Abstract
Invasion of two PNA strands to double-stranded DNA is one of the most promising methods to recognize a predetermined site in double-stranded DNA (PNA = peptide nucleic acid). In order to facilitate this ‘double-duplex invasion’, a new type of PNA was prepared by using chiral PNA monomers in which a nucleobase was bound to the α-nitrogen of N-(2-aminoethyl)-d-lysine. These positively charged monomer units, introduced to defined positions in Nielsen's PNAs (poly[N-(2-aminoethyl)glycine] derivatives), promoted the invasion without impairing mismatch-recognizing activity. When pseudo-complementary nucleobases 2,6-diaminopurine and 2-thiouracil were bound to N-(2-aminoethyl)-d-lysine, the invasion successfully occurred even at highly G–C-rich regions [e.g. (G/C)7(A/T)3 and (G/C)8(A/T)2] which were otherwise hardly targeted. Thus, the scope of sequences available as the target site has been greatly expanded. In contrast with the promotion by the chiral PNA monomers derived from N-(2-aminoethyl)-d-lysine, their l-isomers hardly invaded, showing crucial importance of the d-chirality. The promotion of double-duplex invasion by the chiral (d) PNA monomer units was ascribed to both destabilization of PNA/PNA duplex and stabilization of PNA/DNA duplexes.
INTRODUCTION
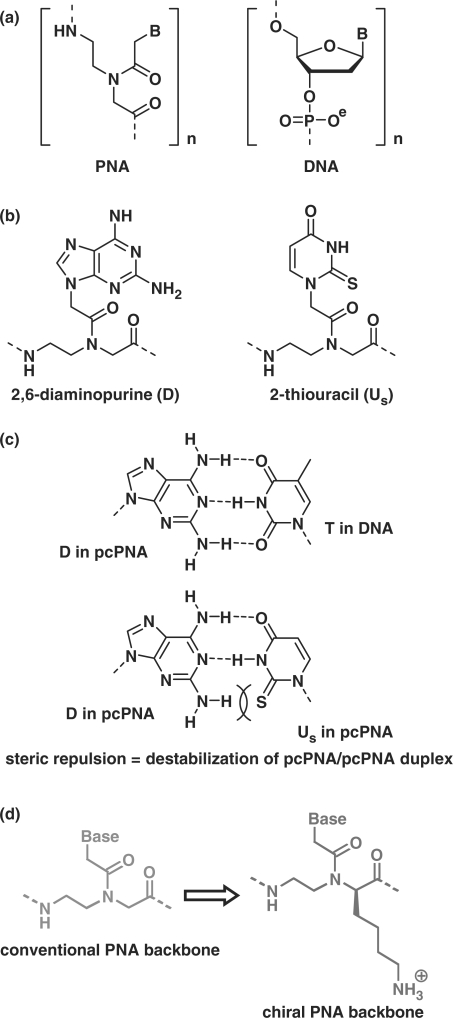
Much interest of chemists and biochemists has been focused on the developments of molecules that selectively bind to a defined site in DNA and regulate its biological functions (1–8). Peptide nucleic acid (PNA), developed by Nielsen et al. (9), bears nucleobases on poly[N-(2-aminoethyl)glycine] backbone (Figure 1a) and forms duplexes with DNA or RNA according to the Watson–Crick rule (10). These duplexes are in general more stable than those of the corresponding DNA, due to the absence of electrostatic repulsion between the two strands. Furthermore, PNA is resistant to nucleases and peptidases (11), and thus highly potent for various applications in vivo and in vitro (12–28). Various PNA derivatives have also been developed (29–31). One of the important characteristics of PNA is its invasion to double-stranded DNA. When two PNA strands are complementary with both strands of DNA at the target site and at the same time the formation of PNA/PNA duplex is suppressed by some factors, these PNA strands can invade the DNA (note that they are complementary with each other). The most successful method for this double-duplex invasion is to use pseudo-complementary PNA (pcPNA) in which nucleobases adenine (A) and thymine (T) in conventional PNA are replaced with modified bases 2,6-diaminopurine (D) and 2-thiouracil (Us) as shown in Figure 1b (32). The invasion is efficient, since the duplex between two pcPNA strands is destabilized by steric repulsion between the 2-amino group of D and the 2-S atom of Us (Figure 1c).
Figure 1.
(a) Chemical structures of PNA and DNA. (b) Pseudo-complementary bases 2,6-diaminopurine (D) and 2-thiouracil (Us) used in pcPNA. The D–Us pair is destabilized by steric repulsion between the 2-amino group of D and the 2-S atom of Us, although D–T and A–Us pairs are sufficiently stable, as shown in (c) (32). (d) Chiral PNA units from N-(2-aminoethyl)-d-lysine, used for the promotion of double-duplex invasion in place of conventional PNA units.
Upon double-duplex invasion, the structure and other physicochemical properties of DNA at the target site are site-selectively changed, indicating unique applications of this method for various purposes. For example, enzymatic functions of restriction enzymes and methylase were inhibited by pcPNAs invading to the corresponding sites (33,34). Double-duplex invasion was also used to prepare artificial restriction DNA cutters for site-selective scission of double-stranded DNA (35–37). Hot spots for the scission, formed by pcPNAs, were selectively hydrolyzed by Ce(IV)/EDTA complex. However, the efficiency of double-duplex invasion by conventional pcPNAs is rather strongly dependent on the target sequence, and gradually decreases with increase in G/C content at the target site [e.g. (G/C)4(A/T)6 > (G/C)5(A/T)5 > (G/C)6(A/T)4] (32). The main reason is that the duplexes between two PNA strands, which involve many G–C pairs, are unfavorably stable and cannot be dissociated for the invasion (no pseudo-complementary bases for G and C are available). Invasion to (G/C)7(A/T)3 sites and still more G–C-rich regions have never been reported. These factors impose a limitation to the applications of this unique and elegant technique, since only A–T-rich sequences can be chosen as the target site.
In this article, we present a new strategy to widen the sequence-versatility of double-duplex invasion [a preliminary communication: (38)]. Positive charges are incorporated to predetermined positions of pcPNA strands by using chiral PNA monomers (39–42) in which D or Us is bound to the α-nitrogen of N-(2-aminoethyl)-d-lysine (Figure 1d). With this chiral pcPNA, G/C-rich sites [e.g. (G/C)7(A/T)3 and (G/C)8(A/T)2] are successfully targeted, and the scope of available sequences is notably expanded. Effects of chiral PNA monomer units bearing conventional nucleobases A and T are also presented. Furthermore, the roles of chiral PNA monomer units in the promotion of double-duplex invasion are discussed in terms of the stability of PNA/PNA and DNA/PNA duplexes in the solutions.
MATERIALS AND METHODS
Materials
PNA strands involving chiral PNA monomer units were synthesized by the method described previously (43). On a (4-methylbenzhydryl)amine resin (from ABI), Nα-Fmoc-protected submonomer (derived from d- or l-N-(2-aminoethyl)lysine) was first connected, and then the nucleobase was attached to the Nα-atom by using the corresponding carboxymethylnucleobase. The carboxymethylnucleobase of 2-thiouracil (Us) was prepared according to the literature (32). In order to introduce the chiral PNA units of 2,6-diaminopurine (D), the carboxymethylnucleobase [synthesized as described in the literature (44)] was activated with 3-hydroxy-1,2,3-benzotriazin-4(3H)-one and N,N′-dicyclohexylcarbodiimide for 2 h at room temperature in N-methyl-2-pyrrolidinone. After filtration using ULTRAFREE-MC 5.0 µm filter unit (from Millipore), the resultant filtrate was added to the resin and incubated for 2 h at 45°C. These procedures were repeated for further three times and then unreacted amino groups were capped by acetic anhydride. For the synthesis of conventional PNA units and pcPNA, Boc-protected monomers were used in place of the submonomer. All the PNA strands were purified by reversed-phase HPLC and characterized by MALDI-TOFMS (Shimadzu, KOMPACT MALDI II or Bruker, AutoFLEX; Supplementary Table 2). Water was deionized by MILLIPORE WATER PURIFICATION SYSTEM, and sterilized immediately before use.
Double-stranded DNAs having the target invasion site (DNA1 and DNA2) were prepared from pBR322 plasmid by PCR amplification. To introduce a mutation into substrate DNA, mutated double-stranded DNA was amplified from pBR322 by overlapping PCR using mutated primers. This PCR product was inserted into the corresponding site of pBR322 by a conventional method using restriction enzymes and a ligase. After cloning from JM109 (Toyobo), substrate DNA for invasion assay was prepared by PCR amplification from the resultant mutated plasmid DNA.
Evaluation of efficiency of strand invasion
The mixture of double-stranded DNA and PNA strands was incubated for 1.5 h at 50°C and pH 7.0 (5 mM HEPES buffer; [NaCl] = 0 M). Then, loading buffer containing bromophenol blue (0.05%) and glycerol (30%) in 0.5× TBE buffer was added, and the mixture was subjected to 5% nondenaturing polyacrylamide gel electrophoresis at 20°C. The bands were stained with GelStar (from Cambrex), and analyzed on a FUJIFILM FLA-3000G imaging analyzer.
Measurements of Tm values
The Tm values were determined by monitoring the absorbance at 260 nm with the temperature ramp 1.0°C/min. Measurement conditions: [DNA] = [each PNA] = 2 μM at pH 7.0 (5 mM HEPES buffer). Unless noted otherwise, no NaCl was added to the mixtures.
RESULTS AND DISCUSSION
Introduction of positive charges to PNA strands using chiral PNA monomers
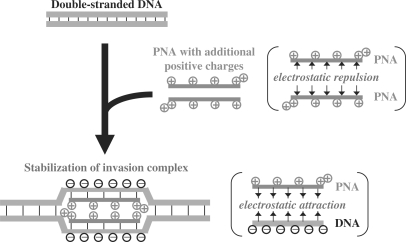
The overall concept of the present study is schematically depicted in Figure 2. The formation of PNA/PNA duplex is hampered by both (i) electrostatic repulsion between the positive charges on the chiral PNA units and (ii) intra-strand steric hindrance induced by the lysine side chains (39,40). On the other hand, PNA/DNA duplexes are stabilized by electrostatic attraction between the positive charges of chiral PNA and the negative charges of DNA.
Figure 2.
Scheme for the promotion of double-duplex invasion by introducing positive charges to PNA strands.
All the PNA strands used in this study are 10-mers bearing one lysine residue at the N-terminus and another lysine residue at the C-terminus (Supplementary Table 1). Accordingly, the target sites for double-duplex invasion in substrate DNA (presented in Supplementary Figure 1) are 10-bp sequences. The ratio of two PNA strands in the invasion mixtures is always 1:1. The units from the chiral monomers are underlined and distinguished from either conventional PNA or conventional pcPNA units (e.g. A versus A and Us versus Us). In the figures, these chiral PNA units are shown in red (with a plus sign), whereas conventional PNA units are in blue. Unless noted otherwise, all the chiral PNA monomers were derived from N-(2-aminoethyl)-d-lysine.
Invasion to G/C-rich sites by chiral PNA
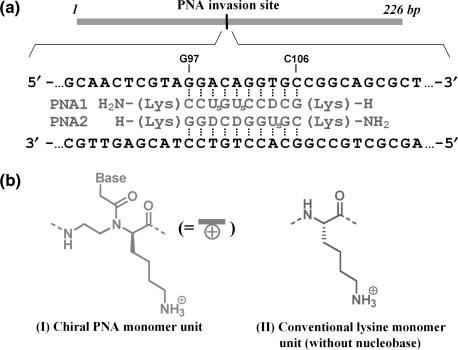
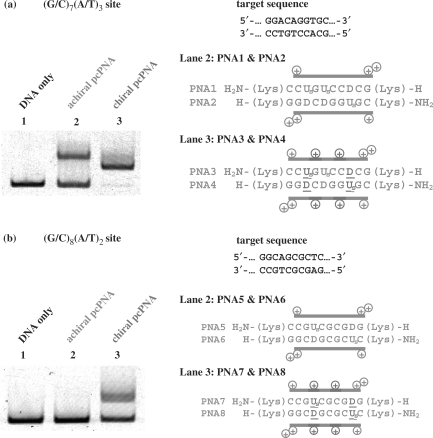
First, we studied on double-duplex invasion to highly G/C-rich regions, which are hardly targeted by conventional pcPNA. Typical sequences at double-duplex invasion site are shown in Figure 3a. The PNA strands are complementary with G97-C106 of a 226-mer double-stranded DNA (DNA1 in Supplementary Figure 1). This target site for invasion (10 bp) is composed of seven G–C Watson–Crick pairs and three A–T pairs [(G/C)7(A/T)3 site]. This highly G–C-rich region is strongly resistant to double-duplex invasion. In invasion experiments, the mixture was incubated at 50°C and pH 7.0 for 1.5 h, and the efficiency of strand invasion was analyzed at 20°C by gel-shift assay on 5% nondenaturing polyacrylamide gel (Figure 4a). In PNA1 and PNA2, conventional pcPNA units, in which D or Us is bound to α-nitrogen of N-(2-aminoethyl)glycine, were used as the counterparts of T and A in DNA strands, respectively [(G/C)7(D/Us)3 additive was formed]. The sequences at the invasion site are presented in the right-hand side of Figure 4a. As shown in lane 2, double-duplex invasion of this ‘achiral pcPNA’ was inefficient, and only half of the DNA was converted to the invasion complex (the upper band in the gel). In PNA3/PNA4, two of these three D–Us pairs in PNA1/PNA2 were replaced with chiral D–Us pairs (D or Us on N-(2-aminoethyl)-d-lysine). In the resultant (G/C)7(D/Us)2(D/Us)1 additive, positive charges are incorporated to the predetermined sites in the PNA strands (see the plus signs in red). The invasion of this ‘chiral pcPNA’ was highly effective and there remained almost no free DNA in the mixture (lane 3). Note that the invasion conditions are exactly the same as employed in lane 2.
Figure 3.
(a) The DNA substrate and PNA strands used in Figure 4a, and (b) the structures of chiral PNA monomer unit (I) and conventional lysine monomer unit (II).
Figure 4.
Strand invasion in G–C-rich regions by pcPNA strands involving chiral PNA monomers; (a) invasion site = (G/C)7(A/T)3 and (b) (G/C)8(A/T)2. Lane 1, DNA only (control); lane 2, conventional pseudo-complementary D–Us's on N-(2-aminoethyl)glycine; lane 3, chiral D–Us's on N-(2-aminoethyl)-d-lysine. Invasion conditions: (i) [DNA1] = 5 nM and [PNA (each strand)] = 50 nM at pH 7.0 (HEPES buffer) and 50°C for 1.5 h; (ii) [DNA1] = 5 nM and [PNA (each strand)] = 30 nM. The gel-shift assay was performed at 20°C.
The target sequence in Figure 4b involves eight G–C pairs and two A–T pairs (G108–C117 of the 226-mer DNA in Figure 3a) and is still less favorable for double-duplex invasion (32). In PNA5/PNA6, two conventional D–Us pairs [on N-(2-aminoethyl)glycines] were incorporated [(G/C)8(D/Us)2) additive]. However, no measurable invasion occurred at this (G/C)8(A/T)2 site as expected (lane 2). The steric repulsion in these two D–Us pairs was too small to destabilize the PNA5/PNA6 duplex sufficiently and allow the invasion. Accordingly, in PNA7/PNA8, these two D–Us pairs were further replaced with two chiral D–Us pairs [on N-(2-aminoethyl)-d-lysines]. As shown in lane 3, this chiral pcPNA [(G/C)8(D/Us)2 additive] efficiently invaded the DNA. The conversion for the invasion monotonously increased with increasing concentration of the PNA strands, and was around 50% when [PNA (each strand)] = 30 nM and [DNA] = 5 nM (see Supplementary Figure 2). Usefulness of chiral PNA monomers for the strand invasion has been further substantiated. Invasion to (G/C)9(A/T)1 site (5′-CCGGCAGCGC-3′/3′-GGCCGTCGCG-5′) using (G/C)9(D/Us)1 additive (PNA17/PNA18 in Supplementary Table 1) was also attempted. However, no invasion was observed under the conditions employed.
Promotion of the invasion by the chiral PNA monomers was also evident at 37°C. However, the invasion was rather slow and required about 1 h to complete. Invasion of achiral pcPNAs was still slower (several fold) than that of chiral pcPNA, and detailed analysis was difficult. Accordingly, all the invasion assays in this article were achieved at 50°C, where the double-duplex invasion of chiral PNA was completed within a few minutes.
Invasion to A/T-rich region by chiral PNA
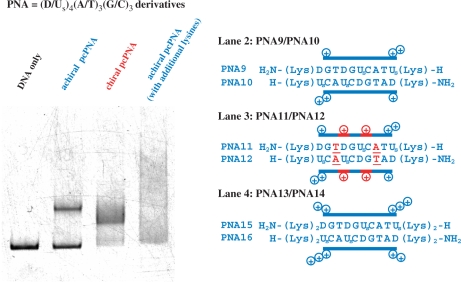
When the target site is rich in A–T pairs, conventional pcPNA [D or Us nucleobase on N-(2-aminoethyl)glycine] eminently achieves double-duplex invasion. However, it has been found that nonpseudo-complementary chiral PNA units [A or T nucleobase on N-(2-aminoethyl)-d-lysine] also promote the invasion. In Figure 5, the PNA strands are complementary with T175-A184 [(G/C)3(A/T)7 site] of a 408-mer double-stranded DNA (DNA2 in Supplementary Figure 1). In PNA9/PNA10, four D–Us pairs on N-(2-aminoethyl)glycine) were incorporated to PNA strands [(G/C)3(D/Us)4(A/T)3 additive]. In spite of destabilization of PNA9/PNA10 duplex by steric hindrance in these four D–Us pairs, the double-duplex invasion occurred only inefficiently (lane 2). Under the same conditions, however, the PNA11/PNA12 combination effectively invaded the DNA, and almost all the DNA was converted to the invasion complex (lane 3). In these PNA strands, two of three A/T pairs in PNA9/PNA10 were replaced with two chiral A/T pairs [(G/C)3(D/Us)4(A/T)2(A/T)1 additive].
Figure 5.
Promotion of invasion to (G/C)3(A/T)7 site by introducing chiral PNA monomers to the pcPNA strands. In lane 3, two conventional A–T pairs (in blue) of the PNA in lane 2 are replaced with chiral A–T pairs (in red). In lane 4, simple lysine residues (Figure 3b, II) were bound to the termini of the pcPNAs used in lane 2 so that the net positive charges (+10) were the same as those in lane 3. Invasion conditions: [DNA2] = 10 nM and [PNA (each strand)] = 200 nM at pH 7.0 (HEPES buffer) and 50°C for 1.5 h. The gel-shift assay was performed at 20°C.
All these notable promotions of invasion by the chiral PNA monomers cannot be ascribed to simple incorporation of positive charges to the PNA strands. Thus, the combination of PNA15 [H2N-(Lys)2DGTDGUsCATUs(Lys)2-H] and PNA16 [H-(Lys)2UsCAUsCDGTAD(Lys)2-NH2] never invaded the DNA (lane 4 of Figure 5). Note that this PNA15/PNA16 combination has the same sequence and the same net charges (totally +10) as the PNA11/PNA12 combination used in lane 3. The only difference between them is whether these positive charges are derived from chiral PNA monomers bearing nucleobases [(I) in Figure 3b] or simple lysine monomers used for conventional peptide synthesis [(II)]. Upon mixing PNA15/PNA16 with the 408-mer DNA together, notable aggregation took place, probably due to formation of polyion complexes, and the strand invasion complex was hardly formed. In these PNA strands, the positive charges are flanking at the ends of strands, and rather freely interact with the negative charges of the DNA in a less controlled manner. In order to promote the strand invasion, steric constraints must be introduced to the middle of PNA strands together with positive charges.
Crucial requirement of d-enantiomers of chiral PNA monomers for the promotion of strand invasion
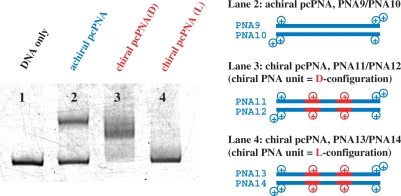
The d-chirality of PNA monomers is critical here. In Figure 6, the PNA11/PNA12 combination [(G/C)3(D/Us)4(A/T)2(A/T)1 additive] was prepared by using chiral PNA monomers, which were derived from either N-(2-aminoethyl)-d-lysine or its l-isomer. When two of three A–T pairs in the PNA strand were chiral PNA monomer units from N-(2-aminoethyl)-d-lysine [(G/C)3(D/Us)4(A(D)/T(D))2(A/T)1 additive, PNA11/PNA12], the strand invasion was highly efficient (lane 3). With the use of the chiral PNA monomers from N-(2-aminoethyl)-l-lysine (PNA13/PNA14), however, the invasion hardly occurred under the same conditions (lane 4). The invasion efficiency of this l-isomer was even lower than that in lane 2 where conventional PNA monomers [A and T on N-(2-aminoethyl)glycine] were used (the PNA9/PNA10 combination).
Figure 6.
Effect of chirality of chiral PNA monomers on the efficiency of invasion. Lane 1, DNA only (control); lane 2, conventional A–T [on N-(2-aminoethyl)glycine] in place of A–T; lane 3, A(D)–T(D) = A–T on N-(2-aminoethyl)-d-lysine (PNA11/PNA12); lane 4, A(L)–T(L) = A–T on N-(2-aminoethyl)-l-lysine (PNA13/PNA14). Invasion conditions are the same as described in Figure 5.
Mismatch recognition of chiral PNAs in double-duplex invasion
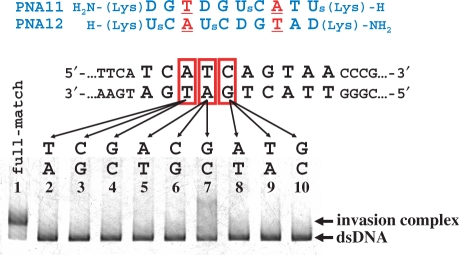
The mismatch-recognizing ability of positively charged chiral pcPNAs is investigated in Figure 7. Here, a single mutation was systematically introduced to the target site in DNA, and the concurrent change in invasion efficiency was assayed. The PNA11/PNA12 combination used in lane 1 is completely complementary with the target site in DNA2, and the invasion is accordingly efficient (this result is the same as observed in lane 3 of Figure 5). In lanes 2–4, the A–T base pair in the original DNA was converted to T–A, C–G and G–C pairs, respectively, and the corresponding mismatches were introduced between the DNA and the PNA additive. In all the cases, no double-duplex invasion was detected. Similarly, the invasion almost completely disappeared when another base pair in the DNA was changed to noncomplementary pair (lanes 5–10). Apparently, the chiral pcPNAs clearly distinguish the mutation at only one DNA base pair in the target site. Their mismatch-recognizing ability is never impaired even when the positive charges introduced should show notable electrostatic interactions with the DNA.
Figure 7.
Effects of mismatch on the strand invasion of chiral pcPNAs into DNA. One of three base pairs in the target site (shown in red squares) was changed to another base pair. Invasion conditions are the same as described in Figure 5.
Fidelity of the present modified PNA additive in the recognition of designated site of DNA was also confirmed by DNaseI foot printing assay (see Supplementary Figure 2). One of the two DNA strands was labeled by FAM. In the absence of chiral pcPNAs, notable cleavage by DNaseI was observed throughout the strand of substrate DNA. In the presence of PNA11/PNA12, however, the target invasion site (5′-TCATCAGTAA-3′) was considerably protected from the digestion. On the other hand, the enzymatic digestion of other portions of the DNA was less affected by the PNA additive.
Origin of high invasion activity of chiral PNA
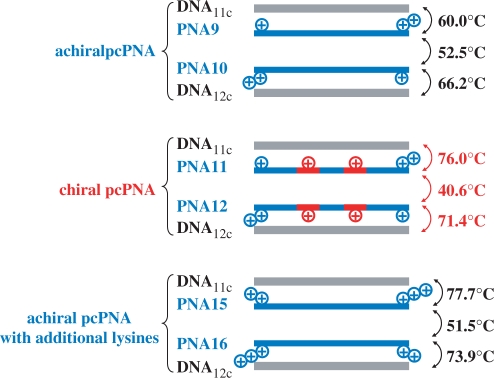
The primary governing factors of the stabilities of invasion complexes are (i) energy gain obtained by the formation of two PNA/DNA duplexes, (ii) energy loss due to competitive formation of the duplex between two PNA strands which are complementary with each other and (iii) energy loss induced by decomposition of the DNA/DNA duplex at the invasion site (Figure 2). Quantitative information on the contributions of the factors (i) and (ii) was obtained by using the melting temperatures (Tm) of these duplexes (Figure 8). It was found that the Tm of PNA11/PNA12 duplex involving two A–T pairs (40.6°C) is notably lower than those for PNA9/PNA10 duplex (52.5°C; two A–T pairs in place of the two A–T pairs) and for PNA15/PNA16 duplex (51.5°C; four additional lysine residues and two A–T pairs in place of the two A–T pairs). Thus, the PNA11/PNA12 duplex shows overwhelmingly larger invasion activity than the other two combinations (Figure 5), mainly because it is less stable due to the electrostatic repulsion between the positive charges on the chiral PNA monomer units A and T. Consistently, the Tm of this duplex gradually elevated with increasing concentration of NaCl, and attained 45.3°C when [NaCl] = 0.5 M (the Tm of the PNA9/PNA10 duplex was little affected by NaCl concentration). This Tm is still lower than that of the PNA15/PNA16 duplex, which is consistent with further destabilization induced by the side-chains of the chiral monomer units in two PNA strands. The intra-strand steric repulsion hampers the strands to take the correct conformations required to form the most stable PNA/PNA duplex. On the other hand, the Tm values for the duplexes of both PNA11 and PNA12 with the corresponding complementary DNA (DNA11c and DNA12c: 10 bp) are 76.0°C and 71.4°C, respectively. These values are much higher than the values for PNA9/DNA11c and PNA10/DNA12c duplexes (Tm = 60.0°C and 66.2°C, respectively), but rather close to those of the DNA duplexes of PNA15 and PNA16 (Tm = 77.7°C and 73.9°C). The stabilities of these PNA/DNA duplexes are mostly governed by the number of positive charges on the PNAs, reflecting the importance of electrostatic attractive interactions. Apparently, the strand invasion of the strands involving chiral PNA monomer units is highly efficient, because (I) the PNA/PNA duplex is destabilized both by the electrostatic repulsion between the positive charges of the PNAs and by the intra-strand steric clash of the lysine side-chains, and (II) the PNA/DNA duplexes are stabilized by the attraction between these positive charges and the negative charges of DNA. The proposal in Figure 2 has been substantiated. Detailed analysis on the double-duplex invasion of PNA1-8 to G/C-rich sites was not successful, since the Tm values of the PNA/PNA and PNA/DNA duplexes in these invasion mixtures were too high.
Figure 8.
Melting temperatures of PNA/PNA duplexes and PNA/DNA duplexes used in Figure 5. The chiral PNA units are shown in red (the plus signs refer to the positions of positive charges in the PNA strands).
The remarkable enantioselectivity in Figure 6 is also attributable to the differences in the stabilities of the corresponding PNA/DNA duplexes. The PNA11/DNA11c duplex (Tm = 76.0°C) is far more stable than the PNA13/DNA11c duplex (Tm = 65.3°C). As shown previously (40), the d-isomers can more easily adopt right-handed helical structures, required for the formation of PNA/DNA duplexes (42), due to more favorable position of the lysine side-chain. On the other hand, the l-isomers favor left-handed helical structures. When chiral PNAs containing l-monomers are forced to assume right-handed helix for PNA/DNA duplex formation, their side-chains are placed in ‘wrong’ positions, giving rise to strong intra-strand steric clash. The thermodynamic stability of the PNA/DNA duplex, which is one of the crucial driving forces for the double-duplex invasion, is too small. Accordingly, chiral PNAs from the d-monomers form more stable duplexes with the complementary DNA.
CONCLUSION
When chiral PNA monomer units, in which a nucleobase is bound to the α-nitrogen of N-(2-aminoethyl)-d-lysine, are introduced to PNA strands containing pseudo-complementary nucleobases, these strands show a very efficient double-duplex invasion to double-stranded DNA. Conventional (achiral) pseudo-complementary PNAs hardly invaded under the same conditions. This promotion of double-duplex invasion by the chiral monomers is notable irrespective of the target DNA sequence. Mismatch recognition of the chiral pcPNAs is so strict that their invasion never occurred even when only 1 bp at the target site in the DNA was changed to another. Both the electrostatic interactions by the positive charges of the PNA strands and the spatial arrangement of the lysine side-chains therein stabilize the PNA/DNA duplex and destabilize the PNA/PNA duplexes, and both factors are important for the promotion of invasion. Interestingly and importantly, only the chiral PNA monomers of d-forms effectively promote the strand invasion. This remarkable enantioselectivity is primarily ascribed to the fact that the DNA duplexes of the d-isomers are more stable than those of the corresponding l-isomers. The d-configurations of the chiral monomer units allow the lysine side-chains to fit in a right-handed helix, and thus stable PNA/DNA duplexes are formed. On the other hand, the lysine side-chains in the l-isomers are placed less suitably for a right-handed helix, leading to decreased stabilities of the PNA/DNA duplexes. Thus, the l-monomer units cannot promote the strand invasion. The achiral pcPNA strands bearing positively charged lysine residues at their termini are far less effective for the invasion than the chiral pcPNAs of d-forms.
By attaching pseudo-complementary nucleobases to N-(2-aminoethyl)-d-lysine units in PNA, double-duplex invasion is successfully achieved when target sequence (10 bp) involves two A–T pairs or more. According to statistic calculation, the probability that there exists no A–T pair in 10-bp sequence is (1/2)10, whereas the probability for the presence of one A–T pair in 10-bp sequence is 10 × (1/2)10. Thus, almost 99% of the 10-bp sequences in DNA should be in principle covered by the present strategy. Promotion of strand invasion by chiral PNA monomers should be applicable to various applications. These attempts, as well as further improvements for in vivo applications, are now under way in our laboratories.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was partially supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology, Japan and by the Global COE Program for Chemistry Innovation. This work was also supported by a grant from Italian MIUR (PRIN 2005). Funding to pay the Open Access publication charges for this article was provided by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Willis B, Arya DP. Major groove recognition of DNA by carbohydrates. Curr. Org. Chem. 2006;10:663–673. [Google Scholar]
- 2.Dhanasekaran M, Negi S, Sugiura Y. Designer zinc finger proteins: tools for creating artificial DNA-binding functional proteins. Acc. Chem. Res. 2006;39:45–52. doi: 10.1021/ar050158u. [DOI] [PubMed] [Google Scholar]
- 3.Pindur U, Jansen M, Lemster T. Advances in DNA-ligands with groove binding, intercalating and/or alkylating activity: chemistry, DNA-binding and biology. Curr. Med. Chem. 2005;12:2805–2847. doi: 10.2174/092986705774454698. [DOI] [PubMed] [Google Scholar]
- 4.Vazquez ME, Caamano AM, Mascarenas JL. From transcription factors to designed sequence-specific DNA-binding peptides. Chem. Soc. Rev. 2003;32:338–349. doi: 10.1039/b206274g. [DOI] [PubMed] [Google Scholar]
- 5.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 6.Wemmer DE. Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct. 2000;29:439–461. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- 7.Segal DJ, Barbas CF. Design of novel sequence-specific DNA-binding proteins. Curr. Opin. Chem. Biol. 2000;4:34–39. doi: 10.1016/s1367-5931(99)00048-4. [DOI] [PubMed] [Google Scholar]
- 8.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 10.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 11.Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchardt O, Sonnichsen SH, Nielsen PE. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 12.Kaihatsu K, Janowski BA, Corey DR. Stability of peptide nucleic acids in human serum and cellular extracts. Chem. Biol. 2004;11:749–758. [Google Scholar]
- 13.Nielsen PE. PNA technology. Mol. Biotechnol. 2004;26:233–248. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 14.Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Adv. Drug Deliv. Rev. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 15.Ray A, Nordén B. Peptide nucleic acid (PNA): its medical and biotechnical applications and promise for the future. FASEB J. 2000;14:1041–1060. doi: 10.1096/fasebj.14.9.1041. [DOI] [PubMed] [Google Scholar]
- 16.Janowski BA, Kaihatsu K, Huffman KE, Schwartz JC, Ram R, Hardy D, Mendelson CR, Corey DR. Inhibiting transcription of chromosomal DNA with antigene peptide nucleic acids. Nat. Chem. Biol. 2005;1:210–215. doi: 10.1038/nchembio724. [DOI] [PubMed] [Google Scholar]
- 17.Ren B, Zhou JM, Komiyama M. Straightforward detection of SNPs in double-stranded DNA by using exonuclease III/nuclease S1/PNA system. Nucleic Acids Res. 2004;32:e42. doi: 10.1093/nar/gnh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komiyama M, Ye S, Liang X, Yamamoto Y, Tomita T, Zhou JM, Aburatani H. PNA for one-base differentiating protection of DNA from nuclease and its use for SNPs detection. J. Am. Chem. Soc. 2003;125:3758–3762. doi: 10.1021/ja0295220. [DOI] [PubMed] [Google Scholar]
- 19.Doyle DF, Braasch DA, Simmons CG, Janowski BA, Corey DR. Inhibition of gene expression inside cells by peptide nucleic acids: effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry. 2001;40:53–64. doi: 10.1021/bi0020630. [DOI] [PubMed] [Google Scholar]
- 20.Lee R, Kaushik N, Modak MJ, Vinayak R, Pandey VN. Polyamide nucleic acid targeted to the primer binding site of the HIV-1 RNA genome blocks in vitro HIV-1 reverse transcription. Biochemistry. 1998;37:900–910. doi: 10.1021/bi972197m. [DOI] [PubMed] [Google Scholar]
- 21.Taylor RW, Chinnery PF, Turnbull DM, Lightowlers RN. Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat. Genet. 1997;15:212–215. doi: 10.1038/ng0297-212. [DOI] [PubMed] [Google Scholar]
- 22.Perry-O’Keefe H, Yao X-W, Coull JM, Fuchs M, Egholm M. Peptide nucleic acid pre-gel hybridization: an alternative to Southern hybridization. Proc. Natl Acad. Sci. USA. 1996;93:14670–14675. doi: 10.1073/pnas.93.25.14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norton JC, Piatyszek MA, Wright WE, Shay JW, Corey DR. Inhibition of human telomerase activity by peptide nucleic acid. Nat. Biotechnol. 1996;14:615–619. doi: 10.1038/nbt0596-615. [DOI] [PubMed] [Google Scholar]
- 24.Bonham MA, Brown S, Boyd AL, Brown PH, Bruckenstein DA, Hanvey JC, Thomson SA, Pipe A, Hassman F, Bisi JE, et al. An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res. 1995;23:1197–1203. doi: 10.1093/nar/23.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen PE, Egholm M, Buchardt O. Sequence-specific transcription arrest by peptide nucleic acid bound to the DNA template strand. Gene. 1994;149:139–145. doi: 10.1016/0378-1119(94)90422-7. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence specific inhibition of DNA restriction enzyme cleavage by PNA. Nucleic Acids Res. 1993;21:197–200. doi: 10.1093/nar/21.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen PE, Egholm M, Buchardt O. Evidence for (PNA)2/DNA triplex structure upon binding of PNA to dsDNA by strand displacement. J. Mol. Recogn. 1994;7:165–170. doi: 10.1002/jmr.300070303. [DOI] [PubMed] [Google Scholar]
- 28.Dragulescu-Andrasi A, Zhou P, He GF, Ly DH. Cell-permeable GPNA with appropriate backbone stereochemistry and spacing binds sequence-specifically to RNA. Chem. Commun. 2005:244–246. doi: 10.1039/b412522c. [DOI] [PubMed] [Google Scholar]
- 29.Kumar VA, Ganesh KN. Conformationally constrained PNA analogues: structural evolution toward DNA/RNA binding selectivity. Acc. Chem. Res. 2005;38:404–412. doi: 10.1021/ar030277e. [DOI] [PubMed] [Google Scholar]
- 30.Pokorski JK, Witschi MA, Purnell BL, Appella DH. (S,S)-trans-Cyclopentane-constrained peptide nucleic acids. a general backbone modification that improves binding affinity and sequence specificity. J. Am. Chem. Soc. 2004;126:15067–15073. doi: 10.1021/ja046280q. [DOI] [PubMed] [Google Scholar]
- 31.Rapireddy S, He G, Roy S, Armitage BA, Ly DH. Strand invasion of mixed-sequence B-DNA by acridine-linked, γ-peptide nucleic acid (γ-PNA) J. Am. Chem. Soc. 2007;129:15596–15600. doi: 10.1021/ja074886j. [DOI] [PubMed] [Google Scholar]
- 32.Lohse J, Dahl O, Nielsen PE. Double duplex invasion by peptide nucleic acid: a general principle for sequence-specific targeting of double-stranded DNA. Proc. Natl Acad. Sci. USA. 1999;96:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Protozanova E, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Pseudocomplementary PNAs as selective modifiers of protein activity on duplex DNA: the case of type IIs restriction enzymes. Nucleic Acids Res. 2003;31:3929–3935. doi: 10.1093/nar/gkg450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izvolsky KI, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. Sequence-specific protection of duplex DNA against restriction and methylation enzymes by pseudocomplementary PNAs. Biochemistry. 2000;39:10908–10913. doi: 10.1021/bi000675e. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Uehara A, Watanabe A, Aburatani H, Komiyama M. Chemical-reaction-based site-selective DNA cutter for PCR-free gene manipulation. Chem. Biochem. 2006;7:673–677. doi: 10.1002/cbic.200500402. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto Y, Uehara A, Tomita T, Komiyama M. Site-selective and hydrolytic two-strand scission of double-stranded DNA using Ce(IV)/EDTA and pseudo-complementary PNA. Nucleic Acids Res. 2004;32:e153. doi: 10.1093/nar/gnh151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto Y, Miura K, Komiyama M. Site-specific scission of lambda phage genomic DNA by Ce(IV)/EDTA-based artificial restriction DNA cutter. Chem. Lett. 2006;35:594–595. [Google Scholar]
- 38.Yamamoto Y, Yoshida J, Tedeschi T, Corradini R, Sforza S, Komiyama M. Highly efficient strand invasion by peptide nucleic acid bearing optically pure lysine residues in its backbone. Nucleic Acids Symp. Ser. 2006;50:109–110. doi: 10.1093/nass/nrl054. [DOI] [PubMed] [Google Scholar]
- 39.Sforza S, Haaima G, Marchelli R, Nielsen PE. Chiral peptide nucleic acids (PNAs): helix handedness and DNA recognition. Eur. J. Org. Chem. 1999:197–204. [Google Scholar]
- 40.Sforza S, Corradini R, Ghirardi S, Dossena A, Marchelli R. DNA binding of a D-lysine-based chiral PNA: direction control and mismatch recognition. Eur. J. Org. Chem. 2000:2905–2913. [Google Scholar]
- 41.Tedeschi T, Sforza S, Dossena A, Corradini R, Marchelli R. Lysine-based peptide nucleic acids (PNAs) with strong chiral constraint: control of helix handedness and DNA binding by chirality. Chirality. 2005;17:S196–S204. doi: 10.1002/chir.20128. [DOI] [PubMed] [Google Scholar]
- 42.Menchise V, De Simone G, Tedeschi T, Corradini R, Sforza S, Marchelli R, Capasso D, Saviano M, Pedone C. Insights into peptide nucleic acid (PNA) structural features: the crystal structure of a D-lysine-based chiral PNA-DNA duplex. Proc. Natl Acad. Sci. USA. 2003;100:12021–12026. doi: 10.1073/pnas.2034746100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sforza S, Tedeschi T, Corradini R, Ciavardelli D, Dossena A, Marchelli R. Fast, solid-phase synthesis of chiral peptide nucleic acids with a high optical purity by a submonomeric strategy. Eur. J. Org. Chem. 2003:1056–1063. [Google Scholar]
- 44.Haaima G, Hansen HF, Christensen L, Dahl O, Nielsen PE. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6-diaminopurine. Nucleic Acids Res. 1997;25:4639–4643. doi: 10.1093/nar/25.22.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.