N.D. |
| SCO4261-3h | SCO4261, possible response regulator | CCC | 1g | Non-coding | 43 | N.D. | aA | 0.088 | Repression |
| C. Class III: operators of complex structure | |||||||||
| SCO1393-4 | SCO1394, possible glycosyl hydrolase | (E)CC | 2 | Non-coding | 30 | N.D. | A0 | 0.049 | Activation |
| SCO2262-3 | SCO2262, possible oxidoreductase | ECC | 2 | Coding | 70 | N.D. | 0A | 0.013 | Repression |
| SCO2465-6 | hrdA, principal sigma factor paralogue | ECC | 2 | Coding | 80 | N.D. | 0A | 0.052 | Repression |
| SCO5447 | SCO5447, putative metalloprotease | ECCE | 3 | Non-coding | 214 | N.D. | A0 | 0.26 | Activation |
| SCO7697 | SCO7697, possible secreted phytase | CCE(E) | 3g | Coding | 122 | N.D. | A0 | 0.001 | Activation |
| SCO4228-9 | phoU, regulator of the PHO response | CCEUES | 2f | Coding | 109 | 38 | A0 | 0.143 | Activationf |
| SCO4228-9 | phoRP, PHO two-component system | CCEUES | 2f | Non-coding | 63 | 65 | A0 | <0.000 | Activationf |
| SCO2878 | SCO2878, unknown | CCCEUES | 2-3 | Coding | 42 | N.D. | AD | <0.000 | Activation |
| SCO1196 | SCO1196, putative secreted protein | CCC[2]ES | 2 | Non-coding | 83 | N.D. | A0 | <0.000 | Activation |
| SCO1845 | pitH2, phosphate transporter | ES[1]ESEUCC[2]ESi | 4i | Coding | 48 | 3 | 00 | >0.25 | Activation/ repressioni |
| SCO2068 | phoD, phospholipase | CCC[8] | 4e | Non-coding | 10 | 10 | 00 | >0.25 | Activation/ repressione |
| ES[1]EEe | Coding | ||||||||
| D. Unclassified operators | |||||||||
| SCO4878-9 | SCO4878, putative glycosyltransferase | 4 DRusj | 1g | Coding | 40 | N.D. | 00 | >0.25 | N.D. |
| SCO4878-9 | SCO4879, hypothetical protein | 5 DRusj | 1 | Coding | 30 | N.D. | A0 | 0.001 | Activation |
aEach letter symbolizes a DRu and denotes the functional class. Class ‘C’ comprises DRus that form the core of the operator; ‘E’ comes from ‘extension’, ‘EU’ from ‘extension unstable’ and ‘ES’ from ‘extension support’; see text for a complete explanation. The DRus that originated the information matrix are represented by underlined letters. In two operators, brackets enclose possible DRus that were not protected in the footprinting assays, but help to explain the number of EMSA complexes. Numbers in square brackets are the separation in base pairs between contiguous DRus.
bTranscription profiles of the response to phosphate limitation. The first letter summarizes the transcription response of the S. coelicolor wild-type strain, and the second, the response of the ΔphoP mutant. ‘A’ means augmented expression, ‘D’ means decreased expression and ‘0’, no detected change (P > 0.1). Data obtained from Rodríguez-García et al. (15).
cThese P-values refer to the interaction contrast (or contrast 5; see Rodríguez-García et al. (15) for details). This comparison quantifies the differences between the response to phosphate limitation of the S. coelicolor wild type and that of the mutant ΔphoP strain. Low P-values support the regulatory role of PhoP.
dThe type of the transcriptional control was inferred from the transcription profile or from the reference provided. In the cases of bidirectional promoter regions, which genes are controlled was decided from the transcriptomic results; exceptionally, there is no data for SCO2263, because no probe was present in the microarrays used.
eAccording to Apel et al. (28).
fAccording to Sola-Landa et al. (23).
gAccording to Rodríguez-García et al. (15).
hThe ORF SCO4262 was annotated as doubtful (6). Since it lies overlapping the putative promoter region of SCO4261, we considered that this ORF is not real.
iAccording to Santos-Beneit, F. unpublished data.
jThe fragment that contains both operators of the SCO4878–SCO4879 intergenic region, showed a unique complex in EMSA experiments. Since it is not possible to dissect the contribution of each operator to the mobility shift, only the number of DRus is indicated.
Promoter activity
The activity of promoters in S. coelicolor M145 and S. coelicolor ΔphoP (15) strains was measured using the reporter xylE gene in pIJ4083 plasmid, coding for a catechol-2,3-dioxygenase (30). Strains were grown in R5 medium with 40 µM phosphate and samples were taken at different times, as described previously (23).
EMSA and footprinting analysis
DNA–PhoPDBD interaction was analysed by the electrophoretic mobility shift assays (EMSA). The promoters were excised from the pBluescript plasmid by digestion with restriction endonucleases and labelled at both ends with digoxigenin with DIG Oligonucleotide 3′-End Labeling Kit, 2nd Generation (Roche Applied Science). The conditions for DNA–protein binding and detection were described previously (23). DNase I footprinting assays were performed by the fluorescent-labelling procedure (31), using the GST-PhoPDBD protein (1.06 μM) as described by Sola-Landa et al. (23).
RESULTS
Expression of phosphate-controlled phoU, pstS and phoRP genes requires PhoP: positive regulation
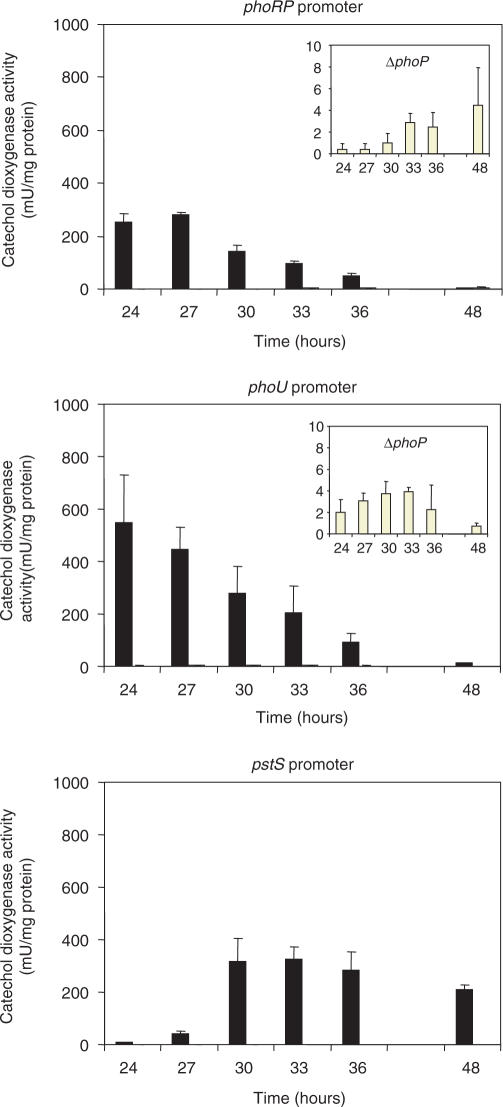
The availability of a deletion ΔphoP mutant of S. coelicolor (15) allowed us to compare the expression of three phosphate-regulated genes phoU, pstS and the phoRP cluster in the parental strain S. coelicolor M145 and the mutant ΔphoP under identical phosphate-limited conditions (for optimal expression of those genes). Expression of these three genes, as shown by time-course quantifications of the catechol oxygenase reporter activity (Figure 1), did not occur in the ΔphoP mutant under conditions favouring gene expression. Only traces of expression of phoRP and phoU were found in the ΔphoP mutant (Figure 1, insets) and no expression at all was observed for pstS. These results prove that the PhoP-mediated control of these genes is exerted by a positive mechanism, i.e. binding of PhoP to the PHO boxes (23) is strictly required for expression of these genes.
Figure 1.
Promoter activity in S. coelicolor M145 (dark bars) and ΔphoP mutant (open bars) of phoRP, phoU and pstS using the xylE gene (encoding a catechol dioxygenase) as reporter, growing in R5 liquid medium with 40 µM phosphate. The activity in ΔphoP mutant is also represented in a larger scale (insets) for easier visualization. Vertical lines on top of each bar indicate SD values. No activity was detected with pstS promoter in the ΔphoP mutant.
Two DRUs are required for PhoP binding
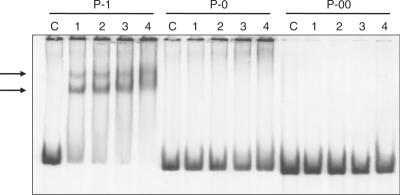
Using the native pstS promoter as model (containing two DRus), we constructed different tailored promoters by directed mutagenesis. In a first step, we introduced two restriction targets, for the endonucleases Acc65I and SpeI, obtaining the native P-1 promoter as a portable cassette. Later, we replaced the PHO box by new sequences with only one DRu (P-0 promoter) or without any (P-00 promoter), preserving the −35 sequence. When binding of PhoPDBD to these promoters was analysed by EMSA, results showed that both DRu are necessary for PhoPDBD binding (Figure 2). While there is a clear binding of PhoPDBD to the native promoter (forming two complexes with different mobility), no binding was observed in the P-0 (one DRu) or in the P-00 promoter (without any DRu) under the same reaction conditions.
Figure 2.
Analysis by EMSA of P-1 (native pstS promoter), P-0 and P-00 promoters (with deletions of one or two DRus, respectively) using increasing concentrations of GST-PhoPDBD protein. Lane C, control without protein; 1, 2, 3 and 4 reactions with 3.12, 6.25, 12.5 and 25 pmol of protein, respectively. The two shift bands of the pstS P-1 promoter are indicated by arrows. Note that there is no band in the gels with P-0 or P-00 promoters.
Selection of candidate target promoters for PhoP regulation
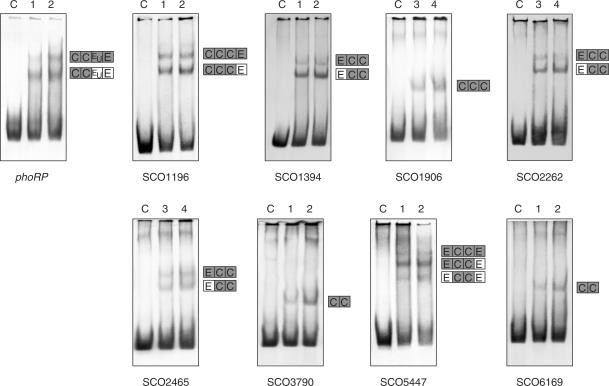
Using an informatic matrix made with the sequence of PHO boxes of pstS, phoRP-phoU (23), phoA, phoD (28) and pitH2 (Santos-Beneit,F., unpublished data) promoters, we analysed the genome of S. coelicolor, as indicated in Materials and Methods section. Several previously uncharacterized genes were found to possess sequences related to the proposed PHO box in their promoter region. Inspection of the candidate target sequences, analysis of the microarray data (15) for signs of PhoP-dependent regulation and concordance of the predicted gene functions with a possible role in phosphate metabolism were considered to choose candidates for experimental analysis. Twenty promoters were amplified by PCR and analysed by EMSA (see Figure 3 for those promoters showing positive binding). These include, in addition to promoters already studied (23,28), SCO0254 (encoding a hypothetical protein), SCO0908 (an unknown protein), SCO1196 (a putative secreted protein), SCO1356 (a putative iron–sulphur protein), SCO1394 (a possible glycosyl hydrolase), SCO1854 (a possible integral membrane protein), SCO1906 and SCO3790 (putative phosphatases different from phoA, phoC, phoD), SCO1993 (an unknown protein), SCO2262 (a possible oxidoreductase), SCO2620 (a probable cell division trigger factor), SCO4451 (a probable export protein), SCO5447 (a putative neutral zinc metalloprotease), SCO6169 (a possible regulatory protein), SCO6722 (a putative regulator), dnaB (SCO3911), encoding a probable replicative DNA helicase, hrcA (SCO2555), encoding a heat-inducible transcriptional repressor, hrdA (SCO2465), encoding a RNA polymerase principal sigma factor, frr (SCO5627), encoding a ribosome recycling factor, redQ (SCO5887) and redP (SCO5888); these last two genes are involved in undecylprodigiosin (red) biosynthesis and constitute an example of secondary metabolite biosynthesis genes that might be directly (or indirectly) regulated by PhoP.
Figure 3.
Analysis by EMSA of the binding of GST-PhoPDBD to several promoters studied in the present work. Lane C, control without protein; 1, 3.12 pmol of protein; 2, 6.25 pmol; 3, 12.5 pmol; 4, 25 pmol of protein. The phoRP promoter was used as control. Only promoters giving positive shifts are shown. Promoter names are indicated under the picture. In the right side of every shift, a scheme is represented explaining the probable DRus structure in the bands detected. C, E and EU correspond with the different types of DRu, Core, Extension and Unstable (see text for explanation). Boxes presumably bound to PhoP are in grey and free boxes (not bound to PhoP) are in white.
Relevance of nucleotides at positions 1 and 2 of a DRu: changing unmatching nucleotides to the consensus sequence restores PhoPDBD binding
Considerable differences in PhoPDBD affinity with these sequences were observed and in some of them no clear binding (as evidenced by EMSA) was detected after repeated gel shift experiments. In some cases, we observed a lack of binding even with promoters having DRus sequences relatively well conserved, but differing in specific nucleotides from the 11-nt consensus.
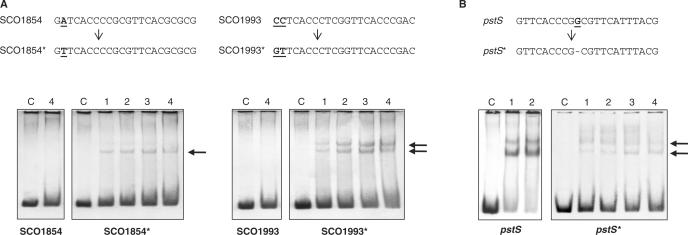
Two of these promoters (SCO1854 and SCO1993) were directly mutated and not matching nucleotides were replaced by the consensus ones (an A to a T in the second position and a CC to GT in the first and second position, respectively; Figure 4A). In both cases, the replacement resulted in a positive binding, confirming that the lack of binding was due to these specific nucleotides. The T in the second position of the direct repeat of SCO1854 and GT in the positions 1 and 2 (they were CC in the native promoter of SCO1993) proved to be essential for DNA binding of PhoPDBD to those direct repeats.
Figure 4.
(A) Analysis of binding of PhoPDBD to the SCO1854 and SCO1993 promoters. The native promoters were mutated (top) and the unmatching bases were replaced by consensus ones. Changed bases are shown in boldface and underlined. The new promoters are named with the same SCO number and an asterisk. Lower panels, EMSA using GST-PhoPDBD protein. Lane C, control without protein; 1, 3.12 pmol of protein; 2, 6.25 pmol; 3, 12.5 pmol; 4, 25 pmol of protein. Note that at least a shift band (indicated by arrows) can be seen in mutated promoters, even with the lowest protein concentration (lane 1), whereas no band is detected in the wild-type promoters with the highest concentration (lane 4). (B) Analysis of the native pstS promoter and a mutant promoter containing a 10-nt DRu. The base deleted between the two DRus is shown in boldface and underlined (top). The new promoter is named pstS*. Note the drastic reduction in binding to the mutant promoter.
DRus of 10 nt and 11 nt
Each DRu was proposed to be 11-nt long. This fits well with the known number of nucleotides per turn of the double helix in the B-DNA (10.6 bp per turn) (helical pitch). However, the Z-DNA structure contains 10-nt per helix turn. We studied the functionality of operators consisting of one DRu of only 10 nt and another of 11 nt. A nucleotide belonging to the low-conserved region (position 10) of the first DRu of the pstS operator was deleted and PhoPDBD binding was compared with that of wild-type promoter by EMSA (Figure 4B). Results showed that PhoPDBD is able to bind the shorter 10-nt sequence, but with a very low efficiency, supporting the model of a DRu with 11 nt, although the number of nucleotides protected by PhoPDBD must take into account the bending of the promoter DNA.
Footprinting identification and structure of the PhoP-binding sites
Because some DRus are poorly conserved, promoters with positive binding were analysed by footprinting to know the exact number of DRus and their organization in every one. In this study, we also included promoters of SCO0034 (encoding an unknown protein), SCO2878 (unknown protein), SCO4261 (a possible response regulator), SCO4878-SCO4879 (putative glycosiltransferase; hypothetical protein, respectively) and SCO7697 (a putative phytase) genes, that have been shown to be recognized by PhoPDBD (15). The nucleotide sequences protected from DNase digestion by PhoPDBD are shown for each promoter in Figure S1 (Supplementary Data).
We searched for DRus in the protected regions using the information theory and programs (see Materials and Methods section). The results of this search led to the discovery of a complex structure of the PhoPDBD-binding sites. Thus, the number of DRus varied from a minimum of two to a maximum of five tandemly arranged DRus in SCO2878 and in SCO4878-9 promoter regions. As occurs in the phoRP and phoD operators, several protected regions contained DRus poorly conserved. Finally, in the SCO1196 protected region, a well-conserved DRu is 2-nt apart from three consecutive DRus (Figure S1).
The analysis of the DRu organization together with the EMSA results allowed us to classify the promoters in three different types. The first class includes promoters with only two DRus, like pstS (23), phoA (28), SCO3790 and SCO6169. This is the simplest structure and the repeats are well conserved (Table 1, panel A). In EMSA analysis of those promoters, generally a unique band was detected; the exception of P-1 promoter will be discussed later. The second class is formed by operators with three conserved DRus (Table 1, panel B). As occurs with the first type of promoters, only one complex was detected in EMSA. Therefore, both these classes of promoters can be defined as composed of DRus that form a firm binding site: each DRu is occupied by a PhoPDBD monomer and the full site is bound at the same time. In contrast, the promoters of the third class showed a complex DNA–protein stoichiometry.
Based on our findings, it is proposed that two (e.g. phoD) or three (e.g. SCO1196) conserved DRus form the core of the binding site and are bound by PhoPDBD at the first stage (denoted by C letters, for core, in Table 1, panel C). At higher protein concentrations, the length of bound sequence is extended to adjacent DRus. This conclusion is supported by the information content analysis (see below). A PhoP cooperative oligomerization on the DNA has been proven for the Bacillus subtilis and E. coli PhoP orthologues (see Discussion section).
Information content analysis of the nucleotide sequences
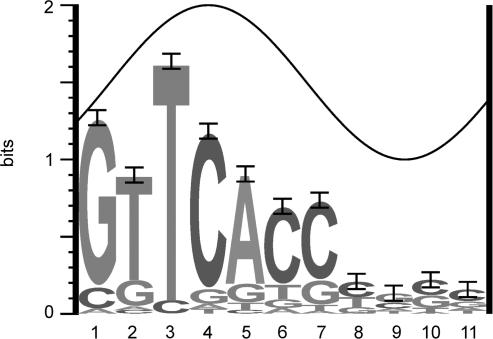
The initial information-based model of the binding site (model 0) was refined taking into account the newly discovered DRus. This model (model I, see Materials and Methods section) was built from 37 DRus that form the core of the selected binding sites (Table 1). The sequence logo (25) that depicts the binding site is shown in Figure 5. The conservation of the first seven positions is clear and remarks the importance of these bases to establish specific protein–DNA contacts, whereas the last four positions contribute with little information. A similar pattern of conservation is found in the repeats that form the PHO boxes in other bacteria (see Discussion section). As proteins usually bind DNA from one face, it is expected that binding-site positions with conservation higher than 1 bit are located at the major groove (26). Thus, it is likely that PhoPDBD contacts bases 1, 3 and 4 through the major groove of the DNA (Figure 5). A similar result has been obtained in E. coli, where the third and fourth bases are directly contacted by PhoB (32).
Figure 5.
Sequence logo of the direct repeats of 11 nt that form the PhoP-binding-site unit. This logo corresponds to model I that comprises 37-DRus of Streptomyces coelicolor (see Materials and Methods section). The height of each letter is proportional to the frequency of the base, and the height of the letter stack is the conservation in bits at that position (25). Note the high conservation of the first seven positions of the DRu. Error bars are shown at the top of the stacks. The total information (Rsequence) for the DRu is 8.08 ± 0.61 bits. The sine wave represents the accessibility of a face of the DNA (B-form, 10.6 bases of helical pitch) with the major groove centred at position 4. C in position 1 or an A or a C at position 2 have a clear negative effect on PhoP binding.
The sequence walkers method displays, for individual sequences, the contribution of each base to the conservation of the binding site (29). The walkers, as well as the individual information contents of the DRus, or Ri (27), allowed us to analyse the requirements of PhoPDBD for DNA recognition. Class I operators are composed of two DRu; i.e. one PHO box. As shown above, this is the simplest operator structure. The sequences of these sites are well conserved, as reflected by high Ri values and by the fact that the sequence GTTCAY is present in seven out of eight DRus (the exception is the first DRu of phoA, Figure 6A). In the cases of class II operators, at least one of the three DRus is well conserved (Ri > 8 bits; this is the Rsequence, Figure 5). The complex class III operators can be seen as composed of a core of two or three DRus associated with other DRus that are bound at higher PhoPDBD concentrations. Those core DRus show the same features than the DRus of class I and II operators (all designated ‘C’ in Table 1). These features are: (i) the sites are made up with at least one DRu with Ri > 8 bits, except phoA; (ii) the DRus are consecutive (null separator); (iii) the information content of the full sites (sum of individual Ri values) have a mean of 18.1 ± 1.29 bits (standard error) for pairs, and 19.5 ± 2.11 bits for trios; i.e. both sets have similar information content.
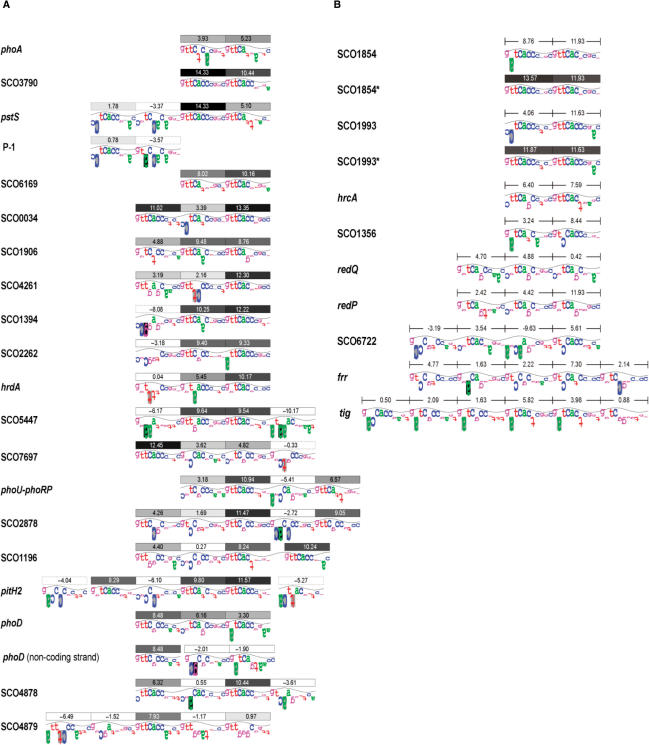
Figure 6.
(A) Individual information analysis of the PhoP-binding sites, identified in this and previous works, using the sequence walker method (29). Each binding site is formed with DRus of 11 nt, which are marked by boxes on its top. Each box contains the individual information (Ri) in bits, and is filled with tones of grey in a scale of 10% steps. Thus, the box of a DRu with a Ri value in the range of 90–100% of the highest possible Ri content (14.63 bits, the Ri of the consensus sequence), are coloured in black; boxes of Ri ≤ 1.46 bits, are shown in white. Hypothetical DRu, which were not comprised in the footprinting-protected area, are represented by boxes with dashed lines. The height of the letters is the information content in bits, and represents the contribution of each base to the conservation of the sequence. Letters extending downward represent unfavourable contacts. The walker limits are 2 bits, which is also the top of the sine wave, and −3 bits at the bottom; 0 bits is at the middle of the walker. If a base does not occur in the set of sequences that forms the model (model I), a black box is given. A grey box indicates that the upside-down letter extends beyond the lower limit. For simplicity, in this figure the sine wave has a periodicity of 11 bases. (B) Analysis of the promoter-region sequences that resembled PhoP-binding sites, but failed to bind PhoP. The mutagenized SCO1854* and SCO1993* sequences are shown below the wild-type sequences. Information scores (bits) of the sequences are indicated.
Nine of the fragments that were initially amplified for EMSA tests failed to contain true binding sites despite of showing certain sequence conservation. The comparison of walkers from true and false sites provides new rules to evaluate potential new sites and highlights the importance of key positions. Firstly, an adenine base at position 2 (A2) seems to impede the binding. This is demonstrated, at least for two-DRus sites, with the mutagenesis of the SCO1854 fragment (Figure 4). The replacement of an A for a consensus T makes the sequence capable of binding PhoPDBD. This would also explain the lack of binding of SCO1356 and tig sites (Figure 6B). Secondly, two-DRus sites appear to require a high conservation at the first six positions in both repeats. This is the case of C1C2 and C1G6 on the first DRu of SCO1993 and hrcA, respectively. Indeed, in three-DRus sites, always the first and third DRu contain a G; a C1 is found only in middle DRus. This observation explains the lack of binding to the redQ sequence (Figure 6B). This sequence and that of redP contain three DRus. The most remarkable features are the lack of conservation in the first positions and the presence of G6 in every DRu. Finally, the sequences of SCO6722, frr and tig, despite of containing several potential DRu, lack core sites. That is, they lack two or three DRu-like sequences that accomplish the features of operator cores.
DISCUSSION
Different pathways known to be regulated by inorganic phosphate availability respond distinctly to different phosphate concentrations (20). Some genes are induced when the concentration of phosphate in the growth medium falls below 5 mM; others, when the concentration falls below 1 mM or less. For instance, the expression of the pstS gene is strongly induced when the concentration of phosphate falls below 0.1 mM (15). Although other regulators could also be involved in the control of the expression of a given gene, its different sensitivity to phosphate availability seems to rely on the affinity of the response regulator PhoP for its cognate-binding sequences (PHO boxes). These were identified as tandem reiterations of direct repeats of 11 nt with the consensus sequence GG/TTCAYYYRG/CG where Y is a pyrimidine and R is a purine (23). The truncated version of PhoP was used because it has been shown previously that PhoPDBD binds more efficiently to promoters than PhoP (23,28). In addition, it has also been shown in other microorganisms that the DBD domain binds in a ‘constitutive’ manner similar to the binding of phosphorylated full protein. Maris et al. (33) used a truncated version of NarL in crystal structure studies because this protein protects exactly the same region and has a longer half-life than the phosphorylated full-length protein.
Proof that the GT nucleotides in the first and second position of the direct repeat are essential was obtained by directed mutation of the first DRu in the promoters of SCO1854 and SCO1993 that were not recognized initially by PhoPDBD. Changes of the GA and CC nucleotides (respectively) to the consensus GT in those positions allowed binding of PhoPDBD leading to the formation of one or two shift bands in the EMSAs. These results confirmed early observations on the importance of the six initial nucleotides, particularly those in positions 1 and 2, in the binding of PhoPDBD. A similar pattern of relevance of the first nucleotides in the PHO boxes has been found in E. coli, B. subtilis, Synechocystis, Campylobacter jejuni, or Corynebacterium glutamicum (34–38).
A significant number of S. coelicolor genes have been found to contain PHO boxes, in agreement with the results of the transcriptomic and proteomic analyses (15). However, there is a considerable variability in the number and organization in the DRus of PHO boxes in those phosphate-regulated genes.
One DRu is not sufficient for clear binding of PhoPDBD. At least two DRus are needed for proper binding of PhoPDBD to a PHO box, since removal of one direct repeat was sufficient to prevent binding of PhoPDBD to a PHO box in the pstS promoter. This result is supported by the lack of binding to SCO1854, SCO1993 and hrcA promoters, which have a good conserved DRu, but lack a second DRu with a high score.
The basic structure of the 11-nt repeat is important since deletion of 1 nt (the G at position 10 in the 11-nt repeat of the well-known pstS promoter) (23) resulted in a drastic reduction of binding of PhoPDBD, as shown by the barely visible formation of PhoPDBD–promoter complex in the shift assays. This result is consistent with the recognition of two direct repeats, at a distance that allows interaction with a PhoPDBD dimer on the same face of the DNA double helix. A similar hypothesis was made for the interaction of the response regulators PhoB and PhoP, from E. coli and B. subtilis, respectively, with the PHO boxes (6 conserved + 5 variable nucleotides) (32,35). In E. coli, the structure of PhoB in complex with its target DNA sequence revealed that PhoB has a winged-helix fold such that helix α3 penetrates in the major DNA groove and a β hairpin wing interacts with the minor groove via the conserved Arg219 (Arg215 in S. coelicolor PhoP). When only two DRus are present, the conservation of the consensus sequence is very important. This property can explain the discrepancy seen with phoA (SCO2286), activated by PhoP (28) but with an AA profile in microarray analysis (Table 1, panel A). The time selected for the phosphate shiftdown experiment was chosen on the basis of pstS gene expression (15), having a promoter with two highly conserved DRus. phoA also has two DRus, but with a lower score, probably requiring a higher concentration of PhoP.
When there are three, four of even more DRus, some of these sequences act as core (C) sequences binding PhoPDBD with high affinity and this allows the subsequent binding of further PhoPDBD molecules, even if those DRus are less conserved.
Three mechanisms explain the sequential binding of PhoPDBD to class III operators. Firstly, the operators of SCO1394, SCO2262, hrdA, SCO7697 and SCO5447 contain one or two DRus, named E (extension), with poor sequence conservation adjacent to the core site (Table 1, panel C). Their Ri values are low or negative (Figure 6A). As the information content can be correlated with the binding energy (a positive Ri corresponds to a negative change of free energy) (27,29), it is proposed that protein–protein interactions stabilize the binding of the PhoPDBD monomer to E repeats. The fast-migrating (lower size) complexes detected in EMSA, should be the result of two PhoPDBD monomers bound to the CC core sites; the slowest complexes should correspond to the full operator-bound complex. This explains the two complexes detected with SCO2262-3 and hrdA fragments. Similarly, the three complexes of SCO5447 and SCO7697 fragments (Figure 3; Table 1, panel C) can be explained by a second E direct repeat of still lower conservation (−6.2 versus −10.2 bits in SCO5447; 4.8 versus −0.3 bits in SCO7697; Figure 6A).
A second mechanism can be illustrated with the previously characterized operator of phoU-phoRP. It is composed of four DRus: the first two repeats form the core (CC), the third DRu has −5.4 bits of information and was named EU (for extension unstable) and the last DRu, named ES (for extension support) is well conserved (Table 1, panel C; Figure 6A). The lack of sufficient conservation of the EU repeat may prevent the binding of PhoPDBD only to this repeat whereas a binding would take place at both EU and ES repeats together. Therefore, the two detected EMSA complexes should correspond to two protein monomers bound to the core (CC) and four monomers bound to the full site, respectively (CCEUES; Figure 3). The same mechanism is proposed for the SCO2878 operator (Table 1, panel C). The third mechanism involves a gap of 1 or 2nt between the core site and the extension repeat. This gap might act in the same way as an EU repeat; i.e. deterring the PhoPDBD cooperative binding on DNA. This effect can be quantified by means of the flexible site information equation (39):
where GS(d) is the gap surprisal for the spacing d. The GS(d) term was calculated from the distribution of gap values (44 gaps of 0 nt, two gaps of 1 nt and two gaps of 2 nt; deduced from Table 1), using the gap surprisal equation (39). It results that a gap between two consecutive DRus subtracts 6.4 bits from the site Ri value (both 1- and 2-nt gaps have the same penalty since both have the same frequency). This amount is similar to the Ri of EU-type repeats (Figure 6A). It follows that the corresponding repeats ES must add information higher than 6.4 bits to obtain a positive Ri value, as it occurs in SCO1196, phoU-RP and SCO2878-binding sites (Figure 6A).
Finally, two putative DRus could have been created in the constructed P-1 promoter when the new restriction sites were introduced. These sequences, located immediately upstream of the DNase-protected site core, would explain the two complexes detected in EMSA (Figures 2 and 4). It is also possible that the wild-type sequences are bound at high regulator concentrations, since they have a sequence similar to the newly introduced one (Figure 6A).
The arrangement of DRus is even more complex in some cases. The operators of pitH2 and phoD are especially complex and appear to use the three mechanisms described above. Furthermore, the phoD operator lies in both strands (Table 1, panel C) (28). Analysing the protected sequences, we observed that in some operators the two DRus are not strictly adjacent but are separated by 1 or 2 nt. This is the case of SCO1196 and the previously reported phoD (28) and pitH2 (Santos-Beneit,F., unpublished data). In these cases, as in the case of phytase of B. subtilis (40), some of the PHO boxes may overlap with the −10 region. At low (or null) PhoP concentration (thus, in ‘high’ phosphate), this site is poorly or not occupied, whereas at high PhoP concentration (thus, in low phosphate), this site acts as a cis-acting negative regulatory site by preventing binding of the RNA polymerase. This seems to happen rarely, since our results and those of E. coli and B. subtilis clearly indicate that most PhoP-regulated genes are activated by this response regulator (this study; 28).
The pho regulon described in this article is of interest as a model for other Streptomyces species that are known producers of secondary metabolites with valuable biological activities. Knowledge of the mechanism of phosphate control provides insight into how to increase the production of those secondary metabolites and even to unlock cryptic secondary metabolites (20,41). Moreover, we cannot dismiss that a gene could be regulated by another regulator (probably a repressor) in addition to the activator PhoP. This putative repressor might be abundant and active in Pi sufficiency and absent/inactive in Pi limitation. In consequence, at a given Pi concentration, the expression of a gene would result from the relative abundance/DNA-binding abilities of the positive and negative regulatory factors.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
A.K.A. received a fellowship of the FPU Program of the Ministry of Education and Science, Madrid, Spain. We acknowledge the excellent technical help of B. Martín, J. Merino, A. Casenave and B. Aguado. This work was funded by ‘Comisión Interministerial de Ciencia y Tecnología’ (BIO2003-01489, BIO2006-14853-C02-01); ‘Ministerio de Educación, Ciencia y Tecnología’ (GEN2003-20245-C09-01) and ‘European Union’ (ACTINOGEN LSHM-CT-2004-005224). Funding to pay the Open Access publication charges for this article was provided by the ACTINOGEN project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Berdy J. Bioactive microbial metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Demain AL. Pharmaceutically active secondary metabolites of microorganisms. Appl. Microbiol. Biotechnol. 1999;52:455–463. doi: 10.1007/s002530051546. [DOI] [PubMed] [Google Scholar]
- 3.Bentley R. Microbial secondary metabolites play important roles in medicine; prospects for discovery of new drugs. Perspect. Biol. Med. 1997;40:364–394. doi: 10.1353/pbm.1997.0009. [DOI] [PubMed] [Google Scholar]
- 4.Martín JF, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Ann. Rev. Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 5.Yanai K, Murakami T, Bibb M. Amplification of the entire kanamycin biosynthetic gene cluster during empirical strain improvement of Streptomyces kanamyceticus. Proc. Natl Acad. Sci. USA. 2006;103:9661–9666. doi: 10.1073/pnas.0603251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James D, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 7.Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl Acad. Sci. USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martín JF, Casqueiro J, Liras P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 2005;8:282–293. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- 10.Recio E, Colinas A, Rumbero A, Aparicio JF, Martín JF. PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem. 2004;279:41586–41593. doi: 10.1074/jbc.M402340200. [DOI] [PubMed] [Google Scholar]
- 11.Mapplestone RA, Stone MJ, Williams DH. The evolutionary role of secondary metabolites – a review. Gene. 1992;115:151–157. doi: 10.1016/0378-1119(92)90553-2. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser D, Losick D. How and why bacteria talk to each other. Cell. 1993;73:873–885. doi: 10.1016/0092-8674(93)90268-u. [DOI] [PubMed] [Google Scholar]
- 13.Martín JF, Demain AL. Control of antibiotic synthesis. Microbiol. Rev. 1980;44:230–251. doi: 10.1128/mr.44.2.230-251.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lounes A, Lebrihi A, Benslimane C, Lefebvre G, Germain P. Regulation of spiramycin synthesis in Streptomyces ambofaciens: effects of glucose and inorganic phosphate. Appl. Microbiol. Biotechnol. 1996;45:204–211. doi: 10.1007/s002530050671. [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-García A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martín JF. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics. 2007;7:2410–2429. doi: 10.1002/pmic.200600883. [DOI] [PubMed] [Google Scholar]
- 16.Asturias JA, Liras P, Martín JF. Phosphate control of pabS gene transcription during candicidin biosynthesis. Gene. 1990;93:79–84. doi: 10.1016/0378-1119(90)90139-i. [DOI] [PubMed] [Google Scholar]
- 17.Slater H, Crow M, Everson L, Salmond GP. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 2003;47:303–320. doi: 10.1046/j.1365-2958.2003.03295.x. [DOI] [PubMed] [Google Scholar]
- 18.Dekeva ML, Titus JA, Strohl WR. Nutrient effects on anthracycline production by Streptomyces peucetius in a defined medium. Can. J. Microbiol. 1985;31:287–294. doi: 10.1139/m85-053. [DOI] [PubMed] [Google Scholar]
- 19.Mendes MV, Tunca S, Antón N, Recio E, Sola-Landa A, Aparicio JF, Martín JF. The two-component phoR-phoP system of Streptomyces natalensis: inactivation or deletion of phoP reduces the negative phosphate regulation of pimaricin biosynthesis. Metab. Eng. 2007;9:217–227. doi: 10.1016/j.ymben.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Martín JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J. Bacteriol. 2004;186:5197–5201. doi: 10.1128/JB.186.16.5197-5201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sola-Landa A, Moura RS, Martín JF. The two-component PhoR-PhoP system controls both primary metabolism and secondary metabolite biosynthesis in Streptomyces lividans. Proc. Natl Acad. Sci. USA. 2003;100:6133–6138. doi: 10.1073/pnas.0931429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghorbel S, Kormanec J, Artus A, Virolle MJ. Transcriptional studies and regulatory interactions between the phoR-phoP operon and the phoU, mtpA, and ppk genes of Streptomyces lividans TK24. J. Bacteriol. 2006;188:677–686. doi: 10.1128/JB.188.2.677-686.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sola-Landa A, Rodríguez-García A, Franco-Domínguez E, Martín JF. Binding of PhoP to promoters of phosphate-regulated genes in Streptomyces coelicolor: identification of PHO boxes. Mol. Microbiol. 2005;56:1373–1385. doi: 10.1111/j.1365-2958.2005.04631.x. [DOI] [PubMed] [Google Scholar]
- 24.Torriani-Gorini A. The Pho regulon of Escherichia coli. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms. Washington, D.C.: ASM Press; 1994. pp. 1–4. [Google Scholar]
- 25.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider TD. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 27.Schneider TD. Information content of individual genetic sequences. J. Theor. Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 28.Apel AK, Sola-Landa A, Rodríguez-García A, Martín JF. Phosphate control of phoA, phoC and phoD gene expression in Streptomyces coelicolor reveals significant differences in binding of PhoP to their promoter regions. Microbiology. 2007;153:3527–3537. doi: 10.1099/mic.0.2007/007070-0. [DOI] [PubMed] [Google Scholar]
- 29.Schneider TD. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK, pp. 439–510: The John Innes Foundation; 2000. [Google Scholar]
- 31.Rodríguez-García A, Ludovice M, Martín JF, Liras P. Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol. Microbiol. 1997;25:219–228. doi: 10.1046/j.1365-2958.1997.4511815.x. [DOI] [PubMed] [Google Scholar]
- 32.Blanco AG, Sola M, Gomis-Ruth FX, Coll M. Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure. 2002;10:701–713. doi: 10.1016/s0969-2126(02)00761-x. [DOI] [PubMed] [Google Scholar]
- 33.Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SM, Kopka ML, Schröder I, Gunsalus RP, Dickerson RE. Dimerization allows DNA target site recognition by the NarL response regulator. Nat. Struct. Biol. 2002;9:771–778. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- 34.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J. Mol. Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 35.Liu W, Hulett FM. Comparison of PhoP binding to the tuaA promoter with PhoP binding to other Pho-regulon promoters establishes a Bacillus subtilis Pho core binding site. Microbiology. 1998;144:1443–1450. doi: 10.1099/00221287-144-5-1443. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki S, Ferjani A, Suzuki I, Murata N. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 2004;279:13234–13240. doi: 10.1074/jbc.M313358200. [DOI] [PubMed] [Google Scholar]
- 37.Wosten MM, Parker CT, van Mourik A, Guilhabert MR, van Dijk L, van Putten JP. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 2006;62:278–291. doi: 10.1111/j.1365-2958.2006.05372.x. [DOI] [PubMed] [Google Scholar]
- 38.Schaaf S, Bott M. Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 2007;189:5002–5011. doi: 10.1128/JB.00121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shultzaberger RK, Bucheimer RE, Rudd KE, Schneider TD. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- 40.Makarewicz O, Dubrac S, Msadek T, Borriss R. Dual role of the PhoP approximately P response regulator: Bacillus amyloliquefaciens FZB45 phytase gene transcription is directed by positive and negative interactions with the phyC promoter. J. Bacteriol. 2006;188:6953–6965. doi: 10.1128/JB.00681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Challis GL, Hopwood DA. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl Acad. Sci. USA. 2003;2:14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.