Abstract
The human AP-endonuclease (APE1/Ref-1), an essential multifunctional protein, plays a central role in the repair of oxidative base damage via the DNA base excision repair (BER) pathway. The mammalian AP-endonuclease (APE1) overexpression is often observed in tumor cells, and confers resistance to various anticancer drugs; its downregulation sensitizes tumor cells to those agents via induction of apoptosis. Here we show that wild type (WT) but not mutant p53 negatively regulates APE1 expression. Time-dependent decrease was observed in APE1 mRNA and protein levels in the human colorectal cancer line HCT116 p53(+/+), but not in the isogenic p53 null mutant after treatment with camptothecin, a DNA topoisomerase I inhibitor. Furthermore, ectopic expression of WTp53 in the p53 null cells significantly reduced both endogenous APE1 and APE1 promoter-dependent luciferase expression in a dose-dependent fashion. Chromatin immunoprecipitation assays revealed that endogenous p53 is bound to the APE1 promoter region that includes a Sp1 site. We show here that WTp53 interferes with Sp1 binding to the APE1 promoter, which provides a mechanism for the downregulation of APE1. Taken together, our results demonstrate that WTp53 is a negative regulator of APE1 expression, so that repression of APE1 by p53 could provide an additional pathway for p53-dependent induction of apoptosis in response to DNA damage.
INTRODUCTION
The mammalian AP-endonuclease (APE1) is a ubiquitous and remarkably multifunctional protein. It plays a central role in the base excision repair (BER) pathway for damaged bases induced by reactive oxygen species and alkylating agents, and for abasic (AP) sites generated after excision of oxidized and alkylated bases by DNA glycosylases (1–3). As an endonuclease, APE1 cleaves the AP site to generate 3′-OH and 5′-deoxyribose phosphate termini, and the 3′-OH terminus is utilized by a DNA polymerase, usually DNA polymerase β, for repair synthesis in mammalian cells (4). APE1 was independently identified as a reductive activator of the c-Jun transcription factor in vitro, and named Ref-1 (5). Subsequently, several other transcription factors (including p53, hypoxia-inducible factor and NF-kB) were also found to be activated by APE1 (6–8). In addition, APE1 directly acts as a trans-acting factor by binding to negative Ca2+ response elements (nCaRE) during Ca2+-dependent repression of the human parathyroid hormone and renin genes (9–11).
Given its multiple functions, it is not surprising that APE1 expression is regulated in vivo in a complex manner. We were one of the first to show the activation of APE1 gene by oxidative stress; several other laboratories subsequently confirmed our observation (12–14). Moreover, based on the identification of the nCaREs in the APE1 promoter itself, we suggested that APE1 regulates its own expression (15). While cell cycle-dependent expression of APE1 was observed in NIH3T3 cells (16), the basal expression of APE1 is variable. The increased APE1 level is often observed in tumor cells including prostate, osteosarcomas, lung and cervical carcinomas compared to normal tissues (17–20). In other types of cancer, subcellular localization of APE1 is altered relative to that in normal tissues (21–23). Tumor cells often overexpress APE1 and are resistant to chemotherapeutic drugs and ionizing radiation (19,24,25). In contrast, siRNA-mediated downregulation of APE1 induced apoptosis of many tumor cell types and enhanced cell sensitization to ionizing radiation and chemotherapeutic agents (26–29). We recently showed that APE1 inactivation-induced apoptosis in mouse embryo fibroblasts (MEF) conditionally nullizygous for endogenous APE1, an effect that could be prevented by ectopic expression of human APE1 (29). Using a complementation assay, we also showed that both the repair and transcription regulatory functions of APE1 are required to prevent apoptosis of MEF (29). Thus, elucidating the molecular mechanisms controlling APE1 expression has profound implications for cancer therapy from both basic and clinical perspectives.
The tumor suppressor gene p53, dubbed ‘the guardian of the genome’, encodes a sequence-specific transcription factor that activates a variety of cellular genes in response to DNA damage, hypoxia and other stress signals (30,31). p53 is one of the most commonly mutated genes in human cancers; 50% of all human cancers lack the wild type (WT) p53 gene allele, and these tumors respond poorly to chemotherapy (32,33). In response to DNA damage or other cellular stresses, p53 is activated and induces cell cycle arrest or apoptosis, depending on the severity of the damage and the context of the cell cycle, and thus helps to maintain genomic stability and prevents cancer (34). Once activated, p53 has the ability to arrest cell cycle by transactivation of Waf-1/p21, 14-3-3δ etc., or induce cells to undergo apoptosis by both transcription-dependent and -independent mechanisms (35–37). p53 activates many downstream target genes involved in the mitochondrial signaling pathway during apoptosis including BAX, PUMA and the death receptor (38–40). In addition, p53 has been shown to downregulate many genes, including cell survival genes such as survivin, bcl-2, IGF1R and DNA repair gene O6-methylguanine-DNA-methyltranferase (MGMT) with the net result of facilitating apoptosis induction (41–46).
Potential link between APE1 expression and p53 status in various tumors and their resistance to chemotherapeutic drugs have been documented (24,47). Despite several studies showing the involvement of APE1 and p53 in drug resistance induction in tumor cells, the question of whether p53 regulates APE1 expression after genotoxic stress has not been addressed. Elucidating the direct role of p53 on APE1 gene expression in human cells should be of major clinical significance, and a clear understanding of the mechanisms by which p53 controls APE1 expression could help in effective use of chemotherapeutic agents in the treatment of tumors expressing WT or mutant p53. In this study, we show that activation of p53 after genotoxic stress downregulates APE1 expression in HCT116 p53(+/+) human colon carcinoma cells, but not in the p53 null mutants derived there from. Transgenic expression of WTp53 in p53 null cells downregulate both endogenous APE1 level and expression of a reporter whose activity is dependent on APE1 promoter activity. We also present evidence that p53 is associated with the endogenous APE1 promoter in vivo, and that interference of specificity protein (Sp1) binding to the APE1 promoter by WTp53 and subsequent recruitment of histone deacetylase (HDAC) to the promoter could explain downregulation of APE1 by WTp53.
MATERIALS AND METHODS
Cell culture and treatment
The human colorectal adenocarcinoma lines, HCT116 p53(+/+) with WTp53 and p53 null HCT116 p53(−/−) (a gift from Dr B. Vogelstein, Johns Hopkins University School of Medicine) were grown in McCoy's 5A (Gibco Life Technologies, Carlsbad, CA, USA) medium supplemented with 10% fetal bovine serum (Sigma-Aldrich, St Louis, MO, USA), 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco BRL, Carlsbad, CA, USA) in a 5% C02 incubator at 37°C. Cells were treated with 200 nM camptothecin (CPT) (Sigma, St Louis, MO, USA) and/or 30 µM pifithrin-α (PFT-α) (Sigma, St Louis, MO, USA).
Plasmids and luciferase assays
Generation of APE1 promoter–reporter plasmids containing DNA fragments (−4800/+65) and (−1800/+65) of the 5′ regulatory region was described earlier (15). Other reporter plasmids with various lengths of the APE1 5′ regulatory region were generated by PCR and cloned into the luciferase reporter vector pGL3 (Promega, Madison WI, USA) using standard procedures. HCT116 p53(−/−) cells were cotransfected with 500 ng of the APE1-reporter plasmids and expression plasmids for WT or mutant [Val143Ala (V143A) and L22G, T23S] p53 or equivalent amounts of empty vector using LipofectAMINE 2000 (Invitrogen, Life Technologies, Carlsbad, CA, USA), according to the manufacturer's instructions. At 48 h after the transfections, cells were lysed with the reporter lysis buffer (Promega), and the luciferase activity in the cell lysates was measured in a luminometer (AutoLumant LB593, Berthold, Oak Ridge, TN, USA) by the luciferase assay kit (Promega, Madison, WI, USA) according to the manufacturer's protocol. The luciferase activity was normalized to the amount of protein in the lysate.
Preparation of total cell extract and western blot analysis
HCT116 cells were lysed in a lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% TritonX-100, and protease inhibitor cocktail (Roche, Nutley NJ, USA) as described earlier (48). Whole-cell extracts (25 µg) were subjected to 12.5% SDS–PAGE and transferred to a nitrocellulose membrane (Trans-Blot 0.2 µm, Bio-Rad, Hercules, CA, USA). Western immunoblotting analyses with mouse monoclonal antip53 antibody (sc-DO-1, dilution 1:200, Santa Cruz Biotechnology, CA, USA), rabbit polyclonal antiAPE1 (dilution 1:2000), or rabbit monoclonal antiβ-actin antibody (Sigma, 1:1000 dilution) were carried out using enhanced chemiluminescence assay (ECL kit, Amersham, London, UK).
RNA isolation and RT-PCR analysis
Total RNA was extracted using Rneasy Mini kit (QIAGEN) according to the manufacturer's protocol. For quantitative, real-time RT-PCR, one step RT-PCR was performed with 100 ng of cellular RNA for both the target gene and an endogenous control in singleplex tubes using TaqMan one-step RT-PCR master mixture reagent kit (P/N 4309196). TaqMan MGB probes (FAMTM dye-labeled) for APEX1 gene and 18s rRNA (VICTM-dye labeled probe) TaqMan® assay reagent (P/N 4319413E) for endogenous control (assay-on-DemandTM (P/N 4331182) were used. The cycling parameters in an ABI 7000 (Applied Biosystem, Foster City, CA, USA) thermal cycler were as follows: reverse transcription at 48°C for 30 min. AmpliTaq activation 95°C for 10 min, denaturation 95°C for 15s, and annealing/extension 60°C for 1 min. The amount of target was calculated after normalization to the 18s RNA as an endogenous control.
Chromatin immunoprecipitation (ChIP) assay
HCT116 p53(+/+) and HCT116 p53(−/−) cells were treated with CPT at final concentration of 200 nM for 6 h unless otherwise indicated. Cells were washed twice with ice-cold PBS, fixed with 1% formaldehyde for 10 min at RT, washed twice with cold PBS, and harvested in cell scraping solution (1X PBS and 0.5 mM PMSF). Pellets were collected by centrifugation at 1200 rpm for 10 min at 4°C and resuspended in ice-cold lysis buffer (Active Motif, ChIP-IT, Carlsbad, CA, USA, supplemented with 0.5 mM PMSF and 0.5 mM protease inhibitor cocktail), and then incubated on ice for 30 min. The DNA fragments were digested with mung bean nuclease to an average size of 200–500 bp, as empirically estimated by agarose gel electrophoresis of the digest. Immunoprecipitations were performed with antip53 (DO-1), IgG (as a negative control) and antiSp1 (as a positive control) antibodies. After sequential washing, DNA–protein complexes were eluted with elution buffer (1% SDS, 0.1 M NaHCO3, 0.01 mg/ml herring sperm DNA). The crosslinks were reversed by heating at 65°C overnight, and treatment with proteinase K (0.17 µg/µl) for 1 h, and the DNA then isolated using a column (Active Motif, ChIP-IT) according to the manufacturer's guidelines. Recovered DNA was suspended in 50 µl of TE. PCR amplification of DNA was carried out with diluted aliquots using appropriate primers to generate PCR products spanning +24 to −368 bp and −118 to −253 bp of the APE1 promoter region. The PCR products were separated by 1.5% agarose electrophoresis in Tris-borate-EDTA buffer and stained with ethidium bromide.
Electrophoretic mobility shift assay (EMSA)
Electrophoretic mobility shift analyses were performed as described earlier (38) with some modifications. The 5′ 32P-labeled oligo 5′-AGAGAGGGAGGCGAGGCTAAGCGTCTCCGTCACGT-3′ (potential p53-binding site on APE1 promoter) and 5′-TTG AAC ATG TCC CAA CAT GTT GA-3′ containing the p53 consensus sequence from the human p21 promoter were annealed with appropriate complementary stands to generate duplex oligos. The DNA (50 fmol) was then incubated with p53 or nuclear extracts (5 μg) from HCT116 p53(−/−) cells for 20 min at 25°C in a buffer containing 40 mM HEPES-KOH, pH 7.5, 50 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 10% glycerol, 1 μg of poly dI-dC (Sigma). After electrophoresis in non-denaturing 5% polyacrylamide gels in Tris-borate buffer at 4°C, the gels were dried and exposed to PhosphorImager (Molecular Dynamics, Piscataway, NJ, USA) and analyzed using IMAGEQUANT software.
RESULTS
WTp53 downregulates APE1 mRNA and protein levels
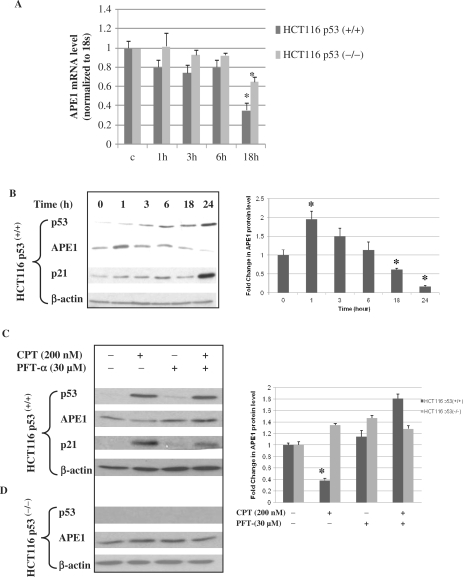
We examined the effect of WTp53 on APE1 expression in HCT116 p53(+/+) and HCT116 p53(−/−) cell lines. To activate p53, these cells were treated with CPT, a DNA topoisomerase I inhibitor, and total RNA was isolated at the indicated times for quantitation of APE1 mRNA levels by real-time RT-PCR. We observed a significant decrease in the APE1 mRNA level at 18 h after treatment with CPT with simultaneous activation of WTp53 in HCT116 p53(+/+) cells (Figure 1A). Treatment with CPT also reduced APE1 mRNA level in p53-negative HCT116 p53(−/−) cells, although not to the same extent as in the p53(+/+) cells indicating a relationship between the p53 status and the downregulation of APE1 expression. Treatment with CPT also induced the p21 level in HCT116 p53(+/+) cells concomitant with enhanced p53 levels (Figure 1B). Activation of the p21 gene was used to monitor in vivo function of p53. Although a transient increase in the APE1 protein level was observed soon after (1 h) CPT treatment, consistent with the decreased APE1 mRNA level, the APE1 polypeptide level was also significantly reduced in HCT116 p53(+/+) cells at 18 h after treatment with CPT (Figure 1B, right panel). We further confirmed the role of p53 by treating cells with the water soluble p53 inhibitor PFT-α, which blocked activation of p53-regulated genes, including cyclin G, p21/Waf-1 and mdm2, and also inhibits apoptosis (49–51). Treatment with PFT-α attenuated APE1 repression after CPT treatment in p53(+/+) cells (Figure 1C), while it had no effect on the APE1 level in p53(−/−) cells (Figure 1D). Taken together, these results suggest that downregulation of APE1 expression is p53-dependent.
Figure 1.
Repression of APE1 gene expression in HCT116 p53(+/+) cells after CPT treatment. (A) Real-time RT-PCR analysis of APE1 mRNA from HCT116 p53(+/+) and HCT116 p53(−/−) cells at indicated time points after CPT treatment. Results correspond to mean ± SD from three separate experiments. (B) After treating HCT116 p53(+/+) cells with CPT (200 nM), as described under Materials and Methods section, total lysates of cells harvested at the indicated times were analyzed for p53, APE1 and p21 levels by western blotting. β-actin was used as the loading control. Right panel, graphical representation of APE1 protein level (normalized to β-actin) at indicated time points after CPT treatment. Results correspond to mean ± SD from three separate experiments. (C and D) HCT116 p53(+/+)or p53(−/−) cells were treated with CPT (200 nM) for 24 h in the presence or absence of the p53 inhibitor PFT-α and total lysates were analyzed for p53, APE1 and p21 levels by western blotting. β-actin was used as the loading control. Right panel, graphical representation of APE1 protein level (normalized to β-actin) after treatment with CPT or PFT-α. Results correspond to mean ± SD from three separate experiments; *P < 0.05.
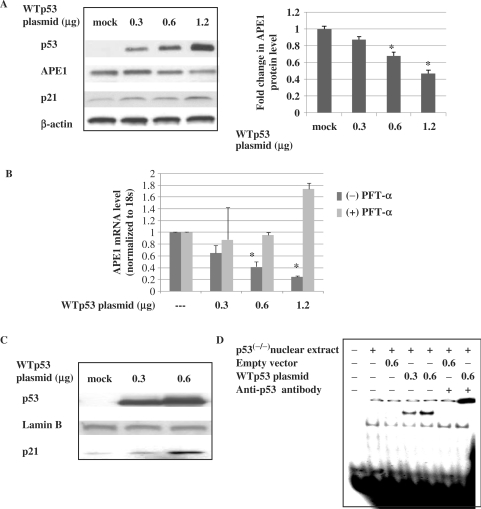
To further confirm that WTp53 acts as a negative regulator of APE1 expression, we investigated the effect of WTp53 overexpression on the APE1 protein and mRNA levels in HCT116 p53 null cells. Various amounts of expression plasmid for WTp53 were used for transfection of p53 null cells, and the expression of p53 and p21 was confirmed by western analysis of the cell extracts (Figure 2A). Overexpression of p53 decreased APE1 mRNA and protein levels in a dose-dependent manner (Figure 2A and B). To confirm that ectopic p53 was transcriptionally active, we measured the p21 levels in nuclear extracts of p53-transfected cells (Figure 2C), and its binding to double-stranded oligonucleotides containing the p53-binding motif of the p21 promoter. Figure 2C shows predominant localization of p53 in nuclear fraction of cells after ectopic expression of WTp53. EMSA of the same extract confirmed the presence of WTp53 which formed a shifted complex with the p53 cis sequence (Figure 2D). Together, these results further confirm that p53 acts as a repressor of APE1 expression.
Figure 2.
Decreased APE1 protein and mRNA levels after ectopic expression of WTp53 in p53 null cells. (A) Total cell extracts were analyzed for p53, APE1 and p21 levels by western blotting. β-actin was used as the loading control. Right panel, graphical representation of APE1 protein level (normalized to β-actin) after transfection with various amounts of WTp53 expression plasmid. Results correspond to mean ± SD from three separate experiments. (B) Real-time RT-PCR analysis of APE1 mRNA levels in HCT116 p53(−/−)cells transfected with various amounts of WTp53 expression plasmid and incubated with PFT-α or vehicle for 48 h. Results correspond to mean ± SD from three separate experiments. (C) Western analysis of p53 and p21 levels in nuclear fraction in p53 null cells after ectopic expression of WTp53. Lamin B was used for nuclear extracts loading control. (D) EMSA of nuclear extracts used in (C) using 32P-labeled duplex oligo from p21 promoter containing consensus p53-binding sequence; *P < 0.05.
APE1 promoter activity is repressed by WTp53, but not by mutant p53
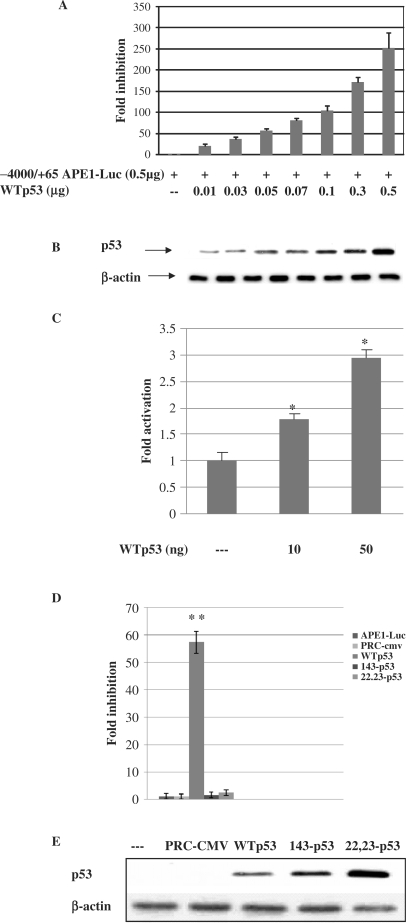
To determine whether p53-mediated downregulation of APE1 occurred at the promoter level, we examined the effect of p53 overexpression on APE1 promoter-dependent luciferase activity in a transient reporter expression assay. We cotransfected an APE1 promoter luciferase reporter construct (−4000/+65 APE1-Luc, containing the APE1 promoter sequence from −4000 to +65 bp) and the WTp53 expression plasmid into HCT116 p53(−/−) cells. Ectopic expression of WTp53 induced a dose-dependent decrease in APE1 promoter activity (up to 250-fold compared to the vector control, Figure 3A). Increase in p53 expression was confirmed by western analysis of the same cells extract with antip53 antibody (Figure 3B). Because p53 is an activator of the p21 gene, we simultaneously examined the effect of ectopic expression of p53 on p21 promoter-dependent luciferase activity. The increase in p21 promoter-dependent luciferase activity with increasing amounts of input p53 plasmid indicated that the inhibitory effect of p53 on APE1 promoter was not due to general inhibition of transcription (Figure 3C). Moreover, we observed that CMV promoter-dependent β-galactosidase and thymidine kinase promoter-dependent renilla-luciferase (pRL-TK, Promega) levels were also increased due to overexpression of WTp53 for unknown reasons (data not shown). We thus could not therefore use these reporter plasmids for normalizing transfection efficiency. In any case, our results indicate that the p53-dependent repression is APE1 promoter specific, and not a general phenomenon. We also examined the effects of two p53 point mutants on APE1 promoter activity namely, V143A, a missense mutation which inactivates p53's sequence-specific DNA binding, and a Leu22Gln Trp23Ser (22,23), double mutant which retains the ability to bind p53-cis sequences but lacks transcriptional activation ability (52). Overexpression of WTp53 significantly decreased luciferase activity under the control of the APE1 promoter (reaching up to ∼60-fold with 50 ng of WTp53), while the mutants had no significant inhibitory effect (Figure 3D). Western analysis of p53(−/−) cells lysates transiently transfected with p53 expression plasmids revealed that the p53 mutant polypeptides were more stable than the WT protein, and were present at higher levels (2- to 3-fold, Figure 3E). These results indicate that despite these higher levels, mutant p53 could not inhibit the APE1 promoter activity.
Figure 3.
Inhibition of APE1 promoter activity by WT but not mutant p53. (A) p53(−/−) cells were transiently transfected with (0.5 μg) of APE1 promoter–luciferase plasmid (−4500/+65) and various amounts of WTp53 expression plasmid or control vector. Luciferase activity was measured at 48 h after transfection and normalized for the amount of protein. The mean ± SD of five independent experiments performed in duplicate is shown. (B) Western analysis of the p53 level in the transfected cell extracts used in (A). (C) p53(−/−) cells were cotransfected with (0.5 μg) of p21 promoter–luciferase plasmid and WTp53. Luciferase activity was measured 48 h after transfection and normalized for the amount of protein. (D) p53(−/−) cells were transiently transfected with (0.5 μg) of APE1 promoter–luciferase plasmid (−4500/+65) and 0.05 μg of expression plasmid encoding WTp53 or mutant p53 (V143A) (22,23) or equivalent amount of empty vector. Other details are given above. (E) Western analysis of WT and mutant p53 in cells lysates used in (D); *P < 0.05; **P < 0.001.
Identification of p53-responsive cis element(s) in APE1 promoter
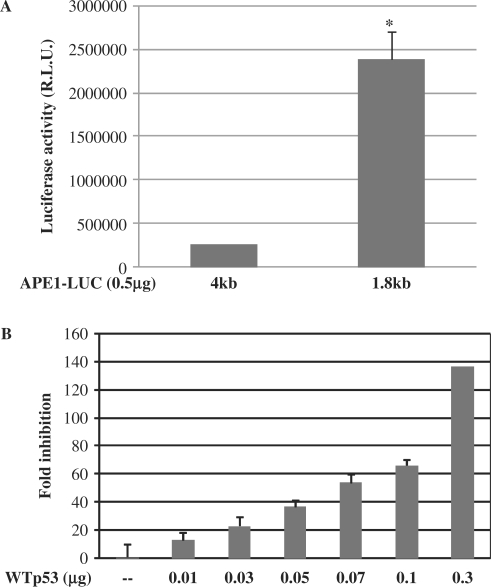
To identify the cis element(s) responsible for p53-mediated downregulation of the APE1 promoter, we carried out promoter deletion analysis. We have shown earlier that the APE1 promoter contains multiple negative regulatory and enhancer elements, including two nCaRE-B elements upstream (−4000 to −1800 bp) of the basal promoter (15). To test whether p53-induced repression of APE1 promoter activity is mediated through the cis elements located in this region, we cotransfected HCT116 p53(−/−) cells with WTp53 and the reporter plasmid containing −1800 to +65 bp (−1800/+65 APE1-Luc) of the APE1 promoter. Deletion of the promoter sequence from −4000 to −1800 bp significantly increased the basal promoter activity (∼9-fold, Figure 4A), confirming our earlier observation about the presence of negative regulatory elements within this sequence (15). Furthermore, ectopic expression of WTp53 caused a decrease in the promoter (−1800/+65 APE1-Luc) activity in a dose-dependent manner (Figure 4B).
Figure 4.
Effect of WTp53 on APE1 promoter reporter activity. (A) Basal promoter activity of APE1 promoter luciferase constructs containing −4500 or −1800 bp of APE1 promoter sequences linked to the luciferase gene. (B) p53(−/−) cells were transiently transfected with 0.5 μg of APE1 promoter–luciferase plasmid (−1800/+65) and WTp53 or control vector as before. Luciferase activity was measured 48 h after transfection as before; *P < 0.05.
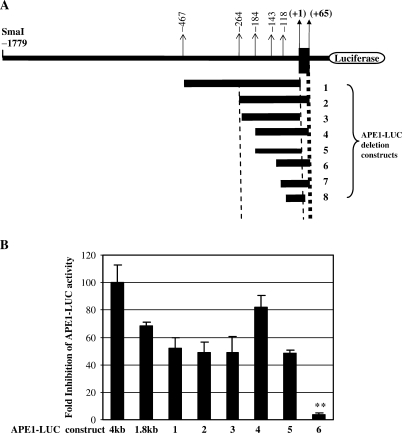
We then carried out detailed mapping of the p53 regulatory sequence by using a series of 5′ promoter deletion constructs. DNA fragments of the 5′ regulatory region (shown in Figure 5A) were cloned upstream of the luciferase coding region and their promoter activity was determined by cotransfection as before. All deletion constructs except 7 and 8 showed basal promoter activity that was abolished by deleting residues −143 to −118 (Figure 5A and B) indicating that the sequence upstream of −118 is required for basal promoter activity. Although deletion of −4000 to −1800 decreased p53-mediated downregulation of promoter activity by ∼1.5-fold, luciferase activity of all the deletion constructs was significantly downregulated after cotransfection with WTp53 (Figure 5B). However, elimination of the sequence from −184 to −143 (construct number 6, Figure 5A) abolished the inhibitory effect of p53 (Figure 5B), indicating that the cis element involved in p53-dependent repression is localized within this sequence. Furthermore, deletion of the +65 bp region downstream to the transcription start site (construct number 5, Figure 5A) had no effect on p53-mediated repression, indicating that these sequences are not required for repression (Figure 5B). Thus the p53-responsive cis elements appeared to be located within the proximal region (−184 to −143 bp) of the APE1 promoter.
Figure 5.
(A) Schematic diagram of the APE1 promoter 5′ regulatory region showing the location of DNA fragments cloned upstream of the luciferase-coding region. The nucleotides are numbered 5′(−) and 3′(+) from the transcription start site (+1). (B) p53(−/−) cells were cotransfected with expression plasmid for WTp53 (0.1 μg) or empty vector and individually for APE1 promoter–reporter plasmids (0.5 μg) 1–8, as indicated in (A). Luciferase activity was measured 48 h after transfection and normalized for the amount of protein. Fold inhibition was calculated as the ratio of luciferase activity from empty vector transfected versus p53-transfected cells; the results represent the mean ± SD in five independent experiments performed in duplicate; *P < 0.001.
Lack of direct binding of p53 to the APE1 promoter in vitro
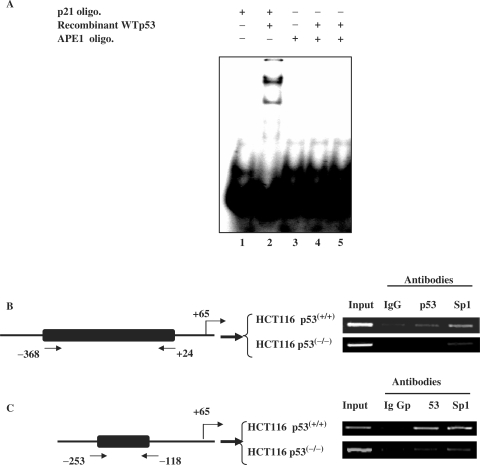
To identify the putative p53-responsive cis element(s) in the 41 bp (−184 to −143) APE1 promoter sequence, we performed a computer search for potential transcription factor binding sites. Putative binding sites were identified one each for Sp1 and upstream factor (USF); however, no consensus p53-binding site could be shown even allowing for two mismatches. Nevertheless, we tested for a potential binding site for p53 in this 41 bp sequence by EMSA using a recombinant p53 polypeptide. The p53 cis element present in the p21 promoter was used as a positive control. As shown in Figure 6A, purified p53 can bind to the p21 promoter (lane 2), but not to the APE1 promoter sequence (lanes 4 and 5), indicating a lack of direct p53 binding to the APE1 promoter in vitro.
Figure 6.
ChIP assay for in vivo association of p53 with APE1 promoter. (A) EMSA of purified p53 (60 ng, lane 4; 120 ng, lane 5) using 32P-labeled duplex oligo corresponding to the bases (−184 to 142) from the APE1 promoter sequence or containing consensus p53-binding sequence from the p21 promoter (lane 2) as a control. (B) p53(+/+) cells were treated with CPT for 9 h, the protein–DNA was crosslinked, and ChIP assays performed as described in Materials and Methods section. Immunoprecipitated APE1 promoter with the indicated antibody was amplified with primers as described in Materials and Methods section. Left panel, schematic diagram of the APE1 promoter showing the relative position of PCR primers used in the ChIP assays.
Recruitment of p53 to the APE1 promoter is induced by cellular stress
Because p53 could exert its repressor activity via interaction with other transcription factors bound to their cognate promoters, we used ChIP assay to test whether p53 is associated with the APE1 promoter in vivo (41). p53(+/+) and p53(−/−) HCT116 cells were treated with CPT for 6 h, and subsequently with 1 mM disuccinimidyl glutarate, which produces protein–protein crosslinks, followed by further incubation with 1% formaldehyde to produce protein–DNA crosslinks. The chromatin fractions were isolated and fragmented by digestion. After immunoprecipitation with antibody to p53 or Sp1, ChIP assays were then performed as described in Materials and Methods section. The amount of immunoprecipitated APE1 promoter sequence was quantitated for each sample by PCR analysis with primers for amplification of the APE1 promoter region from −368 to +24 bp. Figure 6B shows significant enrichment of this sequence in the immunocomplex with p53 antibody, indicating that p53 is associated with the APE1 promoter in vivo in HCT116 p53(+/+) cells. We performed appropriate control experiments to validate our results. Thus, no significant enrichment of the APE1 promoter sequence was observed by the PCR assay in the p53 immunoprecipitate from chromatin fraction of p53(−/−) cells (Figure 6B). Similarly, no significant enrichment of the APE1 promoter sequence was observed in chromatin extracts from either cell line when non-specific IgG was used for immunoprecipitation (Figure 6B). An earlier study identified overlapping Sp1-binding sites within this APE1 promoter sequence (53). As expected, significant enrichment of the APE1 promoter sequence was observed in the Sp1-specific immunocomplexes from both cell lines (Figure 6B), indicating that Sp1 is constitutively associated with the APE1 promoter in the chromatin fraction of both p53(+/+) and p53(−/−) cells. To establish that p53 selectively binds to this (−184 to −143 bp) sequence, we used appropriate primer sequences to amplify the region corresponding to −253 to −118 bp of the APE1 promoter. Figure 6C shows that this sequence was selectively enriched in the p53 immunocomplex from the p53(+/+) but not p53(−/−) HCT116 cells. This confirms binding of p53 to this sequence in vivo. These results provide strong evidence that both p53 and Sp1 are associated with the −253 to −118 bp region in the human APE1 promoter in vivo.
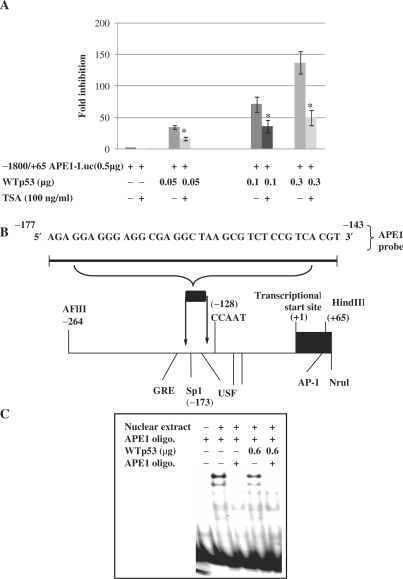
Although there is no evidence for a p53-specific cis element in the APE1 promoter, the presence of p53 in the APE1 promoter complex as indicated by our ChIP assay strongly supports the idea that p53 is recruited to the APE1 promoter via binding to some other trans-acting factors. p53 has been shown to interact with several factors of the basal transcription machinery, e.g. TATA-binding Protein (TBP) or the basal transcription factor Sp1, and also to recruit mSin3A/HDAC repressor complex to the promoter to inhibit transcription (41). Consistent with this, p53-mediated repression of several promoters has been shown to be reversed with the HDAC inhibitor, tricostatin A (TSA) (54,55). To test possible involvement of HDAC in p53-mediated APE1 repression, we used the −1800/+65 APE1-Luc promoter–reporter plasmid which showed the highest promoter activity. At 24 h after cotransfection of HCT116(−/−) cells with the APE1 promoter–reporter and WTp53 plasmids, the cells were treated with 100 ng/ml TSA. Figure 7A shows that TSA reversed p53-mediated repression of luciferase to a significant extent, suggesting that the repression involves p53-dependent recruitment of HDAC to the APE1 promoter.
Figure 7.
Effect of TSA on p53-mediated repression of APE1 promoter activity. (A) p53(−/−) cells were transiently transfected with 0.5 μg of APE1 promoter–luciferase plasmid (−1800/+65) and increasing amounts of expression plasmid encoding WTp53 or equivalent amounts of empty vector. Twenty-four hours after transfection cells were treated with or without TSA (100 ng/ml) and luciferase activity was measured 48 h after transfection and normalized for the amount of protein. The mean ± SD of five independent experiments performed in duplicate is shown. (B) Sequence of the DNA probes for EMSA assay with APE1 promoter. Schematic diagram of the APE1 promoter showing some of the putative transcription factor binding sites (Sp1; USF; glucocorticoid response element (GRE); activator protein 1 (AP-1); and CCAAT box-like sequence). (C) EMSA with nuclear extracts of p53(−/−) cells transfected with empty vector or WTp53 expression plasmid using 32P-labeled duplex oligo corresponding to bases −177 to −143 from APE1 promoter; *P < 0.05.
P53-induced APE1 repression is mediated by interference with Sp1 binding to the APE1 promoter
Identification of one Sp1 site within the putative p53 response element in the APE1 promoter raised the possibility that p53 is recruited by Sp1 to the APE1 promoter complex for inhibition (Figure 7B). ChIP analysis clearly demonstrated association of Sp1 and p53 in the APE1 promoter complex (Figure 6B). p53 has been shown to bind Sp1 and interferes with its binding to the cis element for Sp1-mediated transcription (56–58). To test this possibility, we performed EMSA with APE1's Sp1-binding sequence. Formation of specific protein–DNA complexes suggested the binding of Sp1 to the oligo (Figure 7C), which was competed with unlabeled oligo containing WT but not a mutated Sp1 oligo sequence. These data confirm the specificity of Sp1 binding to this sequence. Furthermore, a significant decrease in binding was observed with nuclear extract from p53-overexpressing HCT116(−/−) cells, suggesting that WTp53 interferes with Sp1 binding to the promoter.
DISCUSSION
APE1 is often overexpressed in tumor cells compared to its basal level in many untransformed cells lines. Downregulation of APE1 was found to sensitize tumor cells to chemotherapy with induction of apoptosis (26,27,29). Thus, elucidating the molecular basis for variable APE1 expression is important from both basic and clinical perspectives. Although several studies have shown that p53 participates in DNA repair processes, and stimulates BER via its direct interaction with APE1 and DNA polymerase β, whether p53 regulates APE1 level itself was not shown before (59). This report provides the first evidence that p53 downregulates APE1 expression. We have shown that the levels of both APE1 mRNA and polypeptide were decreased after CPT treatment in HCT116 p53(+/+), but not in the isogenic HCT116 p53(−/−) cells. This effect on HCT116 p53(+/+) cells was specific, and not a consequence of general effects of genotoxic or other types of stress, because the same treatment did not affect APE1 expression in p53 null cells. This thus indicates direct relationship between the p53 status and APE1 downregulation. Reduced expression of both endogenous APE1 and APE1 promoter-dependent luciferase activity by exogenous p53 in unstressed p53 null cells further confirmed that p53 downregulates APE1 expression. However, ectopic expression of WTp53 downregulated the episomal APE1 promoter to a much greater extent than the endogenous gene which was somewhat unexpected. It is possible that the APE1 promoter in the chromatinized plasmid is more sensitive to the effect of p53 than the folded conformation in cellular chromatin. Furthermore, the endogenous APE1 gene could be regulated by binding of transcription factors to cis elements absent in the episomal promoter. The V143A p53 mutant lacking DNA-binding activity did not downregulate the basal APE1 promoter as much as did WTp53, which strongly suggests that p53's DNA-binding ability is a prerequisite for its repressor activity. However, the L22G, T23S double mutant with normal DNA-binding activity, but lacking trans-acting activity (52), was also unable to repress the APE1 promoter, which suggests that both DNA binding and transactivation abilities of p53 are required for the repression. Western analysis showing that p53 mutants were more stable than the WTp53 protein eliminated the possibility that the former's inability to repress the APE1 promoter was due to reduced stability.
Using functional analysis of nested deletion mutants, we mapped the basal APE1 promoter to a −143 bp segment 5′ to the transcription start site. We also observed that the candidate p53-responsive cis element(s) were located in the −184 to −143 bp segment. However, sequence analysis revealed no apparent p53-binding sites within this segment. Moreover, the recombinant p53 polypeptide failed to bind to an oligo of this sequence (−184 to −143). At the same time we have provided direct evidence from ChIP assays that p53 was bound to the −253 to −118 bp segment sequence in vivo. Together, these data suggest that APE1 repression by p53 is not mediated via direct cis element binding, but rather involves p53's indirect recruitment to the promoter by other transcription factors. p53 has been shown to repress many other promoters that lack p53-specific cis elements, indicating that the mechanisms of p53-mediated repression are varied and complex (41,58,60,61). Such mechanisms could include (i) interference with transcriptional activators, (ii) interference with the basal transcription machinery and (iii) compaction of the chromatin structure at the promoter sites by recruitment of HDACs and other histone-modifying enzymes (41).
The exact mechanism by which p53 represses the APE1 expression is not clear. We have shown that candidate p53-responsive element(s) were located in the −184 to −143 bp segment that includes one Sp1 site. An earlier study using in vitro DNase I footprinting analysis with purified Sp1 polypeptide showed a distinct footprint spanning −169 to −148 bp in the APE1 promoter (53). Consistent with this, our ChIP assay showed constitutive Sp1 binding within this region in both p53(+/+) and p53(−/−) cells, indicating that the Sp1 site is functional in vivo. Moreover, we observed that Sp1 remains bound to the APE1 promoter after p53 activation by CPT, suggesting that both p53 and Sp1 can simultaneously occupy the same promoter region. It thus appears that repression may be achieved through the physical interaction of p53 with Sp1. Consistent with this possibility, p53 has been shown to bind Sp1, inhibiting Sp1-mediated transcription (56–58). However, in P53-dependent repression of the telomerase reverse transcriptase, insulin receptor, VEGF and POLD1 genes, p53 does not inhibit DNA binding by Sp1, but rather interferes with its ability for transactivation (57,58,60–63). We also observed that overexpression of p53 decreased DNA binding by Sp1. On the other hand, treatment with HDAC inhibitor TSA partially abolished p53-mediated repression of the APE1 promoter indicating HDAC recruitment to the promoter site. Thus the mechanism of p53-mediated repression of the APE1 gene appears to be multifaceted.
Our findings are intriguing because p53 acts as a pro-apoptotic factor in the cellular response to stress, whereas APE1 is a pro-survival protein whose DNA repair function is essential for protecting cells from oxidative damage (28,29). However, it is possible that the tumor suppressor function of p53 is mediated in part by transcriptional repression of antiapoptotic genes such as APE1. Thus the cell could use p53 to downregulate APE1 expression in response to severe DNA damage, thereby promoting apoptosis. Consistent with this idea, several earlier studies showed reduction of APE1 expression during apoptosis in myeloid leukemia cells and in neurons after transient global cerebral ischemia in rats (64,65). Additional studies are needed to unravel the physiological significance of such downregulation.
Our study also sheds light on the potential link between APE1 expression and p53 status in different tumors with their resistance to chemotherapeutic drugs. Although only few published studies have directly focused on the potential link between APE1 expression and p53 status in various tumors, an inverse relationship between APE1 nuclear expression and p53 nuclear accumulation has been reported in head and neck cancer (24,47). It is interesting to speculate that tumors with mutated p53 also have high APE1 levels and develop resistance to therapy, while induction of WTp53 in other tumors will reduce their APE1 levels, leading to enhanced therapeutic index of efficacy of the anticancer drugs. Thus the p53-mediated suppression of APE1 expression could offer a potential approach for sensitizing tumor cells to drugs.
ACKNOWLEDGEMENTS
We acknowledge the help of Dr D. Bocangel for this study and we thank Dr D. Konkel for critically editing the manuscript. This work was funded by National Institute of Health (R01 ES08457, R01 CA53791 to S.M. and CA98664 to T.I.), American Heart Association grant (AHA#0565008Y to K.B.). Amira Zaky was supported by a Government of Egypt scholarship for research at University of Texas medical Branch. Funding to pay the Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 2.Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic. Biol. Med. 2002;33:15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- 3.Mitra S, Izumi T, Boldogh I, Bhakat KK, Chattopadhyay R, Szczesny B. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair. 2007;6:461–469. doi: 10.1016/j.dnarep.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair. 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xanthoudakis S, Curran T. Identification and characterization of Ref-1, a nuclear protein that facilitates AP-1 DNA-binding activity. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jayaraman L, Murthy KG, Zhu C, Curran T, Xanthoudakis S, Prives C. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 1997;11:558–570. doi: 10.1101/gad.11.5.558. [DOI] [PubMed] [Google Scholar]
- 7.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 8.Ziel KA, Campbell CC, Wilson GL, Gillespie MN. Ref-1/Ape is critical for formation of the hypoxia-inducible transcriptional complex on the hypoxic response element of the rat pulmonary artery endothelial cell VEGF gene. FASEB J. 2004;18:986–988. doi: 10.1096/fj.03-1160fje. [DOI] [PubMed] [Google Scholar]
- 9.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J. Biol. Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 10.Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J. Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- 12.Ramana CV, Boldogh I, Izumi T, Mitra S. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc. Natl Acad. Sci. USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosch S, Fritz G, Kaina B. Apurinic endonuclease (Ref-1) is induced in mammalian cells by oxidative stress and involved in clastogenic adaptation. Cancer Res. 1998;58:4410–4416. [PubMed] [Google Scholar]
- 14.Tell G, Pellizzari L, Pucillo C, Puglisi F, Cesselli D, Kelley MR, Di Loreto C, Damante G. TSH controls Ref-1 nuclear translocation in thyroid cells. J. Mol. Endocrinol. 2000;24:383–390. doi: 10.1677/jme.0.0240383. [DOI] [PubMed] [Google Scholar]
- 15.Izumi T, Henner WD, Mitra S. Negative regulation of the major human AP-endonuclease, a multifunctional protein. Biochemistry. 1996;35:14679–14683. doi: 10.1021/bi961995u. [DOI] [PubMed] [Google Scholar]
- 16.Fung H, Bennett RA, Demple B. Key role of a downstream specificity protein 1 site in cell cycle-regulated transcription of the AP endonuclease gene APE1/APEX in NIH3T3 cells. J. Biol. Chem. 2001;276:42011–42017. doi: 10.1074/jbc.M106423200. [DOI] [PubMed] [Google Scholar]
- 17.Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, Koch M. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin. Cancer Res. 2001;7:824–830. [PubMed] [Google Scholar]
- 18.Puglisi F, Aprile G, Minisini AM, Barbone F, Cataldi P, Tell G, Kelley MR, Damante G, Beltrami CA, Di Loreto C. Prognostic significance of Ape1/ref-1 subcellular localization in non-small cell lung carcinomas. Anticancer Res. 2001;21:4041–4049. [PubMed] [Google Scholar]
- 19.Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster RS, Einhorn LH, Kelley MR. Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res. 2001;61:2220–2225. [PubMed] [Google Scholar]
- 20.Xu Y, Moore DH, Broshears J, Liu L, Wilson TM, Kelley MR. The apurinic/apyrimidinic endonuclease (APE/ref-1) DNA repair enzyme is elevated in premalignant and malignant cervical cancer. Anticancer Res. 1997;17:3713–3719. [PubMed] [Google Scholar]
- 21.Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 22.Moore DH, Michael H, Tritt R, Parsons SH, Kelley MR. Alterations in the expression of the DNA repair/redox enzyme APE/ref-1 in epithelial ovarian cancers. Clin. Cancer Res. 2000;6:602–609. [PubMed] [Google Scholar]
- 23.Kakolyris S, Kaklamanis L, Giatromanolaki A, Koukourakis M, Hickson ID, Barzilay G, Turley H, Leek RD, Kanavaros P, Georgoulias V, et al. Expression and subcellular localization of human AP endonuclease 1 (HAP1/Ref-1) protein: a basis for its role in human disease. Histopathology. 1998;33:561–569. doi: 10.1046/j.1365-2559.1998.00541.x. [DOI] [PubMed] [Google Scholar]
- 24.Koukourakis MI, Giatromanolaki A, Kakolyris S, Sivridis E, Georgoulias V, Funtzilas G, Hickson ID, Gatter KC, Harris AL. Nuclear expression of human apurinic/apyrimidinic endonuclease (HAP1/Ref-1) in head-and-neck cancer is associated with resistance to chemoradiotherapy and poor outcome. Int. J. Radiat. Oncol. Biol. Phys. 2001;50:27–36. doi: 10.1016/s0360-3016(00)01561-3. [DOI] [PubMed] [Google Scholar]
- 25.Freitas S, Moore DH, Michael H, Kelley MR. Studies of apurinic/apyrimidinic endonuclease/ref-1 expression in epithelial ovarian cancer: correlations with tumor progression and platinum resistance. Clin. Cancer Res. 2003;9:4689–4694. [PubMed] [Google Scholar]
- 26.Robertson KA, Hill DP, Xu Y, Liu L, van Epps S, Hockenbery DM, Park JR, Wilson TM, Kelley MR. Down-regulation of apurinic/apyrimidinic endonuclease expression is associated with the induction of apoptosis in differentiating myeloid leukemia cells. Cell Growth Differ. 1997;8:443–449. [PubMed] [Google Scholar]
- 27.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 28.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol. Cell. 2005;17:463–470. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 29.Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Two essential but distinct functions of the mammalian abasic endonuclease. Proc. Natl Acad. Sci. USA. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin S, Levine AJ. The p53 functional circuit. J. Cell Sci. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 31.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 33.Steele RJ, Lane DP. P53 in cancer: a paradigm for modern management of cancer. Surgeon. 2005;3:197–205. doi: 10.1016/s1479-666x(05)80041-1. [DOI] [PubMed] [Google Scholar]
- 34.Perry ME, Levine AJ. Tumor-suppressor p53 and the cell cycle. Curr. Opin. Genet. Dev. 1993;3:50–54. doi: 10.1016/s0959-437x(05)80340-5. [DOI] [PubMed] [Google Scholar]
- 35.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 36.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol. Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 37.Chan TA, Hwang PM, Hermeking H, Kinzler KW, Vogelstein B. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 2000;14:1584–1588. [PMC free article] [PubMed] [Google Scholar]
- 38.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 39.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 40.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 41.Ho J, Benchimol S. Transcriptional repression mediated by the p53 tumour suppressor. Cell Death Differ. 2003;10:404–408. doi: 10.1038/sj.cdd.4401191. [DOI] [PubMed] [Google Scholar]
- 42.Harris LC, Remack JS, Houghton PJ, Brent TP. Wild-type p53 suppresses transcription of the human O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1996;56:2029–2032. [PubMed] [Google Scholar]
- 43.Grombacher T, Eichhorn U, Kaina B. p53 is involved in regulation of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) by DNA damaging agents. Oncogene. 1998;17:845–851. doi: 10.1038/sj.onc.1202000. [DOI] [PubMed] [Google Scholar]
- 44.Mirza A, McGuirk M, Hockenberry TN, Wu Q, Ashar H, Black S, Wen SF, Wang L, Kirschmeier P, Bishop WR, et al. Human survivin is negatively regulated by wild-type p53 and participates in p53-dependent apoptotic pathway. Oncogene. 2002;21:2613–2622. doi: 10.1038/sj.onc.1205353. [DOI] [PubMed] [Google Scholar]
- 45.Prisco M, Hongo A, Rizzo MG, Sacchi A, Baserga R. The insulin-like growth factor I receptor as a physiologically relevant target of p53 in apoptosis caused by interleukin-3 withdrawal. Mol. Cell. Biol. 1997;17:1084–1092. doi: 10.1128/mcb.17.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srivenugopal KS, Shou J, Mullapudi SR, Lang FF, Jr, Rao JS, Ali-Osman F. Enforced expression of wild-type p53 curtails the transcription of the O(6)-methylguanine-DNA methyltransferase gene in human tumor cells and enhances their sensitivity to alkylating agents. Clin. Cancer Res. 2001;7:1398–1409. [PubMed] [Google Scholar]
- 47.Herring CJ, West CM, Wilks DP, Davidson SE, Hunter RD, Berry P, Forster G, MacKinnon J, Rafferty JA, Elder RH, et al. Levels of the DNA repair enzyme human apurinic/apyrimidinic endonuclease (APE1, APEX, Ref-1) are associated with the intrinsic radiosensitivity of cervical cancers. Br. J. Cancer. 1998;78:1128–1133. doi: 10.1038/bjc.1998.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S. Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 50.Murphy PJ, Galigniana MD, Morishima Y, Harrell JM, Kwok RP, Ljungman M, Pratt WB. Pifithrin-alpha inhibits p53 signaling after interaction of the tumor suppressor protein with hsp90 and its nuclear translocation. J. Biol. Chem. 2004;279:30195–30201. doi: 10.1074/jbc.M403539200. [DOI] [PubMed] [Google Scholar]
- 51.Marsolais D, Cote CH, Frenette J. Pifithrin-alpha, an inhibitor of p53 transactivation, alters the inflammatory process and delays tendon healing following acute injury. Am. J. Physiol. 2007;292:R321–R327. doi: 10.1152/ajpregu.00411.2005. [DOI] [PubMed] [Google Scholar]
- 52.Lin J, Chen J, Elenbaas B, Levine AJ. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 53.Harrison L, Ascione AG, Wilson DM, III, Demple B. Characterization of the promoter region of the human apurinic endonuclease gene (APE) J. Biol. Chem. 1995;270:5556–5564. doi: 10.1074/jbc.270.10.5556. [DOI] [PubMed] [Google Scholar]
- 54.Murphy M, Hinman A, Levine AJ. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 56.Borellini F, Glazer RI. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J. Biol. Chem. 1993;268:7923–7928. [PubMed] [Google Scholar]
- 57.Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol. Cell. Biol. 1999;19:1279–1288. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webster NJ, Resnik JL, Reichart DB, Strauss B, Haas M, Seely BL. Repression of the insulin receptor promoter by the tumor suppressor gene product p53: a possible mechanism for receptor overexpression in breast cancer. Cancer Res. 1996;56:2781–2788. [PubMed] [Google Scholar]
- 59.Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20:914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanaya T, Kyo S, Hamada K, Takakura M, Kitagawa Y, Harada H, Inoue M. Adenoviral expression of p53 represses telomerase activity through down-regulation of human telomerase reverse transcriptase transcription. Clin. Cancer Res. 2000;6:1239–1247. [PubMed] [Google Scholar]
- 61.Zhang L, Yu D, Hu M, Xiong S, Lang A, Ellis LM, Pollock RE. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 2000;60:3655–3661. [PubMed] [Google Scholar]
- 62.Li B, Lee MY. Transcriptional regulation of the human DNA polymerase delta catalytic subunit gene POLD1 by p53 tumor suppressor and Sp1. J. Biol. Chem. 2001;276:29729–29739. doi: 10.1074/jbc.M101167200. [DOI] [PubMed] [Google Scholar]
- 63.Im HJ, Pittelkow MR, Kumar R. Divergent regulation of the growth-promoting gene IEX-1 by the p53 tumor suppressor and Sp1. J. Biol. Chem. 2002;277:14612–14621. doi: 10.1074/jbc.M109414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raffoul JJ, Banerjee S, Singh-Gupta V, Knoll ZE, Fite A, Zhang H, Abrams J, Sarkar FH, Hillman GG. Down-regulation of apurinic/apyrimidinic endonuclease 1/redox factor-1 expression by soy isoflavones enhances prostate cancer radiotherapy in vitro and in vivo. Cancer Res. 2007;67:2141–2149. doi: 10.1158/0008-5472.CAN-06-2147. [DOI] [PubMed] [Google Scholar]
- 65.Walton M, Lawlor P, Sirimanne E, Williams C, Gluckman P, Dragunow M. Loss of Ref-1 protein expression precedes DNA fragmentation in apoptotic neurons. Brain Res. Mol. Brain Res. 1997;44:167–170. doi: 10.1016/s0169-328x(96)00291-4. [DOI] [PubMed] [Google Scholar]