Abstract
Many in vitro and in vivo applications for transgenesis require co-expression of heterologous genes. The use of internal ribosome entry sites (IRESs) in dicistronic expression vectors enables the expression of two genes controlled by one promoter in target cells or whole organisms. Here we describe the expansion of IRES exploitation to generate multicistronic vectors capable of expressing multiple reporter genes, especially to improve the fluorescence yield of autofluorescent reporter gene products such as green fluorescent protein (GFP). We found that the increase in fluorescence output of GFP is proportional to the number of IRES-GFP repeats in the multicistronic vector. At least four genes can be expressed from a multicistonic vector by using tandem IRES elements, with no significant alteration of the expression level of the cap-dependent translated gene. Moreover, gene expression under the control of multiple IRES element has no effect on the posttranscriptional regulation through 3′-untranslated regions (3′UTR). Thus, endogenous gene expression and regulation, especially those controlled by weak promoters, can be analyzed with our IRES-dependent polycistronic reporter gene expression system.
INTRODUCTION
In 1988, Pelletier and Sonenberg (1) as well as Jang et al. (2) demonstrated that the internal ribosome entry site (IRES), originally found in eukaryotic viruses, initiate ribosome binding and translation, in a cap-independent manner. Since then, bicistronic vectors in which the first gene is translated in a cap-dependent manner and the second one in an IRES-dependent manner have been widely used for in vitro and in vivo applications in a variety of experimental settings, from cultured cells and transgenic animals to gene therapy. For example, to link two genes transcribed from a single promoter, one being the gene of interest, and the other a selectable marker or a reporter gene (3–6). Furthermore, IRES-linked expression of fluorescent proteins has been used as reporter in vivo enabling to monitor the expression of target genes without deleting them (7–13).
However, there are limitations using IRES-linked reporter genes. Gene expression downstream of IRES is expressed at lower level as compared with the gene upstream of IRES (14–17), and fluorescence output using IRES-linked green fluorescent protein (GFP) expression is decreased or even absent compared with that found in GFP-fused proteins (18,19). Thus, the opportunity to expand IRES exploitation to generate multicistronic reporter vector capable of expressing multiple genes would open up new possibilities. For example, multiple IRES-enhanced GFP (EGFP) sequences linked in tandem could be used to improve the fluorescence yield. This can be valuable, especially for in vivo targeting of weakly expressed genes with IRES-controlled fluorescent marker.
Although some investigations have employed tagging proteins of interest using multicistronic constructs with multiple cloning sites (MCSs) flanking IRES elements (20–25), no quantitative and systematic studies of this approach have been performed yet. For example, (i) the quantity of the fluorescence output of a fluorescent protein expressed under the control of multiple tandem-IRES compared to single IRES is unknown; (ii) the effect of multiple tandem-IRES on the cap-dependent expressed gene is unclear and (iii) the influence of multiple tandem-IRES on the posttranscriptional regulation of gene expression is also unknown. In the present article we have addressed these questions.
MATERIALS AND METHODS
Vector construction
All vectors were constructed based on the vector backbone pIRES2-EGFP (BD Biosciences Clontech, Palo Alto, CA, USA; Catalog #6029-1) that contains the immediate early promoter of cytomegalovirus (PCMV IE) followed by the IRES of the encephalomyocarditis virus (ECMV), an MCS, the enhanced GFP (EGFP) coding sequence and the SV40 polyadenylation signals (SV40pA) downstream of the EGFP.
The vectors were generated as follows. The luciferase coding sequence was amplified from pGL3-basic (Promega) using the forward primer CGCCTCGAGATGGAAGACGCCAAAAACATAAAG containing XhoI restriction site and the backward primer CGCGAATTCTTACACGGCGATCTTTCCGCCCTTC containing EcoRI restriction site. The product was purified, double digested with XhoI and EcoRI and inserted between the XhoI and EcoRI sites of pIRES2-EGFP. This construct was named pLuc-1x(IRES2EGFP)-SV40pA. The construct pLuc-2x(IRES2EGFP)-SV40pA was created as follows: the IRES2EGFP sequence was amplified from pIRES2EGFP plasmid using the forward primer CGCGCGGCCGCCCGCGGGCCCGGGATCCG containing NotI restriction site and the backward primer CGCGCGGCCGCGATATCTTACTTGTACAGCTCGTCCATGCCGAG containing NotI and EcoRV restriction sites. The insertion of the EcoRV restriction site allows cloning of further fragments downstream of IRES2EGFP, as described subsequently. The PCR-product was purified, digested with NotI and inserted into NotI site, between EGFP sequence and SV40pA, of pLuc-1x(IRES2EGFP)-SV40pA. To generate pLuc-3x(IRES2EGFP)-SV40pA, IRES2EGFP sequence was amplified from pIRES2EGFP using the forward primer CGCGATATCCCGCGGGCCCGGGATCCG and the backward primer CGCGATATCTTACTTGTACAGCTCGTCCATGCCGAG, both containing EcoRV restriction sites. The PCR-product was purified, digested with EcoRV and inserted into the created EcoRV site, between the second EGFP sequence and SV40pA, of pLuc-2×(IRES2EGFP)-SV40pA.
To generate the constructs with the interleukin-10 (IL-10)-3′ untranslated region (UTR) instead of SV40pA, we amplified a 832 base pair (bp) long IL-10-fragment located between the 6th and 838th bp behind the stop codon from mouse genomic DNA which enclose the 3′UTR with the forward primer GCGTCTAGAGTGCAGTGTGTATTGAGTCTGCTGG containing XbaI restriction site and the backward primer GCGCTTAAGCCGAGCTCAACCCCTTCCTGGAG containing AflII restriction site. The SV40pA containing sequence was removed from the constructs pLuc-nx(IRES2EGFP)-SV40pA (n = 1, 2 or 3) by digestion with XbaI and AflII restriction sites. The XbaI/AflII IL10-3′UTR fragment was then inserted the XbaI and AflII of the pLuc-nx(IRES2EGFP) constructs. For the generation of the IL10-3′UTR containing constructs, to prevent Dam methylation of the Xba I sites, the constructs passed through a dam−E. coli mutant (ATCC 47045).
Cell culture and transfection
Prior to transfection, human embryonic kidney (HEK) 293 cell line were seeded in 6-well plates at 5 × 105 cells per well in RPMI 1640 medium with l-glutamine (0.3 g/l), 10% fetal bovine serum (FCS), 10 mM HEPES and penicillin (100 U/ml)–streptomycin (100 μg/ml) (Gibco/Invitrogen). The cells were allowed to attach overnight. A total of 0.9 pmol of each generated construct were transfected separately into HEK 293 using jetPEI™ transfection reagent (Qbiogene Molecular Biology). Selection with G418 (1 mg/ml) was started 48 h after transfection. Transfectants were obtained after 4–6 weeks and maintained under continued selection (0.4–1 mg/ml G418).
Flow cytometry analysis
Cells were washed twice with phosphate buffered saline (PBS) (Gibco/Invitrogen) and harvested by using 1×trypsin ethylene diamine tetra-acetic acid (trypsin–EDTA) (Gibco/Invitrogen). Equal cell numbers were analysed for EGFP fluorescence by a fluorescence-activated cell sorter (FACS, BD FACSCanto). Background fluorescence was determined using non-transfected cells. A total of 10 000 events were collected using list-mode format for each experiment. Cells were gated on EGFP-expressing population. Data were acquired and analyzed using FlowJo-8.4.6 Software (Tree Star Inc; San Carlos, CA, USA). The mean values (represent the peak integrals) of fluorescence for gated EGFP-expressing populations were determined.
Western blot analysis
Equal numbers of cells (5–10 × 106) were washed twice in ice-cold PBS, resuspended in lysis-buffer (50 mM Tris–Hcl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 2% chaps or 1% Triton X-100) and incubated on ice for 15–30 min. Cell debris was removed by centrifugation at 14 000g, 4°C for 10 min and the supernatants were transferred into new tubes. The protein concentration was determined by using BCA Protein Assay Kit (Pierce). Equal amounts of protein lysates were subjected to 12% SDS–PAGE under reducing conditions and transferred to nitrocellulose (NC) membrane by semi-dry electroblotting. The NC-membrane was blocked (PBS; 5% dry milk powder; 0.1% Tween-20) for 1–2 h at room temperature and incubated overnight at 4°C with anti-GFP or anti-actin antibodies (Santa Cruz Biotechnology). The antibodies were used in a final concentration of 0.4 µg/ml. Bands were visualized with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology) and enhanced chemiluminescence substrate. All antibodies were diluted in PBS, 5% FCS and 0.1% Tween-20.
Preparation of total RNA and cDNA synthesis
Total RNA was extracted from cells using the RNA MasterPure RNA Purification kit (Biozym). Contaminating genomic DNA was removed by treatment of the RNA preparations with Baseline-Zero DNase (Biozym). mRNA was stored at −80°C. cDNA was generated from total RNA using the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas), according to manufacturer's instructions. One microgram total RNA was used for each reaction and synthesis was primed by Oligo(dT)18 primers. For negative controls, identical set-ups of reverse transcriptase-polymerase chain reaction (RT-PCR) reactions except reverse transcriptase were performed. This negative control was performed to exclude products amplified from a potential genomic DNA contamination. One microliter of cDNA product was used for a subsequent PCR using 2×BioMix Red (Bioline) in a final volume of 30 μl. The expression level of the multicistronic vectors was detected after 30 amplification cycles using the following primer for luciferase: the forward primer, Luc-F: GGCGCTCCCCTCTCTAAGGA and the backward primer, Luc-B: GGGGTGTTGGAGCAAGATGG. As a loading control, the housekeeping gene, hypoxanthine-guanine-phosphoribosyltransferase (HPRT), was amplified using the primer set: HumHPRT-F: CAGCCCTGGCGTCGTGATTAGTGATG and HumHPRT-B: GGGAACTGATAGTCTATAGGCTCATAGTGC.
Preparation of genomic DNA and Southern blot analysis
Cells were seeded on 6-well plates and cultured until reaching complete confluence (∼5 × 106 cells). Supernatants were then aspirated and 1.5 ml lysis buffer [100 mM Tris–HCl (pH 8.5); 0.2% SDS; 200 mM NaCl; 5 mM EDTA (pH 8.0); 100 μg/ml Proteinase K (freshly added)] was added to each well. After incubation at 37°C for at least 8 h, equal volume (1.5 ml) of isopropanol was added to each well and the plates were incubated on a shaker at 300 r.p.m. and RT for at lest 3 h, until the DNA became visible. DNA were fished with pipette tip, transferred into 150 µl TE-buffer [10 mM Tris–HCl (pH 8); 1 mM EDTA (pH 8)] and incubated at 55°C overnight to allow the dissolution of DNA. The DNA was stored at room temperature (RT) until use. Forty microliter from each DNA sample was digested with 50 U EcoRI (Fermentas) overnight. The digested DNA samples were then fractionated on 0.7 agarose gel in 1×TBE (89 mM Tris–borate, 89 mM boric acid, 2 mM EDTA, pH 8). The gel was treated successively with depurination solution (0.2 M HCl), denaturing buffer (0.5 M NaOH; 1.5 M NaCl) and neutralization buffer (0.5 M Tris–HCl; 1.5 M NaCl; pH 7.2) for ∼45 min, respectively. The gel was subsequently blotted overnight to a positive charged Nylon membrane (Schleicher and Schuell) using 20× SSC (3 M NaCl; 0.3 M tri-Sodium Citrate 2-hydrate; pH 7.2). Cross-linking of the DNA to the membrane was induced by irradiation with a UV lamp for 50 s. The membrane was incubated with hybridization solution [50% Formamide; 5× SSC; 10 mM Tris–HCl (pH 7.5); 1% SDS; 5× Denhardts (2% Ficoll 400; 2% Polyvinylpyrrolidone K30; 2% BSA); 10% Dextransulfate; 100 μg/ml salmon sperm DNA] for 1 h. α32P-dATP-labeled luciferase probe was then added to the hybridization solution and incubated overnight at 42°C. The luciferase probe was labeled using HexaLabel & trade DNA Labeling Kit (Fermentas) according to manufacturer's instructions. The blot was subsequently washed two times with wash-buffer 1 (2× SSC; 0.1% SDS) for 10 min at RT, then two times with wash buffer 2 (0.5× SSC; 0.4% SDS) for 20 min at 70°C. Radioactive signals were detected with Fuji Film Scanner FLA-3000 and analyzed with Aida Image Analyser v.4.00 software.
Luciferase assay
Equal numbers of cells were harvested, washed with PBS, resuspended in luciferase-lysis buffer (Luc-Buffer: 80 mM K2HPO4; 20 mM KH2PO4; 2 mM EDTA; 1% TritonX-100; pH adjusted to 7.8 with 1 M KOH; 1 mM DTT was added freshly before use) and incubated on ice for 15–30 min. Cell debris was removed by centrifugation at 14 000g, 4°C for 10 min and the supernatants were transferred into new tubes. The protein concentration was determined by using BCA Protein Assay Kit (Pierce). Prior to luciferase assay, bovine serum albumin (BSA) was added to a final concentration of 0.5%. The cell lysates were stored at −20°C. Equal amounts of protein lysates (10–60 µg) were transferred into a white microtiter plate (volumes were adjusted to 50–100 µl with Luc-Buffer/0.5% BSA). One volume of Luciferase Assay Reagent (20 mM Tricine; 1.07 mM (MgCO3)4Mg(OH)2.5H2O; 0.1 mM EDTA; pH adjusted to 7.8 with 1 M HCl; 33.3 mM DTT; 270 µM Li3-Coenzyme A; 470 µM d-luciferin; 530 µM Mg-ATP) was added to the cell lysate and chemiluminescence was measured immediately with a MicroLumatPlus LB96V microplate luminometer (EG&G Berthold). All experiments were performed independently at least three times.
RESULTS
Improvement of the fluorescence yield by expressing EGFP under the control of multiple tandem-IRES
To test whether an increase in fluorescence output of EGFP can be achieved by expressing EGFP from a multicistronic vector using the IRES of the ECMV in tandem, three constructs, named pLuc-nx(IRES2-EGFP)-SV40-pA (n=1, 2 or 3), were generated which contain one, two or three IRES-EGFP repeats (Figure 1A), respectively and stably expressed in HEK 293 cells.
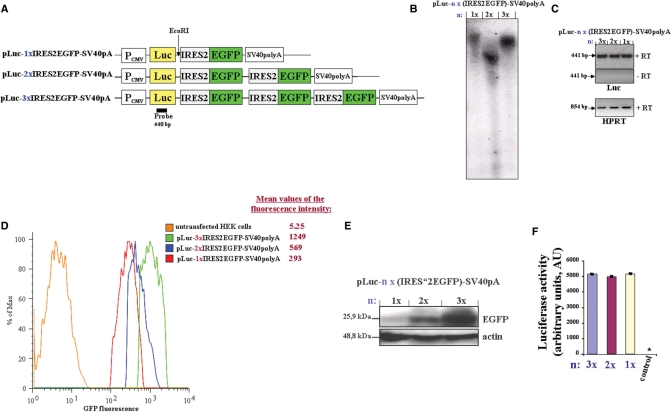
Figure 1.
(A) Schematic representation of the first three constructs used in the studies. The expression vectors were constructed as described in ‘Materials and methods’ section. The EcoRI restriction site and luciferase probe used for Southern blot analysis are shown. The constructs pLuc-nx(IRES2-EGFP)-SV40-pA (n = 1, 2 or 3) were stably transfected in HEK293 cells. (B) Southern blot analysis shows a single insertion of each construct into the genome of the generated three cell lines. (C) The expression level of the multicistronic RNA from the different constructs was analyzed by RT-PCR for luciferase (Luc). (+RT) and (−RT) refer to PCR product from first-strand cDNA synthesis reaction performed from Baseline-Zero DNase-treated total RNA with (+RT) or without (−RT, negative control) reverse transcriptase, respectively. As a loading control, RT-PCR of the housekeeping gene, HPRT, was used. (D) The fluorescence emission of EGFP were analyzed by flow cytometry (using BD FACSCanto). Shown are histograms gated on EGFP-positive cells. The mean values (peak integrals) of fluorescence for the gated EGFP-expressing cells are indicated. (E) Equal amounts of whole cell lysates were separated by SDS–PAGE and the expression level of EGFP were analyzed by western blot using EGFP-antibody. Actin immunostaining is shown as the loading control. (F) Equal amounts of whole cell lysates (10–60 µg) were analyzed for luciferase activity. Control: non transfected HEK293 cells, *, value <50 AU. Experiments were repeated at least three times and similar results were obtained. PCMV, cytomegalovirus promoter; Luc, luciferase gene; SV40polyA, Simian virus polyadenylation site.
To determine the copy number of inserted constructs, genomic DNA was isolated from the HEK 293 cell lines, digested with EcoRI and subjected to Southern blot analysis using a luciferase probe. The pLuc-nx(IRES2-EGFP)-SV40-pA constructs contain only one EcoRI restriction site, directly downstream of the luciferase gene (Figure 1A). The Southern blot analysis showed a single insertion of each construct into the genome of the cell lines (Figure 1B). Furthermore, the analysis of mRNA expression by RT-PCR showed no significant difference in the expression level of the three constructs (Figure 1C), as well no loss in the integrity of the mRNA, at least for the tetracistronic vector (data not shown). Thus, the fluorescence output of EGFP will be dependent on the number of IRES-EGFP repeats in each construct and on their translation efficiency and not on different genomic integration efficiency.
Flow cytometry analysis of the established three cell lines revealed a linear amplification of spectral yield of EGFP (Figure 1D). The mean fluorescence intensity for EGFP (Figure 1D and data not shown) showed that the increase in fluorescence output is proportional to the number of IRES-EGFP repeats. Western blot analysis of whole cell lysates from those cell lines confirmed the increase of EGFP-expression (Figure 1E).
Multiple tandem-IRES do not affect the upstream cap-dependent gene expression
A gene encoding luciferase was inserted right after PCMV and upstream of the first IRES into the three constructs (pLuc-nx(IRES2-EGFP)-SV40-pA; Figure 1A) in order to quantitatively analyze cap-dependent gene expression. If the IRES-dependent translations would interfere with the cap-dependent translation, the luciferase activity should be reciprocated proportional to the number of inserted IRES-EGFP repeats. Equal amounts of whole cell lysates from the three cell lines were collected and analysed for luciferase activity. Interestingly, the luciferase reporter activities were similar for the three different constructs (Figure 1F) indicating that multiple tandem-IRES do not interfere with cap-dependent gene expression. Since the cap-dependent luciferase activity is not affected by the number of IRESs, it can be used as an internal normalization control to eliminate experimental variability, including variations in transfection efficiency.
Multiple tandem-IRES do not influence the posttranscriptional regulation through sequences in the 3′UTRs
The ‘adenylate/uridylate (AU)-rich elements’ (AREs) are well-known RNA sequences in 3′UTRs, which regulate the stability of the mRNA and hence the expression level of the corresponding protein (26). We therefore asked whether the presence of several exogenous IRES sequences in the 3′ region of mRNA would interfere with the 3′UTR function.
To test this, we replaced the RNA-stabilizing SV-40polyA by the IL-10-3′UTR of the pLuc-nx(IRES2-EGFP)-SV40-pA constructs (Figure 2A). IL-10-3′UTR is known to accelerate the degradation of IL-10-mRNA (27). We used a 832 bp long mouse IL-10-fragment spanning the 6th and 838th bp downstream of the stop codon, which include the 3′UTR sequence.
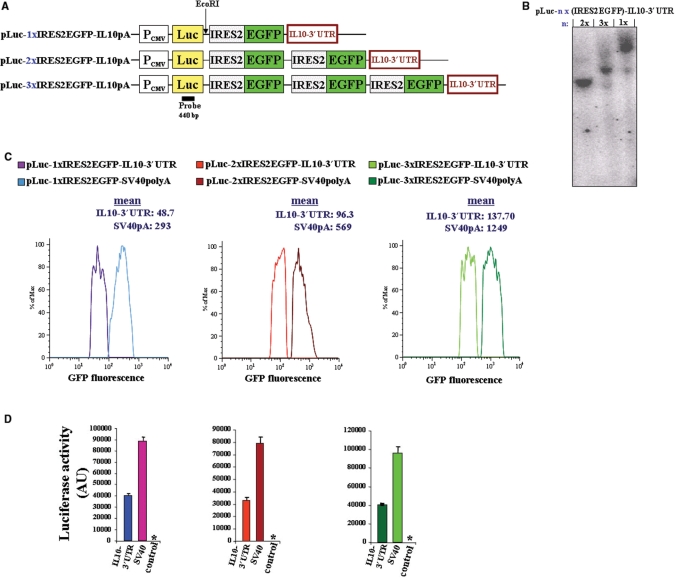
Figure 2.
(A) Schematic representation of the pLuc-nx(IRES2-EGFP)-IL10-3′UTR constructs (n = 1, 2 or 3). The expression vectors were constructed as described in ‘Materials and methods'section. The EcoRI restriction site and luciferase probe used for Southern blot analysis are shown. The constructs were stably transfected in HEK293 cells. (B) Southern blot analysis shows a single insertion of each construct into the genome of the generated three cell lines. (C) Comparison of the fluorescence emission of EGFP between SV40polyA and IL10-3′UTR-containing constructs were determined by flow cytometry. Shown are histograms gated on EGFP-positive cells. The mean values (peak integrals) of fluorescence for the gated EGFP-expressing cells are indicated. (D) Equal amounts of whole cell lysates from all six cell lines expressing the indicated construct were analyzed for luciferase activity. Control, nontransfected HEK293 cells; *, value <15 AU. PCMV, cytomegalovirus promoter; SV40polyA, Simian virus polyadenylation site.
To determine the copy number of inserted constructs, genomic DNA was isolated from the HEK 293 cell lines, digested with EcoRI and subjected to Southern blot analysis using a luciferase probe. Similar to the pLuc-nx(IRES2-EGFP)-SV40-pA expressing cell lines, the Southern blot analysis showed a single insertion of each construct into the genome of the pLuc-nx(IRES2-EGFP)-IL10-3′UTR expressing cell lines (Figure 2B). Thus, the expression level of the different constructs will be dependent on posttranscriptional regulation through the sequences in the 3′-untranslated region.
Flow cytometry analysis of the cell lines stably expressing multicistronic vectors containing IL-10-3′UTR showed decreased fluorescence intensity compared to SV40polyA containing constructs (Figure 2C). The insertion of the luciferase gene upstream of the IRES allowed a second readout system for gene expression monitoring luciferase reporter activities. Exchange of SV-40polyA for an IL-10-3′UTR, results in the loss of luciferase activities by more than 50% (Figure 2D). This is in good agreement with results by Powell et al. (27). Thus, multiple tandem-IRES do not interfere with the posttranscriptional regulation of gene expression through sequences in the 3′UTR.
The proportional increase in fluorescence intensity of EGFP is independent of the posttranscriptional regulation by 3′ UTR
With the pLuc-nx(IRES2-EGFP)-SV40-polyA constructs the increase in fluorescence output was proportional to the number of IRES-EGFP repeats (Figure 1D). We wondered whether similar result can be reached with the IL10-3′UTR containing constructs, which show an altered expression regulation compared to SV40polyA containing constructs (Figure 2).
The analysis of mRNA expression by RT-PCR showed no significant difference in the expression level of the three different constructs (Figure 3A). Furthermore, the Southern blot analysis showed a single insertion of each construct into the genome of the pLuc-nx(IRES2-EGFP)-IL10-3′UTR expressing cell lines (Figure 2B). Thus, the fluorescence output of EGFP will be dependent on the number of IRES-EGFP repeats and on their translation efficiency and not on different genomic integration efficiency.
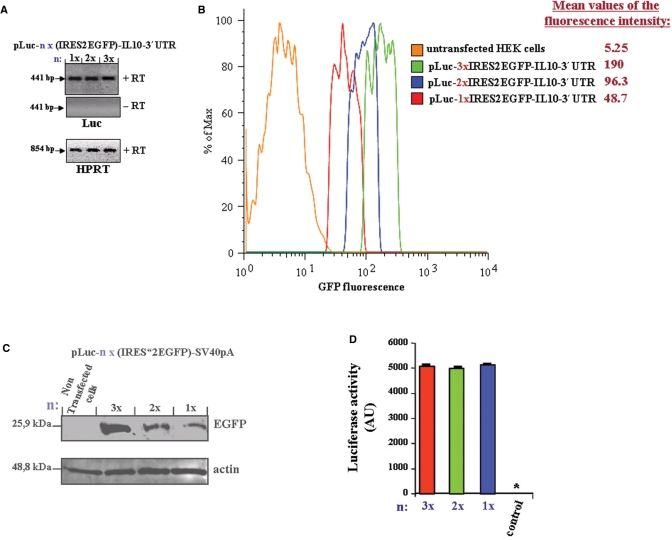
Figure 3.
Stably transfected HEK293 cells with the constructs pLuc-nx(IRES2-EGFP)-IL10-3′UTR, containing one (n = 1), two (n = 2) or three (n = 3) IRES-EGFP repeats were established and (A) The expression level of the multicistronic RNA from the different constructs was analyzed by RT-PCR. (+RT) and (−RT) refer to PCR product from first-strand cDNA synthesis reaction performed from Baseline-Zero DNase-treated total RNA with (+RT) or without (−RT, negative control) reverse transcriptase (RT), respectively. As a loading control, RT-PCR of the housekeeping gene, HPRT, was used. (B) The fluorescence emission of EGFP from the three cell lines were analyzed by flow cytometry. Shown are histograms gated on EGFP-positive cells. The mean values (peak integrals) of fluorescence for the gated EGFP-expressing cells are indicated. (C) Equal amounts of whole cell lysates were separated by SDS–PAGE and the expression level of EGFP were analyzed by western blot using EGFP-antibody. Actin immunostaining is shown as the loading control. (D) Equal amounts of whole cell lysates from all three cell lines were analyzed for luciferase activity. Control, nontransfected HEK293 cells; *, value <15 AU.
Flow cytometry analysis of the established HEK293 cell lines, expressing multicistronic vectors containing IL10-3′UTR and one, two or three IRES-EGFP repeats, respectively, showed a proportional increase in fluorescence output, dependent on the number of IRES-EGFP repeats (Figure 3B). Western blot analysis of whole cell lysates from those three cell lines confirmed the increase of EGFP-Expression (Figure 3C).
Furthermore, the luciferase reporter activities that are cap-dependent, were similar between the cell lysates from the three cell lines (Figure 3D).
Thus, independent of the posttranscriptional regulation through sequences in the 3′-UTR, the increase in fluorescence output of EGFP is proportional to the number of IRES-EGFP repeats, without altering the cap-dependent gene expression.
DISCUSSION
Viral IRES elements can be fused with target genes resulting in the coexpression of multiple proteins from the same mRNA (3–6,28). However, cistrons downstream of IRES are often much less efficiently translated than those directly following the promoter (14–16 and Bouabe, H., unpublished data), and subsequently IRES-directed gene expression using nonenzymatic fluorescent reporters, such as EGFP, result in reduced or even absent fluorescence emission (18,19,29,30). If gene expression from weak promoter should be monitored, the IRES-linked fluorescent reporter gene is usually not ideal.
Here we report a technology that allows improvement of reporter gene activity using multiple IRES-EGFP sequences linked in tandem. Di-, tri- and tetra-cistronic vectors were constructed to express simultaneously luciferase (Luc) gene in a cap-dependent and one, two or three EGFP genes in an IRES-dependent manner, respectively. We used an EMCV-IRES (BD Biosciences Clontech). Flow cytometry and western blot analysis demonstrated linear amplification of fluorescence signal and expression level of EGFP, respectively, which were proportional to the number of IRES-EGFP repeats. Thus, to compensate the IRES-dependent decreased expression, several copies of a gene of interest can be expressed from a multicistronic vector by using tandem IRES elements.
Our results also indicate that all three cistrons following the IRESs are equally translated. Thus, there is no significant influence of each of the IRES-mediated translation on the translation efficiency mediated by the other IRES. Similar results are reported by other observations using di- and tri-cistronic vectors (31,32). Thus the translations of all three EGFP cistrons downstream of IRESs may occur independent of each other.
Translation of capped cellular mRNAs is initiated by recruitment of ribosomes to the 5′end of mRNAs via eukaryotic translation initiation factors (eIF)4E, eIF4A and eIF4G. Translation initiation on most IRES-containing mRNAs, such as EMCV mRNA, requires the same canonical eIFs that are required for translation of capped mRNAs, except for eIF4E (33). It is suggested that IRES containing EMCV mRNA competes with capped cellular mRNA for the recruitment of eIF4G and subsequently for translation (34). This raises the question, whether multiple tandem-IRES in multicistronic RNA affect the cap-dependent gene expression.
Recently, a generated knock-in mouse model, where an IRES-luciferase-IRES-EGFP cassette was inserted into the UTR of the endogenous transcription factor forkhead box P3 (Foxp3) locus, resulting in attenuated expression of the cap-dependent translated endogenous Foxp3 gene (35). The authors assumed that four AREs found dispersed in the luciferase complementary DNA caused the destabilization of the Foxp3 mRNA and subsequently reduced Foxp3 expression.
However, under our experimental conditions, whether one, two or three IRES-EGFP cassettes are inserted downstream of the Luc-gene, the cap-dependent luciferase expression remains similar. Thus, multiple tandem-IRES do not affect the cap-dependent gene expression. In line with our results, Hennecke et al. (36) reported that cap-dependent translation from bicistronic mRNAs remains comparable to monocistronic expression. Nevertheless, it remains to be elucidated, whether this is also valid for other cell types, and in particular for cells under stress conditions, which are known to modulate the availability of eIF4E for cap-dependent translation (34,37)
Posttranscriptional events are central to the regulation of eukaryotic gene expression [reviewed in (38,39)]. They act on the modulation of mRNA stability and/or translation efficiency via the binding of heterogeneous nuclear ribonucleoproteins (hnRNPs) to specific RNA sequences, which are often present in the 3′-UTR (40–42). The AREs are the best-known RNA sequences of the 3′UTR (43). They can bind diverse proteins that either stabilize the mRNA, such as HuR, or accelerate their degradation, such as AUF1 protein and Tristetraprolin (26,44,45), thereby enhancing or decreasing the corresponding translation. Thus, considering on one hand several exogenous IRES sequences present in the 3′ region of the mRNA, which recruit and bind diverse proteins and ribosome complexes, and on the other hand that cellular 3′UTR and IRES are modulated, in part, by common RNA-binding proteins, like polypyrimidine tract-binding protein (PTB) (5,46,47), it seems very likely that multiple tandem-IRES in an mRNA may influence its posttranscriptional regulation through 3′UTR.
We therefore compared the effect of multiple IRESs on the 3′UTR-mediated posttranscriptional regulation by inserting an IL-10-3′UTR as RNA-destabilizing fragment, instead of the SV-40polyA in the pLuc-nx(IRES2-EGFP) constructs. Whether one, two or three IRES-EGFP cassettes are inserted upstream of the 3′UTR, the IL10-3′UTR retains its ability to decrease the expression level of EGFP and luciferase. Thus, multiple tandem-IRES do not affect the posttranscriptional regulation of gene expression through sequences in the 3′UTRs.
Several studies have established that the IRES element of EMCV is functional in a variety of cell types and in whole animals. Using gene targeting strategies several reporter mice have been generated, where the expression of a gene of interest is linked via IRES element with EGFP. These mice enable us to determine which cells express the target gene in vivo (7–13). However, there are several limitations using IRES-linked EGFP knock-in mice. Only some of them are mentioned subsequently. First, IRES-dependent gene expression ranged between 20% and 50% that of the cap-dependent gene (18 and Bouabe, H., unpublished data). Second, in contrast to transgenic animals with several copies of a transgene supplied by retroviruses or microinjection, there are only one or two copies of the IRES-EGFP reporter gene in the genome of knock-in mice generated by gene targeting, thus the IRES-dependent decreased expression cannot be compensated for by high copy number of the transgene. Third, the expression level of the EGFP reporter is dependent on the cell- and tissue-types, as well as on the promoter capacity of the target gene. For these reasons and in view of EGFP's limit of detection that is approximately 105 molecules/cell (48), care should be taken regarding the decreased capacity of IRES-dependent EGFP reporter gene expression. It is very likely that IRES-linked expression of EGFP would not mark all cells expressing the target gene, especially that expressed under weak promoter. Subsequently, the reporter mice would provide incomplete gene expression profile.
In this study, we provide a reporter system that can help to overcome these limitations, at least to some extent, by using multiple IRES-EGFP sequences linked in tandem. Multiple tandem-IRES can enhance the reporter gene activity with no significant effect on the expression regulation of the target gene upstream of IRES. Thus, they perform essential prerequisites allowing their use for in vivo studies.
To evaluate the multiple tandem-IRES-based reporter system in vivo, we used the gene targeting strategy to integrate multicistronic vectors in a specific genomic site of embryonic stem (ES) cells. The reporter mice that we are generating from these modified ES-cell lines, will allow study of the expression behavior of IRES-mediated polycistronic reporter system in vivo.
Finally, in biochemical and cellular studies, a defined expression level of the genes of interest is required. Overexpression can lead to aggregation or to modified function and/or subcellular localization of the protein of interest. IRES-dependent polycistronic gene expression opens up the possibility, to titrate the protein expression level by expressing the gene of interest under the optimal IRES-tandem repeats.
ACKNOWLEDGEMENTS
We are grateful to Dr Cemalettin Bekpen for critical reading of the manuscript. Hicham Bouabe was supported by the DFG Graduate College “Infection and Immunity”. Funding to pay the Open Access publication charges for this article was provided by Max von Pettenkofer Institut, Germany.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 2.Jang SK, Krausslich HG, Nicklin MJH, Duke GM, Palmenberg AC, Wimmer E. A Segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mountford PS, Smith AG. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet. 1995;5:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallardo HF, Tan C, Sadelain M. The internal ribosomal entry site of the encephalomyocarditis virus enables reliable coexpression of two transgenes in human primary T lymphocytes. Gene Ther. 1997;10:1115–1119. doi: 10.1038/sj.gt.3300506. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Salas E. Internal ribosome entry site biology and its use in expression vectors. Curr. Opin. Biotechnol. 1999;5:458–464. doi: 10.1016/s0958-1669(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 6.Attal J, Theron MC, Houdebine LM. The optimal use of IRES (internal ribosome entry site) in expression vectors. Genet. Anal. 1999;15:161–165. doi: 10.1016/s1050-3862(99)00021-2. [DOI] [PubMed] [Google Scholar]
- 7.Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a Bicistronic IL-4 Reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- 8.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;7:2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- 9.Im W, Kim H, Yun D, Seo SY, Park SH, Locksley RM, Hong S. Cytokine reporter mouse system for screening Novel IL12/23 p40-inducing compounds. Mol. Cells. 2005;20:288–296. [PubMed] [Google Scholar]
- 10.Mohrs K, Wakil AE, Killeen N, Locksley RM, Mohrs M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity. 2005;23:419–429. doi: 10.1016/j.immuni.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. PNAS. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhardt RL, Hong S, Kang SJ, Wang Z, Locksley RM. Visualization of IL-12/23p40 in vivo reveals immunostimulatory dendritic cell migrants that promote Th1 differentiation. J. Immunol. 2006;177:1618–1627. doi: 10.4049/jimmunol.177.3.1618. [DOI] [PubMed] [Google Scholar]
- 13.Kamanaka M, Kim ST, Wan YY, Sutterwala FS, Lara-Tejero M, Galan JE, Harhaj E, Flavell RA. Expression of interleukin-10 in intestinal lymphocytes detected by an interleukin-10 reporter knockin tiger mouse. Immunity. 2006;25:941–952. doi: 10.1016/j.immuni.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman RJ, Davies MV, Wasley LC, Michnick D. Improved vectors for stable expression of foreign genes in mammalian cells by use of the untranslated leader sequence from EMC virus. Nucleic Acids Res. 1991;16:4485–4490. doi: 10.1093/nar/19.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houdebine LM, Attal J. Internal ribosome entry sites (IRESs): reality and use. Transgenic Res. 1999;3:157–177. doi: 10.1023/a:1008909908180. [DOI] [PubMed] [Google Scholar]
- 16.Dirks W, Wirth M, Hauser H. Dicistronic transcription units for gene expression in mammalian cells. Gene. 1993;2:247–249. doi: 10.1016/0378-1119(93)90569-o. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Aran J, Gottesman MM, Pastan I. Co-expression of human adenosine deaminase and multidrug resistance using a bicistronic retroviral vector. Hum. Gene. Ther. 1998;9:287–293. doi: 10.1089/hum.1998.9.3-287. [DOI] [PubMed] [Google Scholar]
- 18.Mizuguchi H, Xu Z, Ishii-Watabe A, Uchida E, Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol. Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Zhan X, D’Costa J, Tanavde VM, Ye Z, Peng T, Malehorn MT, Yang X, Civin CI, et al. Lentiviral vectors with two independent internal promoters transfer high-level expression of multiple transgenes to human hematopoietic stem-progenitor cells. Mol. Ther. 2003;7:827–838. doi: 10.1016/s1525-0016(03)00104-7. [DOI] [PubMed] [Google Scholar]
- 20.Fussenegger M, Mazur X, Bailey JE. pTRIDENT, a novel vector family for tricistronic gene expression in mammalian cells. Biotechnol. Bioeng. 1998;1:1–10. doi: 10.1002/(sici)1097-0290(19980105)57:1<1::aid-bit1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 21.Fussenegger M, Moser S, Bailey JE. pQuattro vectors allow one-step multigene metabolic engineering and auto-selection of quattrocistronic artificial mammalian operons. Cytotechnology. 1998;7:229–235. doi: 10.1023/A:1008014706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fussenegger M, Moser S, Mazur X, Bailey JE. Autoregulated multicistronic expression vectors provide one-step cloning of regulated product gene expression in mammalian cells. Biotechnol. Prog. 1997;6:733–740. doi: 10.1021/bp970108r. [DOI] [PubMed] [Google Scholar]
- 23.Moser S, Schlatter S, Fux C, Rimann M, Bailey JE, Fussenegger M. An update of pTRIDENT multicistronic expression vectors: pTRIDENTs containing novel streptogramin-responsive promoters. Biotechnol. Prog. 2000;5:724–235. doi: 10.1021/bp000077r. [DOI] [PubMed] [Google Scholar]
- 24.Azzouz M, Martin-Rendon E, Barber RD, Kyriacos A, Mitrophanous KA, Carter EE, Rohll JB, Kingsman SM, Kingsman AJ, et al. Multicistronic lentiviral vector-mediated striatal gene transfer of aromatic L-amino acid decarboxylase, tyrosine hydroxylase, and GTP cyclohydrolase I induces sustained transgene expression, dopamine production, and functional improvement in a rat model of Parkinson's disease. J. Neurosci. 2002;23:10302–10312. doi: 10.1523/JNEUROSCI.22-23-10302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li T, Zhang J. Stable expression of three genes from a tricistronic retroviral vector containing a picornavirus and 9-nt cellular internal ribosome entry site elements. J. Virol. Methods. 2004;2:137–144. doi: 10.1016/j.jviromet.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 26.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol. Life Sci. 2001;2:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell MJ, Thompson SA, Tone Y, Waldmann H, Tone M. Posttranscriptional regulation of IL-10 gene expression through sequences in the 3′-untranslated region. J. Immunol. 2000;1:292–296. doi: 10.4049/jimmunol.165.1.292. [DOI] [PubMed] [Google Scholar]
- 28.Fussenegger M, Bailey JE, Hauser H, Mueller PP. Genetic optimization of recombinant glycoprotein production by mammalian cells. Trends Biotechnol. 1999;1:35–42. doi: 10.1016/s0167-7799(98)01248-7. [DOI] [PubMed] [Google Scholar]
- 29.Ward CM, Stern PL. The human cytomegalovirus immediate-early promoter is transcriptionally active in undifferentiated mouse embryonic stem cells. Stem Cells. 2002;5:472–475. doi: 10.1634/stemcells.20-5-472. [DOI] [PubMed] [Google Scholar]
- 30.Chung S, Andersson T, Sonntag KC, Bjorklund L, Isacson O, Kim KS. Analysis of different promoter systems for efficient transgene expression in mouse embryonic stem cell lines. Stem Cells. 2002;2:139–145. doi: 10.1634/stemcells.20-2-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu J, Musco ML, Grace MJ. Three-color flow cytometry analysis of tricistronic expression of eBFP, eGFP, and eYFP using EMCV-IRES linkages. Cytometry. 1999;1:51–59. [PubMed] [Google Scholar]
- 32.Li J, Menzel C, Meier D, Zhang C, Dubel S, Jostock T. A comparative study of different vector designs for the mammalian expression of recombinant IgG antibodies. J. Immunol. Methods. 2007;318:113–124. doi: 10.1016/j.jim.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell Biol. 1996;12:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell Biol. 2005;23:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;7129:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 36.Hennecke M, Kwissa M, Metzger K, Oumard A, Kröger A, Schirmbeck R, Reimann J, Hauser H. Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res. 2001;16:3327–3334. doi: 10.1093/nar/29.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;7025:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 38.Hazelrigg T. The destinies and destinations of RNAs. Cell. 1998;4:451–460. doi: 10.1016/s0092-8674(00)81613-x. [DOI] [PubMed] [Google Scholar]
- 39.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;5740:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 40.Blaxall BC, Pende A, Wu SC, Port JD. Correlation between intrinsic mRNA stability and the affinity of AUF1 (hnRNP D) and HuR for A+U-rich mRNAs. Mol. Cell Biochem. 2002;232:1–11. doi: 10.1023/a:1014819016552. [DOI] [PubMed] [Google Scholar]
- 41.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 2003;3:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 42.Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;4:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;11:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 44.Malter JS. Identification of an AUUUA-specific messenger RNA binding protein. Science. 1989;4930:664–666. doi: 10.1126/science.2814487. [DOI] [PubMed] [Google Scholar]
- 45.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 46.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;18:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 47.Martinez-Salas E, Ramos R, Lafuente E, Lopez de Quinto S. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 2001;5:973–984. doi: 10.1099/0022-1317-82-5-973. [DOI] [PubMed] [Google Scholar]
- 48.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]