Abstract
The selective and potent inhibition of mitochondrial translation in Saccharomyces cerevisiae by pentamidine suggests a novel antimicrobial action for this drug. Electrophoresis mobility shift assay, T1 ribonuclease footprinting, hydroxyl radical footprinting and isothermal titration calorimetry collectively demonstrated that pentamidine non-specifically binds to two distinct classes of sites on tRNA. The binding was driven by favorable entropy changes indicative of a large hydrophobic interaction, suggesting that the aromatic rings of pentamidine are inserted into the stacked base pairs of tRNA helices. Pentamidine binding disrupts the tRNA secondary structure and masks the anticodon loop in the tertiary structure. Consistently, we showed that pentamidine specifically inhibits tRNA aminoacylation but not the cognate amino acid adenylation. Pentamidine inhibited protein translation in vitro with an EC50 equivalent to that binds to tRNA and inhibits tRNA aminoacylation in vitro, but drastically higher than that inhibits translation in vivo, supporting the established notion that the antimicrobial activity of pentamidine is largely due to its selective accumulation by the pathogen rather than by the host cell. Therefore, interrupting tRNA aminoacylation by the entropy-driven non-specific binding is an important mechanism of pentamidine in inhibiting protein translation, providing new insights into the development of antimicrobial drugs.
INTRODUCTION
Pentamidine, an aromatic diamidine, has a long history of involvement in the treatment of human protozoan infections (1), and is currently still the clinical drug of choice against African trypanosomiasis, antimony resistant leishmaniais and Pneumocystis carinii pneumonia (PCP) (2–4). PCP is an opportunistic infection in patients with acquired immune deficiency syndrome (AIDS) and other immunocomprised states (5). The selective accumulation of pentamidine and other diamidines by the pathogen, rather than the host cell, is a major reason for the drug selectivity; furthermore, acquired resistance is frequently the result of changes in the transmembrane transport of the drug (6,7). Despite its broad profile of antimicrobial activity, the use of this drug is limited because of its side effects, including nephrotoxicity (1,8). The incidence of kidney toxicity with parenteral pentamidine therapy is quite high, although the damage from this side effect is reversible (9,10).
Several mechanisms have been suggested for pentamidine action against different microbes, but the precise mechanism of its action and its major macromolecular targets have not been entirely elucidated (11–16). Pentamidine has been found to precipitate with DNA, RNA and nucleotides (17). However, only its action in binding to DNA has received a large attention in the past decades (18,19). It has been shown that pentamidine selectively binds to the AT-rich region of duplex DNA (20–22), but the antimicrobial effect of pentamidine and its derivatives does not correlate well with their DNA-binding activity (23). Some proteins may be targets of pentamidine as well: The drug inhibits the in vitro activities of topoisomerase from P. carinii and African trypanosomes (11,13). In addition, pentamidine inhibits PRL (Phosphatase of Regenerating Liver) activity and the growth of WM9 human melanoma tumors in nude mice, coincident with the induction of tumor cell necrosis (24). However, the activity of TcPRL-1 from Trypanosoma cruzi was not affected by pentamidine (25). Pentamidine also demonstrates its activity in selectively modifying ubiquitin (26).
Pentamidine inhibition of RNA function began to draw attention in the last decade. It was first shown to strongly inhibit self-splicing of the nuclear group I introns present in the rRNA genes of P. carinii (27,28), in the 26S rRNA gene of Candida albicans (29) and in the protein coding genes of the mitochondrial genome S. cerevisiae (30). The mechanism of pentamidine inhibition of group I self-splicing has been attributed to its binding of the catalytic intron RNA and its alteration of the intron folding (31). Pentamidine was also unexpectedly shown to specifically and potently inhibit the mitochondrial translation in S. cerevisiae at a concentration three orders of magnitude lower than erythromycin, while keeping cytoplasmic translation unaffected. The underlying mechanism of this important finding is not clear yet.
The majority of antibiotic classes that are used in the clinical setting target the ribosomes to inhibit pathogen translation (32). Appreciation of the high-resolution ribosome structures and their complexes with antibiotics suggests that many naturally occurred antibiotics inhibit bacterial translation through intimate contacts with the ribosomal RNA (33,34). Aminoglycoside antibiotics that specifically bind to the ribosomal acceptor site of the 16S rRNA cause misreading of the genetic code and inhibit translocation in gram-negative and gram-positive bacteria (35). Both rings I and II of the aminoglycoside are important for its binding (36–38). Aminoglycosides also inhibit the activity of most known ribozymes at concentrations much higher than that of the translation (35). The strength of the interaction is dominated by electrostatics, with the positively charged aminoglycosides displacing metal ions and binding to specific sites on the structured RNAs (35).
In contrast, the aminoglycosides tobramycin, neomycin B, and another type of antibiotic purpuromycin have been shown to inhibit the aminoacylation reaction at the level of tRNA charging (39–41). Purpuromycin inhibits protein synthesis in both prokaryotic and eukaryotic cell-free systems (42,43), and it is evident that the inhibition of translation is through the specific inhibition of aminoacyl–tRNA formation (41). Neomycin B inhibits the phenylalanylation of E. coli tRNAPhe; one neomycin molecule is bound to the upper part of the anticodon stem in the yeast tRNAPhe–aminoglycoside crystal complex (40). Tobramycin is a potent and specific inhibitor of the aspartylation reaction (39).
In this report, we show that pentamidine non-specifically binds to tRNAs at concentrations much higher than the specific binding of the known antibiotics mentioned above. The non-specific interaction is primarily driven by hydrophobic interactions, likely due to the insertion of the aromatic rings into paired helices of tRNAs, in contrast to the main role of electrostatic interactions in the specific binding of many antibiotics to rRNA and tRNA. Pentamidine binding has been shown to disrupt the tRNA secondary structures and to mask the anticodon loop of the tRNA tertiary structure in the absence and presence of magnesium respectively, effects which are highly consistent with its inhibition of the tRNALeu aminoacylation and in vitro translation.
MATERIALS AND METHODS
Materials
The E. coli tRNALeu (anticodon GAG) and LeuRS (2100 U/mg) used in this study were provided by Prof. En-Duo Wang's lab (Shanghai Institute of Biological Sciences) (44). In each gel mobility shift assay and footprinting experiment, tRNA was labeled at the 5′ end by incorporation of [γ-32P] ATP (3000 Ci/mmol; PerkinElmer) (45). L-Leucine, ATP, tetrasodium pyrophosphate and dithiothreitol (DTT) were purchased from Sigma (USA). L-[14C] Leucine (50 µCi/ml) and tetrasodium [32P] pyrophosphate were products of Amersham Biosciences.
Gel mobility shift assays
The purified 5′-radiolabeled tRNA (∼0.3 nM) was denatured in 10 mM Tris–HCl (pH 7.5) at 70°C for 5 min and then slowly cooled to room temperature. An aliquot of tRNA was incubated at 37°C for 20 min in 10 μl gel shift buffer containing varying concentrations of pentamidine, 100 mM of Tris–HCl (pH 7.5), 30 mM KCl and 0, 1 mM, 5 mM, 9 mM or 12 mM MgCl2 as indicated. To determine the effect of pH on pentamidine binding, the following buffers at a concentration of 100 mM were used: sodium cacodylate for pH 6.0 and 6.5, Tris–HCl for 7.0, 7.5 and 8.5. Samples were then chilled on ice and analyzed by electrophoresis on 20% non-denaturing polyacrylamide gels in 1× TB buffer (0.045 M Tris–borate) at 4°C with a voltage gradient of 12–15 V/cm. The gels were exposed to Phosphor screens that were scanned and analyzed using a variable scanner Typhoon 9200 (Amersham Pharmacia biotech). Data were plotted using the program GraphPad Prism 4.0.
T1 ribonuclease footprinting
The 5′-end-labeled tRNA was cleaved by ribonuclease T1 and hydroxyl radical to probe the interaction between pentamidine and tRNA. T1 footprinting was performed as described previously (45). An aliquot of the pre-denatured tRNA was incubated in 18 μl gel shift buffer containing the desired concentrations of pentamidine at 37°C for 20 min. RNase T1 (2 μl of 10 U/μl) was then added to initiate the T1 cleavage reaction at 37°C for 10 s, and stopped by adding 20 µl of phenol–chloroform (1:1) and vigorous vortexing. After centrifugation, 16 µl of supernatant was removed for electrophoresis on 12% polyacylamide/7 M urea sequencing gels. Control lanes were digested with the indicated buffer or enzymes under denaturing conditions.
Isothermal titration calorimetry (ITC)
ITC measurements were carried out at 37°C using a VP-ITC calorimeter (MicroCal, Northampton, MA) with stirring at 400 rpm. In a typical experiment, 1.43 ml of 0.01 mM total yeast tRNAs purchased from Sigma (USA) was titrated with 10 mM of pentamidine (28 injection of 5 μl each). The buffer contained 10 mM Hepes (PH 7.5) and 300 mM NaCl with or without 5 mM magnesium. To correct for the dilution and mixing effects, a series of control injections was carried out in which pentamidine was injected into the buffer alone. The heat signal of this control was then subsequently subtracted from the raw data for each injection experiment. MicroCal ORIGIN software supplied with the VP-ITC calorimeter was used for data acquisition and analysis.
Aminoacylation experiments
The tRNALeu aminoacylation tests were assessed as described previously (46), and performed at 37°C in a 20 µl reaction mix containing 100 mM Tris–HCl (pH 7.8), 30 mM KCl, 12 mM MgCl2, 4 mM ATP, 0.1 mM EDTA, 0.5 mM DTT, 0.1 mM [14C] leucine (50 μCi/ml), 1 μM E. coli LeuRS, 3.85 μM tRNALeu (pre-denatured) and the indicated concentrations of pentamidine. The mix except for LeuRS was incubated at 37°C for 10 min and LeuRS was then added to initiate the aminoacylation reaction. After a 30 min of reaction, the samples were applied to Whatman Grade 3 qualitative filter paper, precipitated with 5% trichloroacetic acid (TCA), washed with 5% TCA and ethanol, and analyzed by scintillation counting.
Adenylate formation assay
The [32P] PPi-ATP exchange activity of E. coli LeuRS was assayed in 100 mM HEPES–KOH (pH7.8), 10 mM MgCl2, 10 mM KF, 4 mM ATP, 2 mM [32P] pyrophosphate, 1 mM L-Leucine, and 4 nM LeuRS (47). The analysis was carried out similar as in the aminoacylation reactions, except that 4 min of reaction was run after adding LeuRS.
In vitro translation assays
An assay of protein synthesis in vitro was performed using Flexi rabbit reticulocyte lysate (Promega) according to the manufacturer's instruction with the following modifications: Luciferase control RNA (1 mg/ml) was mixed with reticulocyte lysate on ice and then separated into aliquots for translation at 30°C for 90 min in a final reaction volume of 5 μl containing 20 μM amino acid complete mixtures, 70 mM KCl, varying concentrations of pentamidine and 1.35 mM, 2.5 mM or 3.5 mM magnesium acetate. Then 1 μl of the luciferase translation reaction was removed for bioluminescence assay using a Luminometer TD-20/20 (Turner Designs). Data were plotted using Graphpad Prism. Control reactions were performed in the same condition except that pentamidine was added after the translation.
RESULTS
Pentamidine binds to tRNA cooperatively
Although pentamidine binds to group I ribozyme RNA and inhibits ribozyme function, we found no evidence that this compound, at a concentration of up to 1 mM, binds to single-strand RNA, DNA oligomers and double-strand DNA using similar measuring conditions (data not shown). This result indicated that pentamidine has a lower binding affinity with DNA and non-structured RNA, which is consistent with a model that pentamidine inhibition of the translation in yeast mitochondria occurs through its binding with the two major classes of structured RNAs, tRNA and rRNAs. In this study, we aimed to explore if pentamidine interacts with tRNA; and if it does, how this interaction impacts the function of tRNA charging and translation.
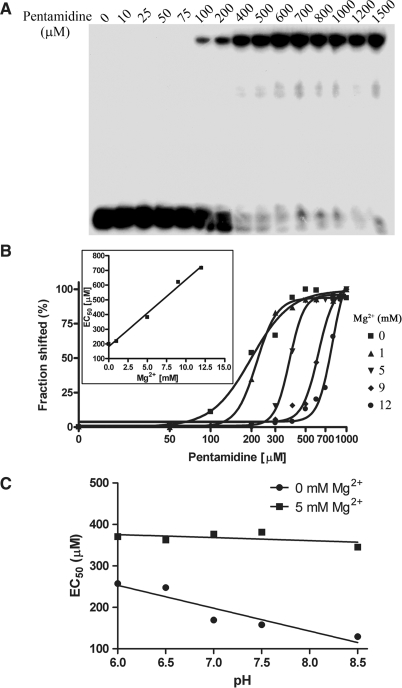
Native PAGE gel analysis was used to examine if pentamidine binds to pure tRNA molecules (Figure 1) in vitro. As shown in Figure 2A, a substantial shift of tRNALeu to a slower mobilized RNA species was observed at 100 μM of pentamidine in the absence of magnesium, and 400 μM of the drug saturated the binding, suggesting that pentamidine binds to tRNA efficiently. Interestingly, the binding affinity was dose-dependently decreased by magnesium (Figure 2B and Table 2). This finding is consistent with the previous conclusion that the charge interaction between pentamidine and structured RNA contributes to its binding with the group I intron ribozyme (28,31). Consistently, pentamidine binding was increased with pH in the absence of magnesium, displaying the contribution of electrostatic interaction between pentamidine and tRNA; while this change was not observed in the presence of 5 mM Mg2+ that presumably overrides the electrostatic interaction imposed by pentamidine (Figure 2C). It should be noted that the majority of the pentamidine-bound tRNA remained in the gel wells and did not run into the gel, probably because of the neutralization of the negative charge of tRNA by multiple pentamidine binding.
Figure 1.
Structures of E. coli tRNALeu (A) and pentamidine (B). The secondary structure of E. coli tRNALeu showing main tertiary interactions is revised from (46); Numbering of nucleotides as well as nomenclature of modified nucleotides are according to Sprinzl et al. (52).
Figure 2.
Gel retardation assay of pentamidine binding to tRNALeu. A fixed concentration of 32P-labeled tRNALeu was incubated with the indicated concentrations of pentamidine in the absence magnesium (A), different concentrations of magnesium (B) at pH 7.5, and different pH in the absence and presence of 5 mM magnesium (C). The shifted bands were quantified by PhosphorImager analysis and plotted. The EC50s and Hill coefficients are listed in Table 2. White misting precipitation started to appear when pentamidine concentration reached 1 mM. The shifted fraction of input tRNA was calculated and plotted against the logarithm of pentamidine concentrations by fitting to a Hill equation Y = Bottom + (Top − Bottom)/(1 + 10[(Log EC50 − X) × Hillslope]). EC50 indicates the effective pentamidine concentration causing 50% of tRNA to shift. The inserts in (B) show the results of plotting EC50s against different magnesium concentrations.
Table 2.
Hill analysis results of different experiments
| Mg2+ (mM) | Hill coefficient | EC50 (μM) |
|---|---|---|
| Native gel analysis | ||
| 0 | 3.1 | 158.3 |
| 1 | 5.8 | 218.7 |
| 5 | 8.1 | 381.4 |
| 9 | 7.4 | 621.0 |
| 12 | 8.3 | 785.1 |
| T1 footprinting | ||
| 0 | 4.9 | 161.3 |
| 12 | 6.3 | 409.8 |
| Leucylation assay | ||
| 12 | 8.2 | 681.5 |
| 12a | 11.7 | 759.8 |
| In vitro translation | ||
| 1.35 | 6.9 | 204.4 |
| 2.5 | 34.5 | 425.4 |
| 3.5 | 14.7 | 470.0 |
Hill analysis is described in the legend to Figure 2, and EC50 indicates the effective pentamidine concentration causing 50% of inhibition.
aIndicates the assay in which tRNA was not pre-denatured.
Figure 2B shows that the drug binds to tRNA in a highly cooperative fashion, and the Hill coefficient indicative of cooperativity was significantly increased by higher concentrations of magnesium, from 3.1 without Mg2+ to 8.3 with 12 mM Mg2+ (Table 2). Clearly, magnesium increases not only the required concentrations of pentamidine to bind to tRNA, but also the binding cooperativity. This indicates that when tRNA folds to a more compacted structure, it becomes less accessible to pentamidine. Furthermore, the drug interacts with folded tRNA in a manner significantly different from tRNA that is not folded.
Pentamidine binding disrupts the tRNA secondary structure and masks the anticodon loop of the tertiary structure
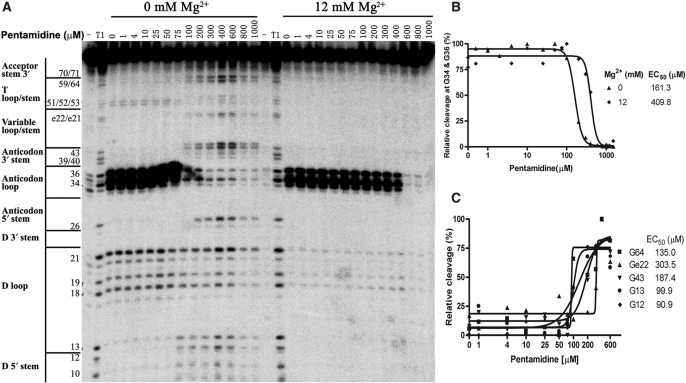
T1 ribonuclease footprinting experiments were then performed to address how pentamidine binding affects the tRNA structure (Figure 3A). RNase T1 ribonuclease specifically cleaves the single-stranded RNA at the 3′ of each guanosine residue and has been used to detect the unpaired guanosine located on the accessible surface of large structured RNAs (45,48,49).
Figure 3.
Pentamidine binding alters the sensitivity of tRNA to ribonuclease T1 cleavage. The radiolabeled tRNA was preincubated with the indicated concentrations of pentamidine and MgCl2 at 37°C for 20 min; ribonuclease T1 was then added to partially digest tRNA and the resulted RNA fragments were fractionated by PAGE on an 12% polyacrylamide/7 M urea gels. One representative gel is shown in (A). Nucleotides and their corresponding locations stems in the tRNA secondary structure are indicated on the left of the gel. Lane T1 indicates tRNA sample partially digested by T1 ribonuclease under denaturing conditions. (B) The protection at G34 and G36 was determined and fit to a Hill equation as in Figure 2. (C) The protection at some other positions including G12 at D stem, G13 at D loop, G43 at anticodon stem, Ge22 at Variable stem and G64 at T stem in the absence of magnesium were also determined and plotted.
In the absence of magnesium, all Gs in the stem regions of tRNALeu were not susceptible to T1 cleavage, indicating the formation of secondary structure. Interestingly, G59 in the TψC loop was resistant to T1 cleavage as well, indicating that this loop might adopt some kind of conformation to prevent G59 from being accessed by T1 ribonuclease. G sites in the D loop and anticodon loop were cleaved by T1 ribonuclease, with the cleavage at G34 and G36 in the GAG anticodon being about 10-fold stronger than that in the D-loop. G37 was not cleaved by T1 ribonuclease, probably because the followed pseudouridine ψ38 alters its conformation to one not recognized by T1. When 12 mM Mg2+ was present, complete protection of Gs in the D loop from T1 ribonuclease attack was evident, consistent with the knowledge that tRNA readily adopts its L-shape tertiary structure at this magnesium concentration. The supersensitive nature of G34 and G36 in the anticodon loop was evident in the absence and presence of magnesium, which has two important implications. First, it is consistent with the requirement for the extreme extended conformation that anticodon bases must adopt to interact with corresponding codon. Second, the tRNA secondary structure might adopt some kind of conformation in the absence of magnesium, resulting in a D loop structure that is much less accessible to T1 cleavage than the extended anticodon loop.
Remarkably, the presence of pentamidine (⩾100 μM) in the tRNA samples containing no magnesium strongly protected G34 and G36 in the anticodon loop from being cleaved by T1, resulting in sensitivity similar to the D-loop, which suggested that pentamidine binding to tRNA reduces T1 accessibility (Figure 3A and 3B). Meanwhile, all Gs in the base-paired stem loops became accessible to T1 ribonuclease, and the accessibility was equivalent to that in the D and anticodon loops (Figure 3A and 3C). These results indicated that pentamidine binding strongly relaxes the preliminary tRNA structure formed in Tris–HCl, and each G residue in the relaxed structure is similarly accessible to T1 ribonuclease. In the presence of 12 mM Mg2+, up to 400 μM pentamidine did not impose an observable effect on the T1-detectable tRNA structure, while the drug at 600 μM nearly completely protected G34 and G36 in the anticodon loop from T1 cleavage, consistent with the sharp increased tRNA-binding ability at the same pentamidine concentration shown in the gel-shift assay (Figure 2). Interestingly, the strong protection at G34 and G36 was not accompanied by an increased cleavage at any other region of tRNA, suggesting pentamidine could not disrupt the tRNA tertiary structure as it did to the secondary structure (no Mg2+). Nevertheless, pentamidine binding very likely alters the tRNA tertiary structure, which is featured by masking its anticodon codon; this binding effect is anticipated to interfere with the tRNA function.
Since T1 ribonuclease is powerless to detect pentamidine binding to the base-paired regions of tRNA, free radical footprinting method detecting the accessibility of the RNA backbone was employed to study the pentamidine-binding sites on tRNALeu (Supplementary Figure S1). Consistent with the model that the tRNA tertiary structure is established by the closing of the D and TψC loops through tertiary interactions, TψC loop and the D loop were drastically protected from the free radical attack in the folded structure. In the absence of magnesium, as low as 10 μM pentamidine significantly protected the tRNA secondary structure from being attacked by free radicals, and ∼50 μM pentamidine protected almost the entire RNA phosphate backbone except for a few sites in TψC stem and the variable stem from free radical cleavage, indicating that pentamidine binding protects the tRNA backbone from being attacked by free radicals. In the presence of magnesium, 400 μM pentamidine was required to reach the near complete protection.
Clearly, free hydroxyl radical footprinting was much more sensitive in detecting the pentamidine–tRNA interaction than was T1 footprinting and gel-shift assay in this study. Hydroxyl radical footprinting was used to study the pentamidine–DNA interaction previously, showing that pentamidine at a concentration up to 100 μM only grants a limited protection to DNA backbone at certain AT-rich regions (50). Obviously, pentamidine protection of tRNALeu from free radical attack completely differed from its DNA protection pattern. The tRNA protection seemed to be equivalently effective at each free radical-accessible site regardless of the presence or absence of magnesium, and the protection pattern was much more effective and quite uniform along the tRNA backbone. This observation strongly suggests that pentamidine binding of tRNA is not sequence-specific or site-specific.
Pentamidine binding is largely driven by non-specific hydrophobic interactions
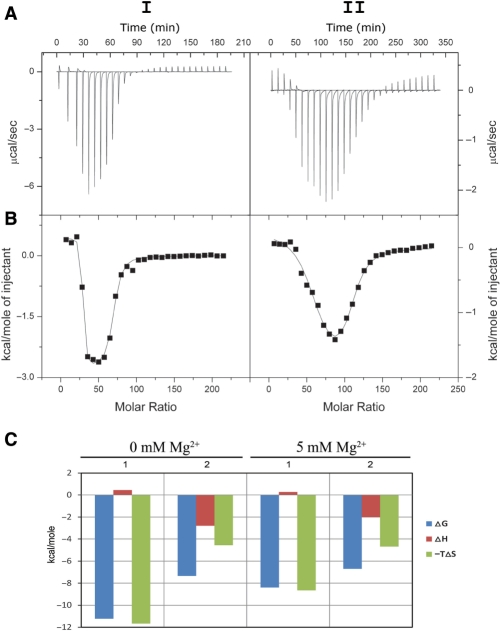
ITC has become an important tool for the direct and reliable measurement of the thermodynamic parameters of the interaction of small molecules with biopolymers (51). This method was used here to directly determine the energetics and stoichiometry of the interaction between pentamidine and tRNA. Figure 4A shows that the ITC profiles for the binding of pentamidine to tRNA with 0 and 5 mM Mg2+ are both biphasic, consistent with the presence of two binding equilibria. Therefore, the ITC results were best fitted by using a model of two sets of binding sites using Origin 7 (Figure 4B). The thermodynamic parameters are presented in Table 1. The enthalpies, entropies and free energies are shown in Figure 4C.
Figure 4.
ITC profiles at 37°C for the titration of pentamidine with a solution of tRNA containing no MgCl2 (I) or 5 mM MgCl2 (II). Each heat burst curve in panel A is the result of an injection of pentamidine into a tRNA solution. The corrected injection heats shown in (B) were derived by integration of the corresponding heat burst curves shown in (A), followed by subtraction of the corresponding dilution heats derived from control titrations of the drug into buffer containing no tRNA. The data points in panel B reflect the corrected experimental injection heats, while the solid lines reflect the calculated fits of the data via a model for two independent sets of binding sites. (C) shows the thermodynamic signatures for the binding events, and 1 and 2 represent the Class I and Class II binding sites, respectively.
Table 1.
Thermodynamic parameters for pentamidine binding to tRNA
| tRNA | Binding site | N | K (M−1) | ΔH (kcal mol−1) | TΔS (kcal mol−1) | ΔG (kcal mol−1) |
|---|---|---|---|---|---|---|
| Without Mg2+ | 1 | 26.0 ± 0.3 | (8.0 ± 5.7)×107 | 0.44 ± 0.05 | 11.66 | −11.23 ± 0.05 |
| 2 | 41.1 ± 0.8 | (1.5 ± 0.3)×105 | −2.79 ± 0.08 | 4.56 | −7.35 ± 0.08 | |
| 5 mM Mg2+ | 1 | 56.4 ± 2.7 | (8.0 ± 4.2)×105 | 0.27 ± 0.08 | 8.65 | −8.39 ± 0.08 |
| 2 | 51.5 ± 4.7 | (5.2 ± 1.5)×104 | −2.00 ± 0.27 | 4.68 | −6.69 ± 0.27 |
Table 1 shows that the binding constant (K) for the first equilibrium (K1) was greater than that for the second (K2). In the absence of magnesium, K1 was 533-fold higher than K2; in the presence of magnesium, K1 was 15-fold higher than K2. The drastically larger K1 as compared to K2 indicates that the first class of binding sites determines the binding affinity of pentamidine with tRNA. Consistent with the hydroxyl radical footprinting result showing the non-specific binding of pentamidine to tRNA, ITC measurements demonstrated evidence for a large number of pentamidine-binding sites on tRNA: In the unfolded tRNA, 26 Class I binding sites and 41 Class II binding sites were present. In the folded tRNA, the Class I and Class II binding sites were increased to 56 and 52, respectively.
Class I binding sites responded to magnesium very sensitively, with K1 decreasing about 100-fold and the binding sites doubling, while Class II binding sites were much less sensitive (Table 1). Figure 4C and Table 1 show that ΔH was a small positive number for Class I binding sites, indicating that enthalpic interactions including hydrogen bond formation, charge interaction and van der Waal's interactions between pentamidine and tRNA were unfavorable. A large positive TΔS indicated entropically favorable binding and suggested that hydrophobic interactions are the main driving force for the Class I binding of pentamidine to tRNA, which is also consistent with the non-specific feature of pentamidine binding.
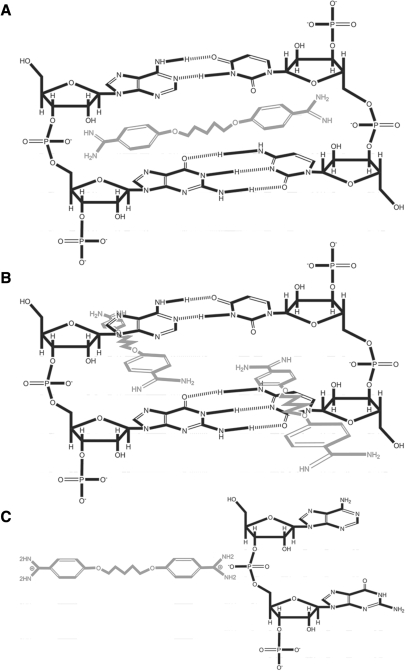
Pentamidine contains two aromatic rings linked by an alkyl chain. Insertion of the aromatic rings into the double helix of tRNA is expected to result in a strong hydrophobic interaction. About 22–30 bp are generally present in the unfolded cloverleaf structure of tRNAs (52); (http://lowelab.ucsc.edu/GtRNAdb/Scere/Scere-align.html). If each pentamidine molecule is inserted between two neighboring base pairs, the average binding sites provided by the total yeast tRNAs should be very close to 26, the number of evident Class I binding sites. Considering that the base-paired helical structures are very flexible in the unfolded tRNAs, it could be very possible for both of the aromatic rings of pentamidine to be inserted into the same neighboring base pairs, with each aromatic ring being stacked with one pair of the neighboring bases (Figure 5A). It is conceivable that this deep insertion can disrupt the hydrogen bonds of each base pair in the tRNA secondary structure, explaining the universal T1 accessibility to all Gs in tRNALeu under the saturated concentrations of pentamidine. The tRNA folds to a more compact structure in magnesium with additional base paires formed between the D loop and the TψC loop. The geometry of the helix of the folded tRNAs is strongly restricted, and insertion of both aromatic rings of the pentamidine into the neighboring base pairs should not be allowed. Because the hydrophobic interaction/entropy change is also the only driving force for pentamidine binding to Class I sites on the folded tRNA, we propose that each pentamidine molecule may form a much weaker hydrophobic interaction with the base paired helix through one of its aromatic rings, consistent with its 100-fold lower binding constant and 2.2 times more binding sites (Figure 5B).
Figure 5.
Proposed binding modes of pentamidine to tRNA. (A) In the absence of magnesium ions, one pentamidine molecule is stacked between 1 bp of the flexible tRNA secondary structure. This insertion should at least partially disrupt the hydrogen bonds between each base pair, and thus blockers are placed between each hydrogen pair of bond donor and receptor to illustrate this possible outcome. (B) In the presence of magnesium, two pentamidine molecules are stacked between base pairs of the compact tRNA tertiary structure, in which only one phenyl group inserts in between the base pairs and the other floats outside because of the structure constraints. (C) Positively charged pentamidine experiences a charge interaction with negatively charged phosphate in the absence and presence of magnesium.
Class II binding was apparently driven by both favorable entropy and favorable enthalpy changes (Figure 4C), consistent with the previous finding that the charge interaction between positively charged pentamidine and negatively charged phosphate in tRNA contributes to their interaction (28). Class II binding sites were not as sensitive to magnesium-mediated tRNA folding as the Class I sites, with K2 decreasing to one third and the binding sites increasing by one fourth. Although the charge attraction between the drug and tRNA obviously did not contribute much to the interaction energetically, the close-distance attraction between differently charged molecules could play a large role in facilitating base stacking interactions (Figure 5C). Therefore, it is very likely that Class I and Class II binding sites on tRNA are close to each other. Consistently, the Class II binding sites numbered 41 and 52 in the absence and presence of magnesium, respectively, which are close to that of the Class I binding sites in the presence of magnesium. It is obvious that not all phosphate units in tRNA can serve as Class II binding sites. It is very possible that only the phosphate backbone of the base-paired regions of tRNAs participates the Class II binding, consistent with the finding that pentamidine did not bind to the single stranded RNA (data not shown). These findings suggested a model in which the charge interaction-driven Class II binding draws pentamidine to a close proximity with tRNA, and then insertion of the aromatic ring of pentamidine into the double helix of tRNA stabilizes the charge interaction by a large entropy change (Figure 5).
Pentamidine binding inhibits tRNALeu aminoacylation
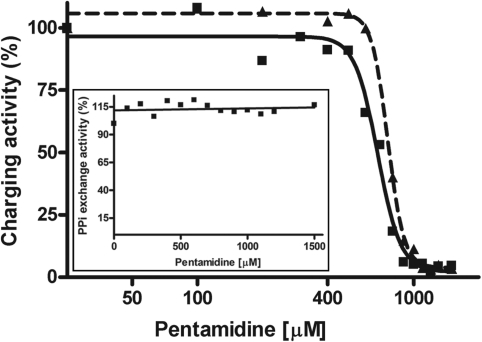
We then asked if the non-specific binding of pentamidine to tRNA affected the tRNA charging process. The ability of tRNALeu to accept [14C] leucine, catalyzed by its cognate tRNA synthetase LeuRS, was studied as a function of increasing concentrations of pentamidine. MgCl2 at 12 mM was included in each reaction because LeuRS-catalyzed tRNALeu charging is very active at this concentration (46). As shown in Figure 6, leucylation of tRNALeu was decreased dose-dependently with increased concentrations of pentamidine (0–1.5 mM). Data were fitted well to the sigmoidal dose-response (variable slope) equation, resulting in an inhibitory concentration and cooperativity of inhibition very similar to those of T1 footprinting and gel-shift analysis (Table 2). These results strongly suggest that the extensive non-specific pentamidine binding converts the tRNALeu to an inactive structure not recognized by LeuRS.
Figure 6.
Pentamidine inhibits aminoacylation of tRNALeu in vitro. The activity of tRNALeu synthetase (LeuRS) in charging E. coli tRNALeu and adenylating L-Leucine were measured at increasing concentrations of pentamidine as described in Materials and Methods. PPi-ATP exchange reaction was used to reflect the adenylation activity of LeuRS. (A) The relative activity of LeuRS to charge tRNALeu at each pentamidine concentration was obtained using the charging activity displayed by the tRNA sample receiving no pentamidine as a control; the relative charging activity was then plotted against pentamidine concentrations as in Figure 2. The solid line (EC50 681.5 μM) and dashed line (EC50 759.8 μM) indicates the experiment performed using tRNA with and without pre-denaturation treatment, respectively. (B) The PPi-ATP exchange activity of LeuRS was plotted against each pentamidine concentration.
During the first step of the aminoacyl–tRNA charging, LeuRS forms an activated leucyl–adenylate using ATP as a substrate in the absence of tRNALeu. Pyrophosphate [PPi] exchange experiments were performed to examine if the adenylation of leucine by LeuRS was interfered by pentamidine. It is showed that pyrophosphate exchange at all pentamidine concentrations from 100 to 1500 μM was at the same level (Figure 6, insert), indicating that pentamidine does not act as a structural analogue of ATP in the aminoacylation reaction and does not interfere with the catalytic activity of LeuRS itself during the amino acid activation step. This conclusion was further supported by the aminoacylation experiment with increasing ATP concentrations, which showed that the inhibitory effect of pentamidine was identical under different ATP concentrations (data not shown).
Pentamidine binding inhibits the in vitro translation
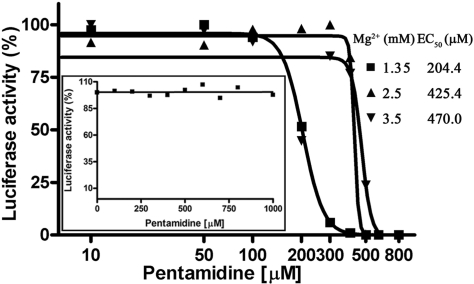
The luciferase mRNA was translated in an in vitro translation system to address whether pentamidine inhibits translation in vitro. Pentamidine dose-dependently reduced the fluorescence emission from the translation reactions, and the increase of magnesium concentration elevated the pentamidine requirement (Figure 7 and Table 2). Pentamidine itself did not quench the fluorescence resulting from luciferase activity (Figure 7, insert). Therefore, the reduced fluorescence intensity was attributed to the inhibition of productive translation, likely due to pentamidine interfering with tRNA structure-related functions such as charging and docking, and possibly rRNA structure-related functions including catalysis and ribosome assembly.
Figure 7.
Pentamidine inhibits proteins synthesis in vitro. Luciferase mRNA was translated in PROMEGA Flexi rabbit reticulocyte lysate reagents with increasing concentrations of pentamidine at different concentrations of magnesium, and the enzymatic activity of the translated luciferase was measured by a luminometer and plotted as in Figure 6. EC50 at each magnesium concentration was listed. Please be noted that the luciferase activity varied greatly with magnesium concentrations; the activity at 1.35 mM, 2.5 mM and 3.5 mM magnesium was 8515.0, 1773.0 and 193.7 counts, respectively. The insert shows the results of the control reactions in which pentamidine was added to each sample after the translation reaction was finished.
DISCUSSION
Pentamidine has been used clinically for over a half of century, and it becomes clear now that pentamidine is selectively accumulated by pathogenic protozoa through high affinity transporters located in the pathogen membranes (6). However, its mechanism of actions inside of cells remains perplexing. Our recent finding that 1 μM pentamidine in the growth media of S. cerevisiae leads to a near complete inhibition of the mitochondrial translation, while up to 25 μM of the drug has no effect on the levels of the cytoplasmic translation, sheds new light on pentamidine targets in vivo (30).
Pentamidine binding to tRNA inhibits tRNA aminoacylation and translation
In this study, we have showed that pentamidine binds to tRNAs at concentrations comparable to that of the Candida group I intron (31). The binding disrupts the tRNA secondary structure formed in the absence of magnesium. In the presence of magnesium, pentamidine uniformly binds to the folded tRNA as well and masks the anticodon loop, but with a greatly reduced affinity. The crystal structure of the complex between leucyl–tRNA synthetase and tRNALeu points that the synthetase recognizes its cognate tRNA depending essentially on tRNA shape rather than base-specific interactions (53). Consistent with this view, pentamidine inhibition of tRNA aminoacylation can be explained by the distorted tRNA structure induced by extensive drug binding. The effects of pentamidine in inhibiting tRNA aminoacylation and in distorting the anticodon loop explain the potent activity of protein translation in vitro and in vivo (Figure 8). Therefore, tRNA is the second class of pentamidine-bound structured RNAs whose biological functions are hindered by pentamidine binding.
Figure 8.
Scheme of the inhibition of tRNA aminoacylation and translation by pentamidine. Magnesium ions help tRNAs fold to their native structures, containing the identity elements that cognate aaRS recognizes. The aminoacylated tRNA then participate in translation. Pentamidine binds to tRNAs non-specifically. If the unfolded tRNAs are bound by pentamidine, the binding disrupts the tRNA secondary structure and the subsequent folding and function. When pentamidine binds to the folded tRNAs, the three dimensional structure of tRNAs is distorted and the anticodon loop is masked, preventing tRNA from being charged by its cognate aaRS, leading tRNA off its functional pathway.
Although the expected targets of pentamidine in yeast are mitochondrial tRNAs while this study has used E. coli tRNALeu and yeast total tRNAs as model. However, the in vitro findings are believed to be in vivo relevant for two reasons: first, the global architecture of tRNAs is highly conserved; second, this study shows that the interaction between pentamidine and tRNA is non-specific to any tRNA sequence. We noticed that about two orders of magnitude higher concentrations of pentamidine are required to inhibit protein translation in vitro (EC50 of 204.4 μM) than that in living S. cerevisae (∼90% inhibition at 1 μM). This large difference can be readily explained by the drug accumulation capability of pathogenic fungi and protozoans. For example, pentamidine is concentrated in trypanosomes up to 1.1 mM after a 3 h incubation with 1 μM of pentamidine (54). Therefore, it is reasonable to predict that the intracellular concentration of pentamidine into mitochondria could reach near millimolar concentrations to inhibit translation.
The mechanism of pentamidine binding to structured RNAs: pentamidine binds to tRNA non-specifically and the binding is driven by hydrophobic interactions
Aminoglycosides represent a major class of translation inhibitors that specifically binds to their targets through electrostatic interactions. In contrast, this study reveals that pentamidine binds to tRNAs non-specifically and primarily through hydrophobic interactions. This non-specific interaction is consistent with its much lower binding affinity compared to that of aminoglycosides.
Two classes of pentamidine-binding sites are present on tRNAs, and each class contains dozens of distinct sites. Class I binding sites are located in the stacked bases of the tRNA double helix, and the negatively charged phosphate backbone provides for Class II binding sites. Remarkably, Class I binding sites determine the binding affinity between pentamidine and tRNA. Hydrophobic interactions are the main driving force for both classes of binding. Although cation competition experiments shown in this study and some previous reports suggest that electrostatic interaction play an role in pentamidine binding of its RNA target (28,30), ITC experiments in this report clearly show that the charge attraction did not contribute much to the binding energetically. We proposed a model in which the flat and rigid phenyl rings of pentamidine are capable of inserting into the stacked bases of RNA double helices, resulting in hydrophobic stacking interactions (Figure 5). The function of multivalent cations in weakening pentamidine's effect can be attributed to the capability of cations in supporting the folding of structured RNAs, which reduces the pentamidine-binding affinity to Class I sites on tRNAs. It is also possible that the charge interaction plays a role in facilitating the base stacking interactions by attracting these two differently charged molecules to a close distance (Figure 5C).
Structured RNAs may be the major molecular targets of pentamidine: implication for developing more effective pentamidine derivatives
Mitochondria are the primary cellular targets of pentamidine in pathogenic eukaryotes and in experimental model eukaryotes (55,56). Resistance to pentamidine in L. mexicana is accompanied by the lack of mitochondrial accumulation of the drug, and pentamidine is consequently excluded from the parasite (57). Mitochondrial translation occurs inside of mitochondria in which the structured RNAs including tRNA and rRNA are present at much higher concentrations than the low-copy DNA genomes. Our finding that pentamidine binding to structured RNAs is governed by non-specific hydrophobic interactions predicts that, in addition to tRNAs, rRNAs are important targets of pentamidine as well. We also found that pentamidine binds to large structured RNAs at a much higher affinity than to DNA and single-stranded RNA (data not shown), possibly because the rigid double helical structure of DNA does not favor pentamidine insertion into the stacked bases and ssRNA lacks base paired helical structure for hydrophobic interaction with pentamidine. The higher binding affinity of pentamidine with RNA over DNA is also supported by the observation that pentamidine concentrations that repress mitochondrial translation have no effect on the steady-state levels of the mitochondrial mRNAs, which are products of transcription from DNA (30). Therefore, we propose that structured RNAs are the dominant binding targets of pentamidine in mitochondria. Importantly, the finding that pentamidine binds to tRNAs and very likely to rRNAs as well, exerting a strong inhibition of mitochondrial translation, explains the pentamidine-induced interruption of many aspects of mitochondrial function and morphology.
Although Trypanosome and Leishmania accumulate pentamidine to a much greater extent than host cells, a certain amount of pentamidine is still expected to be uptaken into host cells. Pentamidine inhibition of translation through binding to tRNAs and possibly rRNAs through a non-specific hydrophobic interaction also explains the side effects of this drug, since pentamidine has no selectivity mechanism to discriminate human and the pathogenic RNAs.
The RNA-binding mechanism of pentamidine revealed in this study shed lights on a target-based approach for designing novel and more specific aromatic diamidines to combat microbial infections through targeting the mitochondrial or kinetoplast translation in pathogenic fungi and protozoans including Pneumocystis carinii, Trypanosome, Leishmania, and Entamoeba histolytica.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We owe thanks to E.-D. Wang (Shanghai Institute of Biological Sciences) for her generosity in providing us tRNALeu and research facilities to perform all of our tRNA charging experiments. Thanks to Prof. Y. Liang for his help of the ITC experiments, and to Prof. J.-G. Wu for providing us Luminometer TD-20/20 equipment for the luciferase activity assay. This work was supported by the Ministry of Education of China (20415), National Natural Science Foundation of China (30330170), and the National Basic Research Program of China (2005CB724604) awarded to Y. Zhang. Funding to pay the Open Access publication charges for this article was provided by National Basic Research Program of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sands M, Kron MA, Brown RB. Pentamidine: a review. Rev. Infect. Dis. 1985;7:625–634. doi: 10.1093/clinids/7.5.625. [DOI] [PubMed] [Google Scholar]
- 2.Goa KL, Campoli-Richards DM. Pentamidine isethionate. A review of its antiprotozoal activity, pharmacokinetic properties and therapeutic use in Pneumocystis carinii pneumonia. Drugs. 1987;33:242–258. doi: 10.2165/00003495-198733030-00002. [DOI] [PubMed] [Google Scholar]
- 3.Apted FI. Present status of chemotherapy and chemopropylaxis of human trypanosomiasis in the Eastern Hemisphere. Pharmacol. Therap. 1980;11:391–413. doi: 10.1016/0163-7258(80)90035-2. [DOI] [PubMed] [Google Scholar]
- 4.Bryceson AD, Chulay JD, Mugambi M, Were JB, Gachihi G, Chunge CN, Muigai R, Bhatt SM, Ho M, Spencer HC, et al. Visceral leishmaniasis unresponsive to antimonial drugs. II. Response to high dosage sodium stibogluconate or prolonged treatment with pentamidine. Trans. Roy. Soc. Trop. Med. Hyg. 1985;79:705–714. doi: 10.1016/0035-9203(85)90199-3. [DOI] [PubMed] [Google Scholar]
- 5.Su TH, Martin WJ., 2nd Pathogenesis and host response in Pneumocystis carinii pneumonia. Annu. Rev. Med. 1994;45:261–272. doi: 10.1146/annurev.med.45.1.261. [DOI] [PubMed] [Google Scholar]
- 6.Bray PG, Barrett MP, Ward SA, de Koning HP. Pentamidine uptake and resistance in pathogenic protozoa: past, present and future. Trends Parasitol. 2003;19:232–239. doi: 10.1016/s1471-4922(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 7.Werbovetz K. Diamidines as antitrypanosomal, antileishmanial and antimalarial agents. Curr. Opin. Investig. Drugs. 2006;7:147–157. [PubMed] [Google Scholar]
- 8.Fairlamb AH. Chemotherapy of human African trypanosomiasis: current and future prospects. Trends Parasitol. 2003;19:488–494. doi: 10.1016/j.pt.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Briceland LL, Bailie GR. Pentamidine-associated nephrotoxicity and hyperkalemia in patients with AIDS. Dicp. 1991;25:1171–1174. doi: 10.1177/106002809102501102. [DOI] [PubMed] [Google Scholar]
- 10.Lachaal M, Venuto RC. Nephrotoxicity and hyperkalemia in patients with acquired immunodeficiency syndrome treated with pentamidine. Am. J. Med. 1989;87:260–263. doi: 10.1016/s0002-9343(89)80147-0. [DOI] [PubMed] [Google Scholar]
- 11.Dykstra CC, Tidwell RR. Inhibition of topoisomerases from Pneumocystis carinii by aromatic dicationic molecules. J. Protozool. 1991;38:78S–81S. [PubMed] [Google Scholar]
- 12.Grady RW, Blobstein SH, Meshnick SR, Ulrich PC, Cerami A, Amirmoazzami J, Hodnett EM. The in vitro trypanocidal activity of N-substituted p-benzoquinone imines: assessment of biochemical structure-activity relationships using the Hansch approach. J. Cell. Biochem. 1984;25:15–29. doi: 10.1002/jcb.240250103. [DOI] [PubMed] [Google Scholar]
- 13.Shapiro TA. Inhibition of topoisomerases in African trypanosomes. Acta Trop. 1993;54:251–260. doi: 10.1016/0001-706x(93)90097-u. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu YK, Weiner AJ, Rosenblatt J, Wong DC, Shapiro M, Popkin T, Houghton M, Alter HJ, Purcell RH. Early events in hepatitis C virus infection of chimpanzees. Proc. Natl Acad. Sci. USA. 1990;87:6441–6444. doi: 10.1073/pnas.87.16.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro TA, Englund PT. Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc. Natl Acad. Sci. USA. 1990;87:950–954. doi: 10.1073/pnas.87.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bornstein RS, Yarbro JW. An evaluation of the mechanism of action of pentamidine isethionate. J. Surg. Oncol. 1970;2:393–398. doi: 10.1002/jso.2930020412. [DOI] [PubMed] [Google Scholar]
- 17.Makulu DR, Waalkes TP. Interaction between aromatic diamidines and nucleic acids: possible implications for chemotherapy. J. Natl Cancer Inst. 1975;54:305–309. [PubMed] [Google Scholar]
- 18.Reddy BS, Sondhi SM, Lown JW. Synthetic DNA minor groove-binding drugs. Pharmacol. Therap. 1999;84:1–111. doi: 10.1016/s0163-7258(99)00021-2. [DOI] [PubMed] [Google Scholar]
- 19.Baraldi PG, Bovero A, Fruttarolo F, Preti D, Tabrizi MA, Pavani MG, Romagnoli R. DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 2004;24:475–528. doi: 10.1002/med.20000. [DOI] [PubMed] [Google Scholar]
- 20.Edwards KJ, Jenkins TC, Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992;31:7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins TC, Lane AN. AT selectivity and DNA minor groove binding: modelling, NMR and structural studies of the interactions of propamidine and pentamidine with d(CGCGAATTCGCG)2. Biochim. Biophys Acta. 1997;1350:189–204. doi: 10.1016/s0167-4781(96)00160-1. [DOI] [PubMed] [Google Scholar]
- 22.Nunn CM, Jenkins TC, Neidle S. Crystal structure of d(CGCGAATTCGCG) complexed with propamidine, a short-chain homologue of the drug pentamidine. Biochemistry. 1993;32:13838–13843. doi: 10.1021/bi00213a012. [DOI] [PubMed] [Google Scholar]
- 23.Tidwell RR, Jones SK, Geratz JD, Ohemeng KA, Cory M, Hall JE. Analogues of 1,5-bis(4-amidinophenoxy)pentane (pentamidine) in the treatment of experimental Pneumocystis carinii pneumonia. J. Med. Chem. 1990;33:1252–1257. doi: 10.1021/jm00166a026. [DOI] [PubMed] [Google Scholar]
- 24.Pathak MK, Dhawan D, Lindner DJ, Borden EC, Farver C, Yi T. Pentamidine is an inhibitor of PRL phosphatases with anticancer activity. Mol. Cancer Therap. 2002;1:1255–1264. [PubMed] [Google Scholar]
- 25.Cuevas IC, Rohloff P, Sanchez DO, Docampo R. Characterization of farnesylated protein tyrosine phosphatase TcPRL-1 from Trypanosoma cruzi. Eukaryot. Cell. 2005;4:1550–1561. doi: 10.1128/EC.4.9.1550-1561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguewa PA, Fuertes MA, Cepeda V, Iborra S, Carrion J, Valladares B, Alonso C, Perez JM. Pentamidine is an antiparasitic and apoptotic drug that selectively modifies ubiquitin. Chem. Biodivers. 2005;2:1387–1400. doi: 10.1002/cbdv.200590111. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Leibowitz MJ. Variation and in vitro splicing of group I introns in rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 1993;21:2415–2421. doi: 10.1093/nar/21.10.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Tidwell RR, Leibowitz MJ. Inhibition of in vitro splicing of a group I intron of Pneumocystis carinii. J. Eukaryot. Microbiol. 1994;41:31–38. doi: 10.1111/j.1550-7408.1994.tb05931.x. [DOI] [PubMed] [Google Scholar]
- 29.Miletti KE, Leibowitz MJ. Pentamidine inhibition of group I intron splicing in Candida albicans correlates with growth inhibition. Antimicrob. Agents Chemother. 2000;44:958–966. doi: 10.1128/aac.44.4.958-966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Bell A, Perlman PS, Leibowitz MJ. Pentamidine inhibits mitochondrial intron splicing and translation in Saccharomyces cerevisiae. RNA. 2000;6:937–951. doi: 10.1017/s1355838200991726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Li Z, Pilch DS, Leibowitz MJ. Pentamidine inhibits catalytic activity of group I intron Ca.LSU by altering RNA folding. Nucleic Acids Res. 2002;30:2961–2971. doi: 10.1093/nar/gkf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutcliffe JA. Improving on nature: antibiotics that target the ribosome. Curr. Opin. Microbiol. 2005;8:534–542. doi: 10.1016/j.mib.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 34.Yonath A, Bashan A. Ribosomal crystallography: initiation, peptide bond formation, and amino acid polymerization are hampered by antibiotics. Annu. Rev. Microbiol. 2004;58:233–251. doi: 10.1146/annurev.micro.58.030603.123822. [DOI] [PubMed] [Google Scholar]
- 35.Walter F, Vicens Q, Westhof E. Aminoglycoside-RNA interactions. Curr. Opin. Chem. Biol. 1999;3:694–704. doi: 10.1016/s1367-5931(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 36.Fourmy D, Yoshizawa S, Puglisi JD. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 1998;277:333–345. doi: 10.1006/jmbi.1997.1551. [DOI] [PubMed] [Google Scholar]
- 37.Fourmy D, Recht MI, Puglisi JD. Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 S rRNA. J. Mol. Biol. 1998;277:347–362. doi: 10.1006/jmbi.1997.1552. [DOI] [PubMed] [Google Scholar]
- 38.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 39.Walter F, Putz J, Giege R, Westhof E. Binding of tobramycin leads to conformational changes in yeast tRNA(Asp) and inhibition of aminoacylation. EMBO J. 2002;21:760–768. doi: 10.1093/emboj/21.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikkelsen NE, Johansson K, Virtanen A, Kirsebom LA. Aminoglycoside binding displaces a divalent metal ion in a tRNA-neomycin B complex. Nat. Struct. Biol. 2001;8:510–514. doi: 10.1038/88569. [DOI] [PubMed] [Google Scholar]
- 41.Kirillov S, Vitali LA, Goldstein BP, Monti F, Semenkov Y, Makhno V, Ripa S, Pon CL, Gualerzi CO. Purpuromycin: an antibiotic inhibiting tRNA aminoacylation. RNA. 1997;3:905–913. [PMC free article] [PubMed] [Google Scholar]
- 42.Rambelli F, Brigotti M, Zamboni M, Denaro M, Montanaro L, Sperti S. Effect of the antibiotic purpuromycin on cell-free protein-synthesizing systems. Biochem. J. 1989;259:307–310. doi: 10.1042/bj2590307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landini P, Corti E, Goldstein BP, Denaro M. Mechanism of action of purpuromycin. Biochem. J. 1992;284(Pt 1):47–52. doi: 10.1042/bj2840047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Wang ED, Wang YL. Overproduction and purifcation of Escherichia coli tRNALeu. Sci. China (Ser. C) 1998;41:225–231. [Google Scholar]
- 45.Xiao M, Leibowitz MJ, Zhang Y. Concerted folding of a Candida ribozyme into the catalytically active structure posterior to a rapid RNA compaction. Nucleic Acids Res. 2003;31:3901–3908. doi: 10.1093/nar/gkg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du X, Wang ED. Tertiary structure base pairs between D- and TpsiC-loops of Escherichia coli tRNA(Leu) play important roles in both aminoacylation and editing. Nucleic Acids Res. 2003;31:2865–2872. doi: 10.1093/nar/gkg382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen JF, Guo NN, Li T, Wang ED, Wang YL. CP1 domain in Escherichia coli leucyl-tRNA synthetase is crucial for its editing function. Biochemistry. 2000;39:6726–6731. doi: 10.1021/bi000108r. [DOI] [PubMed] [Google Scholar]
- 48.Perez-Salas UA, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry. 2004;43:1746–1753. doi: 10.1021/bi035642o. [DOI] [PubMed] [Google Scholar]
- 49.Xiao M, Li T, Yuan X, Shang Y, Wang F, Chen S, Zhang Y. A peripheral element assembles the compact core structure essential for group I intron self-splicing. Nucleic Acids Res. 2005;33:4602–4611. doi: 10.1093/nar/gki770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fox KR, Sansom CE, Stevens MF. Footprinting studies on the sequence-selective binding of pentamidine to DNA. FEBS Lett. 1990;266:150–154. doi: 10.1016/0014-5793(90)81527-u. [DOI] [PubMed] [Google Scholar]
- 51.Doyle ML. Characterization of binding interactions by isothermal titration calorimetry. Curr. Opin. Biotechnol. 1997;8:31–35. doi: 10.1016/s0958-1669(97)80154-1. [DOI] [PubMed] [Google Scholar]
- 52.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tukalo M, Yaremchuk A, Fukunaga R, Yokoyama S, Cusack S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- 54.Carter NS, Berger BJ, Fairlamb AH. Uptake of diamidine drugs by the P2 nucleoside transporter in melarsen-sensitive and -resistant Trypanosoma brucei brucei. J. Biol. Chem. 1995;270:28153–28157. doi: 10.1074/jbc.270.47.28153. [DOI] [PubMed] [Google Scholar]
- 55.Croft SL, Brazil RP. Effect of pentamidine isethionate on the ultrastructure and morphology of Leishmania mexicana amazonensis in vitro. Ann. Trop. Med. Parasitol. 1982;76:37–43. doi: 10.1080/00034983.1982.11687502. [DOI] [PubMed] [Google Scholar]
- 56.Hentzer B, Kobayasi T. The ultrastructural changes of Leishmania tropica after treatment with pentamidine. Ann. Trop. Med. Parasitol. 1977;71:157–166. doi: 10.1080/00034983.1977.11687174. [DOI] [PubMed] [Google Scholar]
- 57.Basselin M, Denise H, Coombs GH, Barrett MP. Resistance to pentamidine in Leishmania mexicana involves exclusion of the drug from the mitochondrion. Antimicrob. Agents Chemother. 2002;46:3731–3738. doi: 10.1128/AAC.46.12.3731-3738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.