Abstract
Expression of the gene cluster icaADBC is necessary for biofilm production in Staphylococcus epidermidis. The ica operon is negatively controlled by the repressor IcaR. Here, the crystal structure of IcaR was determined and the refined structure revealed a homodimer comprising entirely α-helices, typical of the tetracycline repressor protein family for gene regulations. The N-terminal domain contains a conserved helix-turn-helix DNA-binding motif with some conformational variations, indicating flexibility in this region. The C-terminal domain shows a complementary surface charge distribution about the dyad axis, ideal for efficient and specific dimer formation. The results of the electrophoretic mobility shift assay and isothermal titration calorimetry suggested that a 28 bp core segment of the ica operator is implicated in the cooperative binding of two IcaR dimers on opposite sides of the duplex DNA. Computer modeling based on the known DNA-complex structure of QacR and site-specific mutagenesis experiments showed that direct protein–DNA interactions are mostly conserved, but with slight variations for recognizing the different sequences. By interfering with the binding of IcaR to DNA, aminoglycoside gentamicin and other antibiotics may activate the icaADBC genes and elicit biofilm production in S. epidermidis, and likely S. aureus, as a defense mechanism.
INTRODUCTION
Staphylococcus epidermidis is an etiological microorganism that has emerged as one of the most important cause of nosocomial infections, especially for patients who have surgical grafts or implantations in the hospitals. Similar to other bacteria like the sister species S. aureus, it propagates in two alternating habits: plankton and biofilm. Whereas, the plankton form of the bacteria facilitates migration and spreading in the host organism, the biofilm form protects them from host immune system and enhances their ability to defend against antibiotics (1–4). The biofilm of S. epidermidis contains polysaccharide intercellular adhesin (PIA) as a major component (2). Expression of the icaADBC gene cluster (ica for intercellular adhesin) is necessary and sufficient to produce PIA in S. epidermidis (5–7). The genes of icaAD and icaB are involved in polymerization and deacylation reactions; icaC may be responsible for exportation process (8,9). The ica operon is negatively regulated by a repressor encoded by an upstream gene of icaR (10–12). Not much is known about the pathway to inactivate IcaR in stringent environment, which elicits PIA production and biofilm formation, although ethanol-induced and σB-dependent mechanisms have been proposed (13).
IcaR belongs to the tetracycline repressor (TetR) family of proteins, which are involved in a wide variety of gene regulations, either as transcription activators or repressors (14). The crystal structures of three repressors of the TetR family, namely TetR, QacR and EthR, have been reported (15–19). All of the proteins are homodimers. Each monomer contains an N-terminal domain that binds to DNA and a C-terminal domain that is involved in dimerization. Both domains consist entirely of α-helices. Although the overall structures of TetR, QacR and EthR are similar, the amino acid sequences have little homology except in the N-terminal domain. The two N-terminal helices α2 and α3 constitute a helix-turn-helix (HTH) motif. In a homodimer, the two HTH motifs are disposed about the molecular dyad with a separation that matches the distance between the two adjacent major grooves of DNA. Crystal structures of TetR and QacR in complex with DNA revealed the detailed interactions for specific binding to the operators. TetR binds to DNA with a stoichiometry of 2:1 for the protein monomer and double stranded DNA (dsDNA), whereas cooperativity has been reported for QacR and EthR, with ratios to dsDNA of 4:1 and 8:1, respectively (15,17,20). Several drugs have been shown effective in preventing binding of the TetR family proteins to DNA, presumably by modulating the relative disposition of the HTH motifs in the homodimer (16,19,21,22).
The entire DNA sequences of the ica operon of S. epidermidis and S. aureus have been reported (NCBI accession number: NC_002976 and NC_007796). DNaseI foot printing analysis showed that recombinant IcaR binds to a 42 bp region immediately upstream of the icaA gene (11). However, the location and the nature of cooperative binding remain uncertain. Here, in order to study how the ica operon is controlled, IcaR from S. epidermidis was over-expressed in Escherichia coli with a His-tag to facilitate purification. The protein was crystallized and its structure determined by the multi-wavelength anomalous diffraction (MAD) method using a seleno-methionine (SeMet) derivative. The native structure was subsequently determined in a different unit cell by molecular replacement. Furthermore, DNA-binding assays and computer modeling allowed us to elucidate the regulation mechanism of the ica operon. Finally, we observed the inhibitory effect of two aminoglycoside antibiotics, namely, gentamicin and streptomycin, on the IcaR–DNA interactions.
MATERIALS AND METHODS
Cloning, expression and purification
The biofilm-forming strain S. epidermidis ATCC 35984 (RP62A) was obtained from the Food Industry Research and Development Institute in Taiwan. The icaR gene fragment was amplified directly from the S. epidermidis RP62A genome by polymerase chain reaction (PCR) with forward 5% GGAATTCCATATGCACCACCACCACCACCACATGAAAGATAAGATTATTGATAACGC 3′ and reverse 5′ CCCAAGCTTTTATTTTTTTAAAAATACATTTAACAGTG 3′ primers. The PCR product encoding IcaR with an amino-terminal His6 tag was digested with NdeI and HindIII and subsequently cloned into expression vector pET-21a (Novagen). For phasing purposes, a four-residue mutant (L63M, L64M, L143M and L146M) of IcaR was engineered using the QuikChange site-directed mutagenesis kit (Stratagene, CA). Furthermore, by employing the same mutagenesis kit, several site-specific mutants in the DNA binding region of IcaR (L23, K33, A35) were also produced for DNA-binding assay. All DNA constructs were verified by nucleotide sequencing. These constructs were transformed into a non-auxotrophic E. coli strain BL21 (DE3) competent cell for protein expression. The His6-tagged wild-type and mutant IcaR proteins were overexpressed in LB medium containing 100 μg/l ampicillin at 30°C under basal level expression for 2 days. The cells were then harvested by centrifugation at 4000 g for 15 min. The cell pellet was resuspended immediately in the lysis buffer containing 20 mM Tris–HCl, 400 mM NaCl, 30 mM imidazole, pH 7.5. The cell suspension was disrupted by Constant Cell Disruption System (CONSTANT SYSTEM Ltd, UK) and centrifuged at 17 000 g to remove cell debris. The cell-free extract was loaded onto a Ni2+-NTA column, which had been previously equilibrated with lysis buffer. The column was washed with lysis buffer, and the His6-tagged IcaR was subsequently eluted by a linear gradient of imidazole from 30 mM to 500 mM. The purified His6-tagged IcaR was dialyzed three times against 5 l of buffer (20 mM Tris–HCl, pH 8.0), and finally concentrated by 10 kDa cut-off size membrane of Centricon plus-20 (Millipore, MA, USA) for storage at –80°C.
SeMet-labeled IcaR mutant was over-expressed in slightly modified SeMet minimal medium containing 100 μg/l ampicillin at 30°C for 2 days with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) as an inducer (23). The detailed protocol as follows: 200 ml overnight culture of M9 medium (Na2HPO4 6 g/l, KH2PO4 3 g/l, NaCl 0.5 g/l, NH4Cl 1 g/l, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% glucose) of a single transformant was used to inoculate 6 l of fresh M9 medium containing 100 μg/l ampicillin at 37°C until OD 0.6, and then cooled to 30°C. A 120 ml filter-sterilized solution containing 60 mg Fe2(SO4)3 and 60 mg thiamine and 600 mg DL-SeMet was divided equally among the 6 l medium. One hour later IPTG was added to a final concentration of 0.5 mM for 2 days induction.
Purification of the SeMet-IcaR was performed using the same protocol established for native IcaR. About 95 mg/l of native IcaR and 60 mg/l of SeMet-IcaR were obtained, with a purity of greater than 98% as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Full incorporation of SeMet into SeMet-IcaR was determined by mass spectrometry.
Crystallization and data collection
For crystallization, IcaR and SeMet-IcaR solutions were adjusted to 18 mg/ml in 20 mM Tris–HCl pH 8.0 containing 5 mM dithiothreitol (DTT). The sitting-drop vapor diffusion method was employed for crystallization by mixing 3 μl protein solution with 3 μl precipitant solution. The crystals of IcaR were obtained with ∼4–6% (w/v) PEG1500 and ∼1.4–1.8 M NaCl precipitant solution, whereas the crystals of SeMet-IcaR were obtained with ∼12–15% (w/v) PEG1500 and ∼2.0–2.6 M NaCl precipitant solution. High-quality crystals were grown to full size within 1 day at room temperature. The crystals were soaked in a cryoprotectant solution containing 2.5 M NaCl, 150 mM sodium formate, 15% (w/v) PEG1500 and 20% (v/v) glycerol, and then mounted straight from the drop and flash-cooled to 100 K in a stream of cold nitrogen. X-ray diffraction data were collected at SPXF beamline BL13B1 at the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan. All diffraction images were recorded using ADSD Q315 CCD detector and the data were processed and scaled by using the program package of HKL2000 (24). X-ray diffraction data from the native IcaR crystal were collected to 1.9 Å resolution at wavelength λ = 1.0000 Å. MAD data from the SeMet-IcaR crystal were collected to 1.33 Å and 1.45 Å resolution at wavelengths λ = 0.9796 Å (edge) and λ = 0.9641 Å (high remote), respectively. Both native IcaR and SeMet-IcaR crystals belong to the monoclinic space group P21, but with different unit cell dimensions. The data collection statistics are summarized in Table 1. There are four and two IcaR monomers per asymmetric unit in the crystals of native IcaR and SeMet-IcaR, respectively.
Table 1.
Data collection and refinement statistics for the S. epidermidis IcaR crystals
| Crystals | Native-IcaR | SeMet-IcaR | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | |
| Unit cell parameter | |||
| a (Å) | 39.11 | 61.78 | |
| b (Å) | 86.14 | 50.70 | |
| c (Å) | 113.85 | 63.34 | |
| β (°) | 99.15 | 97.09 | |
| Wavelength (Å) | 1.0000 | 0.9796 (edge) | 0.9641 (high remote) |
| Resolution (Å) | 30–1.9 (1.97–1.9)a | 30–1.33 (1.38–1.33) | 30–1.45 (1.5–1.45) |
| Number of reflections | |||
| Observed | 360 641 | 319 708 | 253 970 |
| Unique | 57 448 (5552) | 86 594 (7841) | 67 079 (5977) |
| Completeness (%) | 97.7 (94.4) | 98.0 (89.5) | 98.1 (87.7) |
| Rmerge (%) | 7.5 (35.9) | 5.4 (28.9) | 6.0 (28.5) |
| I/σ (I) | 17.56 (3.5) | 26 (2.9) | 21.7 (2.9) |
| Refinementb | |||
| Number of reflections | 56 361 (5094) | 73 703 (4537) | |
| Rwork (%) | 17.2 (25.0) | 19.0 (29.3) | |
| Rfree (%) | 20.8 (31.5) | 21.8 (29.5) | |
| Geometry deviations | |||
| Bond length (Å) | 0.015 | 0.015 | |
| Bond angles (°) | 1.5 | 1.6 | |
| No. of all protein atoms | 6174 | 3031 | |
| Mean B-values (Å) | 33.6 | 18.4 | |
| No. of water molecules | 740 | 857 | |
| Mean B-values (Å) | 52.7 | 41.0 | |
| Ramachandran plot (%) | |||
| Most favored | 96.7 | 97.3 | |
| Additionally allowed | 3.3 | 2.7 |
aValues in the parentheses are for the highest resolution shells.
bAll positive reflections are used in the refinements.
Structure determination, model building and refinement
Initial phase angles were calculated by employing the program SOLVE (25). Using the MAD data of SeMet-IcaR in the resolution range of 30–1.45 Å. Ten of the 12 Se sites were located in the asymmetric unit. Subsequently, the electron density map was improved using the program RESOLVE (26), and this was followed by automatic tracing of up to 76% of the protein model. Manual building of the remaining model and further refinement were carried out with the program Xtalview (27) and CNS (28) against the 1.33 Å resolution data set of the SeMet-IcaR crystal. The structure of native IcaR was determined by molecular replacement using CNS, with the refined structure of SeMet-IcaR as a search model. For each structure, iterative cycles of model building with Xtalview and computational refinement with CNS were performed, in which 5% reflections were set aside for Rfree calculation (29). The stereochemical quality was assessed with the program PROCHECK (30). The refined structures of IcaR contain a small disordered region of residues 64–70 except the molecule of chain D in the native crystal. Many well-ordered water molecules were also located and included in the models. The refinement statistics are listed in Table 1. The molecular figures were produced by using PyMOL (DeLano Scientific, http://www.pymol.org).
Electrophoretic mobility shift assays (EMSA)
Various single-stranded DNA oligonucleotides, denoted by their lengths (16–28) and a letter for the relative locations (A–G), were purchased from MDBio Inc. (Taiwan). Double-stranded DNA were prepared by annealing complementary oligonucleotides (100 μM each) in 10 mM Tris–HCl, pH 8.0, 20 mM NaCl, heating the reaction to 95°C for 5 min and allowing it to cool to 25°C. A 20 μl binding reaction containing ∼600–2400 pmole of purified recombinant IcaR and 300 pmole of various dsDNA substrates in binding buffer (20 mM Tris–HCl, pH 8.0, 5 mM KCl, 0.1 mM MgCl2, 0.05 mM EDTA, 1 mM DTT and 2 mg/ml BSA) was incubated at room temperature with gentle vortex for 30 min. After incubation, a 10 μl of the reaction was mixed with 2 μl of the sample loading dye and subsequently loaded onto a 1% agarose gel and electrophoresed in 1 × Tris/acetate/EDTA (TAE) at 100 V for 30 min and visualized using ethidium bromide. In the assay for effects of antibiotics on the interaction of IcaR and DNA, 0.5 mg/ml of several antibiotics were pre-incubated with IcaR at room temperature for 30 min and then mixed with the 28E DNA probe, followed by the same procedure as in the other assays. The corresponding molar ratios of antibiotics to IcaR monomer ranged from 3 to 20.
Isothermal Titration Calorimetry (ITC)
For ITC experiments, all samples were prepared with the buffer containing 20 mM Tris, 20 mM NaCl, pH 8.0. Samples were degassed using ThermoVac (MicroCal, MA, USA) for 5 min prior to loading, and all titrations were performed at 25°C. Calorimetric assays were performed using the VP-ITC (MicroCal, MA, USA). The reaction cell (about 1.4 ml) was filled with the degassed solutions. The stirring speed was 300 rpm and the thermal power was recorded every 2 s. ITC titration curves were collected and analyzed using the software Origin 7 package supplied with the instrument. All data obtained were corrected for the heat of dilution of the titrant by subtracting the average of 5–7 data points from the saturated tail. Except the streptomycin–DNA titration, all corrected data were fitted to the one binding site model. In the IcaR–DNA experiments, the dsDNA solution was contained in the cell at a concentration of 2 μM and the titrated IcaR dimer concentration was 36 μM. In the antibiotics–DNA experiments, the dsDNA solution was contained in the cell at a concentration of 7 μM and the titrated antibiotic concentration was 700 μM. In the antibiotics-IcaR experiments, the IcaR solution was contained in the cell at a dimer concentration of 100 μM and the titrated antibiotic concentration was 1 mM. Except the T2U_DNA, all the dsDNA used was the 28E DNA probe as in the EMSA experiments. In the T2U_DNA the key thymidines of 28E DNA were changed to deoxyuridines, with sequences of 5′ ACAACCUAACUAACGAAAGGTAGGTGAA 3′ and 5′ TTCACCUACCUTTCGTTAGTTAGGTTGT 3′.
Computer modeling
Because the IcaR structure is similar to QacR, and so is the DNA binding stoichiometry (1:4, see below), the structure of the QacR–DNA complex from PDB 1JT0 was used as a template to construct a model of IcaR–DNA complex. The 28-bp palindromic DNA sequence of 1JT0 was mutated to the sequence of 28E that was used in the EMSA experiment (see below). Using the program O (31), the new base pairs were positioned as close as possible to match those in the template, whereas the sugar phosphate backbone remained unscathed. The first 50 amino acid residues in the four N-terminal domains of QacR were replaced with those of IcaR (two were omitted), followed by minor adjustments to avoid severe clashes between non-bonded atoms. In some cases the structure of TetR–DNA complex (PDB 1QPI) was also used as a reference. The two dimers of the refined native IcaR structure were then superimposed separately on the two modified QacR dimers, and the C-terminal domains of the former were connected to the corresponding N-terminal domains of the latter, constituting two IcaR dimers bound to the opposite sides of the DNA. Molecular dynamics and energy minimization were subsequently carried out using CNS, with atomic positions of both the protein and the DNA tethered to the originals by applying a moderate harmonic restraint.
Protein Data Bank accession codes
The atomic coordinates and structure factors for the native IcaR crystal and its SeMet derivative have been deposited in the wwPDB with accession numbers of 2ZCM and 2ZCN, respectively.
RESULTS AND DISCUSSION
Features of the IcaR structure
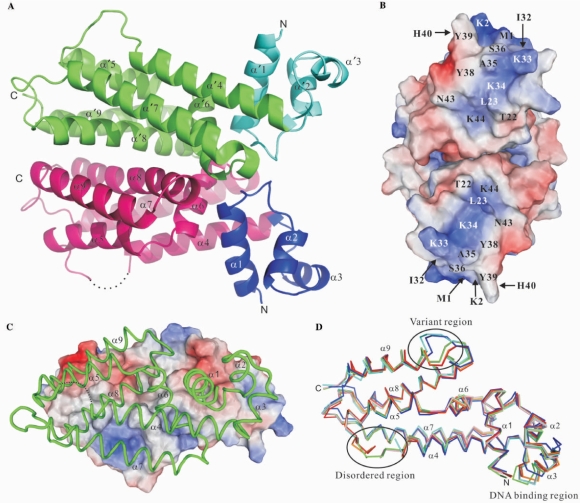
The crystal structures of the native IcaR were refined to 1.9 Å resolution and its SeMet derivative to 1.33 Å, both yielding low R and Rfree values and stereochemical deviations (Table 1). There are four IcaR monomers (A–D) in an asymmetric unit of the former crystal and two (A, B) in the latter, all forming dimers (AB and CD). The N-terminal His-tags were not observed in any of the six monomers. Neither were some loops in the surface regions visible probably due to flexibility. A representative IcaR dimer is shown in Figure 1A. Helices α1–α3 form the N-terminal domain that are supposed to interact with DNA via a HTH motif (α2/α3). This region, expected to be facing the DNA, is densely positively charged in the central patch of surface (Figure 1B). Most are contributed by the helix α3, particularly Lys33 and Lys34. The other helices α4–α9 form the C-terminal domain, which in principle is a large helix bundle that associates with its counterpart to constitute a dimer. Each monomer has a total surface area of about 10 400 Å2, in which 1800 Å2 is buried upon dimer formation. Two-thirds of the interface are contributed by nonpolar residues; the remaining one-third is polar. The charge distribution at the dimer interface is complementary. As shown in Figure 1C, the molecular dyad axis would bring the positively charged surface patch of the front monomer (shown as a worm tracing model) onto the negatively charged surface patch of the back monomer (as a surface). The complementary surface charge distribution about the dyad axis is ideal for efficient and specific dimer formation.
Figure 1.
Structure of IcaR. (A) Overall structure of the IcaR homodimer. The protein structure is shown as a ribbon diagram with the N-terminal domains in blue and cyan, the C-terminal domains in magenta and green, and the disordered region as dotted line. (B) The DNA-binding domains. The electrostatic surface of the dimer is viewed along the dyad axis, after a rotation of ∼90° from (A). The electrostatic surfaces are drawn either blue for positive or red for negative. Possible residues involved in binding DNA are labeled. (C) The dimer interface. One subunit is shown as a surface representation, colored according to the electrostatic potentials as in (B), and another subunit is shown as a worm tracing in green. (D) Superposition of the IcaR models. The polypeptide backbones of six IcaR subunits are shown as stick models. Four subunits from the native crystal are colored in red, pink, orange and green, and two subunits from the SeMet derivative crystal are in cyan and blue.
When the six independent IcaR monomers are superimposed using the program O, the root-mean-squares deviations (RMSD) are 0.42–0.57 Å between 89–149 pairs of Cα atoms with a matching distance criterion of 1.0 Å, except that the monomers A/C and B/D of the native crystal have low RMSD of 0.30 Å and 0.22 Å for 179 and 175 Cα pairs. The two dimers AB and CD are superimposed with an RMSD of 0.33 Å for 362 pairs of Cα. Their positions differ by about half unit cell pseudo-lattice translation along the c-axis, and have almost identical crystal packing environment. With a less stringent criteria of 2.0 Å, the cross-RMSD between the native IcaR dimers (i.e. AB/BA, AB/DC) are comparable to those with the SeMet derivative, ranging from 0.68 Å to 0.96 Å for 300–350 Cα pairs. In general, the C-terminal domain superimposes better than the N-terminal domain, except the loop between helices α8 and α9. Specifically, the first three residues of helix α9 in the SeMet derivative do not have the helical conformation as in the native structure, but form a loop instead (Figure 1D). Large structural alteration is also observed for the DNA binding region, particularly in helix α3 (see Figure S1 in the Supplementary Materials). A maximal displacement of 17 Å is seen for the side chain of Tyr39. Other residues including Tyr38, His40 and Tyr41 also show significant displacements. Although the conformational variation could be a result of crystal packing interactions, it may also represent the intrinsic flexibility of the N-terminal domain.
Despite the difference in the flexible regions, the overall structures of native IcaR and its SeMet derivative are virtually identical. Mutations of the four selected Leu residues to Met did not alter the protein conformation too much (Figure 1D). Actually, SeMet-IcaR also crystallized in the larger unit cell under similar conditions for the native protein.
Cooperative binding to the operator DNA
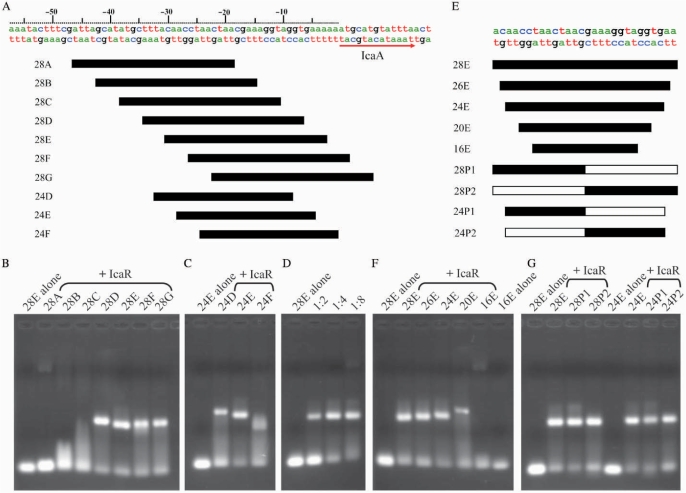
Initial inspection of immediate upstream sequence of the icaADBC genes in the ica operon did not detect obvious pattern of palindrome (Figure 2A). In order to track down the precise location of the IcaR-binding site, and to find out the minimal length of DNA for effective binding (in the hope of growing well-diffracting IcaR–DNA complex crystals), a series of dsDNA segments were designed and tested for IcaR binding by electrophoretic motility shift assay. The experiments began with the 28 bp segments 28A–28G, which ‘walk’ through or span the 52 bp upstream region from –47 to +5 in 4 bp steps. As shown in Figure 2B, the far-upstream segments 28A, 28B and 28C did not bind, whereas the other segments 28D, 28E, 28F and 28G did bind to the IcaR protein, indicating that the operator is located immediately adjacent to the start codon of the icaA gene. Among the latter segments, 28E showed the most significant effect by IcaR on its mobility, suggesting the strongest interaction between them. With the same center as 28E, the shorter 24 bp segment of 24E also showed strong binding, whereas the neighboring 24D and especially 24F appeared to have weaker interactions with IcaR (Figure 2C). Consequently, the core segment of the ica operator that binds to the repressor is centered at –17/–18 from the start codon of icaA gene.
Figure 2.
Electrophoretic mobility shift assay (EMSA) of IcaR binding to the ica operator. (A) Part of the DNA sequence of ica operon of S. epidermidis and diagrams of the dsDNA probes. (B) The EMSA of IcaR binding to different 28-mer dsDNA fragments of the ica operator. (C) The EMSA of IcaR binding to different 24-mer dsDNA fragments of the ica operator. (D) The EMSA of IcaR with different DNA ratio of the 28E probe. The 28E probe was mixed with 1:2, 1:4 and 1:8 molar ratio of IcaR (monomer), respectively. (E) Sequences of the core region of the ica operator and DNA probes. 28P1, 28P2, 24P1 and 24P2 are artificial panlindromic DNA probes of the ica operator. The white bars of the palindromic DNA probes indicate the palindromic sequences to the black bars. (F) The EMSA of IcaR binding to different size probes of the ica operator. (G) The EMSA of IcaR binding to different palindromic probes of the ica operator. A 1:4 molar ratio of DNA and IcaR (monomer) was used in the EMSA experiments except in (D). A 2 mg/mL BSA was included in all EMSA experiments to avoid nonspecific interaction between IcaR and the DNA probes.
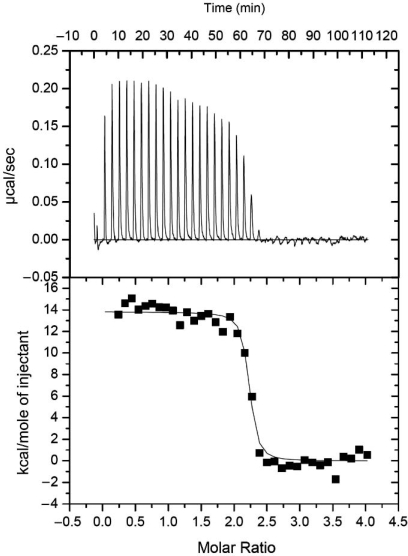
In Figure 2D, the EMSA experiments demonstrated that by incubating the double-stranded 28E DNA with the IcaR monomers at twice the molar equivalent, i.e. with a ratio of 1:2, the mobility of some fraction of the DNA segments was reduced while a significant fraction of the DNA remained unaffected. When the molar ratio was 1:4, all but a small fraction of the DNA was reduced in the gel-shift mobility. These results suggest that IcaR binds to the ica operator by a ratio of 1:4 in a highly cooperative manner. In other words, each operator DNA segment either binds simultaneously to two IcaR homodimers, or it does not bind. Cooperative binding was also observed in the ITC experiments. Here, the stoichiometry was unambiguously determined as two IcaR dimers to one DNA operator (see Figure 3 for details). Similar observations of cooperativity have been reported for QacR, which also binds to DNA with a 1:4 molar ratio, and EthR, which has a ratio of 1:8 (17,20). However, further increase of the molar ratio of dsDNA IcaR to 1:8 did not change the DNA mobility any further (Figure 2D). The length of the 28 bp ica operator DNA core segment also excludes the possibility of binding more than a pair of IcaR dimers. As revealed in the crystal structure of QacR–DNA complex, the two dimeric repressors bind to the DNA on opposite sides, and with overlapping regions of the encompassed base pairs. The cooperativity of the binding is mediated by the protein–DNA–protein interactions, rather than direct interactions between the repressor molecules. We believe that the same DNA-binding mechanism of QacR applies to IcaR as well.
Figure 3.
IcaR–DNA binding determined by ITC. Representative plots from an ITC experiment are shown with raw data in the upper panel and curve fit in the lower panel. The IcaR protein was titrated into the reaction cell containing the 28E dsDNA. Note that the first addition was 3/8 the volume of the other additions. The first two data points were not used in the fitting. Thermodynamic values obtained from the curve fit are: ΔS = 84.3 cal/mol K, ΔH = 13.8 ± 0.2 kcal/mol, KB = 2.0 ± 0.8 × 108 M–1, N = 2.18 ± 0.01. N is the stoichiometry of bound IcaR dimer per dsDNA.
Experiments with different lengths of the DNA segments further confirmed the center position that relates the two IcaR dimers cooperatively bound to the operator. Figure 2E depicts the DNA fragments varying in lengths from 16 to 28 but having the same center. The results showed that the binding strength was weakened significantly when the length was reduced to 20 (Figure 2F). With a length of 16, the DNA virtually did not bind. Thus the minimal length may be less than 24 but more than 20. As shown below, from the computer modeling results, it is most likely 22 bp. In order to increase the homogeneity of the IcaR–DNA complex for crystallization, we tested the binding effectiveness of the palindromes. As seen in Figure 2G, the palindromes based on either upstream or downstream half-site of the operator with length of either 28 or 24 had virtually the same binding strengths as 28E or 24E to the IcaR repressor. These results also supported the correct location of the core operator segment.
Structure comparison and DNA-complex modeling
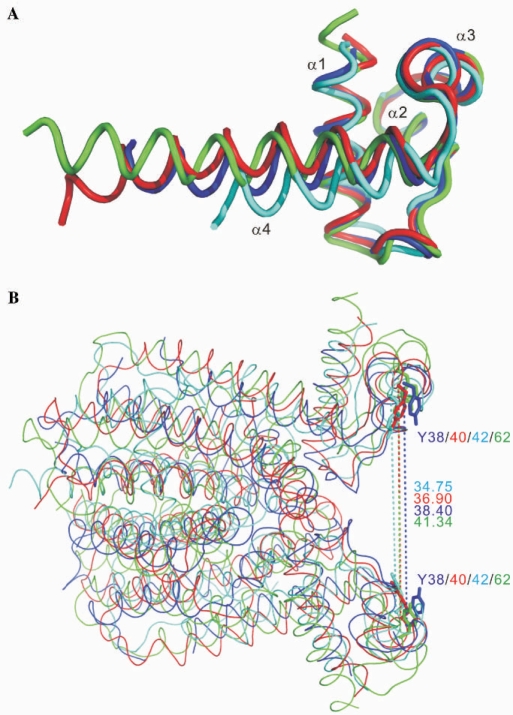
Only the N-terminal DNA-binding domains of the TetR family repressors show significant sequence homology. Excluding monomer B of the SeMet derivative crystal of IcaR, which had a dissimilar conformation of the N-terminal domain due to flexibility (and possibly crystal packing), the RMSD between the monomers of IcaR and three other known repressors TetR, QacR and EthR in the first 45 Cα positions (IcaR) are very small, ranging 0.76–1.35 Å. As shown in Figure 4A, while the first three helices α1–α3 match almost perfectly, the succeeding helix α4 does not (15,17,18). Nevertheless, superposition of the dimeric structures of IcaR, TetR, QacR and EthR, using the program O and a loose matching criterion of 3.0 Å, showed RMSD of 1.9–2.5 Å between 114–160 pairs of Cα atoms, reflecting their similarity in the overall topology (Figure 4B), among which IcaR and QacR showed the closest kinship. The relative dispositions of the HTH motifs are also conserved: the distances between the two Tyr38 Cα atoms in the IcaR dimer and the equivalents in TetR, QacR and EthR, which are supposed to interact with the DNA bases in two adjacent major grooves, have a narrow range of 35–41 Å. Flexibility in the hinge region, probably contributed by the connection to helix α4, should allow each pair of HTH motifs to make specific interactions with its cognate DNA sequence.
Figure 4.
Structural homologues with distinct DNA recognition modes. (A) Superimposition of the three-helix bundle DNA-binding domains. The similarity between IcaR (blue), TetR (cyan), QacR (red) and EthR (green) shows a high degree of structural homology within the DNA-binding domains of these proteins. (B) Comparison of the dimer structures. The dimers of IcaR, TetR, QacR and EthR are superimposed and colored as in (A). The distances between the two Cα atoms of Tyr38 in the IcaR dimer and the equivalents in TetR, QacR and EthR, which are supposed to interact with the DNA bases in two adjacent major grooves, have a narrow range of 35–41 Å.
Attempts to crystallize the IcaR–DNA complex were not successful. Therefore, we resorted to homology modeling to study possible interactions between the IcaR repressor and the ica operator DNA. Because the gel-shift assay showed that cooperative binding to the ica operator involves two IcaR dimers, and because the structure of IcaR is most similar to QacR, we used the crystal structure of QacR–DNA complex (PDB 1JT0) as a template. The 28 bp core region of ica operator as derived from the EMSA results above is shown in Figure 5A, and an overview of the modeled IcaR–DNA complex in Figure 5B. The DNA bases are numbered here according to their relative positions to the center of operator sequence, starting at +1 and –1 in the opposite directions. By analogy with QacR, the binding of IcaR to DNA may principally involve 14 amino acid residues, namely Met1, Lys2, Thr22, Leu23, Ile32, Lys33, Lys34, Ala35, Ser36, Tyr38, Tyr39, His40, Asn43 and Lys44. Most interactions are conserved, including several hydrogen bonds of the polypeptide backbone atoms and those involving the side chains of Thr22, Lys34, Tyr38, Tyr39 and His40 that are identical to those in QacR. In addition to the positive dipole of helix α1, the shorter N-terminus of IcaR may also form an ionic interaction with the phosphate of DNA.
Figure 5.
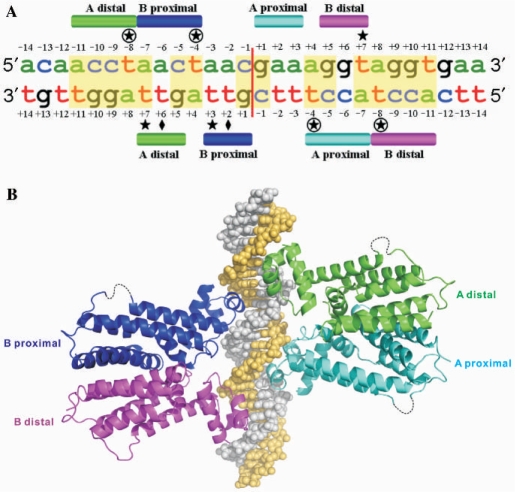
Binding of IcaR to its operator site. (A) Interaction of IcaR with the ica operator. The 28E DNA sequence of ica operator as derived from the EMSA experiment is shown with the bases numbered form –14 to +14. The partial palindromic sequences are shown in yellow background. Approximate locations of the interface with different IcaR monomers are shown as colored bars. Diamonds, circles and stars indicate the locations of the key residues Leu23, Lys33 and Ala35, respectively. (B) The model of IcaR–DNA complex. The model was constructed based on the crystal structure of QacR–DNA complex (PDB 1JT0) as a template. Two IcaR dimers are supposed to bind cooperatively to the operator DNA on opposite sides, in which the binding of dimers involves overlapping DNA sequences. The DNA strands are colored gray and yellow, while the IcaR monomers are colored as in (A).
It is notable that there are three overlapping palindromes in the DNA, as predicted by the dimer of dimers model. One palindrome (central; AGNTAGNT) relates one IcaR dimer to the other, and two (flanking; AGNT) relate the monomers within the two dimers. The central palindrome corresponds to the bases +4 to +11 of both strands in Figure 5A, and each of the two flanking palindromes corresponds to the bases +4 to +7 of one strand and +8 to +11 of another. One of the two Leu23 residues in each IcaR dimer may interact with the methyl group of a thymine base (T+6/T+2 of the complementary strand; Figure 5A). The aliphatic moiety of the side chain of each Lys33 may contact the methyl group of another thymine base (T–8/T–4 of both strands), while the positively charged amino group may interact with a phosphate group. Each side chain of Ala35 may also interact with the same methyl group, as well as one of three thymine bases: T+7 of the coding strand and T+3/T+7 of the complementary strand. The equivalents in QacR are the residues Ser35 and Gly37, respectively. In the QacR–DNA complex structure (PDB 1JT0), Ser35 is hydrogen bonded to the phosphate and Gly37 is in contact with the bases. Pro39 in the TetR–DNA complex (PDB 1QPI), equivalent to Ala35 in IcaR, also interacts with the thymine methyl groups of T–5 in one strand and the opposing T+4 in another (see Figures S2 and S3 in the Supplementary Material). Taken together, the complex model suggests that IcaR may recognize its operator DNA by using the same repertoire of interactions of QacR, with slight variations contributed by the three key residues of Leu23, Lys33 and Ala35. The cooperative mechanism proposed for QacR, in which binding of the first dimer induces conformational change of the DNA that favors the binding of the second dimer (17), may apply to IcaR as well.
The effects of mutations
To investigate the possible roles of residues Leu23, Lys33 and Ala35 in IcaR–DNA recognition and binding, they are substituted with those in QacR, TetR and EthR (Figure S2), and the mutant IcaR proteins were measured for DNA-binding affinity, using ITC. For comparison, the binding constant of wild-type IcaR was also measured (Table 2). In QacR and TetR, the equivalent residue to Leu23 in IcaR is a threonine, which faces an adenine base and forms hydrogen bonds with a neighboring phosphate group. As shown in Table 2, the mutant L23T showed about one-third decrease in affinity, probably because of weaker hydrophobic interactions (see Figure S3 in the Supplementary Material). Surprisingly, the mutant L23V bound strongly to the DNA, with more than 6-fold increase in affinity. Although the binding mode of EthR to its cognate DNA has not been elucidated, in the case of IcaR studied here, the side chain of valine, smaller but also branched, would make better interactions with the methyl group of the thymine base (Figure S3).
Table 2.
Thermodynamic parameters for IcaR–dsDNA, antibiotics–dsDNA and antibiotics–IcaR interactions
| Samples | Binding constants KB (M−1) | ΔH (kcal/mol) | ΔS (cal/mol K) | Relative binding affinity (%) |
|---|---|---|---|---|
| Wild-type and mutated IcaR with 28E dsDNA | ||||
| Wild type | 2.0 ± 0.8 × 108 | 13.8 ± 0.2 | 84.3 | 100 |
| L23T | 1.3 ± 0.4 × 108 | 10.7 ± 0.2 | 72.9 | 65 |
| L23V | 1.5 ± 1.3 × 109 | 14.7 ± 0.2 | 91.2 | 750 |
| K33S | 4.1 ± 0.7 × 107 | 23.5 ± 0.3 | 114.0 | 21 |
| K33E | No binding | |||
| A35G | 3.9 ± 1.2 × 108 | 11.2 ± 0.1 | 76.8 | 195 |
| A35P | 1.0 ± 0.7 × 109 | 15.6 ± 0.2 | 93.5 | 500 |
| Wild-type IcaR with T2U 28E dsDNA | ||||
| T2U_DNA | 1.4 ± 0.3 × 107 | 15.1 ± 0.4 | 83.4 | 7 |
| Antibiotics with wild-type IcaR | Stoichiometry | |||
| Gentamicin | 2.6 ± 1.5 × 104 | −1.3 ± 0.5 | 15.7 | 0.59 ± 0.18 |
| Streptomycin | 8.0 ± 4.1 × 104 | −1.1 ± 0.1 | 18.8 | 0.88 ± 0.08 |
| Kanamycin | Not detectable | |||
| Antibiotics with 28E dsDNA | ||||
| Gentamicin | 8.6 ± 0.9 × 104 | 2.3 ± 0.1 | 30.2 | |
| Streptomycin | Difficult to calculate | |||
| Kanamycin | 1.1 ± 0.2 × 105 | 1.3 ± 0.1 | 27.5 | |
Mutation of Lys33 to serine (K33S) caused an 80% loss of binding affinity, whereas mutation to glutamate (K33E) resulted in complete inactivation of the IcaR repressor (Table 2). The equivalent Ser35 in QacR interacts with a cytidine nucleotide primarily through hydrogen bonding to the phosphate group (Figure S3). In K33S, the short polar side chain may cause unfavorable interaction with the thymine base. In K33E, substituting the positively charged amino group with a negatively charged carboxyl group may disrupt the important salt bridge to the phosphate group. Interestingly, both mutants A35G and A35P showed stronger binding with 2- and 5-fold affinity (Table 2). The glycine residue is more flexible and may provide better accommodation for the DNA, and, as mentioned above for TetR, the proline may also make good interactions with the AT base pairs.
Moreover, because the key two thymine bases (T–8/T–4 of both strands) of the ica operator DNA were implicated in the specificity for IcaR binding, these four thymidines were changed to deoxyuridines and the DNA analyzed by ITC. As shown in Table 2, the binding affinity of IcaR to its cognate DNA was reduced to 7% with the ‘mutant’ T2U_DNA. The T2U_DNA differed from the ‘wild type’ only in the two omitted methyl groups. The structures, including the hydrogen bonds of the AT base pairs, were otherwise identical. Thus, the important roles played by the methyl groups of the key thymines are substantiated.
The effects of antibiotics
As mentioned earlier, IcaR works as a repressor in the negative regulation of the icaADBC genes, whose expression is required for PIA synthesis and biofilm formation (5–7,10–12). However, the mechanism of inactivating IcaR is not clear. In previous studies of TetR and QacR, several drug molecules of diverse chemical nature were shown to bind to the repressor and altered its conformation (16,21,22). The consequence of the binding of drug molecules to DNA was a drastically increased distance between two HTH motifs, which prevented the TetR or QacR dimer from binding to the operator DNA. Recently, it was reported that biofilm formation in bacteria can be induced by aminoglycoside antibiotics (32). Here, to investigate possible effect of drugs on IcaR, six different antibiotics were tested for their potential inhibition of IcaR–DNA binding. These include three aminoglycosides streptomycin, gentamicin and kanamycin; tetracycline, after which the repressor family is named; and two antibiotics often used in laboratory work, ampicillin and chloramphenicol.
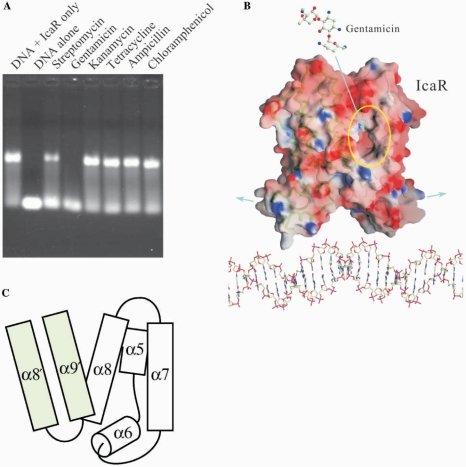
As shown in Figure 6A, only two of the six antibiotics were effective: streptomycin and gentamicin, both belonging to the aminoglycoside class. Gentamicin appeared to inhibit the DNA binding to a significantly greater extent than streptomycin, partially due to the higher molar concentration of gentamicin. Comparison of their chemical structures suggests that gentamicin is more positively charged and less polar than the other antibiotics. Identical results were obtained with the inclusion of spermine, a polycation polyamine, in the EMSA (data not shown), which ruled out nonspecific effect of the positive charges. Furthermore, in order to investigate interactions of IcaR and DNA with the antibiotics, a series of ITC experiments were carried out for gentamicin, streptomycin and kanamycin. As shown in Table 2, gentamicin and streptomycin both bound to IcaR, although not with great strength. The stoichiometry showed further similarity of IcaR to QacR (16). It suggests that only one drug molecule is bound by each dimer, despite the existence of two binding pockets. In contrast, binding to IcaR was not observed for kanamycin, which on the other hand interacted with DNA (see also Figure S4 in the Supplementary Material). Although we cannot exclude the possibility of binding to DNA rather than the repressor, it is also likely that the two aminoglycoside antibiotics interacted with IcaR in a similar way as the drugs with TetR and QacR.
Figure 6.
Gentamicin interrupts IcaR and DNA interaction. (A) The effects of antibiotics to the IcaR binding to ica operator in EMSA experiment. Only two aminoglycosides are capable of interfering the DNA binding, whereas gentamicin inhibited the binding to a significantly greater extent than streptomycin. The other four antibiotics kanamycin, tetracycline, ampicillin and chloramphenicol did not show significant effect on the IcaR–DNA binding. (B) Possible effect of gentamicin on the IcaR binding to ica operator. A negatively charged cavity was observed in each IcaR monomer, near the dimer interface. Binding of gentamicin to this cavity may result in conformation changes that lead to a larger separation of the N-terminal domains. (C) A schematic diagram of the putative antibiotics-binding cavity. The cavity is located at the dimer interface and surrounded by helices α5, α6, α7 and α8 of one monomer as well as α8′ and α9′ of the counter monomer. The α8–α9 loop has different conformations in the two crystal forms (Figure 1D), suggesting some functional flexibility.
Interestingly, a negatively charged cavity was observed in each IcaR monomer. As shown in Figure 6B and 6C, the cavity is located at the dimer interface, surrounded by helices α5, α6, α7 and α8 of one monomer, and flanked by α8′ and α9′ of the counter monomer as well. Inside the cavity, many amino acid residues are non-polar. Helices α5 and α6 are connected with a hinge-like loop, which lies in the back of the cavity. The equivalent connection in QacR contains the three consecutive Tyr91, Tyr92 and Tyr93, which are marked by greatly altered conformations upon drug binding. Structural comparison of IcaR and QacR only shows the identification of Tyr91 in IcaR as an equivalent of Tyr93 in QacR. The adjacent Tyr95 and Tyr152 from helices α6 and α8 of IcaR may be involved in binding the antibiotics. Residues from the counter subunit flank the cavity near its opening. It is worth noting that the loop between helices α8 and α9 in the SeMet-IcaR crystal has a different conformation than that in the native crystal. Flexibility of this lid-like region suggests its participation in the structural change at the dimer interface upon binding antibiotics, which may alter the relative disposition of the two subunits. Possibly with concerted movement of the α5/α6 hinge, the two N-terminal domains would be separated by a larger distance, which prevents the repressor from interacting properly with the operator DNA. Consequently, by interfering with the binding of IcaR to DNA, gentamicin and other antibiotics may elicit biofilm production in S. epidermidis, as a defense mechanism.
CONCLUSION
Understanding biofilm formation would lead to effective means for growth control of microorganisms by suppressing the emergence of obstinate colonies that can be resistant to various treatments for the patients. The sole involvement of the icaADBC genes in PIA synthesis encourages investigation of the regulation mechanism of these genes. Our results of the high-resolution crystal structure of IcaR, the cognate repressor for the ica operon, revealed its kinship to the TetR repressor family. Many of these repressors bind to antibiotics that weaken the interactions with their operator DNA via conformational changes in the protein dimers, resulting in concomitant activation of the genes. We have also shown that two IcaR dimers bind cooperatively to the operator DNA, and that the binding is abolished by gentamicin. Because antibiotics have been implicated in eliciting biofilm formation, cautions should be taken to avoid the use of some antibiotics (e.g. gentamicin) in treating microbial infections (e.g. by Staphylococcus).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
This study was funded by Summit Project of Academia Sinica and the National Science Council (NSC94-3112-B-001-010-Y and NSC95-3112-B-001-015-Y for the National Core Facility of High-Throughput Protein Crystallography at Academia Sinica). Portions of this research were carried out at the National Synchrotron Radiation Research Center (NSRRC), a national user facility supported by the National Science Council, Taiwan. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine. We thank NSRRC of Taiwan, SPring-8 and Photon Factory of Japan, and Advanced Light Source of USA for beam time allocations. We also thank Dr Chris S.-C. Jao for assistance in the ITC experiments. Funding to pay the Open Access publication charges for this article was provided by Academia Sinica.
Conflict of interest statement. None declared.
REFERENCES
- 1.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 2.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, Otto M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6:269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Dobinsky S, Kiel K, Rohde H, Bartscht K, Knobloch JK, Horstkotte MA, Mack D. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 2003;185:2879–2886. doi: 10.1128/JB.185.9.2879-2886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cafiso V, Bertuccio T, Santagati M, Campanile F, Amicosante G, Perilli MG, Selan L, Artini M, Nicoletti G, Stefani S. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin. Microbiol. Infect. 2004;10:1081–1088. doi: 10.1111/j.1469-0691.2004.01024.x. [DOI] [PubMed] [Google Scholar]
- 7.Fluckiger U, Ulrich M, Steinhuber A, Doring G, Mack D, Landmann R, Goerke C, Wolz C. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 2005;73:1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998;273:18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 9.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 10.Conlon KM, Humphreys H, O’Gara JP. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 2002;184:4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jefferson KK, Cramton SE, Gotz F, Pier GB. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 2003;48:889–899. doi: 10.1046/j.1365-2958.2003.03482.x. [DOI] [PubMed] [Google Scholar]
- 12.Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 2004;186:2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 2001;183:2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 16.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural mechanisms of QacR induction and multidrug recognition. Science. 2001;294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- 17.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 2002;21:1210–1218. doi: 10.1093/emboj/21.5.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dover LG, Corsino PE, Daniels IR, Cocklin SL, Tatituri V, Besra GS, Futterer K. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 2004;340:1095–1105. doi: 10.1016/j.jmb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Frenois F, Engohang-Ndong J, Locht C, Baulard AR, Villeret V. Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol. Cell. 2004;16:301–307. doi: 10.1016/j.molcel.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 20.Engohang-Ndong J, Baillat D, Aumercier M, Bellefontaine F, Besra GS, Locht C, Baulard AR. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 2004;51:175–188. doi: 10.1046/j.1365-2958.2003.03809.x. [DOI] [PubMed] [Google Scholar]
- 21.Kisker C, Hinrichs W, Tovar K, Hillen W, Saenger W. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol. 1995;247:260–280. doi: 10.1006/jmbi.1994.0138. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 2002;45:885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 23.Guerrero SA, Hecht HJ, Hofmann B, Biebl H, Singh M. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl. Microbiol. Biotechnol. 2001;56:718–723. doi: 10.1007/s002530100690. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron. Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 27.McRee DE. XtalView/Xfit–A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT. Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr. D. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 31.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 32.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.