Abstract
DNA double-strand breaks (DSBs) induced in the genome of higher eukaryotes by ionizing radiation (IR) are predominantly removed by two pathways of non-homologous end-joining (NHEJ) termed D-NHEJ and B-NHEJ. While D-NHEJ depends on the activities of the DNA-dependent protein kinase (DNA-PK) and DNA ligase IV/XRCC4/XLF, B-NHEJ utilizes, at least partly, DNA ligase III/XRCC1 and PARP-1. Using in vitro end-joining assays and protein fractionation protocols similar to those previously applied for the characterization of DNA ligase III as an end-joining factor, we identify here histone H1 as an additional putative NHEJ factor. H1 strongly enhances DNA-end joining and shifts the product spectrum from circles to multimers. While H1 enhances the DNA-end-joining activities of both DNA Ligase IV and DNA Ligase III, the effect on ligase III is significantly stronger. Histone H1 also enhances the activity of PARP-1. Since histone H1 has been shown to counteract D-NHEJ, these observations and the known functions of the protein identify it as a putative alignment factor operating preferentially within B-NHEJ.
INTRODUCTION
Endogenous cellular processes and exogenous factors such as ionizing radiation (IR) generate in the DNA highly cytotoxic double-strand breaks (DSBs) that undermine genomic integrity. Higher eukaryotes utilize for the majority of DSBs a pathway of non-homologous end-joining (NHEJ) that employs the products of DNA-PKcs, KU70, KU80, LIG4, XRCC4 and Artemis (1,2), as well as the recently characterized factor XLF/Cernunnos (3,4). We will refer here to this pathway as D-NHEJ to indicate its dependence on DNA-PK.
Deficiency in proteins of D-NHEJ compromises rejoining of DSBs in irradiated cells (5–7) and increases DSB misjoining (8), as well as the frequency of chromosomal translocations (9,10). In mice, deficiency of several proteins of D-NHEJ leads to the development of cancer on a p53−/− background (11–13). Chromosomal translocations linking an amplified c-myc oncogene with the IgH locus sequences are frequently observed in these tumors, and the underlying end-joining event makes frequent use of microhomologies (14–16). Defects in components of D-NHEJ have been also implicated in genomic instability (11,17,18), in the formation of soft tissue sarcomas (19), and in the aberrant coding and signal joints formed during V(D)J recombination (20–23). These observations in aggregate, point strongly to an error prone DNA-end-joining pathway that handles DSBs when D-NHEJ is compromised. Because this repair pathway does not show dependence on genes of HRR (7), we proposed that it reflects an alternative form of end joining that functions as backup (B-NHEJ) to D-NHEJ.
The operation of alternative pathways of NHEJ has also been suggested by experiments evaluating joining of ends generated in naked DNA in vitro and transfected into cells for processing. Mammalian cells demonstrate an extraordinary ability to join such transfected DNA, either by direct ligation or by utilizing microhomologies (16,24). Notably, cells deficient in DNA-PKcs (15,25,26), Ku (15,27), XRCC4 (15,27) or DNA ligase IV (15) show high potential of end joining with preferential use of microhomologies (15,27). This microhomology-dependent end joining may overlap partly or completely with B-NHEJ and has been recently shown to be involved in the repair of DNA breaks created during assembly of antigen-receptor genes (28–31). These developments provide solid evidence for the acute biological significance of the backup pathway of DSB repair and implicate it in the chromosomal translocations of lymphoid cancers.
Despite the potential consequences of B-NHEJ function, little is known about the underlying mechanism, its regulation, as well as its integration into the cellular DNA DSB-processing apparatus. Recent work identifies DNA ligase III as a candidate factor in B-NHEJ (32,33) and points to PARP-1 as an additional potential contributor (33,34). Here, we present experiments demonstrating that H1 may be an additional factor contributing to DSB repair as a component of B-NHEJ.
MATERIALS AND METHODS
Cell lines and extract preparation
HeLa cells were grown either as suspension or as monolayer cultures in Joklik's modified MEM (S-MEM) supplemented with 5% bovine calf serum. Experiments were performed either with HeLa nuclear extracts (NE) or with recombinant human DNA ligase IIIβ or recombinant human DNA ligase IV/XRCC4 purified from Sf9 cells (see later). For preparation of cell extracts a 1–30 L HeLa cell suspension was grown in spinner flasks to 0.5–1 × 106 cells/ml and collected by centrifugation. Cells were washed in ice-cold PBS and subsequently in five-packed cell volumes of cold hypotonic buffer (10 mM Hepes, pH 7.5, 5 mM KCl, 1.5 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride, PMSF and 0.5 mM DTT). The cell pellet was resuspended in one volume of hypotonic buffer and, after 10 min in ice, disrupted in a Dounce homogenizer.
For NE preparation 3 M KCl was slowly added to the homogenized cells to a final concentration of 50 mM. The extract was incubated for 10 min on ice and centrifuged for 30 min at 3300 g at 4°C. Supernatant was collected as Cytoplasmic Extract (CE). Nuclear pellet was resuspended in two-packed nuclear volumes (pnv) of low salt buffer (20 mM Hepes, pH 7.9, 20 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.2 mM PMSF and 0.5 mM DTT) and 1 pnv of high salt buffer (10 mM Hepes, pH 7.9, 1.6 M KCl, 1.5 mM MgCl2) was slowly added to a final concentration of 400 mM KCl. Extract was incubated for 30 min at 4°C under gentle rotation and centrifuged for 30 min, 50 000 g at 4°C. The supernatant was collected as NE. NE was dialyzed overnight in dialysis buffer (20 mM Hepes, pH 7.9, 10–20% glycerol, 400 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF and 0.5 mM DTT) before aliquoting, snap freezing and storing at −80°C.
Extract fractionation
Fractionation of DNA-end-joining factors was carried out over a dsDNA-cellulose (Sigma) followed by a Mono-S (Amersham Biosciences) column. Details on these fractionations have been published elsewhere (32). Briefly, fractionation over dsDNA-cellulose was initiated by diluting NE to 300 mM KCl using buffer A (20 mM Hepes pH 7.9, 20% glycerol, 0.2 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT) and loading after equilibration at the same salt concentration. The flow through fraction was designated peak I/II to maintain previous nomenclature. DNA-end-joining activity was eluted at 750 mM KCl (peak III). Active fractions of peak III were pooled, dialyzed against buffer A containing 100 mM KCl and loaded, at a flow rate of 0.5 ml/min onto an 1 ml Mono S HR 5/5 column equilibrated in the same buffer. A linear gradient over 10 column volumes was applied up to 300 mM KCl, followed by three stepwise increases to 500, 750 and 1000 mM KCl, each over 5–10 column volumes. Fractions obtained from all fractionation steps were tested for protein concentration using the Bradford assay, and in vitro DNA-end-joining activity (see later).
Proteins and antibodies
Recombinant human histone H1.2 was purchased from Calbiochem (Darmstadt, Germany). Histone H1 (mixture of different isoforms from calf thymus) was from Alexis Biochemical (San Diego, USA). A mouse monoclonal antibody against histone H1 clone AE-4 was from Acris Antibodies GmbH (Hiddenhausen, Germany). Recombinant human DNA ligase IIIβ and DNA ligase IV/XRCC4 complex was purified from baculovirus infected Sf9 cells as previously described (32,35,36).
Western blot analysis
For Western blotting, proteins were electrophoresed on 12% SDS–polyacrylamide gels, transferred to a nitrocellulose (PVDF membranes were not working well for H1) membrane (Schleicher and Schuell, Dassel, Germany) and probed by the ECL-Plus kit as recommended by the manufacturer (Amersham Biosciences). Signal was detected using the ‘Typhoon’ (GE Healthcare, Freiburg, Germany), or the VersaDoc (Biorad, Munique, Germany).
Mass spectrometry analysis
We used mass spectrometry analysis to identify peptides present in an active fraction of MonoS IIID. For this purpose, proteins were fractionated in a 10% SDS–PAGE gel and stained with Coomassie. Prominent bands were excised and processed for MS analysis at the Medical Proteome Center of Ruhr-University of Bochum, Germany.
DNA-end joining
Supercoiled plasmid pSP65 (3005 bp; Promega) was prepared using CsCl2/EtBr gradients. It was used as a substrate in DNA-end-joining reactions following digestion with SalI to generate linearized DNA. In some experiments, pSP65 was linearized using other restriction endonucleases as indicated. End-joining reactions were performed in 20 mM Hepes–KOH (pH 7.5), 10 mM MgCl2, 80 mM KCl, 1 mM ATP, 1 mM DTT, 25–250 ng of DNA (as indicated) and 0–20 μg of HeLa NE, fractions, purified DNA ligase 3β or purified DNA ligase IV/XRCC4 in a final volume of 20 µl at 25°C for 30 min. Reactions were terminated by adding 2 µl of 0.5% SDS, 2 µl of 0.5 M EDTA and 1 µl of proteinase E (10 mg/ml), then incubated for 1 h at 37°C. DNA from the reaction was loaded on a 0.7% agarose gel and run at 45 V (2 V/cm) for 5 h. Gels were stained in SYBR Gold (Molecular Probes) and scanned in a FluorImager (Typhoon, Molecular Dynamics). For quantification of rejoining the ImageQuant software (Molecular Dynamics) was used to calculate the percent of input plasmid found in dimers and other higher order polymers. The values obtained are included in the figures below the corresponding gels.
For analysis of reaction products by gel-filtration chromatography DNA-end-joining reactions were performed with a 943-bp fragment of pUC19 as substrate. Digestion (500 µl) for 1 h of pUC19 (25 µg) with 50 U Alw44I/ApaLI (MBI Fermentas) generates three fragments (497, 943 and 1246 bp), which were separated in preparative agarose gels after staining with ethidium bromide and collected by electro elution (Biorad, Munique, Germany). Electroeluates were ethanol precipitated, pellets dissolved in TE buffer and the DNA concentration measured in a spectrophotometer (NanoDrop Technologies, Inc., Rockland, USA). Typically, 40 ng of the 943-bp fragment were used in 20 µl end-joining reactions (1 h, 25°C) and products of 25 such reactions were pooled and analyzed by gel filtration. When H1 was present, the reaction mixture was incubated for 10 min at 25°C before adding ATP and ligase IIIβ (20 ng per reaction). As a control one reaction was stopped with 2 µl of 0.5% SDS, 2 µl of 0.5 M EDTA and 1 µl of proteinase E (10 mg/ml) and incubated for 1 h at 37°C while the test reaction was stopped simply by adding DNA-loading buffer.
Gel filtration using a Sephacryl S1000-SF column
Sephacryl S1000-SF (GE Healthcare, Freiburg, Germany) was packed in a HR 10/30 column (GE Healthcare, Freiburg, Germany) giving 290-mm height and 10-mm diameter (∼23 ml packed volume). Chromatography was carried out with 1.5 column volumes of TE buffer containing 80 mM NaCl in an ÄKTA FPLC (GE Healthcare, Freiburg, Germany) and a flow rate of 0.5 ml/min. To calibrate the column pUC19 was digested with PvuII to generate a 322 and a 2364 bp fragment. In addition, λ DNA (48 kb) (Fermentas) was also used.
PARP-1 purification and assays
Recombinant PARP-1 was produced in Sf9 cells using the Baculovirus Constructs, pFastBac1-PARP-1 (kindly provided by Dr Matthew Knight) (37). Small-scale stocks of baculovirus were prepared from monolayer cultures and large-scale stocks from suspension cultures. Sf9 cells were grown as spinner cultures at 27°C in Grace's insect medium (pH 6.1) supplemented with 10% foetal calf serum, Lactalbumin hydrosylate (Sigma), Yeastolate ultrafiltrate(Gibco) and penicillin/Streptomycin. Cells were infected in suspension at multiplicity of infection (MOI) of 2.5 and incubated for 48 h before collection. PARP-1 expression was determined by SDS–PAGE (8%) and Western blotting.
For purification of PARP-1 (38) ∼700 × 106 infected cells were lysed at 4°C with ice-cold homogenization buffer (25 mM Tris–HCl, pH 8.0, 50 mM glucose,10 mM EDTA, 1 mM β-mercaptoethanol, 1 mM PMSF, 0.2% Tween-20, 0.2% Nonidet P-40 (Igepal), 0.5 M NaCl, 2 µg/ml Aprotinin, 1 mM Benzamidine-HCl, 1 µg/ml Pepstatin) and centrifuged. Endogenous DNA was removed by addition of 1 mg/ml salmon sperm protamine sulphate (Biomedicals) followed by centrifugation (20 min, 40 000 rpm, 4°C). Supernatant was precipitated by two steps (0–30% and 30–70%) of ammonium sulphate. The precipitated protein of the second step was dissolved in ice-cold chromatography extraction buffer A (50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 25 mM Na2S2O3, 10 mM 2-mercaptoethanol, 1 mM DTT, 0.1 mM PMSF, 10% glycerol) and dialysed overnight in a buffer containing 50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 25 mM Na2S2O3, 10 mM β-mercaptoethanol, 1 mM DTT, 0.1 mM PMSF, 10% glycerol, 0.25 M NaCl). Protein was loaded on a DNA single-strand cellulose column equilibrated with chromatography buffer A and eluted by a step gradient of 250, 750 and 1000 mM NaCl. PARP-1 eluted at 750 mM NaCl. Purified fractions were used for analysis by SDS–PAGE, protein concentration determination (Bradford) and the measurement of enzymatic activity.
PARP-1 activity was measured with a Western blot assay (39) based on quantification of PARP-1 auto-poly-ADP-ribosylation using an anti-poly-(ADP-ribose) antibody. The reaction mix (20 µl) contains 10× NHEJ buffer (described earlier), 1 mM ATP, 50 ng of Sal-I digested pSP65, 4 mM NAD+ and the indicated amounts of PARP-1, H1.2 or Ligase-3β. The total incubation time was 30 min at 37°C, but Ligase IIIβ and ATP were added after a 10 min pre-incubation. Reactions were stopped by adding 2× SDS loading buffer and sonicated for 15 min at 80°C before running SDS–PAGE (6%) PAGE and blotting onto HyBond-P PVDF membranes (GE Healthcare). For immunodetection, an anti-poly-(ADP-ribose) antibody PAR(m-MAb 10 H), Alexis was used and detected by ECL (GE healthcare).
For dot blot analysis 6 µl aliquots from the above reactions were diluted with SDS and sonicated for 15 min at 80°C. Nitrocellulose membrane (Protran BA 85, 0.45 µm, Schleicher & Schuell, Germany) was activated and 1 µl sample was loaded. The membrane was then incubated in blocking buffer, incubated with the anti-poly(ADP-ribose) antibody and immunodetected as described earlier. Quantification of Western- and Dot-blots was carried out using the ImageQuant (Amersham Biosciences).
RESULTS
Fractionation of nuclear extract identifies a DNA-end-joining-enhancing factor
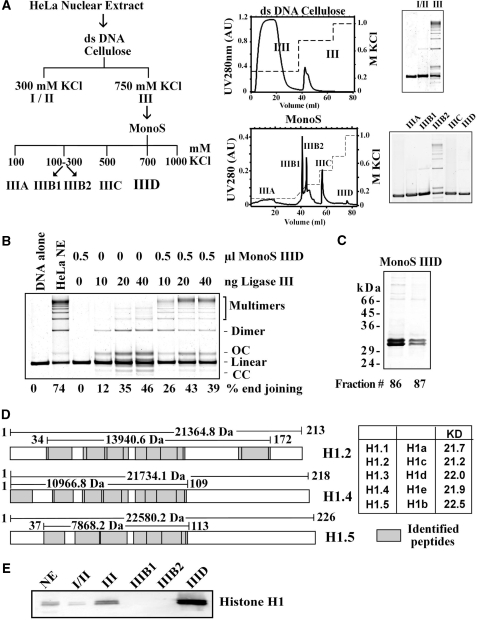
We have previously shown (32) that the DNA-end-joining activity of HeLa nuclear extract can be fractionated according to the scheme outlined in Figure 1A. First, ds-DNA cellulose effectively separates the majority of end-joining activity in the 750 mM NaCl fraction (fraction III). Further fractionation on Mono S generates fraction IIIB2, which contains the majority of DNA-end-joining activity (Figure 1A). Fraction IIIB1 has only limited activity and fraction IIIC is inactive—either by itself or in combination with other fractions [Figure 1A and (32)]. Factors implicated in D-NHEJ, such as DNA ligase IV, DNA-PKcs and Ku are mainly found in IIIB1, whereas DNA Ligase III, PARP-1 and XRCC1 are mainly found in IIIB2 (32). Fraction IIID is inactive by itself, but increases significantly the end-joining activity of IIIB2 suggesting an important role in the reaction (32). Yet, the active factor in IIID remained uncharacterized.
Figure 1.
Identification of H1 as a DNA-end-joining factor. (A) Outline of the nuclear extract fractionation scheme utilized. Extracts were first fractionated over a double-stranded DNA cellulose column followed by further fractionation of active fractions over a Mono S column. Shown are the general fractionation schemes, typical chromatograms for each column, as well as SDS–PAGE gels of peak fractions. (B) Effect of fraction IIID on DNA-end joining catalyzed by purified DNA Ligase IIIβ. Reactions were assembled with the indicated amounts of protein. Shown as control are reactions assembled without extract, or with crude nuclear extract. (C) Coomassie blue-stained SDS–PAGE gel (12%) of fractions 86 and 87 (IIID, 20 µl) from a fractionation over a Mono S column. The two prominent bands detected were excised and subjected to mass spectrometry analysis. (D) Sequest search of peptides characterized by LC/MS-MS from the bands shown in C led to the identification of histone H1 variants H1.4 and H1.5 in the upper band and histone H1.2, H1.4 and H1.5 (74) in the lower band (derived by Flicka sequence analysis). The graph shows the corresponding isoforms and the coverage achieved through the peptides analyzed (grey boxes; lines indicate the size of the covered area and the size of the protein isoform itself). (E) Western blot analysis of the indicated fractions with an anti-histone H1 antibody. Ten micrograms of HeLa nuclear extract (NE) and each 10 µl of fractions I/II and III, IIIB1 and IIIB2 as well as 2 µl of fraction IIID2 were separated on a 12% SDS–PAGE gel and blotted onto a nitrocellulose membrane.
To characterize IIID, we assembled reactions with different amounts of purified recombinant DNA ligase IIIβ in the presence or absence of IIID (Figure 1B). Under the reaction conditions employed the ligase by itself rejoins the plasmid substrate to generate a major band containing open circular plasmid, a minor band with supercoiled plasmid, and several bands containing substrate dimers and multimers. From the percent end-joining results shown in the bottom of Figure 1B it can be seen that fraction IIID at 0.5 µl enhances significantly end joining, particularly at low ligase concentrations, by preferentially enhancing intermolecular joining.
Mass spectrometry analysis identifies histone H1 as the active factor in IIID
SDS–PAGE of IIID shows two prominent bands in close proximity migrating with an apparent molecular weight of 30–32 kDa (Figure 1C). For further characterization, bands were excised, digested, subjected to LC-MS/MS followed by sequential mass spectroscopy (MS/MS spectra) and the results were analyzed with the SEQUEST software. In this way, isoforms of the linker histone H1 could be identified as the major constituents of the bands. Figure 1D graphically shows the coverage achieved by the peptides analyzed. The relatively low peptide sequence coverage at the C-terminus of the H1 isoforms is due to their high lysine and arginine content that leads to the generation of small peptides during trypsin digestion; these small peptides are frequently lost during reverse phase HPLC. Despite this limitation, the content of the two SDS-gel bands could be identified with a high level of confidence. Both gel bands contain the two H1 isoforms, H1.4 and H1.5, while the lower band also contains H1.2. Western blot analysis of IIID using a mouse monoclonal antibody against histone H1 shows bands corresponding to those of SDS–PAGE, in agreement with the mass spectrometry analysis (Figure 1E).
Purified histone H1.2 can fully substitute for fraction IIID
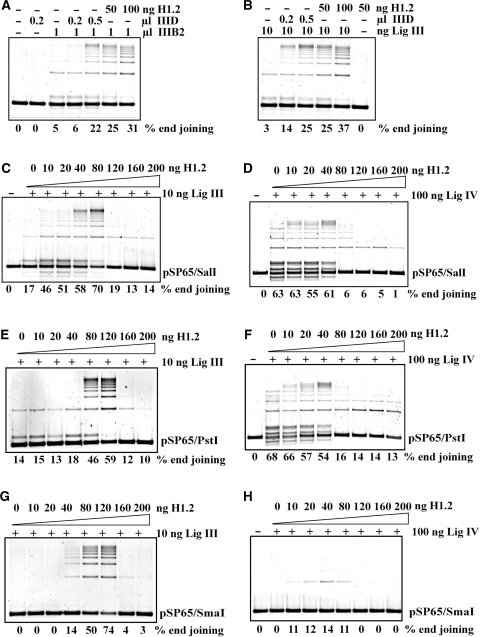
We compared the end-joining activity of fraction IIID with the activity of purified human recombinant H1.2, the most abundant isoform in many cell types (40,41). Recombinant human H1.2 strongly stimulates DNA-end joining when added to reactions assembled with fraction IIIB2 generating an effect practically identical to that of IIID (Figure 2A). Similar results are obtained when fraction IIIB2 is replaced by DNA Ligase IIIβ (Figure 2B), with 50–100 ng H1.2 generating a maximum effect. A mixture of H1 isoforms purified from calf thymus gives similar results (results not shown).
Figure 2.
Characterization of H1 function in DNA-end joining. (A) Comparison of end joining between purified histone H1.2 and MonoS fraction IIID. Reactions were assembled with the indicated amount of protein and end-joining activity evaluated. Note that MonoS IIID, when used alone, does not show detectable DNA-end joining activity. (B) MonoS fraction IIID and histone H1.2 enhance DNA-end joining in reactions assembled with 10 ng purified recombinant DNA ligase IIIβ. Ligase alone has only limited and H1.2 alone (50 ng) shows no detectable DNA-end-joining activity. (C) Histone H1.2 titering in DNA-end-joining reactions assembled with 10 ng DNA ligase IIIβ. The DNA substrate, 50 ng pSP65, was linearized with Sal I to generate ends with 4 nt 5′ overhangs and was incubated with histone H1.2 for 10 min at 25° before adding DNA ligase IIIβ and ATP to start end joining, which was carried out at 25°C for 30 min. (D) Titering of Histone H1.2 in DNA-end-joining reactions assembled with 100 ng DNA ligase IV/XRCC4. Other details as in (C). Reactions were assembled without KCl to increase DNA-end-joining activity (see text). (E) DNA-end joining in reactions assembled with 10 ng DNA ligase IIIβ and different amounts of histone H1.2 using pSP65 plasmid digested with Pst I to generate 4 nt 3′ overhangs. (F) As in (E) but for reactions assembled with 100 ng DNA ligase IV/XRCC4. (G) DNA-end joining in reactions assembled with 10 ng DNA ligase IIIβ and different amounts of histone H1.2 using pSP65 plasmid digested with Sma I to generate blunt ends. Other conditions as in (C). (H) As in (G) but for reactions assembled with 100 ng DNA ligase IV/XRCC4.
Enhancement of DNA-end joining strongly depends on histone concentration
To investigate further the function of histone H1 in DNA-end joining, we studied its concentration dependence in reactions assembled with DNA ligase IIIβ (Figure 2C). Increasing amounts of H1.2 stimulate DNA-end joining and cause a shift in the products from dimers and open circles to multimers. This stimulation reaches a maximum at 80 ng H1.2, but higher concentrations strongly inhibit DNA-end joining with only few dimers forming above 120 ng.
To investigate the ligase specificity of H1-mediated stimulation of end-joining we assembled reactions with purified recombinant DNA ligase IV/XRCC4. Because this ligase shows only a low activity under the conditions optimized for DNA Ligase III, we searched for alternative conditions and noted that omission of KCl from the reaction buffer significantly enhances its activity (42). Reactions assembled in KCl-free buffer show significant end joining that is only slightly stimulated by H1.2 up to 40 ng (Figure 2D). At higher H1.2 concentrations DNA-end joining is rapidly inhibited. This result suggests that the potentiation of end joining by H1 is more pronounced for DNA Ligase III.
Effect of H1.2 on the joining of different DNA ends
All above-described experiments use the pSP65 plasmid linearized with SalI to generate DNA ends with 4 nt 5′ overhangs. To examine the dependence of the H1 effect on the type of DNA ends, pSP65 was linearized with PstI or SmaI to generate 4 nt 3′ overhangs and blunt ends, respectively, and end joining was tested. Reactions assembled with DNA Ligase III and PstI-linearized substrate show an abrupt stimulation of end joining and the formation of multimers at H1.2 concentrations above 40 ng followed by a strong inhibition above 120 ng (Figure 2E). End joining is not detectable in reactions assembled with SmaI-linearized substrate in the absence of H1.2 (Figure 2G). Yet, reactions assembled with 20 ng H1.2 show robust end joining that increases rapidly at 40 ng and reaches a maximum between 80 and 120 ng. At higher H1.2 concentrations an abrupt decrease in DNA-end joining is observed again.
When similar reactions are assembled with DNA ligase IV/XRCC4 and PstI-linearized substrate, only a modest stimulation of end joining occurs up to 40 ng H1.2 (Figure 2F) that is similar to that observed with SalI-linearized substrate (Figure 2D). SmaI-linearized substrate shows no end joining by DNA Ligase IV in the absence of H1.2 (Figure 2H), a marginal stimulation up to 40 ng followed by a clear inhibition at higher concentrations.
The above results in aggregate demonstrate a narrow concentration optimum for stimulation by histone H1 of joining of different types of DNA ends. This stimulation is significantly more pronounced in reactions assembled with DNA Ligase III and may derive from a function of H1 as an alignment factor (43) in end-joining reactions catalyzed by this enzyme.
H1 concentration optimum depends on the amount of substrate DNA
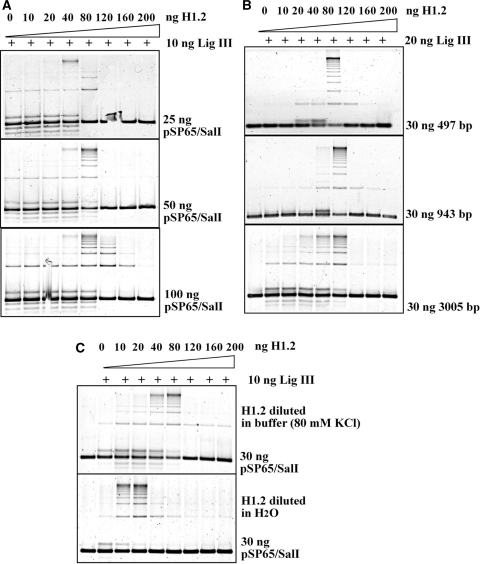
The dramatic increase in multimers among the products of the reactions assembled with H1.2 suggests protein–DNA interactions that fundamentally alter substrate topology. Therefore, we investigated in greater detail the stoichiometry of H1.2 with reference to the substrate DNA. Figure 3A shows the effect of H1.2 on end-joining reactions assembled with 10 ng purified ligase III and different amounts of Sal I-linearized substrate DNA. While the end-joining maximum is achieved at 40 ng H1.2 when using 25 ng substrate, 80 ng is required in reactions assembled with 50 ng substrate. Further increase of substrate amount to 100 ng does not shift the maximum of end joining but reduces the inhibition typically observed at higher H1.2 concentrations. Assuming an even binding of H1 throughout the DNA molecule and considering the relative abundance in the reaction of substrate DNA and H1.2 protein, we estimate that at the end joining maximum there will be one H1 molecule bound for every 13 bp, if all supplied protein is active to bind DNA.
Figure 3.
The effect of histone H1 on DNA-end joining depends upon the amount and the length of the DNA substrate. (A) Titration of the effect of histone H1 on DNA-end joining for reactions assembled with 10 ng DNA Ligase IIIβ and different amounts of Sal I digested substrate DNA as indicated. Other conditions as in Figure 2C. (B) DNA-end joining in reactions assembled with 20 ng DNA Ligase IIIβ and 30 ng substrate DNA of different lengths as indicated. DNA fragments of 497 and 943 bp were prepared by digesting pUC19 with Alw44I. Other conditions as in Figure 2C. Alw44I recognizes the sequence 5′-G|TGCAC-3′ and generates ends with 3′ 4-bp extensions. (C) The choice of solvent modulates the activity of H1.2 in DNA-end joining. The experiments described earlier were carried out with H1.2 dissolved in reaction buffer (see ‘Materials and methods’). Titration of end-joining reactions with H1.2 dissolved in water or reaction buffer and tested in the presence of 10 ng DNA Ligase IIIβ. Other details as in Figure 2C. Note the shift in the maximum of DNA-end joining from 80 to 20 ng.
We also carried out reactions using substrates of different sizes generated by digesting pUC19 with Alw44I (ApaLI) and purifying the 497- and 943-bp fragments. The results summarized in Figure 3B indicate that when reactions are assembled with 30 ng substrate and 20 ng of DNA ligase IIIβ, the optimum in DNA-end joining is always achieved with 80 ng H1.2 regardless of substrate size. Since the abundance of DNA ends is proportionally larger in reactions assembled with small as compared to large substrate molecules, the dependence of the H1 effect on the number of base pairs, rather than the number of DNA ends available is suggested. At 80 ng H1.2, 37, 71 and 226 molecules will be bound on the 497, 943 and 3005 bp (pSP65) DNA molecules, respectively, which is again equivalent to one H1 molecule for every 13 bp.
In the course of our investigations we noticed that the effect of H1 depended strongly on the method of preparation of the working H1 solution. In the above-described experiments, histone H1 was diluted in reaction buffer just before distribution in individual reactions. Notably, when histone H1 was instead diluted in H2O, the end-joining optimum shifted to 10–20 ng (Figure 3C). Other aspects of the H1 effect on end joining remained qualitatively unchanged. Because this preparation protocol increased the apparent activity of the protein for our assays, it was adopted in further experiments.
H1-mediated enhancement of end joining is not due to DNA aggregation
Histones and other DNA-binding factors are known to act as DNA-condensing agents. Such condensation generates molecular crowding that can cause the potentiation of DNA-end joining observed in reactions assembled with H1.2 (44). To examine this possibility we used gel filtration to check the fractionation characteristics of DNA substrate and the products of end-joining reactions carried out in the presence or absence H1.2. We reasoned that high molecular weight aggregates, if forming, they will be unable to enter the column. Therefore, if end joining only occurred in aggregated DNA, high molecular weight products would not pass through the column. The resin used, Sephacryl S-1000, has a separation range between 5 × 105 to 108 Da with an exclusion limit of 20 kb DNA. To accommodate this exclusion limit, end-joining reactions were carried out with the 943-bp fragment of pUC19 (Figure 3B) using 20 ng DNA ligase IIIβ.
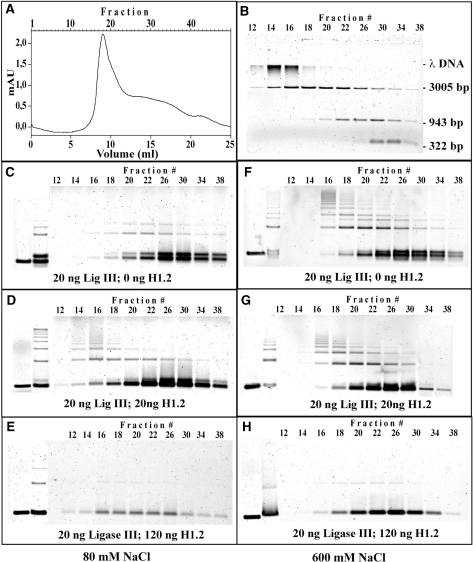
We tested the separation characteristics of the column with a mixture of λ DNA, linearized pSP65 (3005 bp) and the 943- and 322-bp fragments. Figure 4A shows a typical chromatogram and Figure 4B the detection of the DNA fragments in the different fractions. High molecular weight λ DNA is found in the early fractions 14–16, pSP65 substrate size DNA in fractions 14–26, and low molecular weight DNA in fractions 22–34, as expected.
Figure 4.
The effect of histone H1 on DNA-end joining cannot be attributed to H1-induced DNA aggregation. Products of DNA-end joining reactions, carried out as described under ‘Materials and methods’ section in the presence of purified DNA Ligase IIIβ and histone H1.2, were analyzed for possible aggregation by gel filtration (Sephacryl S-1000 SF in HR 10/30, void volume 9.7 ml). (A, B) Calibration of the column with pUC19 fragments, pSP65 and λ DNA. (A) shows a typical chromatogram, whereas (B) the DNA fragments found in the different fractions by agarose gel electrophoresis. The DNA mixture contained 0.45 µg λ DNA, 1 µg linearized pSP65 (3005 bp), 0.72 µg 943 bp and 0.4 µg 322 bp DNA fragments. High molecular weight λ DNA is found in early fractions 14–16, pSP65 substrate size DNA in fractions 14–26, and low molecular weight DNA in fractions 22–34, validating thus the separation potential of the column. (C, D, E) Analysis by gel filtration of substrate and products of DNA-end-joining reactions assembled with 20 ng DNA Ligase III and the indicated amounts of histone H1. Gel filtration was carried out in a buffer containing 80 mM NaCl. (F, G, H) Reactions similar to those shown in C, D and E were analyzed by gel filtration in a buffer containing 600 mM NaCl. Other reaction details as in Figure 2C.
For a test of the products of DNA-end joining, 25 20-µl reactions were allowed to go to completion in the presence of 0, 20 and 120 ng H1.2, were pooled, loaded on the column without adding stop solution and fractionated. Figure 4C, D and E show DNA substrate and product fractionation of reactions assembled with different amounts of histone H1 when 80 mM NaCl were present in the running buffer. Figure 4F, G and H summarize results of identical reactions fractionated in a buffer containing 600 mM NaCl. In reactions carried out in the absence of H1.2 (Figure 4C and F), unrejoined substrate fractionates as expected from the results shown in Figure 4B. The presence of high salt in the running buffer has no effect on the fractionation characteristics (compare Figure 4C with F). In both cases, and in line with their larger size, the products of end joining are found in slightly lower fractions. In reactions carried out in the presence of 20 ng H1.2, substrate is found in the late fractions, as expected and high molecular weight products in fractions 14–18, independently of the presence of high salt in the running buffer (compare 4D with 4G). The unchanged fractionation characteristics of unrejoined DNA substrate and products in reactions assembled with an amount of histone H1 resulting in maximum potentiation of end joining, argue against DNA aggregation. However, histone H1 at higher concentrations does indeed induce substrate aggregation as indicated by the results of reactions assembled with 120 ng H1.2. Under these conditions, aggregation results in extensive loss of substrate DNA and products, which is similar in magnitude in fractionations carried out using either low or high salt buffer (Figure 4E and H). Even low molecular weight material entering the column fractionates aberrantly suggesting gradual release from the aggregate during fractionation. Thus, extensive DNA aggregation can indeed be caused by histone H1, but this effect is not detectable at concentrations stimulating DNA-end joining.
Histone H1 activates PARP-1
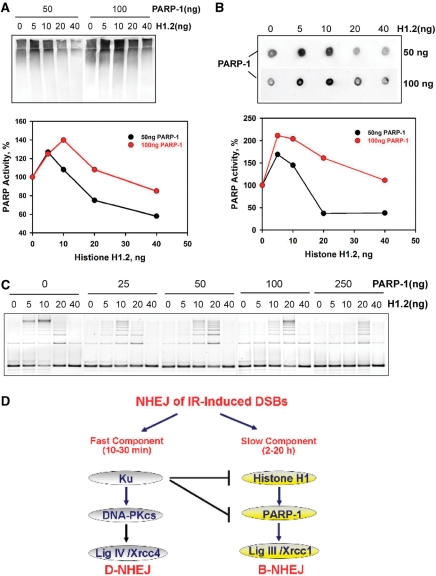
The results presented above implicate histone H1 as an accessory factor of DNA Ligase III and thus as a putative component of B-NHEJ. We inquired, therefore, whether the activity of other candidate components of this pathway is modified by histone H1. End-joining reactions were assembled as described earlier but in the presence of NAD+ and different amounts of PARP-1, and PARP-1 activity measured using an antibody against poly(ADP-ribose). The results in Figure 5A and B show a marked increase in PARP-1 activity with increasing concentration of histone H1, measured either by Western blot (Figure 5A) or by dot blot (Figure 5B), although the increase is of larger magnitude in the dot blot. Thus, a marked stimulation is observed at low concentrations of H1 reaching a maximum at about 10–20 ng and a decline at higher concentrations. The fluctuations in PARP-1 activity observed parallels the fluctuations in DNA-end-joining activity implying a common molecular base (Figure 5C, lanes with 0 ng PARP-1). The effect is PARP-1 concentration dependent and saturates above 100 ng protein (results not shown). These observations point to cooperative interactions between PARP-1 and histone H1, which are compatible with a function in the same repair pathway.
Figure 5.
Histone H1 activates PARP-1. (A) End-joining reactions as described in Figure 2C were assembled with 20 ng DNA Ligase IIIβ, in the presence of NAD+, and with different amounts of histone H1 and PARP-1, as indicated. PARP-1 activity was measured as autopoly(ADP-ribosylation) by Western blotting using an antibody against poly(ADP-ribose). The lower panel shows the quantification of the results shown in the blot. (B) Results from the same reactions used in the Western blot shown in (A) but analyzed by dot blotting. The lower panel shows the quantification of the results shown in the blot. (C) DNA-end-joining activity measured in the reactions used in A and B to measure PARP-1 activity. Reactions were assembled as described in Figure 2C with different amounts of histone H1 and PARP-1 as indicated. (D) The two pathways of NHEJ. D-NHEJ requires DNA-PK and carries out the final ligation step utilizing the LigIV/XRCC4/XLF complex, whereas B-NHEJ uses histone H1 as an alignment factor and carries out end ligation utilizing LigIII/XRCC1 with some contribution from PARP-1.
The enhanced activity of PARP-1 in the presence of histone H1 is not directly reflected by an overall enhancement in end-joining activity (Figure 5C). This is not surprising as the end-joining assay employed here does respond to the presence of PARP-1 (compare 0 ng H1 lanes in Figure 5C). However, other forms of assays have clearly demonstrated an essential role of PARP-1 in B-NHEJ (33). Nevertheless, an enhancement in end joining is notable with increasing PARP-1 concentration in reactions assembled with 20 ng histone H1, which lies beyond the optimum. This suggests that PARP-1 can help to overcome the inhibition mediated by an oversupply of histone H1 and is again compatible with a cooperative function between these two proteins.
DISCUSSION
Histone H1 is an essential component of chromatin
Histone H1 proteins are major structural components of the chromatin fiber with important functions in many chromatin-related activities including DNA repair, although the underlying mechanisms remain largely uncharacterized (45–48). The group of H1 proteins represents the most variable class of the otherwise highly conserved histone proteins, and shows highly dynamic binding to DNA (45–48). Analysis of mice lacking various combinations of histone H1 isoforms suggests a high degree of functional redundancy among them and indicates that the correct overall amount, rather than the correct relative amounts of the various subtypes, is essential for development and survival. Thus, a 2-fold reduction in H1 content is not compatible with normal development, and mutant embryos die in midgestation with multiple defects (49,50).
Histone H1 may inhibit HRR and D-NHEJ
Several reports suggest inhibitory interactions between histone H1 and pathways of DNA DSB repair. Disruption of the Saccharomyces cerevisiae histone H1 homolog, HHO1, has no obvious effect on examined chromatin structure-associated phenotypes, but causes alterations in DNA damage response that are compatible with an inhibitory effect of Hho1 on homologous recombination (51). Despite the beneficial effect on HRR, disruption of HHO1 causes increase in radiosensitivity suggesting disturbances in the balance among DNA repair pathways that reduce the fitness of the organism and its potential to respond to DNA damage (51).
Results in line with an inhibitory role of histone H1 in DNA DSB repair have also been reported in experiments investigating DNA Ligase IV/XRCC4-dependent DNA-end joining in vitro (42). A marked inhibition in DNA-end joining is observed under these conditions, which is partially rescued by H1 phosphorylation through DNA-PK. Since phosphorylation reduces the affinity of histone H1 for DNA binding, the authors propose (42) that phosphorylation by DNA-PK is a mechanism for removing histone H1 from the DNA (52) in preparation for Ku binding and initiation of D-NHEJ. Although Ku has a high enough DNA-end-binding affinity to displace histone H1 from DNA ends (53), DNA-PK-dependent phosphorylation is likely to facilitate this process, but the initial phosphorylation event will need to be catalyzed in-trans by a nearby DSB that already harbors an activated DNA-PK molecule. Overall, these results are in line with the results in Figure 2 that show only a modest increase in DNA Ligase-IV-dependent end joining at low histone H1 concentrations, followed by a strong inhibition at higher concentration. It is likely that the initial small increase in DNA Ligase IV activity observed in our experiments was not seen in the latter study (42) due to the conditions used and the range of histone H1 concentrations studied.
Apparently different results regarding the role of histone H1 in DNA repair have been recently reported in DT40 cells (54). In this model system, only knock-out of H1R, one of the six avian H1 isoforms, increases considerably the sensitivity of cells to the alkylating agent MMS and modestly to IR (54). Since the H1R mutant is not further sensitized to MMS by inactivation of Rad54 and shows a reduction in gene-targeting efficiency and in sister chromatid exchanges, the authors propose that H1R supports HRR. Although this interpretation appears plausible, alternative interpretations not invoking a positive effect of histone H1 on HRR are also possible and are discussed in the following section.
Histone H1 supports DSB repair by backup pathways of NHEJ
As evident from the results presented in the previous section, several observations support the view that histone H1 is a putative component of B-NHEJ. Thus, extract fractionation studies combined with proteomics analysis identify histone H1 as a DNA-end-joining factor (Figure 1). Because the in vitro DNA-end-joining assay used to identify the protein reflects mainly B-NHEJ, the results implicate histone H1 in this pathway of DNA-end joining. This interpretation finds further support by the observation that the DNA-end-ligation activity of ligase III is strongly and relatively specifically enhanced by histone H1, albeit in a narrow range of histone H1 concentrations, which also explains reports of inhibitory action (55,56). Earlier studies also report an enhancement by histone H1 of inter-molecular ligation mediated by either DNA Ligase III, or DNA Ligase IV (57). The results of the latter study are also compatible with a preferential effect on DNA Ligase III, as reflected by a pronounced activation at lower histone H1 concentrations.
How is histone H1 supporting end joining by B-NHEJ? One of the most critical steps in all forms of NHEJ is the alignment of the DNA ends in preparation for the DNA-end-ligation step. Indeed, alignment factors have been considered an important component of NHEJ (43). In D-NHEJ, the Ku/DNA-PKcs complex is thought to play this important role. We propose that histone H1 assumes this function in B-NHEJ. The structured globular domains of the linker histones are known to bind to naked DNA molecules co-operatively by constraining two double helices next to each other and stacking linker histones between them (58,59). Other studies show that histone H1 molecules juxtapose to form end to end polymers (60), and analysis by electron microscopy demonstrates that H1 is capable of assembling DNA in tandem arrangements (57). All these observations are compatible with a function of histone H1 as an alignment factor (43).
Notable is also the observation that in the presence of histone H1, intermolecular ligation is favored over intramolecular ligation, suggesting that histone H1 alters the topology of the DNA substrate. Binding of histone H1 on a linear, naked DNA molecule will force it from a random coil to a more or less-stretched conformation that will take the ends of the molecule apart (47,60,61), thus reducing the probability of intramolecular end joining in an in vitro assay. At the same time, intermolecular interaction between H1 molecules bound to different DNA molecules (see earlier) will facilitate DNA-end alignment and ligation, thus explaining the great increase in intermolecular end joining.
Inhibition of B-NHEJ through a reduction in histone H1 concentration provides an alternative explanation for the results of DT40 presented above (54). Notably, the IR sensitivity of the H1R mutant is further enhanced in a Ku70−/− background. Although the authors attribute this synergistic effect to an involvement of H1R to HRR (see earlier), the expected abrogation of D-NHEJ in the Ku70−/− mutant will also facilitate the function of B-NHEJ. Therefore, compromising B-NHEJ by removing one of its factors, H1R, will lead to radiosensitization. This interpretation is also in line with the observation that abrogation of H1R has no effect on the repair by HRR of I-Sce-I-induced DSBs, and that there is no difference in the response of Rad51 or γ-H2AX after IR (54). Assuming that this alternative interpretation holds, the DT40 system offers genetic evidence for the involvement of histone H1 in B-NHEJ and suggests that different subtypes may have different functions in this process—an effect the in vitro assays employed here or even the mice knockout models cannot reproduce. A separate line of investigation implicates histone H1 in DNA damage response by virtue of its ability to modify chromatin compaction (62).
In line with a role of histone H1 in B-NHEJ are also its effects on PARP-1 activity (63). Thus, an inhibition of DNA ligation has been reported by histone H1 that is mitigated by PARP-1 (64,65). This effect is also observed in our experiments, particularly for inhibitory concentrations of histone H1, suggesting that PARP-1 may have a role in relieving local inhibition of DSB repair by histone H1 (Figure 5C).
Is the observed stimulation of DNA-end-joining specific for the linker histone H1 or can it also be induced by core histones? Experiments along these lines show that a mixture of core histones transiently stimulates DNA-end joining when supplied to reactions similar to those shown in Figure 2. However, the stimulation (results not shown) is only about 30% of that measured for histone H1 and requires slightly higher protein concentrations. This observation together with the fact that in the cell core histones are organized in nucleosomes while the linker histone H1 is available as a monomer that shows highly dynamic binding to chromatin (46) (see earlier), suggests a preferential contribution of the latter to B-NHEJ. On the other hand, the stimulatory activity of core histones may be exploited by the cell to facilitate the joining by B-NHEJ of DSBs induced in DNA directly interacting with the core histone octamer.
Histone H1-induced DNA aggregation
It is well-established that histone H1 binds cooperatively to DNA at the salt concentrations used in the present study. It has also been shown that, together with other nucleoproteins, histone H1 can mediate the generation of large protein–DNA aggregates that separate from aqueous solution (66). Within these aggregates, the DNA-end-joining activity is significantly enhanced (67) raising the question whether the observations in Figures 1–3 reflect such aggregation effects. The gel-filtration experiments carried out to address this question indicate that massive aggregation is only seen at high H1 concentrations and that it is actually associated with inhibition rather than with potentiation of the DNA-end-joining activity. At lower histone H1 concentrations, close to the maximum of the DNA-end-joining activity, such aggregation is not observed and both substrate and products move through the column as expected by their monomeric size. This is in line with results suggesting that at low concentrations histone H1 binds cooperatively to DNA-bridging molecules together, whereas at high concentrations massive aggregates are formed (59–61). The former condition, which is equivalent to intermolecular alignment, is expected to enhance, whereas aggregation will inhibit DNA-end joining.
Interplay of different pathways of NHEJ
Taken together the results presented here identify histone H1 as a putative component of an alternative pathway of NHEJ that operates as a backup to D-NHEJ. Previously published data provide biochemical and genetic evidence for an engagement of DNA ligase III and PARP-1 in this pathway of DSB repair (32,33). Thus, the repair module PARP-1/DNA ligase III/XRCC1 (PLX), hitherto regarded as central in SSB repair (68,69), is implicated in the repair of DSBs and as the results here suggest, it utilizes histone H1 as an alignment factor.
A model for the handling of DSBs by NHEJ is outlined in Figure 5D. When a DSB is induced in the DNA of a repair-proficient cell, the most likely scenario is that Ku will bind the ends replacing bound histone H1 and possibly also other components of chromatin to prepare for the recruitment of DNA-PKcs. DNA-PKcs-dependent phosphorylation events will regulate and coordinate the end-joining process and will culminate with the end ligation catalyzed by the LigIV/XRCC4/XLF complex. Activation of DNA-PKcs is expected to facilitate the removal of histone H1 from chromatin and thus to facilitate the productive functions of Ku at the DNA ends.
If this DNA-PK-dependent pathway of NHEJ is compromised as a result of a defect in one of its components, histone H1 will remain bound and will act as an alignment factor to mediate end joining by LigIII/XRCC1, possibly with the contribution of PARP-1. This pathway operates with slow kinetics and is normally suppressed by D-NHEJ (70). It appears therefore more appropriate to designate this repair pathway as a backup, rather than as an alternative repair pathway. The term alternative implies equal ranking and free choice, both of which do not hold for B-NHEJ.
Several reports implicate inferior backup pathways of DNA-end joining in the phenotypes of mutants of the classical NHEJ pathway. Thus, non-classical pathways of end joining bring together the c-myc and Igh locus and cause B-cell lymphomas in mice with defects in Ku, LIG4 or XRCC4 (11–14,17,18,71). Functionally equivalent pathways may generate the aberrant junctions manifesting chromosome instability in the same mutants. Non-classical pathways of end joining generate the few V(D)J junctions observed in cells with defects in NHEJ (20,21,23) and are implicated in antibody class switching occurring under the same conditions (71). Furthermore, the substantial DNA-end joining observed after exposure to IR of cells with defects in DNA ligase IV, Ku and DNA-PK directly implicates alternative forms of DNA-end joining.
PARP-1/DNA Ligase III-dependent end joining, when utilized, helps the cell to restore its genome and thus presumably to avert cell death. However, its error prone nature causes genomic instability and cancer in the affected organism. The adverse consequences of this repair pathway may derive from inefficient synapsis of the DNA ends, which is probably supported only by histone H1 and microhomologies (15,16) and is therefore inefficient and slow (6,72,73) when compared to the DNA-PK-mediated synapsis. The resulting persistence of DNA ends in the cell can lead to incorrect rejoining and thus to translocations. In addition, it may facilitate DNA-end degradation and thus loss of genetic information. Both phenomena have been described in mutants of the classical pathway of DNA-end joining (see earlier). This microhomology-dependent end joining may overlap partly or completely with B-NHEJ and has been recently shown to be involved in the repair of DNA breaks created during assembly of antigen-receptor genes (28–31). These developments provide solid evidence for the acute biological significance of the backup pathway of DSB repair and implicate it in the chromosomal translocations of lymphoid cancers in systems with compromised D-NHEJ.
ACKNOWLEDGEMENTS
This work was supported by a grant from the DFG. The authors are indebted to Keith Caldecott, Bill Dynan, Steve Jackson and Matthew Knight for reagents. Funding to pay the Open Access publication charges for this article was provided by the DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Ünsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA Ligase IV complex to promote DNA nonhomologous end-Joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche M-C, Sanal O, Plebani A, Stephan J-L, Hufnagel M, le Deist F. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 5.Nevaldine B, Longo JA, Hahn PJ. The scid defect results in much slower repair of DNA double-strand breaks but not high levels of residual breaks. Radiat. Res. 1997;147:535–540. [PubMed] [Google Scholar]
- 6.DiBiase SJ, Zeng Z-C, Chen R, Hyslop T, Curran WJ, Jr, Iliakis G. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000;60:1245–1253. [PubMed] [Google Scholar]
- 7.Wang H, Zeng Z-C, Bui T-A, Sonoda E, Takata M, Takeda S, Iliakis G. Efficient rejoining of radiation-induced DNA double-strand breaks in vertebrate cells deficient in genes of the RAD52 epistasis group. Oncogene. 2001;20:2212–2224. doi: 10.1038/sj.onc.1204350. [DOI] [PubMed] [Google Scholar]
- 8.Rothkamm K, Kuhne M, Jeggo PA, Lobrich M. Radiation-induced genomic rearrangements formed by nonhomologous end-joining of DNA double-strand breaks. Cancer Res. 2001;61:3886–3893. [PubMed] [Google Scholar]
- 9.Martin M, Genesca A, Latre L, Jaco I, Taccioli GE, Egozcue J, Blasco MA, Iliakis G, Tusell L. Postreplicative joining of DNA double-strand breaks causes genomic instability in DNA-PKcs-deficient mouse embryonic fibroblasts. Cancer Res. 2005;65:10223–10232. doi: 10.1158/0008-5472.CAN-05-0932. [DOI] [PubMed] [Google Scholar]
- 10.Virsik-Köpp P, Rave-Fränk M, Hofman-Hüther H, Schmidberger H. Role of DNA-PK in the process of aberration formation as studied in irradiated human glioblastoma cell lines M059K and M059J. Int. J. Radiat. Biol. 2003;79:61–68. [PubMed] [Google Scholar]
- 11.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig NC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Fergson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi JA, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 15.Verkaik NS, Esveldt-van Lange REE, van Heemst D, Brüggenwirth HT, Hoeijmakers JHJ, Zdzienicka MZ, van Gent DC. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 16.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: Role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karanjawala ZE, Grawunder U, Hsieh C-L, Lieber MR. The nonhomologous DNA end joining pathway is important for chromosome stability in primary fibroblasts. Curr. Biol. 1999;9:1501–1504. doi: 10.1016/s0960-9822(00)80123-2. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl Acad. Sci. USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharpless NE, Ferguson DO, O’Hagan RC, Castrillon DH, Lee C, Farazi PA, Alson S, Fleming J, Morton CC, Frank K, et al. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol. Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
- 20.Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Otevrel T, Gao Y, Cheng W-L, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 22.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 23.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins Shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 24.Roth DB, Porter TM, Wilson JH. Mechanisms of nonhomologous recombination in mammalian cells. Mol. Cell. Biol. 1985;5:2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C, Biedermann KA, Mezzina M, Brown JM. Characterization of the DNA double strand break repair defect in scid mice. Cancer Res. 1993;53:1244–1248. [PubMed] [Google Scholar]
- 26.Harrington J, Hsieh CL, Gerton J, Bosma G, Lieber MR. Analysis of the defect in DNA end joining in the murine scid mutation. Mol. Cell. Biol. 1992;12:4758–4768. doi: 10.1128/mcb.12.10.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabotyanski EB, Gomelsky L, Han J-O, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay J-P. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong S-Y, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 30.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131:223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 32.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA Ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 33.Audebert M, Salles B, Calsou P. Involvement of Poly(ADP-ribose) Polymerase-1 and XRCC1/DNA Ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 34.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackey ZB, Niedergang C, Menissier-de Murcia J, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE. DNA Ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of Poly (ADP-ribose) polymerase. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 36.Lee K-J, Huang J, Takeda Y, Dynan WS. DNA Ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system. J. Biol. Chem. 2000;275:34787–34796. doi: 10.1074/jbc.M004011200. [DOI] [PubMed] [Google Scholar]
- 37.Knight MI, Chambers PJ. Production, extraction, and purification of human Poly (ADP-ribose) Polymerase-1 (PARP-1) with high specific activity. Protein Expr. Purif. 2001;23:453–458. doi: 10.1006/prep.2001.1513. [DOI] [PubMed] [Google Scholar]
- 38.Giner H, Simonin F, de Murcia G, Menissier de Murcia J. Overproduction and large-scale purification of the human poly(ADP-ribose) polymerase using a baculovirus expression system. Gene. 1992;114:279–283. doi: 10.1016/0378-1119(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 39.Affar EB, Duriez PJ, Shah RG, Winstall E, Germain M, Boucher C, Bourassa S, Kirkland JB, Poirier GG. Immunological determination and size characterization of poly (ADP-ribose) synthesized in vivtro and in vivo. Biochim. Biophys. Acta. 1999;1428:137–146. doi: 10.1016/s0304-4165(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 40.Th’ng JPH, Sung R, Ye M, Hendzel MJ. H1 family histones in the nucleus: control of binding and localization by the C-terminal domain. J. Biol. Chem. 2005;280:27809–27814. doi: 10.1074/jbc.M501627200. [DOI] [PubMed] [Google Scholar]
- 41.Konishi A, Shimizu S, Hirota J, Takao T, Fan Y, Matsuoka Y, Zhang L, Yoneda Y, Fujii Y, Skoultchi AI, et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks. Cell. 2003;114:673–688. doi: 10.1016/s0092-8674(03)00719-0. [DOI] [PubMed] [Google Scholar]
- 42.Kysela B, Chovanec M, Jeggo PA. Phosphorylation of linker histones by DNA-dependent protein kinase is required for DNA ligase IV-dependent ligation in the presence of histone H1. Proc. Natl Acad. Sci. USA. 2005;102:1877–1882. doi: 10.1073/pnas.0401179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thode S, Schafer A, Pfeiffer P, Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- 44.Bloomfield V, Zimm BH. Viscosity, sedimentaton, et cetera, of ring-and straight-chain polymers in dilute solution. J. Chem. Phys. 1966;44:315–323. [Google Scholar]
- 45.Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 2006;13:305–310. doi: 10.1038/nsmb1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustin M, Catez F, Lim J-H. The dynamics of histone H1 function in chromatin. Mol. Cell. 2005;17:617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 47.Parseghian MH, Hamkalo BA. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem. Cell Biol. 2001;79:289–304. [PubMed] [Google Scholar]
- 48.Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene. 2001;271:1–12. doi: 10.1016/s0378-1119(01)00495-4. [DOI] [PubMed] [Google Scholar]
- 49.Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H10 replacement subtype. Mol. Cell. Biol. 2001;21:7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, Stein A, Woodcock CL, Skoultchi AI. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 51.Downs JA, Kosmidou E, Morgan A, Jackson SP. Suppression of homologous recombination by the saccharomyces cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- 52.Baatout S, Derradji H. About histone H1 phosphorylation during mitosis. Cell Biochem. Funct. 2006;24:93–94. doi: 10.1002/cbf.1293. [DOI] [PubMed] [Google Scholar]
- 53.Roberts SA, Ramsden DA. Loading of the nonhomologous end joining factor, Ku, on protein-occluded DNA Ends. J. Biol. Chem. 2007;282:10605–10613. doi: 10.1074/jbc.M611125200. [DOI] [PubMed] [Google Scholar]
- 54.Hashimoto H, Sonoda E, Takami Y, Kimura H, Nakayama T, Tachibana M, Takeda S, Shinkai Y. Histone H1 variant, H1R is involved in DNA damage response. DNA Repair (Amst.) 2007;6:1584–1595. doi: 10.1016/j.dnarep.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 55.Liang L, Deng L, Chen Y, Li GC, Shao C, Tischfield JA. Modulation of DNA end joining by nuclear proteins. J. Biol. Chem. 2005;280:31442–31449. doi: 10.1074/jbc.M503776200. [DOI] [PubMed] [Google Scholar]
- 56.Blanco MG, Boan F, Gomez-Marquez J. A paradox in the in vitro end-joining assays. J. Biol. Chem. 2004;279:26797–26801. doi: 10.1074/jbc.M402832200. [DOI] [PubMed] [Google Scholar]
- 57.Yamanaka S, Katayama E, Yoshioka K-i, Nagaki S, Yoshida M, Teraoka H. Nucleosome linker proteins HMGB1 and histone H1 differentially enhance DNA ligation reactions. Biochem. Biophys. Res. Commun. 2002;292:268–273. doi: 10.1006/bbrc.2002.6647. [DOI] [PubMed] [Google Scholar]
- 58.Draves PH, Lowary PT, Widom J. Co-operative binding of the globular domain of histone H5 to DNA. J. Mol. Biol. 1992;225:1105–1121. doi: 10.1016/0022-2836(92)90108-v. [DOI] [PubMed] [Google Scholar]
- 59.Thomas JO, Rees C, Finch JT. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 1992;20:187–194. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark DJ, Thomas JO. Salt-dependent co-operative interaction of histone H1 with linear DNA. J. Mol. Biol. 1986;187:569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- 61.Clark DJ, Thomas JO. Differences in the binding of H1 variants of DNA. Eur. J. Biochem. 1988;178:225–233. doi: 10.1111/j.1432-1033.1988.tb14447.x. [DOI] [PubMed] [Google Scholar]
- 62.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang S-M, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kun E, Kirsten E, Mendeleyev J, Ordahl CP. Regulation of the enzymatic catalysis of Poly(ADP-ribose) polymerase by dsDNA, polyamines, Mg2+, Ca2+, histones H1 and H3, and ATP. Biochemistry. 2004;43:210–216. doi: 10.1021/bi0301791. [DOI] [PubMed] [Google Scholar]
- 64.Ohashi Y, Ueda K, Kawaichi M, Hayaishi O. Activation of DNA ligase by poly(ADP-ribose) in chromatin. Proc. Natl Acad. Sci. USA. 1983;80:3604–3607. doi: 10.1073/pnas.80.12.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimmermann SB, Levin CJ. DNA ligase activity on chromatin and its analogs, rejoining of DNA strands in polysine-DNA complexes and in reconstituted chromatins. Biochemistry. 1975;14:1671–1167. doi: 10.1021/bi00679a019. [DOI] [PubMed] [Google Scholar]
- 66.Takahagi M, Tatsumi K. Aggregative organization enhances the DNA end-joining process that is mediated by DNA-dependent protein kinase. FEBS J. 2006;273:3063–3075. doi: 10.1111/j.1742-4658.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 67.Al-Natour Z, Hassan AH. Effect of salt on the binding of the linker histone H1 to DNA and nucleosomes. DNA Cell Biol. 2007;26:445–452. doi: 10.1089/dna.2006.0512. [DOI] [PubMed] [Google Scholar]
- 68.Caldecott KW. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays. 2001;23:447–455. doi: 10.1002/bies.1063. [DOI] [PubMed] [Google Scholar]
- 69.Tomkinson A, Levin DS. Mammalian DNA ligases. Bioessays. 1997;19:893–901. doi: 10.1002/bies.950191009. [DOI] [PubMed] [Google Scholar]
- 70.Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell. Biochem. 2004;92:781–794. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 71.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen H-T, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004;104:14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Zhao-Chong Z, Perrault AR, Cheng X, Qin W, Iliakis G. Genetic evidence for the involvement of DNA ligase IV in the DNA-PK-dependent pathway of non-homologous end joining in mammalian cells. Nucleic Acids Res. 2001;29:1653–1660. doi: 10.1093/nar/29.8.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drabent B, Franke K, Bode C, Kosciessa U, Bouterfa H, Hameister H, Doenecke D. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mamm. Genome. 1995;6:505–511. doi: 10.1007/BF00356166. [DOI] [PubMed] [Google Scholar]