Abstract
In mammals, the expression of 5–10% of genes occurs with circadian fluctuation in various organs and tissues. This cyclic transcription is thought to be directly or indirectly regulated through circadian transcriptional/translational feedback loops consisting of a set of clock genes. Among the clock genes in mammals, expression of the Dbp mRNA robustly oscillates both in vivo and in culture cells. Here, we present circadian enhancer detection strategy using prokaryotic transposon system. The mDbp promoter drives reporter gene expression in robust circadian cycles in rat-1 fibroblasts. To identify the circadian enhancer generating this robust rhythm, we developed a prokaryotic transposon-based enhancer detecting vector for in vitro transposition. Using this system, we identified a strong circadian enhancer region containing the CATGTG sequence in the 5′ flanking region of the mDbp gene; this enhancer region is critical for the ability of the mDbp promoter to drive robust oscillation in living cells. This enhancer is classified as a CANNTG type non-canonical E-box. These findings strongly suggest that CANNTG-type non-canonical E-boxes may contribute, at least in part, to the regulation of robust circadian gene expression. Furthermore, these data may help explain the wider effects of the CLOCK/BMAL1 complex in control of clock output genes.
INTRODUCTION
The mammalian circadian clock is a highly dynamic system that causes 5–10% of genes to be expressed cyclically; this confers near 24-h rhythmicity on behavior, physiology and metabolic processes and allows mammals to adjust to the time of the day (1). The suprachiasmatic nucleus (SCN) of the hypothalamus, which is the master clock in mammals, is entrained by light to keep pace with the day–night cycle autonomic nerve and humoral factors coordinate circadian clocks in peripheral tissues (2,3). Recent studies have also shown that peripheral tissues contain their own self-sustainable circadian clock oscillators (4). Moreover, even cultured fibroblasts contain an active circadian clock, which strikingly resembles the central clock in the SCN (5–8).
Rhythmic transcription of central and peripheral clock output genes is controlled by a molecular oscillator, consisting of interlocked positive and negative transcription/translation feedback loops of a set of clock genes. BMAL1 (MOP3) and CLOCK are basic-helix-loop-helix (bHLH) PAS transcription factors that dimerize and transactivate Period (Per1 and Per2), Cryptochrome (Cry1 and Cry2), and an orphan nuclear receptor, Rev-Erbα, via CACGTG E-box enhancer elements. The PER and CRY proteins act as negative elements by inhibiting the CLOCK/BMAL1 heterodimer, whereas REV-ERBα regulates Bmal1 gene expression via ROR responsive elements (RRE) (2,3,9). Recent reports examining genome-wide gene expression profiles in the SCN and various peripheral organs have shown that hundreds of genes are expressed under circadian regulation systems (10–12). Curiously, most of the cyclically expressed genes differ from organ to organ, which suggests that a much larger number of genes are potential targets of circadian control (13). Thus, each peripheral tissue or organ cyclically expresses a specific set of genes governing tissue-specific functions, such as cholesterol metabolism-related genes in the liver (10). Although it is unquestionably important to study the transcriptional regulatory network connecting the circadian clock and physiology, the findings of micro-array studies are often too indirect to uncover the distinct regulatory mechanisms and pathways regulating circadian gene expression.
To further elucidate the circadian transcriptional regulatory network, we here establish an effective enhancer-detecting method to identify enhancers eliciting circadian gene expression in mammalian cells using an in vitro random transposon system and a high-throughput luciferase-based circadian real-time monitoring system.
The expression of Dbp in the SCN, liver and cultured fibroblast cell lines oscillates very robustly(14,15). In this study, we first showed that a promoter region of the mDbp gene, which contains ∼1.4 kb upstream from the transcriptional start site, generates robust circadian oscillation of luciferase bioluminescence in rat-1 and NIH3T3 fibroblast cells even though no functional circadian enhancer has been reported. Next, we used an enhancer-trap approach based on an in vitro transposon system to efficiently discover unknown circadian enhancers. Using this system, we have identified a circadian enhancer contributing for cyclic transcriptional regulation in the 5′ flanking region of the mDbp gene. This identified enhancer is classified as a non-canonical E-box (CANNTG), which is identical with recently reported as a new circadian enhancer of mDbp promoter (16). Moreover, our observations suggest that CLOCK/BMAL1-mediated transcriptional regulation may influence a much larger set of genes than previously thought; these effects are exerted via not only CACGTG E-boxes, but also non-canonical CANNTG E-boxes.
MATERIALS AND METHODS
Cell culture
Rat-1 fibroblast cells (HSRRB, Osaka, Japan), NIH3T3 fibroblast cells, and COS7 cells (kindly donated by Dr Ishitani, Nagoya University) were cultured in DMEM with 10% FBS and penicillin–streptomycin. For real-time analysis, the medium was changed to phenol-red-free DMEM buffered with HEPES at 24 h after transfection.
Construction of transposons for the enhancer trap
To construct transposons for the enhancer trap, we used the EZ::Tn™ pMOD™-2<MCS> Transposon Construction Vector (EPICENTRE Biotechnologies) (17). We used the PCR-based overlap extension method to generate fragments carrying the minimal CMV promoter and luciferase reporter gene. First, upper fragments carrying the minimal CMV promoter were amplified using pCMVupKpnI (5′-CCGGGGTACCTAGGCGTGTACGGTGGGAG GCCTATATAAGC-3′; the KpnI site is underlined) and pCMVlowOver (5′-CCATGGTGGCAGGCTGGATCGGTCCCGGTGTCTTCTATGG-3′; the overlapping section is underlined) as primers and the pTRE2 Vector (CLONTECH) as a template. Lower fragments carrying the luciferase reporter gene were amplified using pLUCupOver (5′-GGACCGATCCAGCCTGCCACCATGGAAGATGCCAAAAAC-3′; the overlapping section is underlined) and pLUClowBamHI (5′-TTATGGATCCTACCACATTTGTAGAGG TTTTACTTGC-3′; the BamHI site is underlined) as primers and the pGL4.11 Luciferase Reporter Vector (Promega) as a template. The upper and lower DNA fragments were used as templates for a second PCR reaction using pCMVupKpnI and pLUClowBamHI as primers. The resulting 2.2 kb PCR product was digested with KpnI and BamHI and ligated to KpnI- and BamHI-digested pMODTM-2<MCS> Transposon Construction Vector. The kanamycin resistance gene was amplified by PCR using pKmupHindIII (5′-TTCGGAAGCTTC AACAAAGCCACGTTGTGTCTC-3′; the HindIII site is underlined) and pKmlowHindIII (5′-AGCGTAAGCTTCTGCCAGTGTTACAACCAATTAACC-3′; the HindIII site is underlined) as primers and the EZ::Tn™ <KAN-2> Transposon (EPICENTRE Biotechnologies) as a template. The resulting 1.0 kb PCR product was digested with HindIII and ligated to the HindIII-digested vector described earlier to yield the transposon vector for the enhancer trap. This vector was sequenced to confirm that no unexpected mutations were introduced. The larger PshAI fragment from the transposon vector for the enhancer trap was isolated, purified by agarose gel electrophoresis, and used as the transposon for the enhancer trap.
Enhancer trap
The transposon for the enhancer trap was integrated into a vector carrying the DBP promoter using EZ::Tn™ pMOD™ Transposase (EPICENTRE Biotechnologies) according to the manufacture's instructions. One microliter of transposed sample was transformed into Escherichia coli. DH5 alpha (Toyobo, Osaka, Japan), and plasmid DNA was prepared from 48 independent colonies. One microgram of each plasmid was transfected into rat-1 cells and bioluminescence was monitored. Plasmids were sequenced to determine the insertion sites of the transposon.
Plasmids
The Dbp promoter was obtained by PCR from C57BL/6 mouse genomic DNA using pDbpupSacI (5′-AACTCGAGCTCCACGTCCTGATAGTCTGCAC-3′; the SacI site is underlined) and pDbplowBglII (5′-GCTAGATCTGTACCAAGTGGGCGAGTCTC-3′; the BglII site is underlined) as primers. The resulting 1.4 kb PCR product was subcloned into the pGEMT-EAZY vector (Promega) to yield the Dbp promoter vector. In the same way, the mPer2 and mBmal1 promoters were obtained using the primer sets pmPer2upKpnI (5′-AAGGTACCTCTGCCGGCTGTGAGTTGCGCAG-3′; the KpnI site is underlined) and pmPer2lowXhoI (5′-TTCTCGAGACCGCTAGTCCCAGTAGCGCCG-3′; the XhoI site is underlined), or pmBmal1upSacI (5′-GATCGAGCTCGCAGAGTCC GCAACGCAGTGG-3′; the SacI site is underlined) and pmBmal1lowXhoI (5′-GATCCTCGAGCGCACCCGCACTCGGATCCCG-3′; the XhoI site is underlined). Each promoter was then ligated into the pGL4.11 [luc2P] Vector (Promega). To generate a mutated Dbp promoter reporter vector, a QuikChange site-directed mutagenesis kit (Stratagene) was used along with the primers pDbpEbox-mut-up (5′-GCACGCGCAAAGCGGTACCC TTCCCCCTTC-3′; substituted nucleotides are underlined) and pDbpEbox-mut-low (5′-GAAGGGGGAAGGTACCGCTTTGCGCGTGC-3′; substituted nucleotides are underlined). The mutated Dbp promoter reporter vector was sequenced to confirm the presence of the specific mutations and the absence of any unexpected mutation. The Dbp reporter vector was digested with KpnI and religated to generate the Dbp(-462):Luc vector. The mutated Dbp reporter vector was digested with KpnI and religated to generate the Dbp(-410):Luc vector. DNA fragments carrying the non-canonical E-box from the Dbp promoter (−416) were obtained by PCR using pDbpNEKpnIup (5′-AAGAGGTACCCCTCTTGCCCCAC-3′; the KpnI site is underlined) and pDbpNEKpnIlow (5′-ATATGGTACCAATGGGAGGAGGCGCTG-3′; the KpnI site is underlined) as primers and C57BL/6 mouse genomic DNA as template. The resulting 0.1 kb PCR product was digested with KpnI and subcloned into KpnI digested transposon vector for the enhancer trap, described earlier. The reporter vector carrying the non-canonical E-box from the Dbp promoter was sequenced to confirm the presence of the expected DNA fragment and the absence of any unexpected mutations.
Real-time monitoring of bioluminescence
The 2 × 105 rat-1 cells were plated in 35 mm dishes or 5 × 104 cells were plated in 24-well plates. For the 35 mm dishes, rat-1 cells were transfected with 1 μg of each construct using 3 μl of FUGENE 6 transfection reagent (Roche). For 24-well plate assays, cells were transfected with 300 ng of each construct and 0.8 μl of FUGENE 6. After overnight culture, medium was changed to DMEM without phenol-red and containing 100 nM luciferin and 15 mM HEPES. After additional overnight culture, cells were reset with 100 nM dexamethasone and bioluminescence was automatically monitored using a photomultiplier-tube-based bioluminescence monitoring system (18).
Transcriptional assays
Luciferase reporter gene assays were performed in NIH3T3 cells. Cells (2 × 105) were seeded on 6-well plates and transfected the following day. In this study, we used the Dual Luciferase reporter assay system (Promega). Cells in each well were transfected with 50 ng of pGL4.11 vector containing the indicated DNA fragments as described as well as the indicated amounts of expression plasmids for flag-tagged mClock, mBmal1 and HA-mCry1pcDNA3.1. The total amount of DNA per well was adjusted to 1 μg by adding pcDNA3 vector as carrier. Forty-eight hours after transfection, cells were harvested and luciferase activity was determined by luminometer.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as described (19). Briefly, COS7 cells expressing flag-tagged mCLOCK and mBMAL1 were washed with 5 ml of Tris-buffered saline (TBS) and pelleted by centrifugation at 1500 × g for 5 min. The pellet was resuspended in 1 ml of TBS, transferred into a microtube and centrifuged for 5 s. The supernatant was removed and the pellet was dissolved in 400 μl of chilled Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT and 0.5 mM phenylmethylsulfonyl fluoride). The cells were allowed to swell on ice for 15 min, after which 25 μl of 10% NP-40 was added. The tubes were vigorously vortexed for 10 s and centrifuged for 30 s at 15 000 rpm. The nuclear pellet was dissolved in 50 μl ice-cold Buffer C (20 mM HEPES ph 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT and 1 mM PMSF). The nuclear extract was centrifuged at 12 000 rpm for 5 min, and 2 μl of the supernatant was used for each reaction.
The interaction of the BMAL1 and CLOCK proteins expressed in COS7 nuclei with the DNA fragment containing the non-canonical E-box enhancer element of the mDBP promoter was analyzed by EMSAs using the LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL). The 30 bp DNA duplex used in the EMSA experiments consisted of the annealed pair of oligonucleotides E-box F (5′-GCACGCGCAAAGCC ATGTGCTTCCCCCTTC-3′; the non-canonical E-box is underlined), and E-box R (5′-GAAGGGGGAAGCACATGGCTTTGCGCGTGC-3′; the non-canonical E-box is underlined). These two oligonucleotides were biotinylated at their 5′-ends to allow chemiluminescent detection. DNA–protein binding reactions were carried out at room temperature and consisted of nuclear extracts mixed with a reaction buffer composed of 10 mM Tris–HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 50 ng/μl poly (dI–dC), 2.5% glycerol, 10 mM EDTA (pH 8.0) and 0.05% NP-40. 20 fmol of biotin-labeled E-box DNA was then added to the reaction mixture and incubated for 25 min at room temperature. For competition experiments, a 50-fold molar excess of unlabeled duplex DNA fragments corresponding to the E-box site (described earlier) was added to the reaction mixture before addition of the labeled oligonucleotides. For mutant E-box probe, the annealed pair of oligonucleotides E-box mut F (5′-GCACGCGCAAAGCGGTACCCTTCCCCCTTC-3′; substituted nucleotides are underlined), and E-box mut R (5′-GAAGGGGGAAGGTACCGCTTTGCGCGTGC-3′; substituted nucleotides are underlined) were used. Reaction mixtures were analyzed by non-denaturing 6% polyacrylamide gel electrophoresis and transferred to Biodyne B Nylon Membranes (PALL). The biotin-labeled DNA probes were detected by chemiluminescence using the Chemiluminescent Nucleic Acid Detection Module Kit (Pierce). The membranes were exposed to X-ray film for 1–3 min and developed according to the manufacturer's instruction.
Chromatin immunoprecipitation (ChIP) assay and Q-PCR
NIH3T3 cells (American Type Culture Collection) were grown in DMEM (Invitrogen) supplemented with 10% FBS (JRH Biosciences) and antibiotics (25 U/ml penicillin, 25 mg/ml streptomycin; Invitrogen). Cells were plated at 6 × 105 cells in 10 cm dishes 24 h before transfection, and transfected with two plasmids (6 µg each of FLAG tagged BMAL1 and FLAG tagged CLOCK expression vector) with FuGene6 transfection reagent (Roche) according to the manufacturer's instructions. Cells were harvested 48 h after transfection.
ChIP assays with these transfected NIH3T3 cells were performed as previously described (20) with the following modifications. In this report, we used Chromatin Immunoprecipitation Assay Kits (Upstate Biotechnology), anti-Flag M2 antibody (Sigma) and anti-V5 antibody (Sigma). Protein G-Sepharose (Amersham) was used for precipitation of antibody/protein immune complexes. The resulting precipitated DNA was quantified by Q-PCR, which was performed according to the previously described method (21). Absolute DNA abundance was calculated using the standard curve obtained from NIH3T3 cell's genomic DNA. Tbp-5′ region were quantified and used as the internal control.
Primer sequences for Q-PCR of ChIP products
P(Dbp)-forward, 5′-TGAACACTCGGCTCCTTTAAGAG-3′; P(Dbp)-reverse, 5′-GCGTGCTATCGGATGCG-3′; Act-5′-forward, 5′-CCAAGAGGCTCCCTCCACA-3′; Act-5′-reverse, 5′-GTGCAAGGGAGAAAGATGCC-3′; Tbp-5′-forward, 5′-GTTTGGCTAGGTTTCTGCGG-3′; Tbp-5′-reverse, 5′-GAAAGGACCGTTACCTGGGC-3′.
RESULTS
Robust circadian oscillation driven by the mDbp promoter
The Dbp mRNA is expressed with a robust circadian rhythm in the SCN, peripheral tissues and cultured cells (5,22). Recent reports have suggested that a CACGTG E-box in the second intron of the Dbp gene is likely to be responsible for this cyclic mRNA expression (14,15). Although the reported intronic E-box was functional in response to CLOCK/BMAL1-mediated transcriptional activation, it is still unknown whether this intronic E-box is enough to generate robust circadian oscillation.
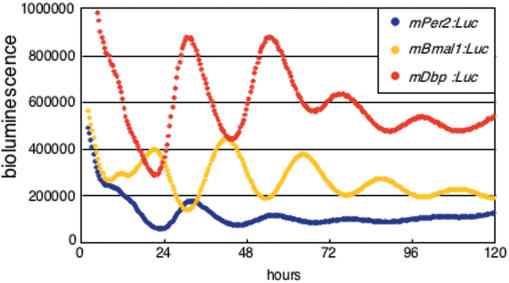
We first focused on the 5′ flanking region of the mDbp gene because many clock genes, including mPers and Bmal1, have circadian enhancers near their transcription start sites (12,23,24). We cloned the mDbp promoter region (−1332 ∼ +83) and fused it upstream of a luciferase reporter gene in the pGL4.11 vector (Promega). This vector was then transfected into rat-1 fibroblast cells, and bioluminescence was monitored. Surprisingly, the mDbp promoter drove luciferase expression with a robust circadian rhythm; the amplitude of this rhythm was larger than that of the oscillation driven by the mPer2 or mBmal1 promoters (Figure 1). These results indicate that the mDbp promoter region contains strong circadian enhancer(s) that are capable of generating robust oscillation of expression.
Figure 1.
Robust circadian oscillation of mDbp:Luc-driven bioluminescence in rat-1 cells. Luciferase reporter constructs driven by the mPer2 promoter (mPer2:Luc, blue), the mBmal1 promoter (mBmal1:Luc, yellow) and the mDbp promoter (mDbp:Luc, red) were transfected into rat-1 fibroblasts and synchronized by treatment with 100 nM dexamethasone. Real-time bioluminescence monitoring showed that all three promoters were able to drive robust circadian oscillation of the reporter gene. The phases of the mDbp:Luc and mPer2:Luc-driven rhythms are quite similar, whereas the mBmal1:Luc driven rhythm is in almost the opposite phase.
Transposon-based enhancer detection for the mDbp promoter
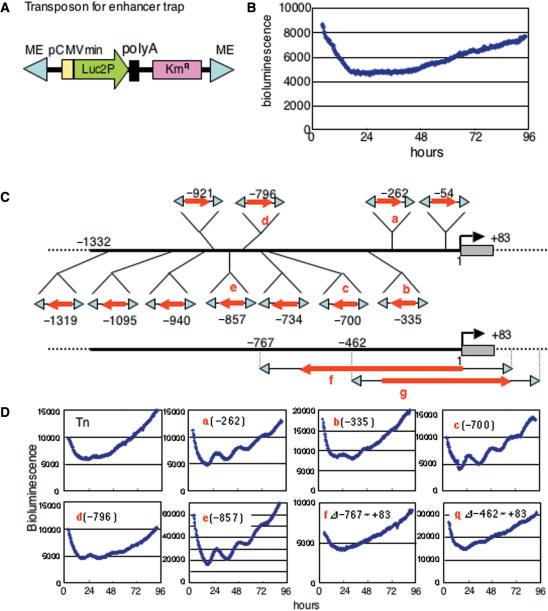
To determine the unknown circadian enhancer(s) in the ∼1.4 kb mDbp promoter region, we developed an enhancer-detection system based on the Tn5 transposon for in vitro tranposition. We constructed a transposon carrying ∼120 bp of minimal CMV promoter fused to a bioluminescence reporter gene and a kanamycin resistance gene (Figure 2A). We analyzed the CMV minimal promoter driven bioluminescence of this trap vector following transfection into rat-1 cells. As expected, the bioluminescence was detectable, constitutive, and showed no circadian intensity changes during several days (Figure 2B).
Figure 2.
Transposon-based in vitro enhancer trap for the mDbp promoter. (A) Structure of the Tn5-based enhancer trap vector. Reduced half-life firefly luciferase (Luc2P) obtained from the pGL4.11 vector (Promega) is used as a reporter driven by the CMV minimal promoter. ME indicates the mosaic end sequence to transpose into the host DNA. (B) Real-time bioluminescence monitoring of rat-1 cells transfected with the enhancer trap vector. Cells are synchronized by treatment with 100 nM dexamethasone. No rhythmic fluctuation was observed over a period of 4 days. (C) The enhancer trap vector was transposed into the ∼1.4 kb fragment of the mDbp promoter cloned into the pGEM T-Easy vector (Promega). After Ampicillin and Kanamycin selection, 13 clones with integration of the trap vector into the mDbp promoter were obtained. Red arrows indicate the direction of the luciferase gene in the trap vector. The position of integration is noted for each red arrow. Clones f and g showed integration with some deletion. (D) Clones with trap vector integrations were transfected into rat-1 cells and analyzed by real-time bioluminescence monitoring. Representative results from clones a to g are shown. Rat-1 cells transfected with clones a, c or e exhibited robust circadian oscillation of bioluminescence. In contrast, rat-1 cells transfected with clones b, d, f or g exhibited only faint oscillation.
The enhancer-detecting transposon vector was randomly integrated into the plasmid carrying the mDbp promoter in vitro, and each plasmid was sequenced to determine the transposon insertion site. Eleven different clones with the transposon integrated into the Dbp promoter region were obtained (Figure 2C). The arrows in Figure 2C indicate the direction of reporter gene expression. These plasmids were then transfected into rat-1 fibroblast cells and bioluminescence was monitored (Figure S1). Representative results, indicated as (a)–(e), are shown in Figure 2D. Interestingly, among these five bioluminescence profiles, (a), (c) and (e) show apparent circadian oscillation, whereas (b) and (d) show only faint bioluminescence fluctuation or no apparent circadian change (Figure 2D). These results suggest that the circadian enhancer(s) in the mDbp promoter may not be active at long distances from the trap vector. More importantly, these results indicate that the putative circadian enhancer is likely to be located between −700 bp (c) and −262 bp (a) of the 5′ flanking region of this gene. In additon, there was no strong circadian enhancer in downstream from the position of −335 bp, because only faint oscillation was seen in −335 bp integrated one (Figure 2Db). Strikingly, one transposed clone (f) in Figure 2C, lacking this region did not show circadian fluctuation of bioluminescence in rat-1 cells (Figure 2D). In addition, a similar transposition, clone (g), lacking −462 to +83 bp, also exhibited almost no apparent circadian oscillation in bioluminescence compared to (a) or (c) (Figure 2D). This strongly suggests that the region between −462 and −335 bp contains the putative circadian enhancer responsible for the robust circadian oscillation of Dbp promoter driven bioluminescence.
The predicted circadian enhancer region in the mDbp promoter contains a non-canonical E-box
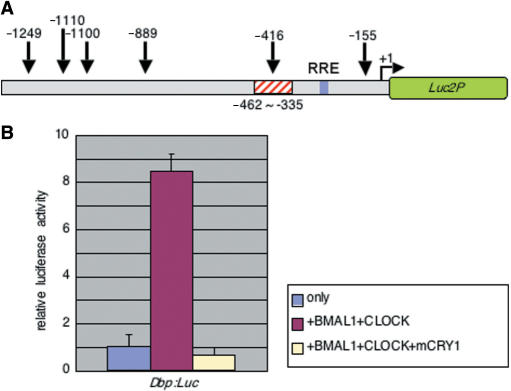
Using the in vitro transposon-based enhancer-trap method, we were able to predict a region that may contain a strong circadian enhancer. Thus, the next step was to identify the exact enhancer sequence responsible for mDbp promoter driven circadian oscillation. Ripperger et al. reported that CLOCK controls the circadian expression of the mDbp gene in vivo (14). Moreover, our findings in this study show that the phase of mDbp promoter driven rhythm is similar to the rhythm driven by the mPer2 promoter and opposite from mBmal1 promoter-driven rhythms (Figure 1). Several studies have indicated that CLOCK/BMAL1 activate transcription via CACGTG E-box enhancers existing in various mammalian clock genes such as mPer1, mCry1, Rev-erb alpha and Dbp. Recent observations have also shown that CLOCK/BMAL1 regulate not only CACGTG E-boxes, but also CACGTT E-box like sequences (25,26). Furthermore, one recent report suggested that a CANNTG non-canonical E-box was able to regulate circadian expression of prolactin (27). Although no CACGTG or CACGTT E-boxes were seen, several CANNTG sequences exist in the mDbp promoter region (Figure 3A). Thus, we investigated the possibility of CLOCK/BMAL1-mediated regulation of the mDbp promoter region we cloned.
Figure 3.
Transcriptional activation driven by the mDbp promoter is upregulated by CLOCK/BMAL1 co-expression. (A) Six CANNTG non-canonical E-boxes are found within the ∼1.4 kb region of the mDbp promoter that is able to drive robust circadian oscillation of transcription. Each number indicates the position of a CANNTG consensus sequence. The red-hashed box represents the location of a putative strong circadian enhancer as predicted by the enhancer-trap assay. (B) Transcriptional assay using the mDbp:Luc reporter vector. Co-expression of flag-tagged mBmal1 and mClock results in a greater than 8-fold up-regulation of promoter activity. Additional expression of HA-mCry1 dramatically suppresses mDbp promoter activity.
We performed transcriptional assays using the ∼1.4 kb mDbp promoter-luciferase (Dbp:luc) reporter construct. Co-transfection of flag-tagged mClock and mBmal1 expression vectors with the Dbp:luc reporter resulted in significant up-regulation of luciferase activity compared to pcDNA3 empty vector co-transfection conditions (Figure 3B). Moreover, additional expression of mCry1 dramatically reduced the CLOCK/BMAL1-mediated transcriptional activity (Figure 3B). These results are consistent with the reported features of canonical E-boxes, and suggest that non-canonical E-box(es) in the mDbp promoter are likely to be functional.
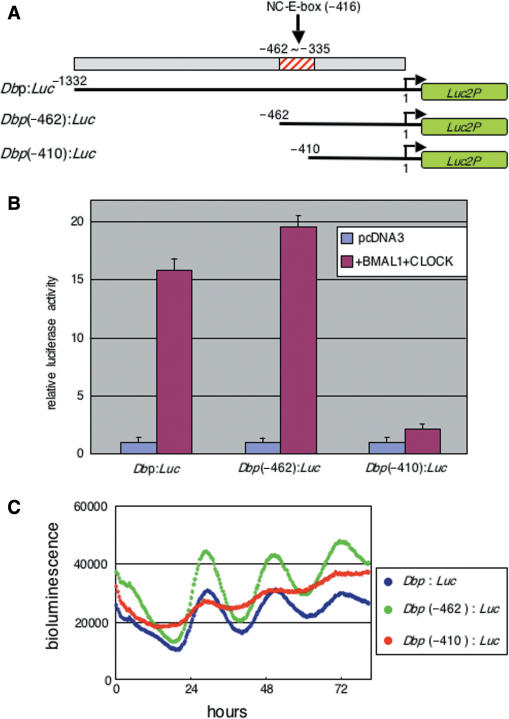
One CANNTG non-canonical E-box sequence was located in the region predicted to contain the putative circadian enhancer as shown (Figures 2 and 3A). To determine whether this non-canonical E-box is the putative enhancer, we generated deletions of the mDbp promoter named Dbp(−462):luc and Dbp(−410):luc (Figure 4A). Strikingly, transcriptional assays showed that deletion of the region containing the non-canonical E-box resulted in a loss of responsiveness to co-expression of the CLOCK/BMAL1 proteins (Figure 4B). More importantly, real-time bioluminescence monitoring showed that the circadian oscillation of Dbp(−410):luc was dramatically reduced, whereas Dbp(−462):luc exhibited robust circadian oscillation similar to that of Dbp:luc (Figure 4C). These results indicate that the predicted circadian enhancer region containing a non-canonical E-box (CATGTG) was actually responsible for transcriptional activation by the CLOCK/BMAL1 heterodimer and for exhibiting robust circadian oscillation of bioluminescence reporter expression. The weak bioluminescence oscillation observed in Dbp(−410):luc transfected cells may be elicited through another non-canonical E-box existing in the position of −155 bp. In addition, an RRE compatible sequence (indicated in Figure 3) in mDbp promoter may be not significantly functional, because the phase of Dbp(−410):luc was still similar to the bioluminescence rhythm driven by other two Dbp promoter luciferase reporter vectors.
Figure 4.
The predicted circadian enhancer region is essential for robust circadian oscillation driven by the mDbp promoter. (A) Deletions of the mDbp promoter: Dbp(−462):Luc contains the whole predicted enhancer region, Dbp(−410):Luc contains only a part of the predicted enhancer region. (B) Transcriptional assay using these reporter constructs. Both Dbp:Luc and Dbp(−462):Luc are strongly up-regulated (16-fold and 19-fold induction, respectively) by co-expression of the CLOCK/BMAL1 proteins, whereas the Dbp(−410):Luc reporter is induced only about 2-fold induction in response to CLOCK/BMAL1 co-expression. (C) The Dbp(−410):Luc-driven bioluminescence rhythm exhibits a dampened oscillation of very low amplitude; in contrast, both Dbp:Luc and Dbp(−462):Luc drive robust circadian oscillation of bioluminescence.
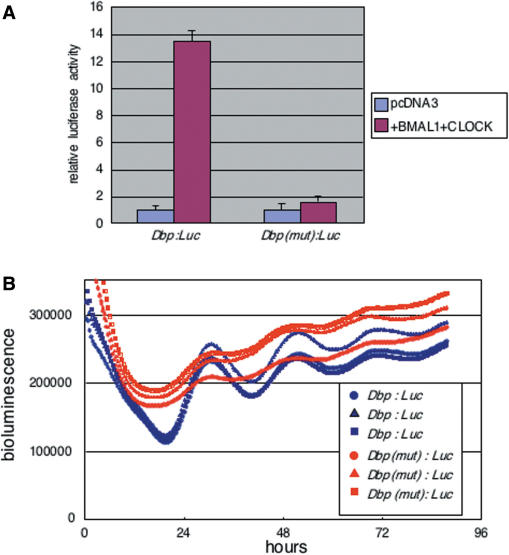
Identification of the non-canonical E-box as an enhancer contributing for robust circadian oscillation driven by the mDbp promoter
To determine whether the non-canonical-E-box (CATGTG: −416 bp from the putative transcription start site of the Dbp promoter) located in the predicted circadian enhancer region is actually functional, we generated a mutation of this non-canonical-E-box in the Dbp:luc reporter [Dbp(mut):luc] using site-directed mutagenesis. The non-canonical E-box CATGTG was substituted with the sequence GGTACC, and transcriptional assays using the mutated promoter construct were performed to examine its responsiveness to CLOCK/BMAL1. The [Dbp(mut):luc] promoter was induced 1.6-fold by CLOCK/BMAL1 co-expression, whereas the wild-type Dbp:luc promoter was induced 13.5-fold (Figure 5A). Subsequently, real-time bioluminescence monitoring was performed using the wild-type and mutated Dbp promoters. Dbp(mut):luc driven bioluminescence showed very weak circadian oscillation, but the amplitude of this oscillation was dramatically lower than that of Dbp:luc-driven bioluminescence oscillation (Figure 5B). These findings are consistent with the results obtained from deletion of this predicted enhancer region (Figure 4C). Therefore, these results strongly suggest that the non-canonical E-box located in the predicted circadian enhancer region was responsible for driving robust circadian transcriptional oscillation from the Dbp promoter. Although mutation of CATGTG E-box sequence resulted in the dramatic damp of promoter driven circadian oscillation, slight circadian rhythm of bioluminescence still remained. This suggested that other weak circadian enhancer(s) in part may contribute the transcriptional rhythm of Dbp gene.
Figure 5.
Mutation of the non-canonical E-box localized in the predicted circadian enhancer region results in loss of robust circadian oscillation. (A) Substitution of the CATGTG non-canonical E-box to GGTACC results in loss of CLOCK/BMAL1-mediated transactivation of the Dbp:Luc reporter. Mutated Dbp:Luc is named Dbp(mut):Luc. Relative luciferase activity is reported as mean + SEM (B) Real-time bioluminescence analysis using wild-type Dbp:Luc (three blue lines) and Dbp(mut):Luc (three red lines) shows a dramatic dampening of circadian oscillation in the case of the mutant. Each line represents one 35-mm dish transfected with Dbp:Luc or Dbp(mut):Luc. Cells were synchronized by treatment with 100 nM dexamethasone and bioluminescence monitoring was performed using the same photomultiplier.
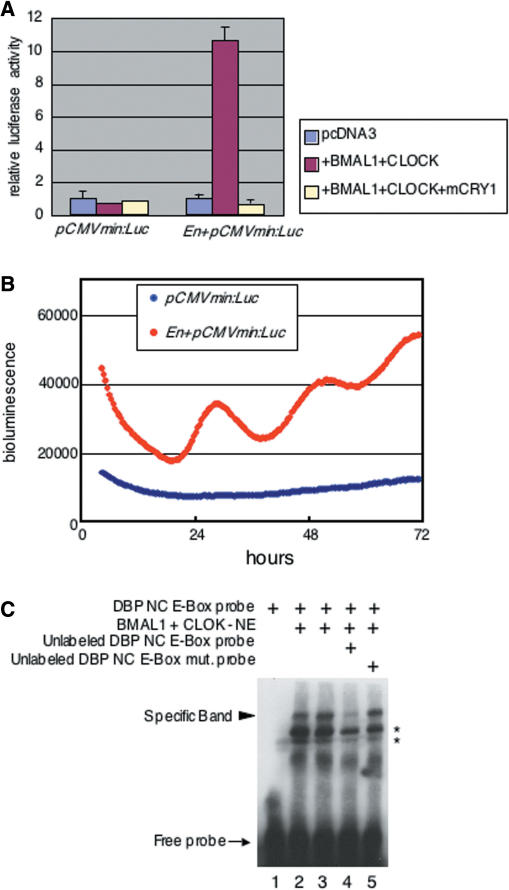
Finally, we investigated the ability of the predicted enhancer region alone to generate exact circadian oscillation of gene expression. We subcloned a 127 bp DNA fragment, containing −466 to −340 bp of the mDbp promoter, upstream of a CMV minimal promoter driven luciferase reporter construct (En + pCMVmin:luc). Transcriptional assays were performed to examine the effects of CLOCK/BMAL1 on this DNA fragment. The relative luciferase activity of En + pCMVmin:luc was up-regulated after CLOCK/BMAL1 co-expression, and this transcriptional activity was suppressed completely by the additional expression of mCRY1 (Figure 6A right). In contrast, no response was observed inform a CMV minimal promoter luciferase reporter without the predicted enhancer DNA fragment (Figure 6A left). Real-time bioluminescence monitoring demonstrated that the predicted enhancer region had the ability to generate circadian gene expression in living rat-1 fibroblast cells (Figure 6B). These results indicated that the non-canonical E-box contained in this enhancer region functioned as a circadian enhancer. Thus, we next confirmed the direct binding of the CLOCK/BMAL1 complex to this non-canonical E-box sequence using electrophoretic gel mobility shift assays (EMSA). When a biotin-labeled 30-bp oligo DNA probe containing the non-canonical E-box (CATGTG) sequence was incubated with nuclear extract from COS7 cells transfected with expression vectors for mClock and mBmal1, a shifted band was observed (Figure 6C lanes 2 and 3). The specific band was abolished by a 50-fold excess of unlabeled probe (Figure 6C lane 4); in contrast, it was not abolished by a 50-fold excess of unlabeled probe containing a mutation of the non-canonical-E-box sequence from CATGTG to GGTACC (Figure 6C lane 5). These results indicate that the CLOCK/BMAL1 complex directly binds the CATGTG non-canonical E-box located in the predicted enhancer region of the mDbp promoter.
Figure 6.
The isolated predicted circadian enhancer region containing the non-canonical E-box elicits distinct circadian oscillation. (A) The isolated predicted circadian enhancer region of the mDbp promoter was subcloned into the pCMVmin:Luc reporter vector, and strong up-regulation of transcriptional activity was observed after co-expression of CLOCK/BMAL1. CLOCK/BMAL1-mediated transcriptional activity was dramatically reduced by the additional expression of HA-mCRY1. In contrast, the pCMVmin:Luc vector without the non-canonical E-box showed no response to co-expression of CLOCK/BMAL1 and HA-mCRY1. (B) Real-time bioluminescence monitoring of pCMVmin:Luc with or without the non-canonical E-box. The pCMVmin:Luc vector exhibited no cyclic fluctuation of bioluminescence over 3 days; in contrast, the En+pCMVmin:Luc vector, which contains the predicted circadian enhancer elicited apparent circadian oscillation of bioluminescence (red line). (C) Electrophoretic gel mobility shift assays (EMSA) indicate direct binding of the CLOCK/BMAL1 complex to the CATGTG non-canonical E-box located in the predicted circadian enhancer region of the mDbp promoter. Shifted bands are detected when labeled non-canonical E-box probe is incubated with nuclear extract from CLOCK/BMAL1 expressing COS7 cells (lanes 2 and 3); incubation with an excess of unlabeled probe results in dramatic reduction of the shifted bands (lane 4). An excess of unlabeled mutated non-canonical E-box probe (CATGTG to GGTACC) does not reduce the shifted band (lane 5). Asterisks indicate non-specific bands.
Interaction of BMAL1/CLOCK with Dbp promoter around identified non-canonical E-box region in NIH3T3 cells
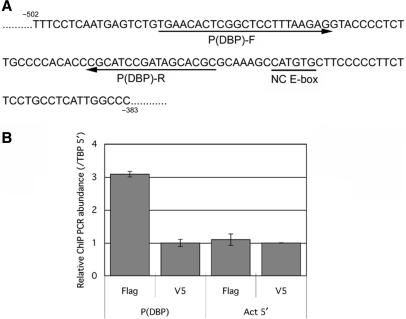
To confirm the identified non-canonical E-box actually functioned in living cells, we performed chromatin immunoprecipitation (ChIP) using flag-tagged BMAL1 and CLOCK expressed NIH3T3 cells. Co-precipitated genomic DNA fragments by anti-flag immunoprecipitation were analyzed by quantitative PCR (Q-PCR) method to detect the identified non-canonical E-box region. Position of primer sequences for Q-PCR and the non-canonical E-box of Dbp promoter region are presented in Figure 7A. Q-PCR analysis revealed that the significant recruitment of flag-tagged BMAL1 and CLOCK around the non-canonical E-box region of Dbp promoter [P(DBP)] was observed, comparing with the beta-actin promoter region (Act5′) as a negative control (Figure 7B). ChIP using anti-V5 antibody for flag-BMAL1 and flag-CLOCK expressed NIH3T3 showed no significant difference between P(DBP) targeted and Act5′-targeted Q-PCR (Figure 7B). These results confirmed that BMAL1/CLOCK actually bound the 5′ flank region of Dbp gene around the identified non-canonical E-box.
Figure 7.
BMAL1/CLOCK binds the endogenous non-canonical E-box region of 5′ flank end of the Dbp gene in NIH3T3 cells. Chromatin immunoprecipitation (ChIP) was performed to analyze the binding of BMAL1/CLOCK with the endogenous non-canonical E-box identified in this study. (A) The position of identified non-canonical E-box is indicated as underline (NC E-box). Arrows below the DNA sequences indicate the primers [P(DBP)-F and P(DBP)-R] to detect co-immunoprecipitated target Dbp promoter region. (B) ChIP is performed using flag-tagged BMAL1/CLOCK expressed NIH3T3 cells. Quantitative PCR (Q-PCR) using P(DBP)-F and P(DBP)-R primers revealed that amplified signals from the precipitants with anti-Flag antibody are significantly stronger than the signals amplified from the precipitants using anti-V5 antibody as a control (left two columns). As a negative control, Q-PCR using specific primers for beta-actin promoter (Act5′) shows that amplified signals from precipitant samples using anti-flag or anti-V5 antibodies both are low and no significant difference between them (right two columns). Error bars indicate standard deviation (SD).
DISCUSSION
In this study, we have presented that we developed a transposon-based in vitro enhancer trapping method, which is a new strategy to identify unknown enhancers contributing to circadian gene expression in living cells. Using this method, we determined that the CATGTG sequence, which can be classified as a CANNTG non-canonical E-box, at position −416 bp in the mDbp promoter functioned as a strong circadian enhancer to elicit robust oscillation of mDbp promoter driven luciferase bioluminescence. These results were compatible with recent study showing that this non-canonical E-box actually functioned as a circadian enhance in vivo (16).
Transposon-based enhancer detection system
In this study, we developed a Tn5 transposon-based in vitro enhancer detection vector to identify unknown circadian enhancer elements. The Tn5 transposon system was originally developed for mutagenesis of bacterial genes in vitro and in vivo (17). However, this system is also useful for examining mammalian genes in vitro when the target DNA fragments are subcloned into a cloning vector. Therefore, we used this transposon system to try to trap putative enhancer elements. Moreover, using short half-life firefly luciferase (from pGL4.11, Promega), we were able to analyze real-time transcriptional activity for several days following transfection of this trap vector into rat-1 cells. Combining the newly generated enhancer trap vector with high throughput real-time bioluminescence monitoring allowed us to detect a highly dynamically controlled circadian enhancer element. As shown in Figure 2, some of the transposons integrated into the mDbp promoter exhibited apparent circadian bioluminescence cycles, whereas others did not (Figure 2). This indicated that the circadian enhancer in the mDbp promoter must localized on the up-stream of the luciferase, and allowed us to identify the CATGTG non-canonical E-box as a strong circadian enhancer driving robust circadian oscillation.
Another interesting point of this study is why these circadian enhancers exhibit ‘positional effect’ or ‘distance effect’. In general, it is known that an enhancer shows its function without any relation of distance and direction. However, our strategy was actually useful to find out the non-canonical E-box enhancer. Although we have not yet had exact answer for this question, one speculation is that the position of BMAL1/CLOCK E-box may be important for expressing its function. Because in many clock genes having E-box on their promoter, the E-boxes reside near transcriptional start sight in many case. The exact answer should be waited for further studies.
This study used transient transfections to analyze the real-time bioluminescence driven by the enhancer detection vector. We were able to predict the circadian enhancer region of the mDbp promoter based on the amplitudes of reporter oscillation observed in trap vector integrated clones; this suggests that our in vitro enhancer trap strategy is a powerful and convenient method to identify unknown enhancers. Furthermore, much microarray data has recently become available in public databases. Using these data, in vitro enhancer trapping may become a much more effective way to understand the regulatory mechanisms of gene expression.
A non-canonical E-box as a strong circadian enhancer
Here, we have identified a CATGTG non-canonical E-box as a strong circadian enhancer driving an mDbp promoter luciferase reporter with clear circadian rhythm. One report recently suggested that a CANNTG non-canonical E-box (CATTTG) in the prolactin promoter is regulated by the CLOCK/BMAL1 complex (27). These results showed that both CACGTG canonical E-boxes and CANNTG non-canonical E-boxes could be important circadian enhancers. On the other hand, our findings raise a new question. The CANNTG sequence is mathematically predicted to appear once every 256 bp in the genome. This means that circadian enhancers may exist widely in the promoters and intronic regions of many genes. Storch et al. reported that hundreds of genes are cyclically expressed in liver and heart; however, most of these genes do not overlap in both organs (11). Other microarray studies have supported the idea of divergent circadian gene expression in a tissue-specific manner (10,12). These studies suggest the possibility that a large number of genes have the potential to exhibit circadian expression under appropriate conditions (13). In addition, our data showed that weakly but apparent circadian oscillation of bioluminescence was still remained even this non-canonical E-box enhancer was mutated (Figure 5B). These results may suggest that other weakly functioned unknown circadian enhancers still existed on the mDbp promoter region or other mechanism (for example, chromatin remodeling) would contribute for circadian gene expression. Another important and interesting question arising from this study is how the circadian system selectively regulates gene expression. Tissue-specific circadian gene expression may be controlled by a combination of enhancer and tissue-specific regulatory factors, such as the genome sequence around the non-canonical E-boxes or chromatin regulatory mechanisms. Further studies are required to address these questions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Sayaka Tagao, Akira Morita and Yukiko Sugisawa for technical support. This work is supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (16021237 and 17650092 to K.Y., Genome Network Project to H.R.U.); an intramural Grant-in-Aid from CDB (to H.R.U.); SAKURA Grant from JSPS (K.Y.); Grant-in-Aid from KANAE Medical Foundation (K.Y.); a Leading Project for Biosimulation (K.Y. and H.R.U.). Funding to pay the Open Access publication charges for this article was provided by Grant-in-Aid from Ministry of Education, Culture, Sports, Science and Technology of Japan (to YU and KY 16GS0315).
Conflict of interest statement. None declared.
REFERENCES
- 1.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Gemonics Hum. Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 3.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 4.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 5.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammlian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 6.Yagita K, Tamanini F, van der Horst G, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 7.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Welsh DK, Yoo S.-H, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr. Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 10.Panda S, Antoch M, Miller B, Su A, Schook A, Straume M, Schultz P, Kay S, Takahashi J, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 11.Storch K-F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 12.Ueda H, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 13.Reddy AB, Wong GKY, O'Neill J, Maywood ES, Hastings MH. Circadian clocks: neural and peripheral pacemakers that impact upon the cell division cycle. Mutat. Res. 2005;574:76–91. doi: 10.1016/j.mrfmmm.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 14.Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Develop. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaguchi S, Mitsui S, Yan L, Yagita K, Miyake S, Okamura H. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 2000;20:4773–4781. doi: 10.1128/mcb.20.13.4773-4781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripperger J, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 17.Steiniger-White M, Rayment I, Renznikoff WS. Structure/function insights into Tn5 transposition. Curr. Opin. Struct. Biol. 2004;14:50–57. doi: 10.1016/j.sbi.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Golden SS, Ishiura M, Johnson CH, Kondo T. Cyanobacterial circadian rhythms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- 19.Schreiber E, Matthias P, Mueller M, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts”, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 reponse element upstream proximal from the interleukin-13 gene locus. J. Biol. Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno K, Kasukawa T, Dauwalder B, Itoh T, Takahashi K, et al. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 2007;21:1687–1700. doi: 10.1101/gad.1552207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi S, Mitsui S, Miyake S, Yan L, Onishi H, Yagita K, Suzuki M, Shibata S, Kobayashi M, et al. The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr. Biol. 2000;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- 24.Preitner N, Damiola F, Lopes-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERB alpha controles circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 25.Yoo S, Ko C, Lowrey P, Buhr E, Song E, Chang S, Yoo O, Yamazaki S, Lee C, et al. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 27.Leclerc G, Boockfor F. Pulses of prolactin promoter activity depend on a noncanonical E-box that can bind the circadian proteins CLOCK and BMAL1. Endocrinology. 2005;146:2782–2790. doi: 10.1210/en.2005-0100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.