Abstract
The RNA import complex (RIC) from the mitochondrion of the kinetoplastid protozoan Leishmania tropica contains two subunits that directly bind to import signals on two distinct subsets of tRNA and interact with each other allosterically. What happens to the tRNA subsequent to its loading on the complex is unknown. A third subunit—RIC9—has intrinsic affinity for both types of tRNA and is essential for import in vivo. Here we show that antibody against RIC9 inhibited the import of both types of tRNA into mitoplasts in vitro, but failed to inhibit the binding of these tRNAs to their respective receptors, indicating that RIC9 acts in a subsequent step. Using photoaffinity crosslinking-immunoprecipitation to detect translocation intermediates, it was observed that tRNA was transferred from its cognate receptor to RIC9, followed by translocation across the membrane and release as free tRNA in the inner compartment. Transfer required elevated temperatures and ATP, but ATP was substituted by acid pH. These tRNA movements were sensitive to uncouplers and inhibitors, suggesting distinct roles of the electrical and chemical components of the proton motive force generated by vectorial proton translocation accompanying ATP hydrolysis. By analysis of partially assembled complexes in L. tropica depleted of various subunits, and in vitro assembly assays, RIC9 was shown to make stable contacts with RIC8A, a tRNA receptor and RIC6, a membrane-embedded component. The results have implications for the mechanism of tRNA import.
INTRODUCTION
Import of nucleus-encoded tRNAs into mitochondria occurs in a large number of species, to compensate for the evolutionary loss of the corresponding mitochondrial tRNA genes (1,2). An extreme example is kinetoplastid protozoa of genera Leishmania and Trypanosoma, in which the mitochondrial genomes are completely devoid of tRNA genes, necessitating the large-scale import of tRNAs from the cytosol to maintain mitochondrial translation (3,4).
The mechanisms of recognition and translocation of the hypercharged tRNA molecules of an awkward (L-shaped) structure across two mitochondrial membranes against a potential gradient are largely unresolved at present. Presumably, import of tRNAs from the cytosolic compartment, would involve (i) the chaperoning of the tRNA to import sites on the outer membrane; (ii) recognition of importable tRNAs, or discrimination against non-imported species; (iii) translocation though the outer membrane; (iv) recognition or discrimination at the inner membrane; (iv) translocation though the inner membrane into the matrix compartment. Significant progress has been recently achieved by the development of in vitro import systems from yeast (5), plants (6) and kinetoplastid protozoa (7–10) and by the application of gene knockdown protocols to identify import factors. So far, these studies indicate similarities as well as differences in the import mechanism in different organisms.
There is general agreement that in all organisms, tRNA import is mediated by protein factors or complexes on the mitochondrial membranes, but some in vitro systems additionally require soluble carrier proteins, while others do not. Both membrane-bound and soluble factors have been recently identified. In Arabidopsis, three different outer membrane-bound tRNA-binding import factors have been described (11), while in yeast, components of the inner and outer membrane protein import complexes have been implicated (12). In the latter system, soluble carrier proteins (lysyl tRNA synthetase and enolase) are involved (13,14). A soluble cytosolic tRNA-binding protein (EF1α) is also required for import into T. brucei mitochondria in vivo but not in vitro (15). Finally, a functional import complex of several proteins has been isolated from L. tropica (see subsequently).
In the L. tropica in vitro import system, as well as in transiently transfected cells, there is evidence for interactions between two different types of importable tRNA at the inner membrane (16). Type I tRNAs are imported efficiently by themselves, whereas import of type II tRNAs is stimulated by type I tRNAs; conversely, type II tRNAs inhibit the import of type I substrates. These two tRNA types differ in the sequence motifs recognized by the import apparatus (17), and interact with distinct receptors (see subsequently). Such allosteric interactions may help to balance the tRNA pool in the matrix, and must be adequately accounted for by any proposed import mechanism.
A combination of biochemical and genetic approaches is being used to define components of the inner membrane-associated import apparatus of L. tropica. A large (∼600 kDa) multi-protein complex (the RNA import complex, or RIC) was isolated from Leishmania mitochondria and shown to be functional for the translocation of tRNAs across artificial (18) or mitochondrial (19) membranes. This complex contains several tRNA-binding proteins and a tRNA-dependent ATPase (18,20). The genes for the major subunits have been identified (21–23). The largest subunit, RIC1, binds type I tRNAs (21) and is essential for the import of this subset in vitro (18) as well as in vivo (21). The other tRNA subset (type II) is recognized by RIC8A (22). Binding of type II tRNAs to RIC8A is positively regulated by the RIC1–tRNA complex, while that of type I tRNAs is inhibited by RIC8A complexed with type II tRNA (18,22). Moreover, in vivo, downregulation of the type I receptor RIC1 results in mitochondrial depletion of both type I and type II tRNAs, whereas import of type I tRNAs is stimulated in cells depleted of the type II receptor RIC8A (21,22), as expected from the operation of the allosteric interactions in vivo.
Not much is known about the events subsequent to the binding of tRNA to the receptors. All the in vitro import systems require ATP for translocation. Additionally, in the L. tropica (24), yeast (12) and plant (6) systems, a membrane potential is also required (as judged by sensitivity of import to potential-dissipating protonophores), although the L. tarentolae system appears to be resistant to these inhibitors (10). There is also clear evidence for the requirement of a membrane potential in T. Brucei in vivo (15). It is possible that, at least in some systems, ATP hydrolysis (mediated in L. tropica by RIC1) results in proton pumping across the membrane, resulting in a proton gradient that drives import (20).
To better define the translocation step, we looked for additional tRNA-binding subunits of the import complex. One such candidate is RIC9, a major RNA-binding component of the purified complex (Chatterjee,S. and S. Adhya,S., unpublished data). RIC9 is the smallest subunit of size 19 kDa. It is encoded by a single gene with partial structural homology to subunit VI (COXVI) of cytochrome c oxidase (complex IV) (23). Antibody against RIC9 detected the presence of a cross-reactive 19 kDa protein in complex IV (23); since no other COXVI-related sequence is observed in the Leishmania genome, this is likely to be a bifunctional protein. Knockdown of RIC9 by expression of the corresponding antisense RNA resulted in depletion of mitochondrial tRNAs and loss of mitochondrial function, suggesting its involvement in import (23). In this report, we have examined the role of RIC9 in the translocation of tRNAs across membranes. The results suggest that RIC9 acts as a transit stop for tRNAs traveling from the receptor to the pore, and that this transient interaction is energized by a proton gradient across the membrane.
MATERIALS AND METHODS
Cloning and expression of RIC9 gene
The PCR amplification of the RIC9 gene from L. tropica genomic DNA has been described (23). The complete gene was inserted into vector pGEX4T-1 (Amersham, Buckinghamshire, UK) and expressed in Escherichia coli BL21 as a glutathione-s-transferase fusion protein. Recombinant RIC9 was cleaved off the fusion protein and gel-purified as described (21). Prior to assay, 200 μl of the eluate (∼3 μg/ml protein, in 0.2% w/v SDS, 0.05M Tris–HCl, pH 7.5, 0.1 mM EDTA, 5 mM DTT, 0.1 mg/ml BSA, 200 mM NaCl) was diluted 5-fold in TETN250 buffer (250 mM Tris–HCl, pH 7.5, 5 mM EDTA, 250 mM NaCl, 1%v/v Triton-X-100 and 2 mg/ml BSA) and kept for 2 h on ice in order to refold the protein. The protein was finally concentrated by ultrafiltration to ∼20 μg/ml (∼1 pmol/μl). The final detergent concentrations in the concentrated protein solution are estimated to be 1% Triton X-100, 0.04% SDS.
Knockdown of RIC9 and other subunits
The RIC9 gene was inserted in the antisense orientation between the HindIII and BamH1 sites downstream of the tetracycline-inducible T7 RNA polymerase promoter in targeting vector pGET (21), to form the recombinant pGET(RIC9AS). Expression host L. tropica 13–90 was transfected with the recombinant carrying a BLE (bleomycin-resistant) marker gene, and phleomycin-resistant colonies were selected. A selected clone was cultured and induced with 10 μg/ml tetracycline for 2 days, resulting in antisense synthesis and growth arrest (23). Mitochondrial analyses were typically carried out on 2 day-induced cultures. Other subunits (RIC6, RIC8A) were similarly knocked down by induction of the appropriate antisense RNAs (23).
Preparation of radiolabeled tRNA
Genes encoding Leishmania tRNAArg (ACG), tRNATrp (CCA), tRNAVal (CAC) and tRNAMet-e (CAU) were PCR-amplified from L. tropica strain UR6 genomic DNA and cloned between the HindIII and BamH1 sites of vector pGEM4Z (Promega), as described (21). Bam H1-linearized plasmids were run-off transcribed in 10 μl reactions containing 40 mM Tris–HCl (pH 7.5), 6 mM MgCl2, 1 mM Spermidine, 1 mM DTT, 0.5 mM each of CTP, ATP and GTP, 0.01 mM UTP, 1 μCi of [α-32P] UTP and 10 units of T7 RNA polymerase at 37°C for 60 min. Each transcript contains a nonanucleotide extension at the 5′-end, derived from the vector polylinker region. 5-Bromouridine (BU)-labeled tRNAs were synthesized under light-limiting conditions in reactions containing, additionally, 20 μM 5-BU triphosphate. Effector RNAs were similarly synthesized except that the amount of [α-32P] UTP in the transcription reaction mix was reduced to one-tenth of the normal, and the UTP concentration was raised from 10 to 250 μM. tRNAs were purified by gel electrophoresis.
Import assay systems
Import was assayed by incubating 32P-labeled tRNAs with intact L. tropica mitochondria, or mitoplasts, or liposomes reconstituted with purified RIC or with mitochondrial extract from induced or tetracycline-induced cultures, adding ribonucleases and analyzing the RNAse-resistant RNA.
Mitoplasts
Mitochondria were prepared by hypotonic lysis of L. tropica promastigotes followed by Percoll gradient centrifugation (25). Mitoplasts were obtained by treating mitochondria with 320 μM digitonin at 4°C to selectively remove the outer membrane (24).
RIC-liposomes
RIC was purified by affinity chromatography from inner mitochondrial membrane fractions of L. tropica (18). Phosphatidyl choline liposomes (50 μg of lipid in 10 μl of buffer DB) were incubated with RIC (100 ng) for 1 h on ice before import assay.
Knockdown extract reconstituted liposomes
Mitochondria were prepared from 2 × 108 promastigotes. Crude mitochondrial fractions (∼400 μg protein) prepared from uninduced or RIC9 knockdown cells were extracted with 40 μl of BAM buffer (50 mM Bis Tris–HCl pH 7.0, 0.75 M aminocaproic acid, 2% w/v dodecyl maltoside), diluted to 200 μl with buffer DB (0.2 M Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 0.1 mM PMSF, 10% v/v glycerol) and concentrated to 30 μl (protein concentration 13 mg/ml) by using a Microcon 10 ultrafilter (Amicon). The ultrafiltration step also serves to remove excess detergent (dodecyl maltoside), change the buffer to the glycerol-containing DB (to facilitate subsequent reconstitution). Before reconstitution, recombinant RIC9 (120 ng in 6 μl, or 20 μg/ml, in 0.2% w/v SDS-containing gel elution buffer) was diluted 5-fold into BAM buffer, kept on ice for 2 h, further diluted to 200 μl with buffer DB and concentrated down to 30 μl by ultrafiltration. Liposomes (50 μg of lipid in 10 μl of dialysis buffer) were incubated with mitochondrial extract (8 μl = ∼100 μg protein) with or without refolded RIC9 (8 ng in 2 μl) for 1 h on ice before import assay (final detergent concentrations: 0.0006% SDS, 0.16% dodecyl maltoside).
RNase protection assay
Mitoplasts or reconstituted liposomes were incubated with 100 fmol of 32P-labeled tRNA (5 nM) and ATP (4 mM) in 20 μl reactions containing buffer BB (10 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 2 mM DTT) for 15 min at 37°C. RNase A (2.5 μg/ml) and RNase T1 (50 units/ml) were added, and incubation continued for an additional 15 min at 37°C. The RNase-protected radiolabeled RNA was recovered by guanidium isothiocyanate extraction and ethanol precipitation and analyzed by urea-6% w/v PAGE (18).
RNA-binding assays
Mitoplasts were incubated with 32P-labeled tRNA (1 nM) in the presence of buffer BB containing 0.1 M KCl on ice for 30 min, washed, and the bound RNA recovered as above. To perform northwestern analysis, blots of recombinant RIC9 were probed with radiolabeled tRNAs (26). Purified and re-folded RIC9 (∼1 pmol/μl, see above) was diluted to ∼50 fmol/μl in buffer DB and 1 μl was incubated with indicated amounts of 32P-labeled tRNA in buffer BB (reaction volume 10 μl, estimated detergent concentrations: 0.05% Triton X-100, 0.002% SDS) in the presence of indicated concentrations of KCl for 30 min on ice. Gel-shift assays were performed on native 5–15% w/v gradient gels (18). Dissociation constants (Kd) were estimated by Scatchard analysis (27). Binding of tRNA to liposomes was assayed by incubating with labeled substrate for 30 min at 4°C in the absence of ATP, followed by washing the liposomes and recovery of the bound RNA.
Photochemical crosslinking and immunoprecipitation
RIC-incorporated liposomes or mitoplasts were incubated with BU-labeled tRNA (1 nM) for 30 min on ice in 20 μl buffer BB containing 0.1 M KCl, and washed with cold buffer DB. The tRNA-loaded vesicles were resuspended in 20 μl of BB containing 4 mM ATP and incubated at 37°C for the indicated times. For time course reactions, the reaction for each time-point was diluted out with cold DB and the vesicles were recovered by centrifugation. The vesicles were then irradiated with 313 nm UV on a transilluminator for 30 min–1 h with cooling, to form RNA–protein crosslinks. SDS was then added to a final concentration of 0.2%. For immunoprecipitation, the crosslinked and dissociated proteins were diluted 20-fold into TETN250 buffer containing a 1 : 50 dilution of polyclonal mouse antiserum, kept on ice for 1 h and the protein–antibody complex recovered by precipitation with Protein-A-Sepharose. Crosslinked complexes or immunoprecipitates were dissociated in SDS–PAGE sample buffer and resolved by 8 M urea-6% w/v PAGE in the presence of 0.05% w/v SDS.
Analysis of mitochondrial complexes
Mitochondria were extracted with BAM buffer as above, and the extracts (∼130 μg protein) subjected to Blue Native (BN) electrophoresis, as described (21). High molecular complexes were visualized by Coomassie staining. This was followed by either western blotting of the resolved complexes, or excision of individual bands and analysis of their subunit compositions by SDS–PAGE (21).
In vitro assembly assay
Liposomes (50 μg lipid) were reconstituted in 20 μl of buffer DB with RIC9-knockdown mitochondrial extract (∼5 mg/ml protein) as described above, in presence or absence of recombinant RIC9 (∼0.4 μg/ml) for 1 h on ice, washed and the membrane-associated complexes were solubilized with BAM buffer. The complexes were run under native (BN–PAGE) or denaturing (SDS–PAGE) conditions, transferred to nitrocellulose membranes and probed with indicated antibodies. Where indicated, mouse polyclonal antiserum (1 : 50) against a specific subunit was added before RIC9.
Protein interaction assay
Recombinant subunits (0.2–0.6 μM) were incubated alone or in combination in 10 μl buffer BB for 1 h on ice, then subjected to native 4–15% gradient PAGE (18), before Coomassie staining or western blotting.
RESULTS
Depletion of RIC9 results in defective import of both type I and type II tRNAs
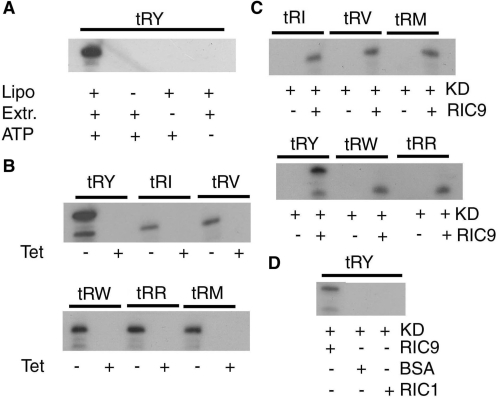
In view of the putative bifunctionality of RIC9, the depletion of mitochondrial tRNAs in vivo upon downregulating RIC9/COX6b could be an indirect consequence of the loss of the respiratory function of this protein, leading to depletion of intracellular ATP. It was therefore pertinent to ask whether the import apparatus was specifically affected in any way. The import function of RIC can be assayed by reconstitution of phospholipid vesicles (liposomes) with purified RIC (18) or mitochondrial extracts (21), followed by detection of ribonuclease-resistant radiolabeled tRNA. Control experiments showed that the import was dependent on the presence of liposomes as well as the extract in the reconstitution mix, and dependent on ATP (Figure 1A). When mitochondrial extracts from uninduced L. tropica antisense transformants were assayed in this way, normal import was observed by RNase protection; in contrast, mitochondrial extracts from RIC9-deficient cells were inactive for import of all the tRNAs tested (Figure 1B). These included type I (tRNATyr, tRNAArg and tRNATrp) as well as type II (tRNAIle, tRNAVal and tRNA) tRNAs. Import was restored by the addition of purified, bacterially expressed RIC9 (Figure 1C), but not by unrelated proteins such as bovine serum albumin (BSA), or other import factors such as RIC1; RIC9 itself was not able to protect the tRNA from RNase digestion (Figure 1D). Thus, the defect in the import apparatus was specifically caused by the deficiency of RIC9.
Figure 1.
Import activity of mitochondrial extracts from RIC9-knockdown Leishmania tropica. (A) Import of tRNATyr into liposomes reconstituted with mitochondrial extracts from uninduced pGET(RIC9AS) transformants of L. tropica 13–90. Indicated combinations of liposomes (50 μg phosphatidylcholine) and mitochondrial extract (5 mg/ml) were incubated together in 20 μl for 1 h at 4°C, then 32P-labeled tRNATyr (3.3 nM) and ATP (4 mM, as indicated) were added, and import assayed by RNase protection. (B) Import of the indicated tRNAs into liposomes reconstituted with mitochondrial extracts from uninduced (−Tet) or RIC9 knockdown (+Tet) cells. In addition to high specific activity import substrate (3.3 nM), 0.33 nM of low specific activity tRNATyr was present as positive effector for type II tRNAs in all cases. (C) Liposomes were reconstituted with mitochondrial extracts from knockdown cells (KD) in the absence or presence of recombinant, refolded RIC9 (0.4 μg/ml), before assay of the indicated tRNAs for import. (D) Liposomes were reconstituted with knockdown mitochondrial extracts in the absence or presence of RIC9, BSA or recombinant RIC1 (0.4 μg/ml each), then assayed for import of tRNATyr, tRY, tRNATyr; tRI, tRNAIle; tRV, tRNAVal; tRW, tRNATrp; tRR, tRNAArg; tRM, tRNAMet-e.
tRNA binding activity of isolated RIC9
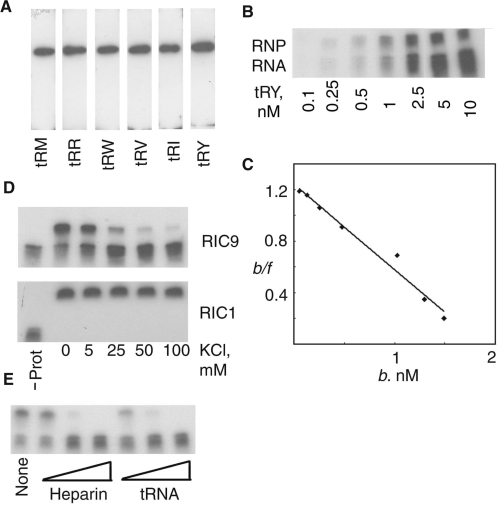
The Leishmania RIC9 gene was expressed in E. coli, the recombinant protein purified and subjected to northwestern analysis using a variety of importable tRNAs as probes. While unrelated proteins such as BSA did not react under these conditions (data not shown), strong signals were obtained with all the tRNAs tested, revealing a general tRNA-binding property of RIC9 (Figure 2A).
Figure 2.
tRNA-binding activity of recombinant RIC9. (A) Northwestern blots of purified recombinant RIC9 (100 pmol) were probed with the indicated 32P-labeled tRNAs (106 c.p.m./ml). (B) Gel-shift assay of refolded RIC9 (5 nM) incubated with indicated amounts of tRNATyr in 10 μl of buffer BB without added salt. (RNP = tRNA–RIC9 complex). (C) Scatchard plot of bound/free (b/f) against bound RNA concentration (b). Data from the gel-shift assay in panel (B). The slope of the best-fit curve = −1/Kd. (D) Effect of KCl on the binding of tRNATyr (1 nM) to 5 nM of recombinant RIC9 (upper panel) or RIC1 (lower panel). (E) Effect of heparin (10, 50 and 100 μg/ml) or E.coli tRNA (1, 5 and 10 nM) on binding of tRNATyr (1 nM) to RIC9 (5 nM).
Gel-shift assays using purified RIC9 revealed a specific tRNA–RIC9 complex (Figure 2B). From Scatchard plots (Figure 2C), the dissociation constant (Kd) of the complex at 4°C in the absence of added monovalent salt was estimated to be 1.5 nM. The interaction was sensitive to salt and to the polyanion heparin, and competed out by an excess of E. coli tRNA (Figure 2D and E). In contrast, binding of tRNA to the import receptor RIC1 was insensitive to KCl up to at least 0.1 M (Figure 2D). Thus, the tRNA-binding activity of RIC9 is non-specific with respect to tRNA sequence and involves mainly ionic interactions.
In the complex, RIC9 is not a tRNA receptor but is required for translocation
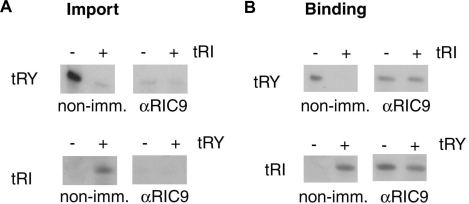
Translocation of tRNA through the import channel is preceded by the binding of the substrate to the appropriate receptor in the import complex. The role of RIC9 in binding and translocation was investigated by immuno-inhibition experiments with wild-type L. tropica mitoplasts. In presence of control (non-immune) serum, import as well as binding of type I tRNATyr were inhibited by low concentrations of type II tRNAIle; in contrast, import and binding of tRNAIle were stimulated by tRNATyr. Anti-RIC9 antibody inhibited the import of type I tRNATyr as well as that of type II tRNAIle (Figure 3A).
Figure 3.
Effect of anti-RIC9 antibody on tRNA import in vitro. (A) Mitoplasts (100 μg protein) from L. tropica strain UR6 were pre-incubated with non-immune or anti-RIC9 serum (1 : 50) for 1 h at 4°C, then incubated for 15 min at 37°C with 4 mM ATP and 5 nM of high specific activity tRNATyr (upper panels) or tRNAIle (lower panels) in the absence or presence of low specific activity effector (0.5 nM) as indicated, and assayed for import by RNase protection. (B) Mitoplasts, pre-incubated as above, were incubated in binding buffer containing 0.1 M KCl with indicated combinations of substrate (1 nM) and effector (0.1 nM) in the absence of ATP for 30 min at 4°C, then washed and the bound RNA recovered.
The same antibody failed to inhibit the binding of tRNATyr to mitoplasts, but in its presence the binding of tRNATyr was no longer inhibited by tRNAIle (Figure 3B). Conversely, in presence of anti-RIC9 antibody, the type II tRNAIle was bound in the absence of tRNATyr (Figure 3B). These results indicate that (i) RIC9 does not directly bind tRNA in the native complex in the absence of ATP; (ii) contacts between RIC9 and the two receptors are altered upon binding of the antibody, leading to the activation of the type II binding site of RIC8A and the loss of contact between tRNA-bound RIC1 and RIC8A and (iii) RIC9 is required for the translocation step.
ATP-dependent transfer of tRNAs from receptors to RIC9
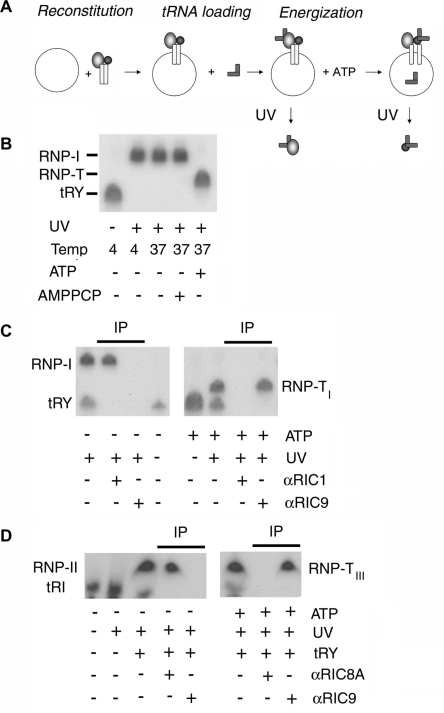
Although RIC9 in isolation has tRNA-binding activity (Figure 2), antibody against this subunit was unable to inhibit tRNA binding to the intact, membrane-bound complex, but inhibits translocation (Figure 3). This indicates either that the tRNA-binding activity of RIC9 is not utilized during import, or that the binding of tRNA to RIC9 in the import complex follows the initial receptor-binding step but precedes the actual translocation. To investigate these possibilities, radiolabeled tRNA containing the photoactivable analog 5-BU was incubated with mitoplasts, or with proteoliposomes reconstituted with purified RIC, under conditions favoring binding or translocation, then crosslinked to the complex, which was subsequently dissociated to its constituent subunits, and the subunit crosslinked to tRNA identified by immunoprecipitation with subunit-specific antibody (Figure 4A). The initial tRNA-binding step occurs at 4°C in the absence of ATP, and under these conditions tRNATyr crosslinks specifically to RIC1 (18).
Figure 4.
ATP-induced transfer of tRNA from import receptors to RIC9. (A) Crosslinking protocol. Liposomes were incubated with purified RIC (25 μg/ml) for 1 h at 4°C, then incubated with 1 nM of 32P- and BU-labeled tRNATyr at 4°C. The resultant tRNA-loaded vesicles were transferred to 37°C in the presence of 4 mM ATP, then UV-irradiated. (B) RIC-liposomes (25 μg/ml) were incubated with 1 nM of BU-labeled tRNATyr at 4°C for 30 min, followed by transfer to 37°C for 5 min in the presence or absence of 4 mM of ATP or AMPPCP as indicated. The reactions were then UV-irradiated and the crosslinked products analyzed by urea–PAGE. (C) RIC-liposomes were incubated with BU-labeled tRNATyr as above, followed by transfer to 37°C in the presence of 4 mM ATP. The resulting complex was UV-irradiated and the crosslinked adduct was immunoprecipitated with anti-RIC9 or anti-RIC1 serum (1 : 50). (D) RIC-liposomes were incubated as above (panel C) with 1 nM of BU-labeled tRNAIle. Low specific activity effector tRNATyr (0.1 nM) was present as indicated. The crosslinked adducts were immunoprecipitated with anti-RIC9 or anti-RIC8A serum (1 : 50). RNP-I, RIC1–tRNATyr complex; RNP-TI, RIC9–type I tRNA complex.
In an initial experiment, tRNATyr, double-labeled with 32P and BU, was incubated with mitoplasts at 4°C in absence of ATP; excess unbound label was washed off; the bound RNA was photocrosslinked to mitoplast protein; and the ribonucleoprotein (RNP) was analyzed by gel electrophoresis. A discrete RNP (designated as RNP-I) was observed, migrating slower than the uncrosslinked RNA (Figure 4A). If, after the initial binding at 4°C, followed by washing off excess label, the incubation temperature was raised to 37°C, there was no change in the mobility of the RNP. To identify the RIC subunit crosslinked to the tRNA, the liposome-bound RIC was dissociated after UV-irradiation, and the RNP immunoprecipitated with subunit-specific antibody (Figure 4C). The RNP of lower mobility formed at 37°C in the absence of ATP was precipitated by anti-RIC1 antibody, as previously observed for the complex formed at 4°C (18), but not by anti-RIC9 (Figure 4C, left). This confirms that RIC9 is not the initial tRNA-binding site in the complex (see above).
On addition of ATP at 37°C, the mobility of the RNP was increased (RNP-TI; Figure 4B). RNP-TI was pulled down by anti-RIC9 but not by anti-RIC1 (Figure 4C, right). The same result was obtained with intact mitoplasts instead of RIC-proteoliposomes (data not shown). Formation of the tRNA: RIC9 complex required hydrolysis of ATP, since the non-hydrolyzable analog βγ-methylene ATP could not substitute for ATP (Figure 4B). This experiment demonstrates that tRNA is transferred from RIC1 to RIC9 upon ATP hydrolysis.
tRNATyr is a Type I tRNA that, through interaction with RIC1, promotes the binding of Type II tRNAs to their cognate receptor, RIC8A (18,22). To examine the movement of Type II tRNAs subsequent to receptor binding, BU-labeled tRNAIle (Type II) was loaded on to the complex in presence of tRNATyr effector, then incubated under different conditions as above, before crosslinking. In the absence of ATP, the crosslinked complex (RNP-II) was immunoprecipitated by anti-RIC8A antibody, as expected (Figure 4D, left). In the presence of ATP, a complex (RNP-TII) of about the same mobility as RNP-II was formed, but this was specifically precipitated by anti-RIC9, but not by anti-RIC8A antibody (Figure 4D, right). Thus, both tRNA types are transferred from their cognate receptors to RIC9.
Stepwise movements of tRNA in presence of ATP
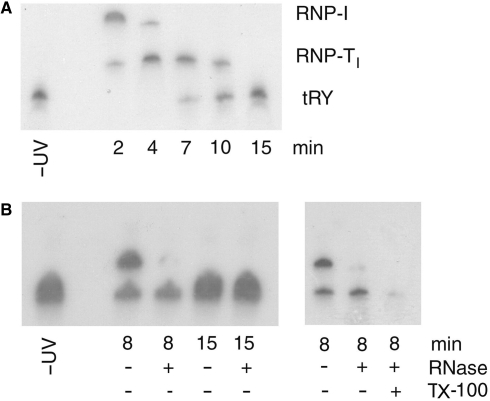
To examine the kinetics of transfer of tRNA from the receptor to RIC9, the BU-labeled tRNATyr-RIC complex on liposomes was incubated for different times at 37°C in presence of ATP, then UV-crosslinked and the RNA adduct analyzed by gel electrophoresis (Figure 5A). Between 0 and 10 min, there was a progressive reduction in the receptor complex RNP-I. The amount of the RIC9 complex (RNP-TI) increased to a peak at ∼4 min, and then declined. Between 7 and 15 min, the formation of a species migrating as free tRNA was observed (Figure 5A).
Figure 5.
Time course of tRNA movement from receptor to import pore. (A) BU-labeled tRNATyr (1 nM) was bound to 25 μg/ml of RIC-liposomes. The bound complex was transferred to 37°C in the presence of ATP and incubated for various times before UV-crosslinking and gel analysis. (B) Left, the crosslinked products obtained upon incubation at 37°C for 8 or 15 min were digested with RNase (2.5 μg/ml RNase A, 50 U/ml RNase T1). Right, the crosslinked product obtained after incubation at 37°C for 8 min was treated with 0.5% v/v Triton X-100 prior to RNase digestion.
The locations of the free and bound tRNA on the liposomes were determined by their RNase sensitivity following the crosslinking step. The RNP-TI complex (as well as the RNP-I complex; data not shown) formed at early and intermediate incubation times was RNase sensitive (Figure 5B), indicating their location of the liposome surface. In contrast, the free tRNA formed at later times was resistant to RNase, but became sensitive upon disruption of the liposomes with detergent (Figure 5B), and therefore represents imported tRNA. Thus, in the presence of ATP, the tRNA is first transferred from the receptor to RIC9 on the membrane surface, and this is followed by its translocation through the membrane.
H+-dependent transfer of tRNA to RIC9
Vectorial membrane-bound ATPases such as the vacuolar ATPase and the F1 ATPase (the ATP synthase acting hydrolytically) act as ATP-driven proton pumps (28). We have previously shown that tRNA-dependent ATP hydrolysis by RIC generates a proton gradient across the phospholipid membrane that is sensitive to protonophores and ionophores, and that acidification of the medium results in import in the absence of ATP, implying the proton gradient as the major driving force (20). Complex-bound RIC1 carries out this reaction (29).
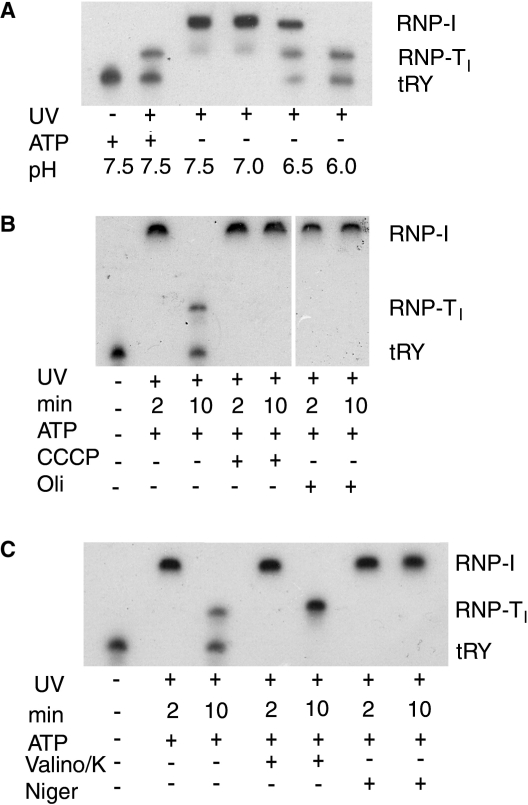
Thus, it was of interest to determine the role of protons in the stepwise transfer of tRNA from the receptor to the import pore. The tRNATyr–RIC complex, bound to liposomes, was incubated at 37°C for 10 min at different pH values in the absence of ATP, then UV-crosslinked and analyzed by gel electrophoresis as before. Between pH 7.0 and 7.5, the tRNA remained bound to RIC1 (as RNP-I), but further lowering of the pH to ∼6 resulted in transfer to RIC9 (RNP-TI) and release as free imported tRNA (Figure 6A), mimicking the situation with ATP at neutral pH. Thus, protons effectively substitute for ATP as the energy source for translocation.
Figure 6.
Role of protons in tRNA translocation. (A) RIC-liposomes (25 μg/ml) were washed and incubated in buffers of indicated pH in the absence or presence of 4 mM ATP, with 1 nM of BU-labeled tRNA. The vesicles containing the bound RNAs were then transferred to 37°C for 10 min and then UV crosslinked. (B) BU-labeled tRNATyr was bound to RIC-liposomes which had been pre-incubated with 50 μM of either m-chlorocarbonylcyanide phenylhydrazone (CCCP) or oligomycin, and incubated at 37°C in the presence of 4 mM ATP for 2 or 10 min. (C) Effect of valinomycin (50 μM) plus 50 mM KCl, or nigericin (50 μM) on translocation. Reaction conditions were as in (B).
In the presence of the protonophore uncoupler CCCP, which dissipates the gradient by equilibrating protons across the membrane, ATP-dependent translocation of tRNA from RIC1 to RIC9 was abolished (Figure 6B). The same step was inhibited by oligomycin (Figure 6B), which does not inhibit RIC1-dependent ATPase but blocks proton-driven import (20), presumably by inhibiting back-flow of protons. These observations indicate the crucial role of protons generated by ATP hydrolysis in tRNA translocation.
Separate roles of the chemical and electrical components of the proton motive force
The electromotive force generated by vectorial transport of protons [the proton motive force (PMF)] is the sum of chemical (ΔpH) and electrical (Δψ) components. The role of these two components can be assessed by the use of ionophores that selectively dissipate either ΔpH or Δψ. In the presence of valinomycin and K+, which neutralizes Δψ by equilibrating K+ across the membrane, the tRNA was transferred from RIC1 to RIC9, but failed to translocate across the pore as free intra-vesicle tRNA (Figure 6C). K+ alone had no effect (data not shown). In contrast, nigericin that carries out electroneutral exchange of H+ with K+ (30), and therefore specifically affects ΔpH, inhibited the transfer of tRNA from RIC1 to RIC9 (Figure 6C). Thus, protons are required to move the tRNA from one protein to another of the membrane surface, whereas the electrical potential drives the tRNA across the membrane.
RIC9 contacts both membrane-embedded and receptor subunits within the import complex
The structure of the RIC is currently unknown. However, transient and stable interactions between the subunits can be revealed by a combination of biochemical and genetic methods. One approach is based on the prediction that the subunits are assembled in an ordered manner. Therefore, knockdown of a given subunit (X) will result in the accumulation of a partially assembled RIC sub-complex containing all the subunits assembled up to X in the assembly pathway, but lacking X as well as all subunits assembled after X. Moreover, addition of purified X to an extract from X-knockdown cells will result in the formation of a complex containing X as well as the other missing subunits.
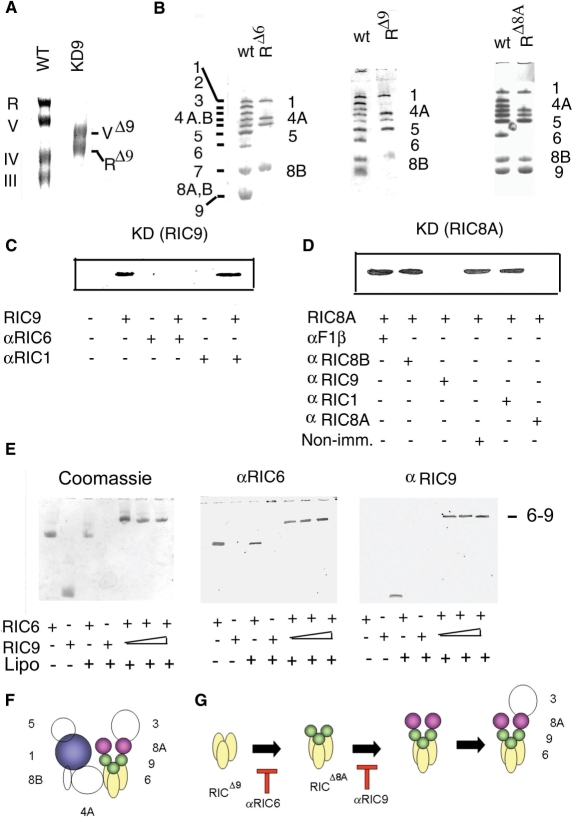
Intracellular RIC9 was depleted by inducible antisense-mediated knockdown of the corresponding mRNA (23). Inner membrane macromolecular complexes in mitochondrial extracts from the growth-arrested cells were resolved by BN electrophoresis (21). As shown previously (21), wild-type cells have four high molecular weight complexes—complex III (ubiquinol cytochrome c reductase), complex IV (cytochrome c oxidase), complex V (F1Fo ATP synthase) and RIC (Figure 7A). Knockdown of RIC9 resulted in the disappearance of complexes III and IV, and the formation of a smaller complex V (VΔ9) lacking the mitochondrion-encoded subunit A6 (data not shown). In normal cells, RIC contains 11 major subunits, including the single copy type I receptor RIC1, two copies of the type II receptor RIC8A and three copies each of RIC6 and RIC9 (23; Figure 7B and F) In RIC9 knockdown cells, a subcomplex of RIC (RICΔ9) was observed, which, on subunit analysis by second-dimension SDS–PAGE, was found to lack subunits 9, 8A and 3 (all nucleus-encoded) as well as the mitochondrion-encoded subunits 2, 4B and 7 (Figure 7B). The absence of organelle-encoded subunits is a general feature of the knockdown of essential import subunits (such as RIC6 and RIC8A; Figure 7B), which leads to depletion of the intramitochondrial tRNA pool, and consequently, to the inhibition of organellar protein synthesis. The lack of subunits 8A and 3 in RICΔ9 indicates that these subunits are assembled after RIC9.
Figure 7.
Interaction of RIC9 with other subunits during complex assembly at the membrane. (A) Effect of RIC9 knockdown on mitochondrial inner membrane complexes. Mitochondrial extracts from uninduced (WT) or tetracycline-induced cultures of L. tropica 13–90 transformed with pGET(RIC9AS) (KD9) were subjected to BN electrophoresis, and the protein complexes visualized by Coomassie staining. The positions of RIC (R) and respiratory complexes V, IV and III in the WT extract are indicated; RΔ9 and VΔ9 are the subcomplexes in knockdown cells derived from RIC and complex V, respectively. (B) Wild-type RIC (wt) and knockdown complexes (RΔX) from cells deficient in subunit X, where X = subunit 6, 8A or 9. (C and D) In vitro assembly assay. Complexes from RIC9 (C) or RIC8A (D) knockdown cells were reconstituted in presence of liposomes and recombinant RIC9 or RIC8A, respectively. Where indicated, antibody (α) against a specific RIC subunit was present during the reconstitution reaction. Liposome-bound complexes were resolved by native (C) or denaturing (D) PAGE, western blotted and probed with anti-RIC9 (C) or anti-RIC8A (D) antiserum. (E) Interaction between RIC6 and RIC9 at the membrane. Recombinant RIC6 (0.2 μM) and RIC9 (0.2–0.6 μM) were incubated alone or in combination with liposomes, which were washed and the liposome-bound proteins resolved by native PAGE. Left, Coomassie stain. Duplicate lanes were blotted on nitrocellulose and probed with anti-RIC6 (middle) or anti-RIC9 (right) serum. (F) Hypothetical model of RIC, showing the nucleus-encoded subunits. Subunit numbering and stoichiometry are as in ref. (23). Subunits 1, 8A, 9 and 6 are highlighted in color. (G) Sequential assembly of subunits 6, 9 and 8A, indicating the steps blocked by specific antibodies. For clarity, the common subunits of the knockdown complexes (1, 4A, 5 and 8B, and the mitochondrion-encoded subunits 2, 4B and 7) are not shown.
A comparative analysis of the subunit compositions of partially assembled sub-complexes in cells depleted for various subunits showed that RICΔ9 was identical to RICΔ6 (the sub-complex in RIC6-depleted cells) except that the former contained RIC6 (Figure 7B). Similarly, RICΔ8A contained all the subunits of RICΔ9 and additionally, RIC9 (Figure 7B). These results indicate that the subunits are assembled in the following order: RICΔ6 + 6 → RICΔ9 + 9 → RICΔ8A + 8A → RICΔ3 (Figure 7G).
To study the interaction between RIC9 and other subunits of the import complex, the RIC9–knockdown complex was incubated with liposomes in the presence of recombinant RIC9. The washed liposomes were then analyzed for the presence of RIC9 by western blot. This showed the presence of RIC9 in the membrane-bound complex (Figure 7C). To identify the subunit in the pre-assembled sub-complex RICΔ9 that directly interacts with RIC9, the reconstitution reaction was carried out after pre-incubating the liposome-bound RICΔ9 with antibodies against the other subunits. Antibody against RIC6 inhibited the binding of RIC9 to the complex, as shown by western blot analysis (Figure 7C). In contrast, antibodies against other subunits, e.g. RIC1, did not prevent assembly of RIC9 (Figure 7C), indicating specificity of the immuno-inhibition. Both antibodies were bound to the complex, as indicated by a gel-shift assay (data not shown). These results illustrate the direct interaction between RIC6 and RIC9.
In a similar experiment, the assembly of recombinant RIC8A to the RIC9–knockdown complex was assayed in the presence of antibodies against various RIC subunits. Western analysis of the liposome-bound complex showed that RIC8A assembly was specifically inhibited by anti-RIC9 antibody (as well as, expectedly, by the self, i.e. anti-RIC8A, antibody (Figure 7D). In summary, these immuno-inhibition experiments confirm the direct, stable interactions between RIC9 and subunits 6 and 8A during assembly of the complex.
RIC6, identical to the iron sulfur protein (ISP) subunit of complex III, is essential for import, and is present in the stoichiometry of 3 mol/mol of RIC (23); the stoichiometry is identical to that of RIC9. A long hydrophobic α-helix in the modeled structure of RIC6 suggests the possibility of membrane insertion. This was tested by incubating the purified subunit with liposomes, separating the liposomes by centrifugation, solubilizing the bound proteins and separating them by native gel electrophoresis. It was observed that RIC6 alone, but not RIC9, was bound to liposomes (Figure 7E). In the presence of both subunits, a larger membrane-bound heterodimer was observed, containing RIC6 as well as RIC9 in a roughly 1 : 1 ratio, as shown by western blotting, and the pattern was not altered by titrating with RIC9 (Figure 7E). Thus, the two subunits interact to form stable membrane-associated heterodimers, but further polymerization of the heterodimers was not observed; this would presumably require one or more additional RIC subunits.
DISCUSSION
In this report, we present evidence that RIC9, the 19 kDa subunit of Leishmania RIC is involved in the translocation of several tRNAs from import receptors through the import pore. (i) RIC9 is an RNA-binding protein with no specificity for tRNA sequence. (ii) tRNA binding by RIC9 involves reversible ionic interactions, as expected of a transport factor interacting transiently with the import substrate. (iii) Antibody against RIC9 inhibits translocation but not binding of tRNA. (iv) Energy-driven movement of tRNA from the receptor to RIC9, followed by delivery to the pore, was observed.
RIC9, and a second subunit, RIC6, are each present in the mole ratio of 3 per complex (13). Experiments on the assembly of RIC showed that RIC6 is tethered to the membrane, where it directly interacts with RIC9, and that RIC9 in turn contacts the type II receptor RIC8A (Figure 7). A reasonable hypothesis is that RIC9 and RIC6 form a hexameric complex that functions in translocation, with RIC9 transferring tRNAs from receptors to the channel constituted by RIC6. Although both the transfer subunit and the channel are activated by the PMF, RIC9 is unlikely to be an integral part of the transmembrane channel, since the RNA bound to it is sensitive to RNAse (Figure 5) and is therefore, exposed to the exterior of the vesicles. The tRNA is released directly from RIC9 into the membrane interior in the free form, since we could not detect any other crosslinked RNP complex prior to the appearance of imported tRNA (Figure 5); however, transient interactions with RIC6 or some other subunit cannot be excluded at this time.
RIC9 is encoded by a single Leishmania gene homologous to COX6b, and is shared by RIC and Complex IV. Thus, in common with RIC1 (21) and RIC8A (22), it appears to be a bi-functional protein with both oxidative phosphorylation and tRNA import activity. A second common feature of these three import components is the presence of an extra domain, or extension, which appears to confer upon the protein an import-related activity. In case of RIC1 (21) and RIC9 (23), a C-terminal extension is present, while the additional domain of RIC8A is at the N-terminus (22). Thus, RIC9 is a further example of evolution of bi-functionalism by domain addition.
The plant and protozoal tRNA import systems share a number of common features such as the involvement of membrane-bound proteins, and the requirement of a trans-membrane potential (1,2). The protozoal RIC9/COX6b is closely related to COX6b from higher plants (23). In Arabidopsis and rice, multiple COX6b genes have been found, which code for long and short versions of the protein (31). The function of the long gene is unknown; it may have been created during evolution by exon addition to the short gene. The long gene from plants resembles Leishmania RIC9/COX6b in having an extra domain or extension (at the N-terminus).
In potato mitochondria, the voltage-dependent anion-selective channel (VDAC) is required for tRNA import (11). VDAC in the isolated state binds tRNA, but in intact mitochondria or in the import complex, the tRNA-binding site is not exposed, since antibodies do not inhibit receptor binding but affect translocation (11). Functionally, these properties resemble those of RIC9, but no sequence similarities between the two proteins are apparent; this may be an example of different proteins having undergone convergent evolution to a tRNA transfer factor.
The tRNA-binding property of recombinant RIC9 is different from that of the previously described import receptors for type I and type II tRNAs (21,22). RIC9 binds both types of tRNA, and binding is sensitive to salt as well as polyanions (Figure 2). In contrast, binding of tRNAs to the import receptors is sequence/structure specific, and insensitive to agents that disrupt ionic interactions. These observations, made with purified recombinant proteins, recall our earlier results with isolated mitochondria showing that import, but not binding, of RNA is salt- and heparin-sensitive (7). In fact, the dose–response curves of RIC9 binding and import with respect to salt and heparin are very similar: monovalent salt inhibits above 0.1 M while heparin inhibits at 10–50 μg/ml.
The present photocrosslinking studies, coupled with the use of inhibitors, show that RIC9-mediated movements of tRNA from import receptors to and through the import pore are sensitive to protonophores, and that acidification substitutes for ATP in the process (Figure 6). Thus, translocation is powered by the PMF generated by hydrolysis of ATP. The contrasting effects of valinomycin and nigericin (Figure 6), which alter different components of the PMF, further suggest that the PMF acts in two ways: protons move the tRNA from the receptor to RIC9, while the tRNA is released from RIC9 into the pore by the membrane potential.
The chemical requirement of protons for the first step suggests protonation of one or more critical residues on the RNA or one of the proteins. Since the transfer from RIC1 to RIC9 is completed within the narrow pH range of 7–6 (Figure 6), it is unlikely that protonation of the phosphate groups of the RNA (pKa ∼ 1–2) or carboxylate groups of the protein (pKa ∼ 5) are involved. On the other hand, histidine residues (pKa ∼ 7 for the imidazole group) are the only protein side chains that would be protonated in this pH range. Hydrogen bonding between histidine residues and specific RNA bases would thus be affected by changes in pH, leading to either destabilization of the RIC1–tRNA complex or stabilization of the RIC9–tRNA complex. RIC1 is a bifunctional form of the α subunit of F1Fo ATP synthase with a C-terminal extension that is involved in tRNA binding (21). The C-terminal 100-residue region contains four of the nine histidine residues of this 574-amino acid protein; the involvement of these residues remains to be tested.
The movement of tRNA from RIC9 through the membrane pore requires the membrane potential (Δψ) established by proton pumping. According to the Nernst equation (32), a pH gradient between 6 and 7 (a 10-fold concentration gradient) would generate a Δψ of ∼60 mV, negative inside. Thus the pore is likely to be voltage-gated. It is interesting to note that RIC6, the putative pore component (see above), is a Fe-S protein (23); whether the paramagnetic Fe2+ ions sense voltage is a possibility worth investigating.
ACKNOWLEDGEMENTS
Supported by CSIR Network Project # NWP0038. S.B. and S.M. received Fellowships from the Council of Scientific and Industrial Research. Funding to pay the Open Access publication charges for this article was provided by the above project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bhattacharyya SN, Adhya S. The complexity of mitochondrial tRNA import. RNA Biol. 2004;1:84–88. doi: 10.4161/rna.1.2.1180. [DOI] [PubMed] [Google Scholar]
- 2.Mirande M. The ins and outs of tRNA transport. EMBO Rep. 2007;8:547–549. doi: 10.1038/sj.embor.7400989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson AM, Suyama Y, Dewes H, Cambell D, Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock K, Hajduk SL. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 1990;265:19203–19215. [PubMed] [Google Scholar]
- 5.Tarassov IA, Entelis NS. Mitochondrially-imported cytoplasmic tRNALys(CUU) of Saccharomyces cerevisiae: in vivo and in vitro targeting systems. Nucleic Acids Res. 1992;20:1277–1281. doi: 10.1093/nar/20.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delage L, Dietrich A, Cosset A, Marechal-Drouard L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol Cell Biol. 2003;23:4000–4012. doi: 10.1128/MCB.23.11.4000-4012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahapatra S, Adhya S. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J. Biol. Chem. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- 8.Yermovsky-Kammerer AE, Hajduk SL. In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol. Cell. Biol. 1999;19:6253–6259. doi: 10.1128/mcb.19.9.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nabholz CE, Horn EK, Schneider A. tRNAs and proteins are imported into mitochondria of Trypanosoma brucei by two distinct mechanisms. Mol. Biol. Cell. 1999;10:2547–2557. doi: 10.1091/mbc.10.8.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubio MAT, Liu X, Yuzawa H, Alfonso JD, Simpson L. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA. 2000;6:988–1003. doi: 10.1017/s1355838200991519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salinas T, Duchene A-M, Delage L, Nilsson S, Glaser E, Zaepfel M, Marechal-Drouard L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:18362–18367. doi: 10.1073/pnas.0606449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarassov I, Entelis N, Martin RP. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J. Mol. Biol. 1995;245:315–323. doi: 10.1006/jmbi.1994.0026. [DOI] [PubMed] [Google Scholar]
- 13.Tarassov I, Entelis N, Martin RP. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Entelis N, Brandina I, Kamenski P, Krasheninnikov IA, Martin RP, Tarassov I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–1620. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouzaidi-Tiali N, Aeby E, Charriere F, Pusnik M, Schneider A. Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. EMBO J. 2007;26:4302–4312. doi: 10.1038/sj.emboj.7601857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharyya SN, Chatterjee S, Adhya S. Mitochondrial RNA import in Leishmania tropica: aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Mol. Cell. Biol. 2002;22:4372–4382. doi: 10.1128/MCB.22.12.4372-4382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goswami S, Chatterjee S, Bhattacharyya SN, Basu S, Adhya S. Allosteric regulation of tRNA import: interactions between tRNA domains at the inner membrane of Leishmania mitochondria. Nucleic Acids Res. 2003;31:5552–5559. doi: 10.1093/nar/gkg773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharyya SN, Chatterjee S, Goswami S, Tripathi G, Dey SN, Adhya S. “Ping-pong” interactions between mitochondrial tRNA import receptors within a multiprotein complex. Mol. Cell. Biol. 2003;23:5217–5224. doi: 10.1128/MCB.23.15.5217-5224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahata B, Bhattacharyya SN, Mukherjee S, Adhya S. Correction of translational defects in patient-derived mutant mitochondria by complex-mediated import of a cytoplasmic tRNA. J. Biol. Chem. 2005;280:5141–5144. doi: 10.1074/jbc.C400572200. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharyya S, Adhya S. tRNA-triggered ATP hydrolysis and generation of membrane potential by the Leishmania mitochondrial tRNA import complex. J. Biol. Chem. 2004;279:11259–11263. doi: 10.1074/jbc.C300540200. [DOI] [PubMed] [Google Scholar]
- 21.Goswami S, Dhar G, Mukherjee S, Mahata B, Chatterjee S, Home P, Adhya S. A bi-functional tRNA import receptor from Leishmania mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:8354–8359. doi: 10.1073/pnas.0510869103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee S, Home P, Mukherjee S, Mahata B, Goswami S, Dhar G, Adhya S. An RNA-binding respiratory component mediates import of type II tRNAs into Leishmania mitochondria. J. Biol. Chem. 2006;281:25270–25277. doi: 10.1074/jbc.M604126200. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee S, Basu S, Home P, Dhar G, Adhya S. Necessary and sufficient factors for import of tRNA into the kinetoplast-mitochondrion. EMBO Rep. 2007;8:589–595. doi: 10.1038/sj.embor.7400979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee S, Bhattacharyya SN, Adhya S. Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J. Biol. Chem. 1999;274:31249–31255. doi: 10.1074/jbc.274.44.31249. [DOI] [PubMed] [Google Scholar]
- 25.Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghosh A, Ghosh T, Ghosh S, Das S, Adhya S. Interaction of small ribosomal and transfer RNAs with a protein from Leishmania donovani. Nucleic Acids Res. 1994;22:1663–1669. doi: 10.1093/nar/22.9.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharyya SN, Mukherjee S, Adhya S. Mutations in a tRNA import signal define distinct receptors at the two membranes of Leishmania mitochondria. Mol. Cell. Biol. 2000;20:7410–7417. doi: 10.1128/mcb.20.19.7410-7417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Futai M, Oka T, Sun-Wada GH, Moriyama Y, Kanazawa H, Wada Y. Luminal acidification of diverse organelles by V-ATPase in animal cells. J. Exp. Biol. 2000;203:107–116. doi: 10.1242/jeb.203.1.107. [DOI] [PubMed] [Google Scholar]
- 29.Goswami S, Adhya S. The α subunit of Leishmania F1 ATP synthase hydrolyzes ATP in presence of tRNA. J. Biol. Chem. 2006;281:18914–18917. doi: 10.1074/jbc.C600089200. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls DG, Ferguson SJ. Bioenergetics 3. San Diego/London: Academic Press; 2001. pp. 17–26. [Google Scholar]
- 31.Ohtsu K, Nakazono M, Tsutsumi N, Hirai A. Characterization and expression of the genes for cytochrome c oxidase subunit VIb (COX6b) from rice and Arabidopsis thaliana. Gene. 2001;264:233–239. doi: 10.1016/s0378-1119(01)00334-1. [DOI] [PubMed] [Google Scholar]
- 32.Dawson A, Klingenberg M, Kramer R. Transport across membranes. In: Darley-Usmar VM, Rickwood D, Wilson MT, editors. Mitochondria: a Practical Approach. Oxford: IRL Press; 1987. pp. 35–78. [Google Scholar]