Abstract
ELAV is a neuron-specific RNA-binding protein in Drosophila that is required for development and maintenance of neurons. ELAV regulates alternative splicing of Neuroglian and erect wing (ewg) transcripts, and has been shown to form a multimeric complex on the last ewg intron. The protein has three RNA recognition motifs (RRM1, 2 and 3) with a hinge region between RRM2 and 3. In this study, we used the yeast two-hybrid system to determine the multimerization domain of ELAV. Using deletion constructs, we mapped an interaction activity to a region containing most of RRM3. We found three conserved short sequences in RRM3 that were essential for the interaction, and also sufficient to give the interaction activity to RRM2 when introduced into it. In our in vivo functional assay, a mutation in one of the three sequences showed reduced activity in splicing regulation, underlining the functional importance of multimerization. However, RRM2 with the three RRM3 interaction sequences did not function as RRM3 in vivo, which suggested that multimerization is not the only function of RRM3. Our results are consistent with a model in which RRM3 serves as a bi-functional domain that interacts with both RNA and protein.
INTRODUCTION
The RNA recognition motif (RRM) is the most common RNA-binding domain, and also one of the most abundant protein domains in eukaryotes (1–3). The RRM consists of about 80–90 amino acids, and has the conserved structure of two α-helices packed against four anti-parallel β-strands. Structural studies have shown that it interacts with RNA through amino acids located on the β-strands (4–9). In addition to RNA-binding, it has been shown that RRMs can also serve as the site of protein–protein interactions. Such a case was initially reported for the interactions between U2 small nuclear ribonucleoprotein particle (snRNP) B″ and A′ proteins (10–13), and later structural studies, including that of other proteins, revealed that the protein–protein interactions often, but not always, occur through amino acids on the α-helices of the RRM (8,14–21). This dual interacting capability of the RRM domain might explain its prevalence.
The ELAV protein family is a conserved RNA-binding protein family found in animals from Caenorhabditis to humans. Its characteristic structure is the three RRMs (RRM1, 2 and 3) with a ‘hinge’ region of about 60–80 amino acids separating the second and third RRMs. Mammals have four members of the family called Hu proteins (HuR, HuB, HuC and HuD) that have been implicated in stabilization and/or translation activation of mRNAs containing AU-rich elements (AREs) (22–24). Drosophila has three ELAV-family proteins (ELAV, Rbp9 and Fne). ELAV is specifically expressed in all neurons in Drosophila, and elav null mutants show embryonic lethal phenotype with developmental defects in the nervous system (25,26). It has been shown that ELAV regulates alternative splicing of Neuroglian (Nrg) and erect wing (ewg) transcripts, by binding to their introns to produce neuron specific isoforms (27–30). Additionally, ELAV has been shown to autoregulate its own message (31–33).
Recently, ELAV has been shown to form a multimeric complex on the ewg intron RNA. It appears that ELAV in solution is mainly in a tetramer-configuration without the substrate RNA, and is assembled into a larger dodecameric complex upon binding to the target RNA (34). Mammalian ELAV family proteins have also been shown to form multimers (35–37). Therefore, it is likely that multimerization is a common feature shared by the ELAV-family proteins, and an important part of the mechanism by which they exert their effect on the bound RNA.
In this study, we show that one of the major sites for ELAV–ELAV interaction activity is located in RRM3 and a small adjacent region of the hinge. Three short sequences in RRM3 are shown to be necessary for the interaction, and also sufficient to give the interaction activity to RRM2 when introduced into it. However, RRM3 requires more than the three short sequences to function in vivo, suggesting that acting as a multimerization domain is not the sole function of RRM3. Our data are consistent with a model in which RRM3 acts as a bi-functional domain that interacts with both RNA and protein.
MATERIALS AND METHODS
Plasmid construction
To construct plasmids for the yeast two-hybrid assay, we used pGBKT7 and pGADT7 vectors (Clontech) for the GAL4 DNA-binding-, and transcription-activation-domain fusions, respectively. Both fusions were N-terminal fusions; i.e. the DNA-binding, or activation domain was fused to the N-terminus of the protein to be tested. The full-length and truncated elav coding sequences were amplified by PCR from an elav cDNA clone, digested with EcoRI and BglII, and subcloned between EcoRI and BamHI sites of pGBKT7 and pGADT7 vectors.
For construction of hinge-RRM1 (eH1) and hinge-RRM2 (eH2) cDNAs, the elav RRM1 and 2 coding sequences were PCR-amplified with primers that have the 3′ end sequence of the hinge region fused to the 5′ region of the RRM1 or 2 coding sequence. The fused hinge sequence would anneal to the corresponding site of the hinge region during the next round of PCR. The secondary PCRs were performed in which eH1 or eH2 were amplified from mixed templates of the first PCR product and pGADT7-eH. The resulting PCR fragments were digested with EcoRI and BglII, and subcloned into EcoRI/BamHI sites of pGADT7.
The mutant hinge-RRM3 constructs (eH3mu1-7) were generated by site-directed PCR mutagenesis. For each mutation, a complimentary pair of primers was designed at the target site to replace a certain RRM3 sequence with the corresponding sequence from RRM2. In the primary PCRs, the 5′ and 3′ parts were amplified separately using one of the complimentary mutagenic primers at one end. The PCRs produced the 5′ and 3′ parts of the constructs overlapping with each other for the length of the mutagenic primers. The 5′ and 3′ parts were joined together by the secondary PCRs in which a mixture of the two primary PCR products was used as a template. The secondary PCR fragments were digested with EcoRI and BglII, and subcloned into EcoRI/BamHI sites of pGADT7.
The eH3mu8 was constructed in a similar way as the other mutant hinge-RRM3 constructs were made, but with three separate primary PCRs; from the 5′ end of the hinge to the first two mutations (ETEE and W), from the first two mutations to the third mutation site (MTNY), and from the third mutation to the 3′ end of RRM2. The three primary PCR fragments were joined together by the secondary PCR with the primers at both ends of the eH3mu8. The secondary PCR fragment was digested with EcoRI and BglII, and subcloned into EcoRI/BamHI sites of pGADT7.
For Rbp9 and fne constructs, the coding sequences of Rbp9 and fne were amplified by PCR from cDNA prepared from adult Drosophila RNA. The amplified Rbp9 coding sequence was digested with BglII and XhoI, and subcloned into BamHI/SalI and BamHI/XhoI sites of pGBKT7 and pGADT7, respectively. The fne coding sequence was digested with EcoRI and BglII, and subcloned into EcoRI/BamHI sites of pGBKT7 and pGADT7.
For the upstream activating sequence (UAS) constructs, PCRs were performed to put a mutant RRM in place of RRM3 in the full length elav cDNA. The pGADT7 with a mutant hinge-RRM insert (eH3mu4, eH3mu8, or eH2) was digested with HpaI, and used as a template together with pGBKT7-eQ12H. With the primers at the 5′ end of elav coding sequence and downstream to the cloning site of pGADT7, the full length elav coding sequences with a mutant RRM3 were amplified. The PCR fragments were digested with EcoRI and XhoI, and subcloned first into EcoRI/XhoI sites of pGADT7, then cut out by BglII and XhoI digestion, and subcloned into BglII/XhoI sites of the pUAST vector (38). Likewise, the control elav coding sequence was excised from the pGADT7-eQ12H3 construct with BglII and XhoI digestion, and subcloned into BglII/XhoI sites of pUAST. All the UAS-elav constructs have hemagglutinin (HA) epitope tag sequence at the N-terminus that is derived from pGADT7.
All the subcloned PCR fragments were fully sequenced to confirm the absence of inadvertent mutations introduced during PCR. See Supplementary Data for all the primer sets used for the PCRs (Table S1) and their sequences (Table S2).
Yeast two-hybrid assay
The Matchmaker Two-Hybrid System 3 (Clontech) was used for the two-hybrid interaction assays. The Saccharomyces cerevisiae Y187 and AH109 strains (Clontech) were transformed with the pGBKT7 and pGADT7 constructs, respectively. To test two-hybrid interactions, Y187 with a pGBKT7 construct and AH109 with a pGADT7 construct were mated, and the resulting diploids were tested for two-hybrid reporter gene activation. Each mating culture was plated on a SD/-Ade/-His/-Leu/-Trp testing plate to examine the activation of both ADE2 and HIS3 reporter genes, as well as on a SD/-Leu/-Trp plate to confirm mating success. Colony formation on the SD/-Ade/-His/-Leu/-Trp plate was scored after 6 days of incubation at 30°C. In all figures, ‘+’ means yeast colony formation indicating interaction between the two proteins, and ‘−’ means no or very few colony formations indicating no or very weak interaction (see Figure 3B for examples of ‘+’ and ‘−’ results). All the constructs were tested to confirm that they did not activate the reporter gene expressions by themselves.
Figure 3.
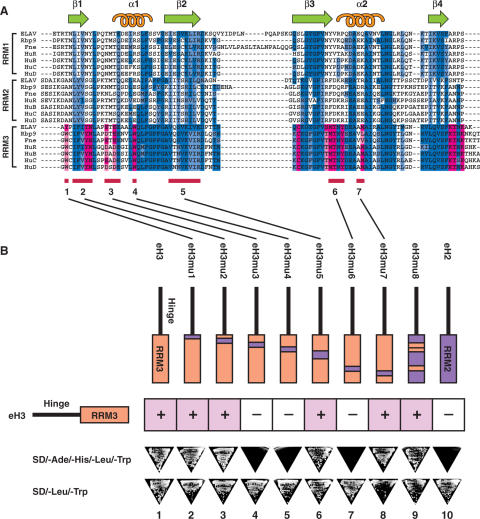
Uniquely conserved sequences in the RRM3 that are responsible for ELAV–ELAV interaction. (A) An amino-acid sequence alignment of RRMs from three Drosophila, and four human ELAV-family proteins; ELAV, Rbp9, Fne, HuR, HuB, HuC and HuD. Amino acids that are conserved among RRM3s but not at the same sites in RRM1 or 2 (RRM3-specific conservations) are indicated by magenta. Conserved amino acids among the RRM3s that are not specific to RRM3 are shown by blue. Darker color indicates identical amino acids to those of ELAV RRM3, and lighter color indicates conservative substitutions. Putative secondary structural elements are shown above. The seven underlined sequences were chosen for the mutational analysis. (B) The effect of mutations on the interaction. The hinge-RRM3 construct was mutated by replacing one of the seven underlined sequences with the corresponding sequence of RRM2. The interactions between the mutants and the wild-type hinge-RRM3 were tested by the two-hybrid assay. In the figure, the constructs above the ‘+’ or ‘−’ box are activation-domain fusions, and the eH3 ‘bait’ construct on the left is a DNA-binding-domain fusion. The loss of interaction activity of the mutants eH3mu3, 4 and 6 (columns 4, 5 and 7) suggests that the mutated amino acids are essential for the interaction. Introduction of the three sequences, 3, 4 and 6, to RRM2 (eH3mu8 construct, column 9) was sufficient to give the interaction activity to the hinge-RRM2. Photographs of representative testing and control plates are shown below.
Immunoprecipitation
For co-immunoprecipitation (co-IP), adult flies of the following genotypes were prepared: elave5; elavDvORF/CyO, elave5; elavDvORF/+; elav−13/+, elave5; elav−13/TM3 Ser, UAS-eQ12H3/+; Hsp70-GAL4/+ and Hsp70-GAL4/+. The elave5 (26) is a null allele of elav, elavDvORF (39) and elav−13 (40) are elav transgenes, UAS-eQ12H3 is HA-tagged elav under the control of UAS, and Hsp70-GAL4 (P{GAL4-Hsp70.PB}; Bloomington Drosophila Stock Center) is GAL4 driven by the Heat shock protein 70 (Hsp70) promoter. A 37°C heat-shock was applied for 30 min to UAS-eQ12H3/+; Hsp70-GAL4/+ and Hsp70-GAL4/+ flies 3 h and 30 min before head-collection. Eighty heads were collected for each genotype and homogenized in 250 μl of immunoprecipitation buffer [IPB; 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 0.1% Triton X-100, Complete Mini Protease Inhibitor Cocktail Tablet (Roche)]. The homogenate was centrifuged, the supernatant was divided into two equal volumes of 110 μl, and the RNase Cocktail (Ambion; 0.1 U RNase A and 4U RNase T1) was added to one (RNase +). Both RNase + and − homogenates were incubated at room temperature (RT) for 30 min. After the incubation, the homogenate was centrifuged again and 100 μl of the supernatant was recovered. The Immobilized Protein A (Pierce Biotechnology) was pre-washed with IPB, then incubated with either the mouse anti-ELAV monoclonal antibody (mAb) 7D (for ELAVDvORF) or the mouse anti-HA mAb F-7 (Santa Cruz Biotechnology; for HA-tagged ELAV) in IPB at 4°C for several hours. About 10 μl of the beads were added to the homogenate after being washed with IPB. The mixture was incubated at 4°C for 1 h and 30 min with gentle rotation. The beads were washed with IPB. Then 10 μl of 2 × SDS sample buffer [100 mM Tris–HCl pH 6.8, 4% sodium dodecyl sulfate (SDS), 2% 2-mercaptoethanol, 20% glycerol, 0.001% bromophenol blue] was added to the beads, and incubated at 95°C for 8 min. The precipitated proteins were separated by SDS polyacrylamide gel electrophoresis and western blotting was probed by the rat anti-ELAV anti-serum.
Germline transformation
Df(1)w; Ki pp Δ2-3/+ embryos were injected with the pUAST constructs (41,42). The germline transformants were recovered based on [w+] eye color, and transgenic lines were established by standard procedures.
Immunostaining
The y w; UnGA; dpp-GAL4 (P{GAL4-dpp.blk1}) (43)/TM6B Tb females were crossed with males of one of the UAS-elav lines. The [Tb+] wandering third instar larvae in the next generation were used for dissection. Wing discs were dissected in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde in PBS for 30 min at RT. The wing discs were washed in 0.3% Triton X-100 in PBS (PBT), blocked in 5% normal goat serum in PBT for 2 h and 30 min at RT, and incubated with primary antibodies of the mouse anti-ELAV mAb 7D and the rabbit anti-GFP polyclonal antibody (Torrey Pines Biolabs) at the dilutions of 1:100 and 1:200, respectively, in PBT for overnight at 4°C. The secondary antibody incubation was carried out with the Cy5-conjugated donkey anti-mouse immunoglobulin antibody (Jackson ImmunoResearch Laboratories) and the fluorescein-conjugated goat anti-rabbit immunoglobulin antibody (Jackson ImmunoResearch Laboratories) at the dilution of 1:200 each in PBT for 2 h and 30 min at RT. The wing discs were mounted with 70% glycerol in PBS after washing with PBT.
Confocal microscopy and image analysis
Immunofluorescent images were acquired by Leica TCS SP2 mounted on Leica DM IRE2 inverted microscope. Intensities of fluorescence were measured by ImageJ software (National Institutes of Health) from average Z-series projection images of the wing discs. The ELAV (Cy5) and GFP (fluorescein) signals in the dpp expression pattern as well as in the background were measured for eight wing discs for each genotype. The background signal was subtracted from the signal in the dpp-pattern to obtain the compensated signal intensity for each disc. The compensated GFP signal was normalized by the compensated ELAV signal for each disc, and the average and confidence interval of the normalized GFP signal was calculated for eight wing discs of each genotype.
RESULTS
All Drosophila ELAV-family proteins interact with each other
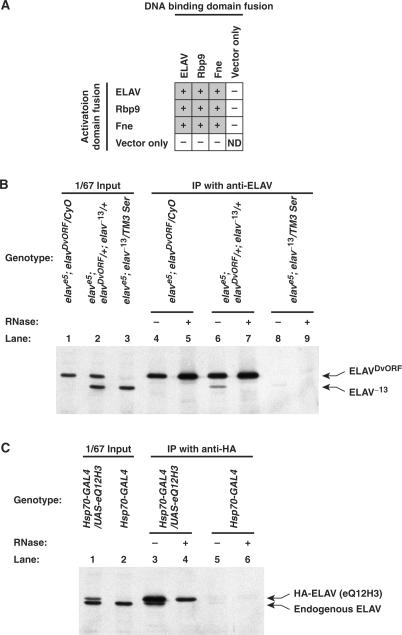
It has been shown that ELAV forms multimeric complexes in the presence or absence of substrate RNA (34), and mammalian Hu proteins form homo- and hetero-multimers (35–37). In our yeast two-hybrid screening of a Drosophila embryonic cDNA library for ELAV-interacting proteins, we identified ELAV itself and Fne, another ELAV-family protein in Drosophila, as the strongest ELAV-interacting proteins (Toba and White, our unpublished data). This suggests that Drosophila ELAV-family proteins also interact with each other as Hu proteins do. To examine interactions among all three Drosophila ELAV-family proteins, we cloned the cDNA of elav, Rbp9 and fne into yeast two-hybrid vectors. The interactions between the proteins in yeast were assessed by looking at colony formation on the testing plate that requires activation of both of the two reporter genes (see Materials and Methods section for details). Interactions were positive in all the combinations (Figure 1A), which suggests that Drosophila ELAV-family proteins may form homo- and hetero-multimers under these conditions.
Figure 1.
(A) Yeast two-hybrid interactions between Drosophila ELAV-family proteins. In the matrix, ‘+’ and ‘−’ indicate positive and negative interactions in the yeast two-hybrid assay, respectively. The results show that all Drosophila ELAV-family proteins, ELAV, Rbp9 and Fne, interact with themselves as well as each other. ‘Vector only’ means empty pGBKT7 and pGADT7 vectors for the DNA biding and activation domain fusions, respectively. ND, not determined. (B and C) ELAV–ELAV interaction is sensitive to RNase treatment. (B) Both ELAVDvORF and ELAV−13 were expressed in the elav null mutant flies. Immunoprecipitation (IP) was performed with the anti-ELAV mAb 7D which does not recognize ELAV−13, and western blot was probed with the anti-ELAV polyclonal antiserum. ELAV−13 was co-IPed with ELAVDvORF (lane 6), and the co-IPed ELAV−13 became undetectable with RNase treatment (lane 7). The elave5; elavDvORF/CyO (lanes 1, 4 and 5) and elave5; elav−13/TM3 Ser (lanes 3, 8 and 9) are negative controls in which ELAVDvORF and ELAV−13 were solely expressed, respectively. (C) HA-tagged ELAV was expressed in the flies with Hsp70-GAL4 driver, and an anti-HA mAb was used for IP. Western blot was probed with the anti-ELAV polyclonal antiserum. The endogenous ELAV was co-IPed with HA-ELAV (lane 3), and the interaction was disrupted by RNase treatment (lane 4). The Hsp70-GAL4 (lanes 2, 5 and 6) is a negative control that does not have the HA-tagged ELAV (UAS-eQ12H3) transgene.
ELAV–ELAV interaction requires RNA
To examine ELAV multimerization in the in vivo environment, we conducted co-IP from Drosophila extracts in two different ways: (i) A protein extract was prepared from flies expressing both the elavDvORF (39) and elav−13 (40) transgenes over the elave5 (null) background. The elavDvORF expresses ELAV of D. viliris that has a larger molecular weight than D. melanogaster ELAV. The ELAV−13 protein has a 13 amino acid deletion in the first RRM that includes the epitope recognized by the monoclonal anti-ELAV antibody. The ELAVDvORF was IP-ed with the monoclonal antibody to see if the ELAV−13 would be co-IP-ed; (ii) The HA-tagged ELAV was expressed in adult Drosophila using the GAL4-UAS system (38), and the HA-ELAV was IP-ed with an anti-HA antibody to examine if the endogenous ELAV would be co-IP-ed with it. In both cases, ELAV protein that was not recognized by the antibody was co-IP-ed with the other form of the protein (Figure 1B lane 6 and Figure 1C lane 3). However, the interactions seemed RNase sensitive since RNase treatment of the protein extract made co-IP-ed proteins undetectable in both cases (Figure 1B lane 7 and Figure 1C lane 4). This result suggests that either the interaction is mediated by RNA, or binding of the RNA allows a conformational change in ELAV that facilitates the protein–protein interaction.
Mapping of ELAV multimerization domains
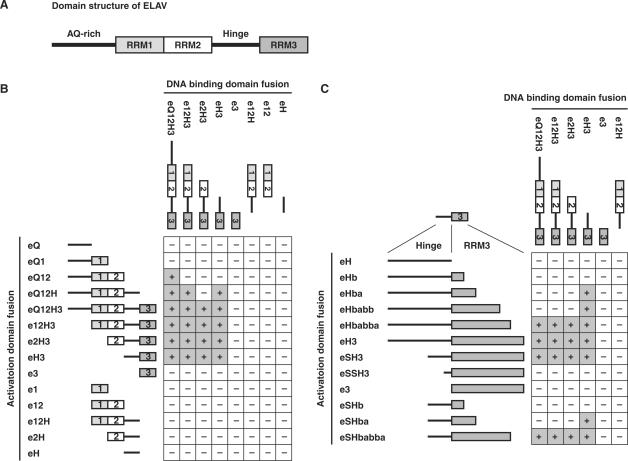
ELAV consists of three RRMs, an N-terminal alanine/glutamine (AQ)-rich region, and a hinge region between the RRM2 and 3 (Figure 2A). To map the region that is responsible for the ELAV–ELAV interaction, we made a series of deletion constructs and tested interactions using the yeast two-hybrid system. Interaction activity required the presence of both the hinge region and RRM3 in all cases but of eQ12 and eQ12H constructs in Figure 2B. The eH3 fragment interacted with itself (Figure 2B). In addition to the hinge-RRM3 region, the N-terminal AQ-rich region seemed to contribute to the interaction since the constructs that lack RRM3 or both hinge and RRM3 (eQ12 and eQ12H in Figure 2B) still retained some interaction activity as long as they had the AQ-rich region (compare eQ12 with e12, eQ12H with e12H in Figure 2B). Our results suggest the involvement of both the AQ-rich and hinge-RRM3 regions in the interaction. We resolved to pursue the activity of the hinge-RRM3 region since it is likely to be a general property of ELAV-family proteins, as the AQ-rich region is not present in any other ELAV-family protein. Additionally, the AQ-rich region is not essential to the vital function of ELAV (44).
Figure 2.
Mapping of ELAV–ELAV interaction activity by the yeast two-hybrid assay. (A) Schematic representation of ELAV structure with the N-terminus to the left. ELAV consists of three RNA recognition motifs (RRMs), the N-terminal AQ-rich region, and a hinge region between the RRM2 and 3. (B and C) Yeast two-hybrid interactions among deletion mutants of ELAV. In the matrices, ‘+’ and ‘−’ indicate positive and negative interactions in the yeast two-hybrid assay, respectively. Schematics of the deletion mutants are shown upper and left sides of each matrix. Boxes labeled with 1, 2 and 3 represent RRM1, 2 and 3, respectively. The bars next to the RRM1 and between RRM2 and 3 represent the AQ-rich and hinge regions, respectively.
We narrowed down the activity within the hinge-RRM3 region with deletion constructs of the hinge-RRM3 fragment. The result suggests that N-terminal two thirds of the hinge region and the C-terminal 18 amino acids of RRM3 are dispensable for the interaction (Figure 2C). The break point of the eSH3 in Figure 2C is the C-terminal end of the sequence required for nuclear localization of the protein (45). The construct eSHbabba in Figure 2C that consists of most of the RRM3 and a C-terminal part of the hinge was the smallest fragment that showed the same interaction activity as the original eH3 fragment.
Three conserved short sequences in RRM3 play a critical role in ELAV–ELAV interaction
To identify amino acids responsible for the interaction in the hinge-RRM3 region, we looked for evolutionary conservation within the region where we mapped the interaction activity. While we found conserved sequences in the RRM3, the sequence in the hinge region was very divergent and we hardly found any conservation within the region where interaction activity was mapped (data not shown). Although it is still possible that the region has important structures for the interaction without apparent sequence conservation, we focused our investigation on the RRM3.
Before proceeding to the analysis of the sequence, we wanted to make sure that it is the amino acid sequence of RRM3 that determines the interaction activity, not just the presence of any RRM after the hinge. The latter could be the case since the general structure of the RRM is quite well conserved. To test the possibility, ELAV RRM1 or 2 were fused to the C-terminus of the hinge, and checked for the interaction by the two-hybrid assay. Our results showed that RRM1 or 2 do not substitute for RRM3, indicating that amino acids unique to the RRM3 are responsible for the interaction (Figure 3B column 10, data not shown).
To find the amino acids responsible for the interaction, we looked for RRM3-specifically conserved residues. Figure 3A shows an amino acid sequence alignment of RRMs from three Drosophila, and four human ELAV-family proteins. RRM3-specific conservations are indicated by magenta, which are defined as the amino acids conserved among RRM3s, but not in RRM1 or 2. Conserved amino acids among the RRM3s that are not specific to RRM3 are indicated by blue. We chose the underlined sequences (numbers 1–7) as the targets for the mutational analysis. The sequence number 5 is not an RRM3-specific conservation, and was chosen as a negative control.
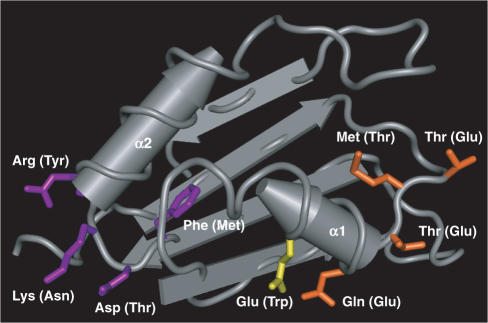
The eH3 construct was mutated by replacing one of the underlined sequences in Figure 3A with the corresponding sequence of ELAV RRM2. The interaction with eH3 was tested by the two-hybrid assay. While mutation in sequences 1, 2, 5 and 7 did not affect the interaction (Figure 3B columns 2, 3, 6 and 8), mutations in sequences 3, 4 and 6 caused loss of interaction (Figure 3B columns 4, 5 and 7), which indicated that the mutated amino acids were essential for the interaction. The three essential sequences 3, 4 and 6 are the ‘ETEE’ sequence adjacent to the putative α-helix 1, the single tryptophan on the α-helix 1, and ‘MTNY’ close to the putative α-helix 2, respectively. Next, we asked if the three sequences are sufficient to provide interaction activity to RRM2 when introduced into the eH2 construct at the corresponding sites. The hinge-RRM2 construct with the RRM3 sequences 3, 4 and 6 (eH3mu8) interacted with eH3 (Figure 3B column 9), demonstrating that the sequences play a critical role in ELAV–ELAV interaction. The examination of expected positions of the identified amino acids in relation to the tertiary structure of the RRM revealed that most of them are exposed to the outside, and thus qualified as candidates for the amino acids that mediate the interaction (Figure 4).
Figure 4.
Expected positions of the three sequences in relation to the tertiary structure of the RRM. The RRM2 part of the crystal structure of HuD associated with c-fos ARE (9; Protein Data Bank ID 1FXL) is shown. The RRM1, linker region between RRM1 and 2, and c-fos ARE RNA are omitted. The corresponding amino acid residues of the three interaction sequences of ELAV RRM3 are shown in orange (sequence 3), yellow (sequence 4) and purple (sequence 6). The labels indicate the identity of the shown residues of HuD RRM2 and that of the corresponding residues of ELAV RRM3 in the parentheses. All the residues are exposed to the solvent except the phenylalanine in the sequence 6. Columnar and flat arrows represent α-helix and β-strand, respectively. The α-helices 1 and 2 are labeled. The image is created by the Cn3D software (National Center for Biotechnology Information).
The three sequences are not sufficient for RRM3 to function in vivo
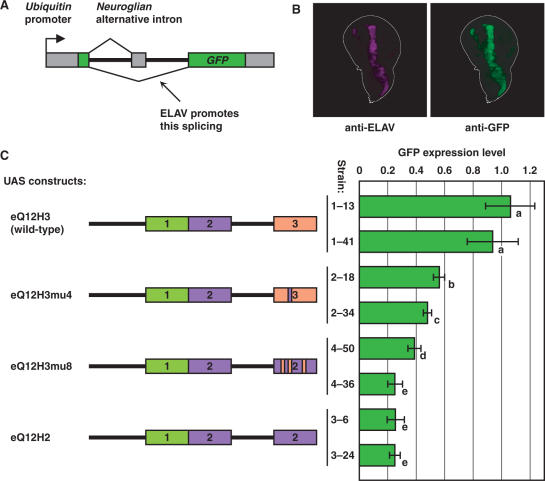
Given that our results indicate RRM3 has a critical role in ELAV–ELAV interaction, it is possible that RRM3 serves mainly as a protein–protein interaction domain, not as an RNA-binding domain. To test the possibility, we examined the in vivo function of ELAV with mutant RRM3 by expressing them using the GAL4-UAS system. We made four UAS-elav constructs: (i) the wild-type control (eQ12H3); (ii) the W419E mutant (eQ12H3mu4) that is presumably interaction defective; (iii) a mutant in which RRM2 replaces RRM3 except for the sequences 3, 4 and 6 (eQ12H3mu8) and (iv) a mutant in which RRM2 replaces RRM3 completely (eQ12H2) (see Figure 5C for schematics). To evaluate the in vivo function of the mutant proteins, we employed a GFP reporter gene (UnGA) whose expression is dependent upon ELAV-regulated alternative splicing of the Nrg alternative intron (46). Although the reporter is transcribed ubiquitously, GFP is expressed only when ELAV promotes the neural splicing of the intron (Figure 5A). GFP expression from UnGA is limited to neurons in the wild-type animals, but when ELAV is ectopically expressed, GFP is also expressed where ectopic ELAV is expressed (46; Figure 5B). We ectopically expressed ELAV in the wing discs of the third instar larvae using the dpp-GAL4 driver (43), and measured the GFP expression level relative to the ectopic ELAV level (see Materials and methods section for details). The right panel of Figure 5C shows induced GFP reporter expression levels by two independent insertion lines of each UAS-elav construct, as fractions of the mean value of the wild-type controls. The W419E mutant showed a reduced level of GFP expression compared to the wild-type construct, suggesting the importance of multimerization for the alternative-splicing regulation (Figure 5C). The mutant in which RRM3 was replaced by RRM2 except for the sequences 3, 4 and 6 functioned no better than the RRM3/2 total replacement mutant (Figure 5C). The result indicates that RRM3 requires more than the three sequences to function in vivo, and implies that multimerization is not the only function RRM3 performs.
Figure 5.
In vivo functional analysis of the RRM3 mutants. (A) Schematic representation of the UnGA reporter gene. The UnGA has the alternatively spliced intron from Nrg whose splicing is regulated by ELAV. It is transcribed ubiquitously, but GFP is expressed only when ELAV promotes the neural splicing of the intron. (B) Ectopic expression of ELAV in the wing disc by the dpp-GAL4 driver leads to the GFP expression from UnGA. Anti-ELAV (left panel) and anti-GFP (right panel) staining of a UnGA/+; dpp-GAL4/UAS-eQ12H3 wing disc are shown. The outline of the disc is shown with a white line. (C) The wild-type control and three mutant forms of ELAV were expressed by the dpp-GAL4 driver. Two independent insertion lines were used for each UAS construct. GFP expression levels in the wing disc were quantified, and normalized by ELAV expression levels. The bars on the right panel show the averaged values of eight wing discs for each genotype, as fractions of the mean value of the wild-type controls. The error bars show the 95% confidence intervals. Bars labeled with different letters (a–e) are significantly different at P < 0.01 in the t-test.
DISCUSSION
Previous studies have suggested that the RRM3 of ELAV-family proteins has a less important role in specific RNA-binding than the other two RRMs. For example, experiments using deletion constructs of Hu proteins have shown that mutant proteins lacking RRM3 still bind ARE efficiently (37,47,48). Yet, evolutionary conservation of the RRM3 sequence is the strongest among the three RRMs, which indicates an important function for RRM3. In this study, we showed that ELAV RRM3 has a central role in multimerization of the protein, and we believe that the multifunctional aspect of RRM3 makes it the most conserved RRM of the protein. It is noteworthy that RRM3 of HuC and HuB has been shown to have a dominant negative effect on neural-phenotype-inducing activity of the proteins (49). An explanation for this effect is competition between the full-length protein and the RRM3 fragment for binding to the RNA substrates, as shown by Gao and Keene (36). Considering our results, however, one can conceive another possible mechanism that the over-expression of the RRM3 fragment interferes with multimerization of the full-length proteins.
Kasashima et al. (35) studied multimerization of Hu proteins. They found that RRM3 of HuC showed the strongest interaction activity in the yeast two-hybrid system when full length HuB was used as bait, basically agreeing with our result. However, when HuC RRM3 was used as bait, a fragment containing both RRM1 and 2 also showed strong interaction activity (35). In our yeast two-hybrid experiment, similar constructs (e12 construct in Figure 2B) did not show interaction with either the full length ELAV or hinge-RRM3 fragment. This discrepancy may come from the presence of an arbitrary threshold in our system; i.e. we evaluate interaction by scoring colony formation of the yeast, and weak interactions may fail to be scored in principle. Therefore, it is possible that RRM1 and 2 also contribute to multimerization. In fact, we found that the RRM1 + 2 fragment showed limited interaction activity when the N-terminal AQ-rich region was added to it (eQ12 construct in Figure 2B). Nonetheless, our results suggest that the interaction activity of RRM3 is at least stronger than that of RRM1 or 2. The three sequence elements in the RRM3 we identified in this study are likely to be important for multimerization of all ELAV-family proteins, since they were identified as conserved sequences among the family.
Structural studies have shown that the RRMs have a well conserved general structure; i.e. four anti-parallel β-strands form a β-sheet, and two α-helices are packed against the β-sheet (6; Figure 4). Studies on RNA-bound forms of RRMs have demonstrated that RRMs interacts with RNA through the amino acids located on the β-sheet, including those in the conserved short consensus sequences ribonucleoprotein (RNP) 1 and 2 by which RRM was initially defined (4,5,7–9). It has been shown that RRMs are also involved in protein–protein interactions. In some cases, the RRM interacts with another RRM (17,19,21), while in other cases, the RRM interacts with non-RRM proteins (8,10–16,18,20). Amino acids on the α-helices are involved in most interactions although detailed mode of interaction varies from one case to another. We identified three sequence elements that are essential for ELAV–ELAV interaction, and also sufficient to give the interaction activity to RRM2 when all of them are introduced into it. All the three elements are estimated to be located on or adjacent to the α-helices when we look at the positions of the corresponding amino acids in other RRMs for which the tertiary structures have been determined (Figure 4). This suggests a model in which the ELAV RRM3 interacts with another RRM3 through the surface opposite to the RNA-binding surface of the domain.
In our co-IP experiment, ELAV–ELAV interaction required the presence of RNA. This observation raises the question as to whether the interaction is based on protein–protein interaction or is mediated by RNA. Although the RNA-mediated interaction model could not be excluded completely, the protein–protein interaction model is preferred for the following reasons: (i) the three identified important sequence elements for the interaction are all located away from the putative RNA binding surface; (ii) the mutations on the putative RNA binding surface (mutants eH3mu2 and 5 in Figure 3B) that potentially alter the binding specificity did not interfere with the interaction. It has also been shown that HuD–HuD interaction is greatly reduced with RNase treatment although not completely abolished (35). We hypothesize that a conformational change of the RRM upon RNA binding ensures efficient protein-protein interaction in both ELAV and HuD cases. Interestingly, a recent study on ARE-binding of HuR has revealed that RRM3 is required for cooperative assembly of HuR oligomers on RNA (37). The observation fits the idea that the initial binding of an ELAV-family protein on a target RNA facilitates the protein-protein interaction between RRM3s.
We find that RRM2 with the three critical sequences allows ELAV interaction in the yeast two-hybrid system, but still fails to support the in vivo function. Thus, additional sequences in RRM3 are also necessary for ELAV function. This is consistent with the previous study that suggests that the RNA binding ability of each RRM, including RRM3, is essential, as transgenes that carry mutations that individually disrupt the RNA binding ability of individual RRMs were unable to provide ELAV function (40). Therefore, it is suggested that ELAV RRM3 is a bi-functional domain that interacts with both RNA and protein.
One of the three critical sequence elements for ELAV–ELAV interaction was the single tryptophan at the position 419 (W419). Coincidentally, two independent temperature-sensitive alleles of elav, elavts1 and elavFliJ2, have TAG and TGA stop codons at the site of W419, respectively (50). In these two mutants, ELAV protein with an apparent normal size is produced, along with a truncated form. It is hypothesized that some form of nonsense suppression occurs in these mutants, and probably a different amino acid substitutes the original W419 (50). Further, it is suggested that the temperature sensitivity of the mutants is due to impaired function of the protein, rather than the thermolability of the protein, because the amounts of either forms of ELAV are unaffected by temperature (50). Since W419E substitution strongly reduced multimerization activity in our yeast two-hybrid experiment, we hypothesize that the amino acid substitution resulted from the nonsense suppression compromise multimerization of the protein, which leads to the temperature sensitive phenotype of the mutants.
ELAV-family proteins have been shown to form multimeric complexes on their target RNA (34–37). It is likely that multimerization is an integral part of the mechanism by which ELAV-family proteins carry out their functions. However, the structural basis of the multimer formation remains unknown. In this study, we showed the importance of RRM3 for ELAV multimerization, and identified three sequence elements that are essential for the interaction. Our results suggest a rough model for a ELAV multimer in which ELAV molecules interact with each other by their RRM3s. Further structural and biochemical studies are required to draw the complete picture of the multimeric complex, and understand the physical mechanisms of RNA processing/stabilization by ELAV-family proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank M. Zhadina for help with the two-hybrid assay, P. Parmenter and E. Dougherty for technical assistance. We thank the Bloomington Drosophila Stock Center at Indiana University for providing fly stocks. The project described was supported by Grant Numbers P01NS044232 and P30 NS045713 from the National Institutes of Neurological Disorders and Stroke and Grant Number S10 RR16780 from the National Institute of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institute of Health. The Open Access funding charges for this paper were waived.
Conflict of interest statement. None declared.
REFERENCES
- 1.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 2.Dreyfuss G, Swanson MS, Pinol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 1988;13:86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- 3.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 4.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 5.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 6.Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 7.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 8.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 10.Bentley RC, Keene JD. Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein particle (snRNP) B” protein and through interactions with U2 snRNP-A' protein. Mol. Cell Biol. 1991;11:1829–1839. doi: 10.1128/mcb.11.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fresco LD, Harper DS, Keene JD. Leucine periodicity of U2 small nuclear ribonucleoprotein particle (snRNP) A' protein is implicated in snRNP assembly via protein-protein interactions. Mol. Cell Biol. 1991;11:1578–1589. doi: 10.1128/mcb.11.3.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B” and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 13.Scherly D, Dathan NA, Boelens W, van Venrooij WJ, Mattaj IW. The U2B” RNP motif as a site of protein-protein interaction. EMBO J. 1990;9:3675–3681. doi: 10.1002/j.1460-2075.1990.tb07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 15.Kielkopf CL, Rodionova NA, Green MR, Burley SK. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001;106:595–605. doi: 10.1016/s0092-8674(01)00480-9. [DOI] [PubMed] [Google Scholar]
- 16.Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S. Crystal structure of the human nuclear cap binding complex. Mol. Cell. 2001;8:383–396. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 17.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 18.Selenko P, Gregorovic G, Sprangers R, Stier G, Rhani Z, Kramer A, Sattler M. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell. 2003;11:965–976. doi: 10.1016/s1097-2765(03)00115-1. [DOI] [PubMed] [Google Scholar]
- 19.Shamoo Y, Krueger U, Rice LM, Williams KR, Steitz TA. Crystal structure of the two RNA binding domains of human hnRNP A1 at 1.75 A resolution. Nat. Struct. Biol. 1997;4:215–222. doi: 10.1038/nsb0397-215. [DOI] [PubMed] [Google Scholar]
- 20.Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 2003;17:971–976. doi: 10.1101/gad.260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu RM, Jokhan L, Cheng X, Mayeda A, Krainer AR. Crystal structure of human UP1, the domain of hnRNP A1 that contains two RNA-recognition motifs. Structure. 1997;5:559–570. doi: 10.1016/s0969-2126(97)00211-6. [DOI] [PubMed] [Google Scholar]
- 22.Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am. J. Hum. Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl Acad. Sci. USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campos AR, Grossman D, White K. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J. Neurogenet. 1985;2:197–218. doi: 10.3109/01677068509100150. [DOI] [PubMed] [Google Scholar]
- 26.Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 27.Koushika SP, Lisbin MJ, White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr. Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- 28.Koushika SP, Soller M, White K. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol. Cell Biol. 2000;20:1836–1845. doi: 10.1128/mcb.20.5.1836-1845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisbin MJ, Qiu J, White K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 2001;15:2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soller M, White K. ELAV inhibits 3'-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 2003;17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borgeson CD, Samson ML. Shared RNA-binding sites for interacting members of the Drosophila ELAV family of neuronal proteins. Nucleic Acids Res. 2005;33:6372–6383. doi: 10.1093/nar/gki942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samson ML. Evidence for 3' untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics. 1998;150:723–733. doi: 10.1093/genetics/150.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samson ML, Chalvet F. found in neurons, a third member of the Drosophila elav gene family, encodes a neuronal protein and interacts with elav. Mech. Dev. 2003;120:373–383. doi: 10.1016/s0925-4773(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 34.Soller M, White K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol. Cell Biol. 2005;25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasashima K, Sakashita E, Saito K, Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 2002;30:4519–4526. doi: 10.1093/nar/gkf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao FB, Keene JD. Hel-N1/Hel-N2 proteins are bound to poly(A)+ mRNA in granular RNP structures and are implicated in neuronal differentiation. J. Cell Sci. 1996;109(Pt 3):579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 37.Fialcowitz-White EJ, Brewer BY, Ballin JD, Willis CD, Toth EA, Wilson GM. Specific protein domains mediate cooperative assembly of HuR oligomers on AU-rich mRNA-destabilizing sequences. J. Biol. Chem. 2007;282:20948–20959. doi: 10.1074/jbc.M701751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 39.Yao KM, White K. Organizational analysis of elav gene and functional analysis of ELAV protein of Drosophila melanogaster and Drosophila virilis. Mol. Cell Biol. 1991;11:2994–3000. doi: 10.1128/mcb.11.6.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisbin MJ, Gordon M, Yannoni YM, White K. Function of RRM domains of Drosophila melanogaster ELAV: Rnp1 mutations and rrm domain replacements with ELAV family proteins and SXL. Genetics. 2000;155:1789–1798. doi: 10.1093/genetics/155.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 43.Staehling-Hampton K, Jackson PD, Clark MJ, Brand AH, Hoffmann FM. Specificity of bone morphogenetic protein-related factors: cell fate and gene expression changes in Drosophila embryos induced by decapentaplegic but not 60A. Cell Growth Differ. 1994;5:585–593. [PubMed] [Google Scholar]
- 44.Yao KM, Samson ML, Reeves R, White K. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol. 1993;24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- 45.Yannoni YM, White K. Domain necessary for Drosophila ELAV nuclear localization: function requires nuclear ELAV. J. Cell Sci. 1999;112(Pt 24):4501–4512. doi: 10.1242/jcs.112.24.4501. [DOI] [PubMed] [Google Scholar]
- 46.Toba G, Qui J, Koushika SP, White K. Ectopic expression of Drosophila ELAV and human HuD in Drosophila wing disc cells reveals functional distinctions and similarities. J. Cell Sci. 2002;115:2413–2421. doi: 10.1242/jcs.115.11.2413. [DOI] [PubMed] [Google Scholar]
- 47.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 49.Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc. Natl Acad. Sci. USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samson ML, Lisbin MJ, White K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics. 1995;141:1101–1111. doi: 10.1093/genetics/141.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.