Abstract
The inherent properties of DNA as a stable polymer with unique affinity for partner molecules determined by the specific Watson–Crick base pairing makes it an ideal component in self-assembling structures. This has been exploited for decades in the design of a variety of artificial substrates for investigations of DNA-interacting enzymes. More recently, strategies for synthesis of more complex two-dimensional (2D) and 3D DNA structures have emerged. However, the building of such structures is still in progress and more experiences from different research groups and different fields of expertise are necessary before complex DNA structures can be routinely designed for the use in basal science and/or biotechnology. Here we present the design, construction and structural analysis of a covalently closed and stable 3D DNA structure with the connectivity of an octahedron, as defined by the double-stranded DNA helices that assembles from eight oligonucleotides with a yield of ∼30%. As demonstrated by Small Angle X-ray Scattering and cryo-Transmission Electron Microscopy analyses the eight-stranded DNA structure has a central cavity larger than the apertures in the surrounding DNA lattice and can be described as a nano-scale DNA cage, Hence, in theory it could hold proteins or other bio-molecules to enable their investigation in certain harmful environments or even allow their organization into higher order structures.
INTRODUCTION
DNA strands are, due to their unique ability to dimerize in a specific manner determined by the rather strict rules of Watson–Crick base paring between adenine and thymine or guanine and cytosine together with its high chemical and physical stability, the ideal molecule for the design of self-assembling structures (1–5) Successful strategies for synthesis of simple synthetic DNA substrates to be used for enzyme analyses (6–8) as well as more complex two-dimensional (2D) DNA structures for both scientific and technological purposes (9–13) have been presented. Assembly of 3D DNA structures, however, has proven more difficult (14–19) and a better understanding of the behavior of DNA structures in 3D is necessary for future progress.
In the building of synthetic DNA structures folding/assembly of one/or more DNA strand(s) is controlled by designing the base sequences of the strand(s) in such a way that the desired structure supports a maximum number of correct base pairs and, hence, will be the energetically favored one (20,21). Although the specific base pairing presents a very precise way of controlling assembly of DNA structures, unwanted side products may be a problem, in particular, when it comes to the assembly of more complex structures. The yield of the desired structure highly depends on the DNA oligonucleotide design and is hampered when the formation of unspecific polymeric structures is only slightly less energetically favorable than the correct structure. The latter possibility should, therefore, be avoided in the sequence design. Moreover, to ensure high stability of the complete structure it is necessary to link the DNA ends covalently after assembly to prevent the structure from disintegrating into the single oligonucleotides of which it consists, upon denaturation. Problems caused by unspecific assembly and/or instability may be an inevitable property of some designs and in fact many 3D DNA structures suffer from one or the other disadvantage, emphasizing the importance of a continued effort within this field. Few designs have been presented in the past fulfilling the important requirements for the successful assembly of 3D DNA nano-structures. Seeman and coworkers (14,15) were the first to complete the building of a cube and a truncated octahedron DNA structure although these were hampered by a low yield (∼1%). Later, an octahedron was elegantly folded with high yield from a single DNA fragment by the Joyce group (16). The design of this structure, however, compromised the possibilities for covalent closure. Goodman et al. (17) synthesized tetrahedrons characterized both by high yield (95%) and covalent closure and most recently, an elegant new approach of using relatively simple building blocks for the efficient assembly of more complex 3D structures including cubic, pentagonal and hexagonal prisms was presented by Aldaye and Sleiman (19).
Here we present a covalently closed and efficiently assembled 3D DNA structure with a complexity exceeding that of the tetrahedron presented by Goodman et al. (17). Our structure is built from eight oligonucleotides and has the connectivity of an octahedron as defined by its double-stranded regions. In other words, the structure has the form of a DNA cage composed of an outer lattice with apertures smaller in diameter than the central spherical cavity which was confirmed by the two very different and complementary techniques cryo-Transmission Electron Microscopy (cryo-TEM) and Small Angle X-ray Scattering (SAXS). Potential applications for the created DNA cage in biochemical science and technology will be discussed.
MATERIALS AND METHODS
Design of oligonucleotides
The sequences of the oligonucleotides were as described in the text, and chosen using a computer program that minimized unwanted base pairing of the following three types: (i) annealing between different oligonucleotides, (ii) annealing between two identical oligonucleotides and (iii) annealing within a single oligonucleotide. This was done under the constraints defined by the desired structure and the different desired melting temperatures for the interactions.
Assembly of the DNA cage
The cage was assembled by mixing OL1–OL8 (purchased from GeneLink) in equimolar amounts in an assembly buffer containing 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg/ml bovine serum albumin. Phosphates were added to the 5′ ends by incubation with T4 polynucleotide kinase for 30 min at 37°C. Subsequently, the sample was heated to 65°C for 10 min and cooled by 0.25°C/min. until the sample reached a temperature of 30°C, where T4 DNA ligase was added. Cooling was continued by 0.25°C/min. until 16°C. For analyses of radiolabeled DNA cage each of the oligonucleotides OL1–OL8 were radiolabeled by incubation with T4 polynucleotide kinase and γ-(32P)ATP for 30 min at 37°C prior to G50-sephadex spin dialysis and EtOH precipitation. The individually radiolabeled oligonucleotides were subsequently dissolved in assembly buffer and mixed with equimolar amounts of the remaining cold phosphorylated oligonucleotides and assembly performed as described above. Hence, 10 pmol of radiolabeled OL1 was mixed with 10 pmol of each of the cold phosphorylated OL2–OL8 and so forth.
Gel-electrophoresis and purification of the DNA cage
The partly and fully assembled cage was analyzed in native or denaturing 5% polyacrylamide gels following standard procedures. Before loading on the denaturing gel formamide was added to the samples and boiled for 2 min to ensure denaturation. The fully assembled cage was purified from a native gel by excision of the band representing the product, followed by soaking the gel slice in elution buffer [500 mM NH4Ac, 10 mM MgAc2, 0.1 mM EDTA (pH 8.0), 0.1% SDS] for 24 h at 37°C and EtOH precipitated. All products were visualized by staining the gels with SyBr Green. For SAXS analysis, an additional purification step was included after elution from the gel slice. The eluted cage was purified on a QIAGEN-tip 500 column (from QIAGEN Plasmid Maxi kit), essentially following the standard procedure from the kit. The cage was eluted from the column in 300μl fractions and EtOH precipitated. The fractions containing the highest concentrations of eight-stranded DNA cage (1–2 μg/μl) were used for SAXS analysis.
Cryo-TEM
Cryo-TEM was performed essentially as described previously (22). A 0.5 μl drop of purified DNA cage (86 nM) was adsorbed on a 300-mesh copper grid covered with perforated polymer film with thin evaporated carbon layers on both sides. The charge of the polymer film was neutral. The cryo preparation was performed in a climate chamber with a temperature of 25°C and a relative humidity of 98–100%. The grid was blotted for 0.5 s with Whatman filter paper #1 and plunge-frozen into liquid ethane held just above its freezing point. The sample was observed at a temperature below −170°C using a Zeiss EM 902A, top-entry electron microscope operating at 80 kV and in zero loss bright-field mode. Images at ∼1 μm under focus were recorded on a BioVision Pro-SM SSCCD camera under low-dose conditions using the analySIS data collection and image processing software. The magnification was ×218 000, resulting in a pixel size of 4.04 Å. The best images of boxed particles were used for 3D reconstruction employing the EMAN program (23). An initial model was generated with 50 particles using the ‘startoct’ option in step 2 of the EMAN program. Cycles of angular refinement were performed with an angular interval of 9° until convergence, with a total of 269 particles (from nine micrographs) contributing to the final average map. Full octahedral symmetry was imposed during the reconstruction. The resolution of the final reconstruction was estimated as 42 Å by the EOTEST subroutine of the EMAN program. The actual resolution of the map may be better than indicated by this number that most probably reflects the rather low number of particles used for the reconstruction. Isodensity maps were viewed and modeled using the UCSF Chimera package (24). A ‘negative’ control reconstruction that employed the initial 50-particle model and randomly boxed regions of the micrographs known not to contain DNA cages did not produce a meaningful map.
SAXS measurements and modeling
SAXS measurements were performed at the SAXS laboratory of the Chemistry Department at the University of Aarhus as described in greater detail in Supplementary Data. The characteristic real-space distance distribution functions p(r) were determined from the scattering data using indirect Fourier transformation (25). This function corresponds to a histogram over all distances between pairs of points within the particle and it gives direct insight into the particle shape and size. To model the DNA-cage structure, we developed a method using rigid body refinement and least-squares procedures. In a first very low-resolution approach, the double-strand regions that form the sides of the cage were modeled as linear arrangement of spheres, and octahedral symmetry was applied to build the full structure. The Debye equation (26) was used for the intensity calculation and a least-squares procedure (27) was used to optimize the structure and fit the experimental data. In the next approach, atomic structures of DNA were used in a model for the cage. To speed up the calculations, we use only the C2* sequence to describe the DNA structure. It was shown that this is a reasonable approximation (See Supplementary Data, Figure S3). The octahedral symmetry was again applied to build the full structure and the ends of consecutive paired DNA sides with 18 bp were connected using linear arrangements of spheres to mimic the bases in this region.
RESULTS
Design of oligonucleotides for the DNA cage
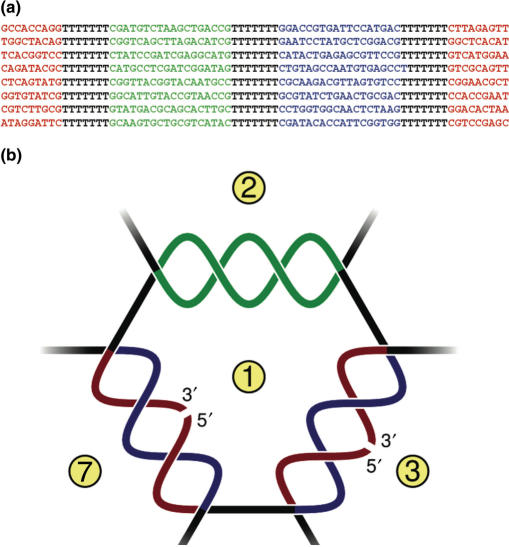
For the current design we used eight 75mer oligonucleotides (OL1–OL8) to build a DNA cage with the connectivity of a truncated octahedron with a theoretical outer diameter of 150–200 Å as calculated from the published dimensions of DNA (28). The sequences of the oligonucleotides are shown in Figure 1a. The oligonucleotides each contain three annealing regions of 18 bases interrupted by seven bases spacer regions. One of the annealing regions (marked in red) is divided in two halves located at each end of the oligonucleotides, while the two other annealing regions (marked in blue or green) are undivided and located internally in the oligonucleotides. During assembly, each of the green regions form specific base pairing with the complementary green region on the partner oligonucleotide with a theoretical annealing temperature of 56°C. The two halves of each of the red regions are brought together by specific base pairing with the complementary blue region on the partner oligonucleotide with a theoretical annealing temperature of 28°C (Figure 1b). This design allowed the DNA structure to assemble simply by heating and cooling a mixture of equimolar amounts of OL1–OL8 with added 5′ phosphates necessary for covalent closure of the structure by subsequent DNA ligation.
Figure 1.
Design of the eight-stranded DNA cage. (a) Sequences of oligonucleotides OL1–OL8. Annealing regions are shown in color and non-annealing spacer regions in black. (b) Schematic illustration of assembly of the strands. The green region of one oligonucleotide base pairs with the green region of a partner oligonucleotide (example: green region of OL1 pairs with green region of OL2). The interrupted red region of one oligonucleotide pairs with the blue region of a partner oligonucleotide. This brings the ends of the first oligonucleotide together allowing covalent closure of the red region by ligation (example: OL1 forms a circle when the red region anneals with the blue region of OL7). Complete annealing of OL1–OL8 allows covalent closure of the structure. Oligo numbers are indicated in yellow circles.
Assembly and gel-electrophoretic analysis of DNA cage
The cage was assembled by mixing, heating and cooling equimolar amounts of 5′-phosphorylated OL1–OL8 as described in Materials and Methods section. When the temperature of the assembly mixture reached 30°C, the cage was covalently sealed by the addition of T4 DNA ligase.
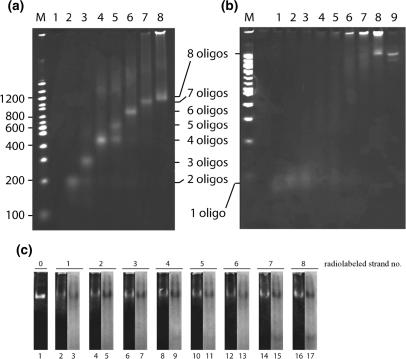
The specific assembly was tested by analyzing the products obtained when annealing and ligating increasing numbers of equimolar amounts of oligonucleotides added in successive order from OL1 to OL8 to the reaction mixture in native or denaturing 5% polyacrylamide gels (Figure 2a and b, lanes 1–8). As evident from Figure 2a adding increasing numbers of oligonucleotides to the assembly reaction resulted in products running mainly as single bands with decreasing mobility in the native gel. This electrophoretic pattern supports the formation of specific products of increasing complexity upon the addition of increasing numbers of oligonucleotides as expected from the sequence design of OL1–OL8. Assembly of OL1–OL5, however, resulted in two products (lane 5) with the faster running product having a mobility corresponding to the mobility of the product obtained when mixing OL1–OL4 (lane 4). Since OL1–OL5 was mixed in equimolar amounts this result suggests that the formation of the five-stranded complex is not as favorable as complexes containing either fewer or more strands. Note, that mixing OL1–OL6 resulted in a single product (lane 6) with a mobility slower than the slowest mobility band in lane 5. The presence of all eight oligonucleotides in the slow mobility product obtained after mixing OL1–OL8 (Figure 2a, lane 8) was confirmed by 5′-radiolabeling each of OL1–OL8 by 32P before mixing them one by one with equimolar amounts of the remaining seven cold phosphorylated oligonucleotides. As evident from Figure 2c, radiolabeling either one of the eight oligonucleotides resulted in a radiolabeled product with a mobility corresponding to the product of lane 8 in Figure 2a (see lane 1, Figure 2c). Moreover, the amount of radiolabeling present in the product corresponded to the amount of total product (as estimated by the SyBr green staining) regardless which of the oligonucleotides that carried the labeling (compare lane 3 with 2, lane 5 with 4 and so on). This result indicates that the product contains equal amounts of all eight oligonucleotides and, hence, represents the fully assembled eight-stranded DNA cage. The presence of a high-mobility radiolabeled band with a mobility corresponding to a single oligonucleotide was observed in some of the lanes (most pronounced in lanes 15 and 17) suggesting that a part of the oligonucleotides remained separate and were not assembled into the eight-stranded cage structure. The corresponding bands could not be observed following visualization by SyBr green staining most probably due to the inability of SyBr green to intercalate and thereby stain single-stranded DNA. When run in a denaturing gel only the mixture of four or more oligonucleotides gave rise to products with a mobility slower than the mobility of single oligonucleotides (Figure 2b, compare lanes 4–8 with lanes 1–3). Since oligonucleotides not linked to each other by covalent bonding will fall apart in a denaturing gel this result is consistent with the predicted assembly dictated by the sequences of OL1–OL8 (Figure 1), where covalent catenanes are predicted to be formed only after annealing of at least OL1–OL4 followed by ligation. Such catenanes lack a well-defined 3D structure, which likely explains the smeared appearance of bands observed in lanes 4–7. The smearing of the bands explains their relatively low intensity. The appearance of a single well-defined band in lane 8 with a mobility slower than the faint and smeared bands in lanes 4–7 lends further credence to the successful specific assembly and covalent closure of the eight-stranded DNA cage. By quantification of this band relative to the products retarded in the gel-slot we estimated an approximate yield of the correctly assembled cage of 30% (this yield estimate corresponds to the result of an estimation based on output versus input amount of DNA as measured spectrophotometrically; data not shown). A band similar to the specific slow-mobility band in lane 8 was observed even after boiling the reaction mixture for 10 min after complete assembly. This was only observed when both T4 polynucleotide kinase and T4 DNA ligase were present during assembly (data not shown) supporting that the band represents a covalently closed product as predicted.
Figure 2.
Gel-electrophoretic analyses of the partly and fully assembled DNA cage. (a) Lanes 1–8 show the results of subjecting assembly reactions containing OL1, OL1 and OL2, OL1–OL3, OL1–OL4, OL1–OL5, OL1–OL6, OL1–OL7 or OL1–OL8 to analysis in a native polyacrylamide gel. Lane M contains a DNA ladder with sizes indicated to the left of the gel picture. (b) Lanes 1–8 and lane M, same as (a), except that samples were analyzed in a denaturing polyacrylamide gel. Lane 9, gel-purified cage. The number of oligonucleotides present in each band is indicated between (a) and (b). (c) Assembly after radiolabeling of each of the oligonucleotides OL1–OL8 one by one. Lanes 2 and 3 represent the product assembled using a radiolabeled version of OL1 with lane 2 showing the result after SyBr green staining and lane 3 being the result of autoradiography of the gel. Lanes 4–17 represent pairwise identical analyses of products assembled with OL2–OL8 being radiolabeled before assembly as indicated above the gel pictures. Lane 1 is gel purified unlabeled DNA cage.
Structural analysis of DNA cage
To determine the 3D structure of the assembled DNA cage we have chosen to use single-particle image reconstruction based on cryo-TEM (22,24,29) and SAXS (30). These two approaches are complementary, with cryo-TEM being a direct space imaging technique looking at individual particles, whereas SAXS is an ensemble technique measuring simultaneously on all particles in a macroscopic volume. The results from cryo-TEM are used as a basis for the modeling that is required for the interpretation of the SAXS data.
For the structural analyses, the fully assembled cage was purified from a native gel by excision of the band representing the product. After resuspension in water, the purity of the sample was confirmed by denaturing gel-electrophoresis (Figure 2b, lane 9). For the SAXS analysis, the eluted DNA cage was further purified on a DNA affinity column.
The cryo-TEM was performed essentially as described previously (23). The best images of boxed particles were used for 3D reconstruction employing the EMAN program (29). The 3D isodensity map is shown in Figure 3a. Overall, the obtained isodensity map is consistent with the structure expected to result from assembly of the eight DNA oligonucleotides. The dimensions of the most dense areas in the reconstructed map (contoured at a σ of −2.28), presumably corresponding to the double-stranded arms, are identical to the dimensions of 18 bp double-stranded DNA as determined by crystallization studies (28) (Figure 3b). From the density map, the outer diameter of the cage can be estimated to 190 Å and the inner diameter to 130 Å, which is in accordance with the expected diameter of 150–200 Å.
Figure 3.
Structure of the eight-stranded DNA cage. (a) 3D isodensity map of the cage generated from single-particle reconstruction based on cryo-transmission electron microscopy imaging. (b) Superimposition of the reconstructed structure and cylinders representing a stretch of 18 bp DNA. The grid surface illustrates the front section of the isodensity map in (a) at a low contour level (−1.69 σ). The higher contour level (−2.28 σ) is shown in the blue solid surface. The dimensions of the pink cylinders (diameter: 2 nm, length: 6 nm) equal the dimensions of an 18 bp DNA helix based on crystal structures of double-stranded DNA (28). The cylinders are shown in a 1:1 scale to the isodensity map. Scale bar: 5 nm.
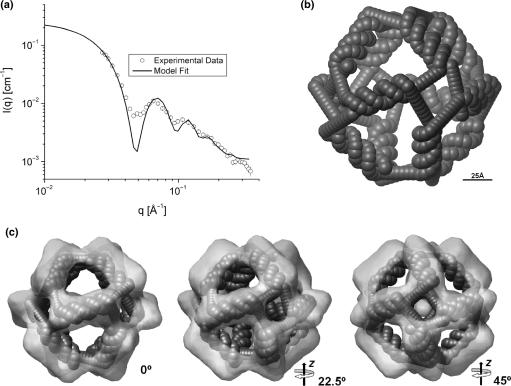
The analysis of the SAXS data using indirect Fourier transformation (25) shows that the sample contains very well-defined hollow particles (see Supplementary Data, Figure S1). The low-resolution modeling in terms of linear arrangement of spheres gave a reasonable agreement with the experimental data considering the simplicity of the model (see Figure S2). The next approach using spheres for representing the C2* sequence of the DNA structure also gave a good fit to the data (Figure 4a) resulting in the model shown in Figure 4b (model in stereo view is shown in Figure S4b). However, combining the cage structure into a trimeric structure improves the fit suggesting that there is weak aggregation in the high concentration sample used for SAXS (Figure S5). The model reveals that the distance from the center of the cage to the center of the double-stranded DNA is 62.5 ± 0.7 Å and the maximum diameter of the model is ∼155 Å, which is in agreement with the expected size. The inner diameter obtained from the SAXS is ∼100 Å and the side of the apertures is around 60 Å in diameter. Interestingly, from the modeling we found that the double-stranded DNA in the sides of the structure are twisted by an angle of 18 ± 5° from the horizontal position. The proposed modeling method thus enabled retrieving of the dimensional parameters of the DNA structure with reasonable accuracy.
Figure 4.
SAXS and 3D model for the DNA cage. (a) Experimental data (points) and model fit (solid lines). (b) Octahedral model obtained from the SAXS data. (c) Overlay of SAXS model and cryo-TEM reconstruction in three different views.
Figure 4c shows the overlay of the data obtained from SAXS and cryo-TEM analyses. The two structures are in very good agreement. The tilting of the double-stranded region as well as the size of the apertures are the same in the two structures. Overall, the structure from cryo-TEM is slightly larger than the SAXS structure, which is most likely caused by the use of a relatively small number of particles in the cryo-TEM analysis.
DISCUSSION
Here we have reported the design of an octahedral DNA cage that assembles from eight individual oligonucleotides with an efficiency of ∼30% as estimated by gel-electrophoretic analysis and spectrophotometric measurements of the amount of output versus input DNA material. The cage could be covalently closed by DNA ligation as proven by its resistance towards thermal and chemical denaturation demonstrated by gel-electrophoretic analysis. Correct assembly was supported by the stepwise assembly of the eight oligonucleotides when mixed successively in assembly buffer as expected from the sequence design and by the presence of all eight oligonucleotides in the product as demonstrated by radiolabeling each of OL1–OL8 one at a time prior to assembly. Further support of correct assembly of the eight oligonucleotides came from structural analyses using two complementary techniques, cryo-TEM and SAXS, both suggesting that the utilized DNA strands, OL1–OL8, form an octahedral cage-like structure with a central cavity of 100–130 Å in diameter surrounded by a lattice with apertures of 60 Å in diameter and an outer ‘cage’ diameter of 155–190 Å. This structure and its dimensions are in agreement with the predictions. The results of cryo-TEM and SAXS analysis showed that the double-stranded arms of the structure were twisted slightly compared to the theoretical model suggesting some topological constraints in the assembled cage. This possibility will be further investigated using computer simulation techniques.
As described, the eight-stranded DNA cage reported here assembles with a yield of around 30%, which is lower than the 95% yield of the four-stranded DNA tetrahedron presented by Goodman et al. (17) and higher than the 1% yields of the 10-stranded cubic and 48-stranded octahedral structures constructed by Seeman and coworkers (14,15). The high yield of the Goodman DNA tetrahedron was ascribed to a two-stage assembly process in which their four DNA oligonucleotides form two pairs at a relatively high temperature, which subsequently join to form the final structure at a lower temperature. This is reminiscent to the procedure used to assemble the cage presented here, where the eight oligonucleotides form four pairs at around 56°C which join to the final structure at around 28°C. This assembly strategy, together with the careful choice of oligonucleotide sequences to avoid mispairings along with the fact that all annealing regions end on G-C basepairs, is probably a contributing factor to our decent yield. The final step in our cage assembly requires the joining of four DNA pairs making it somewhat more complex than the final assembly step of the Goodman tetrahedron where only two pairs join (17). This added complexity of the cage assembly may increase the risk of polymer formation and may be one of the reasons for our yield being around 30% rather than 95%. That our yield is significantly higher than the 1% yields obtained by Seeman and coworkers (14,15) is most likely due to their much more complicated assembly procedure composed of several steps. A DNA octahedron folding from a single strand with a close to 100% yield was presented by Joyce and coworkers (16), and lends further credence to the idea of an inverse relationship between yield and number of steps in the assembly procedure. In addition to the aforementioned factors, the flexible spacer regions composed of seven thymidines in the eight-stranded cage give the individual strands a degree of flexibility to orient themselves relative to their pairing strands, which may improve the yield. In favor of the theory that a certain flexibility in the structure improves the assembly, we find that OL1–OL4 assemble quite efficiently while the annealing of OL5 to the OL1–OL4 complex appears more problematic. OL1–OL4 represents the smallest number of strands giving rise to catenanes that are anticipated to impose some rigidity and may hamper the entering of OL5 to the complex.
The development of complex self-assembling DNA structures are anticipated important factors for future bio- and nano-technological conquests. The dimensions of the DNA cage (having a central cavity larger than the apertures in the surrounding lattice) described in the present study, suggest that it may be able to hold proteins or other bio-molecules in its center, reminiscent to the protein encapsulation demonstrated by Erben et al. (31). Such entrapment may protect proteins/biomolecules from potential harmful components such as inactivating or degrading protein complexes (larger in diameter than 60 Å) in the surroundings while still being able to interact with substrates or ligands small enough to pass the apertures of the lattice. It is easy to imagine how this could be of advantage both in scientific investigations as well as in drug delivery when the conditions necessitate protection of the agent of interest. The possibilities of using the presented DNA cage as such an entrapment device is currently under investigation. Also, linking the octahedral DNA cages together in 2D or 3D networks may enable the strict organization of proteins (or other components) entrapped in the cage center to ease structural analyses. Such networks composed of several cages linked together in an ordered fashion are currently being developed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are thankful to Maria Vinther and Kirsten Andersen for technical assistance. This work was supported by the Carlsberg Foundation, the Danish Research Councils, the Danish Cancer Society, the Novo Nordisk Foundation, the Augustinus Foundations, the Hartmann Foundation, the Aase and Ejnar Danielsen Foundation and the Harboe Foundation. Funding to pay the Open Access publication charges for this article was provided by Danish Research Councils.
Conflict of interest statement. None declared.
REFERENCES
- 1.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Seeman NC. DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct. 1998;27:225–248. doi: 10.1146/annurev.biophys.27.1.225. [DOI] [PubMed] [Google Scholar]
- 3.Seeman NC. DNA enables nanoscale control of the structure of matter. Q. Rev. Biophys. 2005;38:363–371. doi: 10.1017/S0033583505004087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem. Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding B, Seeman NC. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao S, Mao C, Birktoft JJ, Shuman S, Seeman NC. Resolution of undistorted symmetric immobile DNA junctions by vaccinia topoisomerase I. Biochemistry. 2004;43:1520–1531. doi: 10.1021/bi0358061. [DOI] [PubMed] [Google Scholar]
- 7.Andersen FF, Andersen KE, Kusk M, Frohlich RF, Westergaard O, Andersen AH, Knudsen BR. Recombinogenic flap ligation mediated by human topoisomerase I. J. Mol. Biol. 2003;330:235–246. doi: 10.1016/s0022-2836(03)00593-x. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, Tonozuka T, Jayaram M. Mechanism of active site exclusion in a site-specific recombinase: role of the DNA substrate in conferring half-of-the-sites activity. Genes Dev. 1997;11:3061–3071. doi: 10.1101/gad.11.22.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 10.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 11.Malo J, Mitchell JC, Venien-Bryan C, Harris JR, Wille H, Sherratt DJ, Turberfield AJ. Engineering a 2D protein-DNA crystal. Angew. Chem. Int. Ed. Engl. 2005;44:3057–3061. doi: 10.1002/anie.200463027. [DOI] [PubMed] [Google Scholar]
- 12.Rinker S, Liu Y, Yan H. Two-dimensional LNA/DNA arrays: estimation the helicity of LNA/DNA hybrid duplex. Chem. Commun. 2006;25:2675–2677. doi: 10.1039/b603872g. [DOI] [PubMed] [Google Scholar]
- 13.Rothemund PWK. Folding DNA to create nono-scale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 14.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Seeman NC. The construction of a DNA truncated octahedron. J. Am. Chem. Soc. 1994;116:1661–1669. [Google Scholar]
- 16.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 17.Goodman RP, Schaap AT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 18.Erben CM, Goodman RP, Turberfield AJ. A self-assembled DNA bipyramid. J. Am. Chem. Soc. 2007;6:6992–6993. doi: 10.1021/ja071493b. [DOI] [PubMed] [Google Scholar]
- 19.Aldaye FA, Sleiman HF. Modular access to structurally switchable 3D discrete DNA assemblies. J. Am. Chem. Soc. 2007;129:13376–13377. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- 20.Turner DH. Thermodynamics of base pairing. Curr. Opin. Struct. Biol. 1996;6:299–304. doi: 10.1016/s0959-440x(96)80047-9. [DOI] [PubMed] [Google Scholar]
- 21.Sundaralingam M, Ponnuswamy PK. Stability of DNA duplexes with Watson-Crick base pairs: a predicted model. Biochemistry. 2004;43:16467–16476. doi: 10.1021/bi048158+. [DOI] [PubMed] [Google Scholar]
- 22.Frank J. Averanging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy. 1975;1:159–162. doi: 10.1016/s0304-3991(75)80020-9. [DOI] [PubMed] [Google Scholar]
- 23.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 24.Frank J. Classification of macromolecules assemblies studied as “single particles”. Q. Rev. Biophys. 1990;23:281–329. doi: 10.1017/s0033583500005564. [DOI] [PubMed] [Google Scholar]
- 25.Pedersen JS, Hansen S, Bauer R. The aggregation behavior of zinc-free insulin studied by small-angle neutron scattering. Eur. Biophys. J. 1994;22:379–389. doi: 10.1007/BF00180159. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen JS. Small-angle scattering from precipitates: analysis by use of a polydisperse hard-sphere model. Adv. Colloid Interface Sci. 1997;70:171–210. doi: 10.1103/physrevb.47.657. [DOI] [PubMed] [Google Scholar]
- 27.Debye P. Zerstreuung von Rötgenstrahlen. Ann. Phys. Leipzig. 1915;46:809–823. [Google Scholar]
- 28.Premilat S, Albiser G. Conformations of A-DNA and B-DNA in agreement with fiber X-ray and infrared dichroism. Nucleic Acid Res. 1983;11:1897–1908. doi: 10.1093/nar/11.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 30.Kratky O. X-ray small angle scattering with substances of biological interest in diluted solutions. Prog. Biophys. Mol. Biol. 1963;13:105–173. doi: 10.1016/s0079-6107(63)80015-2. [DOI] [PubMed] [Google Scholar]
- 31.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.