Abstract
Ribosome biogenesis in eukaryotes requires the participation of a large number of ribosome assembly factors. The highly conserved eukaryotic nucleolar protein Nep1 has an essential but unknown function in 18S rRNA processing and ribosome biogenesis. In Saccharomyces cerevisiae the malfunction of a temperature-sensitive Nep1 protein (nep1-1ts) was suppressed by the addition of S-adenosylmethionine (SAM). This suggests the participation of Nep1 in a methyltransferase reaction during ribosome biogenesis. In addition, yeast Nep1 binds to a 6-nt RNA-binding motif also found in 18S rRNA and facilitates the incorporation of ribosomal protein Rps19 during the formation of pre-ribosomes. Here, we present the X-ray structure of the Nep1 homolog from the archaebacterium Methanocaldococcus jannaschii in its free form (2.2 Å resolution) and bound to the S-adenosylmethionine analog S-adenosylhomocysteine (SAH, 2.15 Å resolution) and the antibiotic and general methyltransferase inhibitor sinefungin (2.25 Å resolution). The structure reveals a fold which is very similar to the conserved core fold of the SPOUT-class methyltransferases but contains a novel extension of this common core fold. SAH and sinefungin bind to Nep1 at a preformed binding site that is topologically equivalent to the cofactor-binding site in other SPOUT-class methyltransferases. Therefore, our structures together with previous genetic data suggest that Nep1 is a genuine rRNA methyltransferase.
INTRODUCTION
Ribosomes are highly complex macromolecular machines responsible for the synthesis of all cellular proteins and are therefore essential for all living cells. Roughly, 200 000 ribosomes are present in a yeast cell (1) and about 2000 ribosomes need to be synthesized per minute in growing yeast. Ribosomal subunits can be assembled from their components in vitro without the presence of any additional factors albeit with very low efficiency. Efficient ribosome synthesis in vivo requires a plethora of additional assembly factors, especially in eukaryotes where literally hundreds of additional assembly factors have been described (2). Although the structure of ribosomal subunits and complete ribosomes have been determined to high resolution in prokaryotes (3–8) and eukaryotes (9), the maturation of rRNAs, their modification and their assembly with ribosomal proteins during ribosome biogenesis is still under intensive investigation.
In eukaryotes, the 18S rRNA of the small 40S subunit as well as the 25S and the 5.8S rRNAs of the large ribosomal subunit are processed from a 35S rRNA precursor. This precursor assembles with the 5S rRNA and the respective proteins to the 90S pre-ribosome at the nucleolus (10–12), a nuclear substructure that is organized around the rDNA containing chromosomal regions (11). During 35S rRNA processing a number of chemical modifications occurs at the rRNAs which include base modifications and 2′-O-methylations at specific nucleotides (13–15). The basic steps of ribosome biogenesis are conserved within eukaryotes, but the most detailed results have been obtained for Saccharomyces cerevisiae as the model system.
In eukaryotes and also in archaea, which share several steps in rRNA processing with eukaryotes, 2′-O-methylation and pseudouridinylation are usually catalysed by ribonucleoprotein particles (snoRNPs) which consist of snoRNAs and associated proteins. The snoRNAs guide the snoRNPs to the specific modification sites by virtue of their sequence complementarity with the respective rRNA target sequences (16–18). C/D box snoRNAs together with the Nop1 methyltransferase catalyse 2′-O-methylation (19,20) and H/ACA-box sno-RNAs together with pseudouridinkinase Cbf5 catalyse the formation of pseudouridine (21). In addition, dedicated enzymes modify specific nucleotides of the rRNA without the participation of the snoRNPs (22,23).
Recently, the Nep1 protein family, also referred to as Emg1, has been described as essential for ribosome biogenesis (24,25). Nep1 homologues are found in all eukaryotic genomes and in some archaea. It was demonstrated that the yeast and the human Nep1 proteins are localized in the nucleolus. In yeast, a temperature sensitive Nep1-mutant, Scnep1-1ts, showed an increased sensitivity at elevated temperatures to paromomycin, a translation inhibitor that binds to ribosomal RNA. Additionally, pre-rRNA processing of the 18S-rRNA precursor was blocked at sites A0, A1 and A2 in these Scnep1-1ts mutants which indicated that Nep1 is involved in ribosomal biogenesis. Furthermore, S-adenosylmethionine (SAM)—a common cofactor for methyltransferases—as well as overexpression of the SAM2-gene encoding a SAM synthetase restored growth of a Scnep1-1ts mutant at restrictive temperatures. This finding suggested that Nep1 is involved in rRNA methylation (25). Yeast 3-hybrid analysis demonstrated that ScNep1 binds directly to a 6-nt RNA consensus motif (5′-C/UUCAAC-3′) that is found at three positions in the 18S rRNA (nucleotides 1566–1571 near helix 47, nucleotides 1190–1195 near helix 35 and nucleotides 349–354 near helix 35) (26). Interestingly, nucleotides 1566–1571 are in proximity to 2 nucleotides that are modified through methylation (G1572, 2′-OH methylation, and G1575, N7-methylation) whereas nucleotides 1190–1195 include the hypermodified N1-Methyl-3-(3-amino-3-carboxypropyl) pseudouridine (m1acp3-Ψ in position 1191). Genetic analysis demonstrated that the essential phenotype of a ScΔnep1 deletion could be suppressed either by the deletion of snoRNA snr57 which mediates the 2′-OH-methylation of G1572 or by multi-copy overexpression of ribosomal protein Rps19 (26). Taken together, this suggests that Nep1 interferes with a methyltransferase reaction and that Nep1 activity is needed in the early steps of ribosome biogenesis to remove snr57 from the 18S rRNA, which then enables Rps19p recruitment to the 18S rRNA. Therefore, the essential phenotype of a Δnep1 deletion could result from arrested ribosome biogenesis at the Rps19 assembly step.
Despite this wealth of genetic data the actual function of Nep1 is still uncertain. Obvious sequence homologies to other known methyltransferases could not be detected. Only very recently a comprehensive bioinformatics study suggested that the Nep1-family of proteins might share homology with the SAM-dependent SpoU- and TrmD-related methyltransferases (27), the so-called SPOUT-class of methyltransferases (28). To gain further insight into the function of Nep1, we determined the structure of the Nep1 homolog from the archaebacterium Methanocaldococcus jannaschii (MjNep1), which is very similar to both the Nep1 from yeast (26% identity, 52% similarity) and the human Nep1 (26% identity, 47% similarity). We crystallized MjNep1 in three different forms: as the apo-protein, bound to the SAM-analogue S-adenosylhomocysteine (SAH) and bound to the antibiotic and general methyltransferase inhibitor sinefungin. The structures were determined using X-ray crystallography to a resolution of 2.2 Å for the free protein, 2.15 Å for the Nep1–SAH complex and 2.25 Å for the Nep1–sinefungin complex. The structure revealed a fold that is similar for the SPOUT-class of methyltransferases but contains a novel extension of this fold. Conservation of a number of arginine residues indicates a role for this extension in RNA binding. All SPOUT-class methyltransferases that have been functionally characterized to date modify nucleotides in either tRNAs or rRNAs in a site-specific manner by methylation of either ribose 2′-OH-groups, guanine N1 or uridine N3 nitrogens (27).
MjNep1 binds SAH and sinefungin in a preformed binding pocket that is topologically similar to the known cofactor binding sites of other SPOUT-class methyltransferases. The structure for Nep1 presented in this article, together with the previous genetic data therefore suggest that it functions as an 18S-rRNA-methyltransferase.
MATERIAL AND METHODS
Protein overexpression and purification
The gene encoding the full-length M. jannaschii Nep1-protein (aa1-205) was amplified by PCR from M. jannaschii genomic DNA (ATCC 43967D-5) with the appropriate primers and subcloned into the pET11a (Novagen, Madison) overexpression vector. This vector (MjNep1:pET11a) was transformed into Escherichia coli BL21(DE3) cells (Invitrogen). Native protein was overexpressed in ampicillin-containing (100 mg/l) LB-medium after induction with IPTG (250 mg/l) at an OD600=0.8 for 12 h at 24°C. Cells were harvested by centrifugation (5000g, 20 min, 4°C) and resuspended in sonication buffer (25 mM Tris/HCl, pH 7.8, 500 mM NaCl, 2 mM mercaptoethanol). Following sonication, the supernatant was cleared by centrifugation (20 min, 15 000g, 4°C) and the NaCl-concentration adjusted to 250 mM. Nep1 was purified from the supernatant by an ion-exchange step on a SP-Sepharose column using a salt gradient with buffer B (25 mM Tris/HCl, pH. 7.8, 1 M NaCl, 2 mM mercaptoethanol) and a subsequent gel filtration step (Superdex75, HiLoad 16/60, Amersham). Protein purity following this procedure was >95% as judged by SDS–PAGE. MALDI mass spectrometry revealed that the N-terminal methionine was cleaved. For subsequent crystallization trials the protein was exchanged into a buffer containing 20 mM Tris/HCl, 20 mM NaCl and 2 mM mercaptoethanol and concentrated to a final concentration of 15–20 mg/ml using Centricon YM-10 microconcentrators (Millipore).
For labelling the protein with selenomethionine E. coli BL21(DE3) cells transformed with MjNep1:pET11a were grown on a M9-medium supplemented with 75 mg/l l-selenomethionine (Sigma), 50 mg/l l-leucine, l-isoleucine and l-valine and 100 mg/l l-phenylalanine, l-threonine and l-lysine according to van Duyne et al. (29). The purification proceeded as described for the native protein but the mercaptoethanol concentration was raised to 10 mM. The complete replacement of all methionines by selenomethionines was confirmed by MALDI-mass spectrometry.
Crystallization, structure determination and refinement
Screening of the crystallization conditions for the native protein (15 and 10 mg/ml) was carried out at 4°C and 16°C using the hanging drop vapour-diffusion method. Suitable crystals were obtained at 4°C in 100 mM BisTris–propane buffer, pH 9.5, 6–10% polyethylene glycol 400, 30–48% glycerol and 0–400 mM trimethylamine-N-oxide. Crystals of the selenomethionine containing protein in the presence or absence of a 3-fold excess of S-adenosylhomocysteine (Sigma) were grown under similar conditions at 16°C. Crystals of the native protein bound to sinefungin (Sigma) were grown in the presence of a 3-fold excess of sinefungin at 4°C in 100 mM glycine buffer at pH 10.0, 20–24% polyethylene glycol 400, 30% glycerol and 0–200 mM trimethylamine-N-oxide.
Nep1 crystals were flash-cooled in liquid nitrogen prior to data collection at the Berkeley Advanced Light Source beamlines 4.2.2 and 8.2.2. Data for the apo-Nep1 and the Nep1–sinefungin complex were collected at beamline 4.2.2. Data for the selenomethioninyl-Nep1 crystals containing S-adenosyl-homocysteine were collected at beamline 8.2.2 at the absorption peak for selenium (0.9797 Å) as determined by X-ray fluorescence. Nep1–SAH complex diffraction data were processed with HKL-2000 (30). Native Nep1 and Nep1–sinefungin complex data were processed with d*TREK (31).
The structure of Nep1 bound to S-adenosylhomocysteine was determined by the single-wavelength anomalous diffraction (SAD) method implemented in autoSHARP (32). Phases from 14 sites (each of the two monomers in the asymmetric unit contained six SeMet residues and one extra site from a SeMet alternate conformation) were calculated and refined using SHARP (33) followed by density modification using DM (34). Phase calculation prior to density modification resulted in an initial figure of merit of 0.42. ARP/wARP (35) automatically traced 398 out of 410 residues into the modified electron density map and the model was completed by hand using COOT (36). Coordinates were refined against the data using PHENIX (37), including simulated annealing, and alternated with manual rebuilding. Data collection, phasing and refinement statistics are shown in Table 1. Figures were prepared using PyMol (38).
Table 1.
Data collection, phasing and refinement statistics
| Native Nep1 | Nep1 + SAH | Nep1 + Sinefungin | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 84.5, 91.1, 94.0 | 84.9, 91.1, 93.7 | 84.6, 89.6, 94.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength | 1.2400 | 0.9797 | 0.9790 |
| Resolution (Å) | 50–2.2 | 50–2.15 | 50–2.25 |
| Rsyma | 0.064 (0.490) | 0.107 (0.565) | 0.053 (0.468) |
| I/σI | 11.4 (2.6) | 17.5 (3.3) | 15.2 (2.8) |
| Completeness (%) | 100 (100) | 100 (100) | 99.9 (99.9) |
| Redundancy | 7.1 (6.9) | 7.1 (6.1) | 7.0 (6.8) |
| SAD phasing | |||
| Resolution (Å) | – | 50.0–2.15 | – |
| Rcullis | – | 0.67 | – |
| Phasing power | – | 1.55 | – |
| Figure of merit | – | 0.42 | – |
| Refinement | |||
| Resolution (Å) | 50–2.2 | 50–2.15 | 50–2.25 |
| No. reflections | 37 456 | 74 988 | 34 521 |
| Rwork/Rfree | 0.207/0.246 | 0.180/0.221 | 0.215/0.244 |
| No. atoms | |||
| Protein | 3372 | 3377 | 3372 |
| Ligand | 48 (8 glycerol) | 108 (2 SAH, 7 glycerol) | 90 (2 sinefungin, 7 glycerol) |
| Solvent | 254 | 265 | 193 |
| B-factors | |||
| Protein | 48.3 | 36.4 | 51.3 |
| Ligand | 61.0 | 42.5 | 59.1 |
| Solvent | 52.8 | 43.6 | 54.0 |
| RMS deviations | |||
| Bond lengths (Å) | 0.008 | 0.010 | 0.007 |
| Bond angles (°) | 1.123 | 1.198 | 1.019 |
aValues in parentheses are for highest-resolution shell.
Analytical ultracentrifugation
Equilibrium sedimentation analysis was performed with a Beckman Optima XLI ultracentrifuge using interference optics. In a buffer containing 20 mM KPO4, pH 6.5, 20 mM NaCl, the NEP1 protein was sedimented to equilibrium at three loading concentrations (85 μM, 142 μM and 200 μM) and four speeds (19 300, 26 200, 31 600 and 36 200 r.p.m.), producing data between 0 and 30 fringes, corresponding to concentrations ranging between 0 and 9.4 mg/ml (assuming a standard 3.2 fringes/mg/ml for proteins). The data were globally fitted with a fixed molecular weight distribution with 100 terms ranging between 10 and 80 kDa (39). The fit was then analysed with a 10 000 iteration Monte Carlo analysis according to (40).
Yeast three-hybrid RNA-binding assays
Yeast three-hybrid RNA-binding assays were essentially carried out as described previously (26). Briefly, plasmids containing either one (pIII/UUCAAC) or three copies (pIII/3×UUCAAC) of the 5′-UUCAAC-3′ consensus RNA-binding motif that binds to the yeast Nep1 protein were transformed into strain L40coat containing either pGAD-Nep1 (26) encoding the wild-type yeast Nep1-protein or pGAD-Nep1 with the appropriate point mutations. Quantification of β-galactosidase activity in Miller-units was performed as previously described (26 and references cited therein). The given values are averaged activities from two independent transformants.
RESULTS
Structure determination and refinement
The crystal structure of Nep1 from M. jannaschii (205 aa, MW 24 085 KDa) bound to the substrate analogue SAH was determined to 2.15 Å resolution using SAD data collected on a crystal of the selenomethionine (SeMet)-substituted protein grown in the presence of a 3-fold excess of SAH. Subsequently, the structures of the apo-protein and the sinefungin-bound form were determined using the Nep1–SAH-complex structure as an isomorphous starting model and refined to 2.2 Å and 2.25 Å resolution, respectively. The final models include residues 2–205 in all monomers. Mass spectrometry indicated that the N-terminal methionine corresponding to residue 1 is cleaved. Although double anomalous peaks for the selenium atom in each of the two selenomethionine 141 residues of the asymmetric unit were located and used for phasing, only selenomethionine 141 in chain B is modelled with a clear dual conformation. Because the minor conformation of this selenomethionine occupies the binding site for SAH with an occupancy of 0.3, the SAH ligand in chain B was set to a partial occupancy of 0.7.
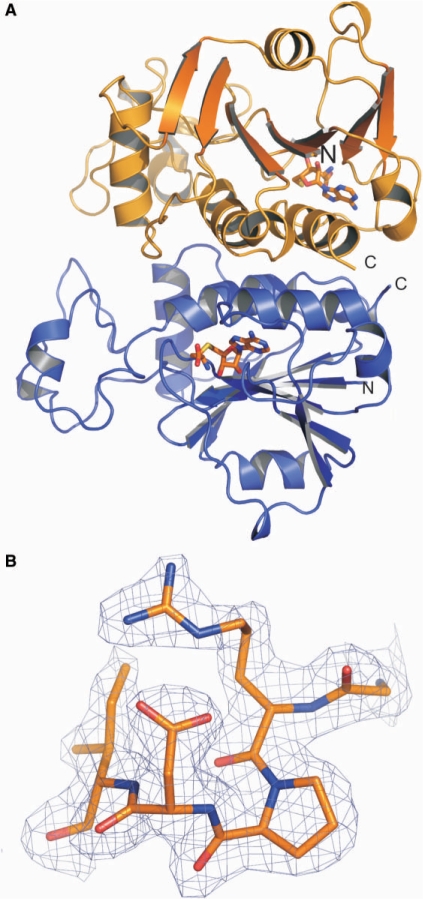
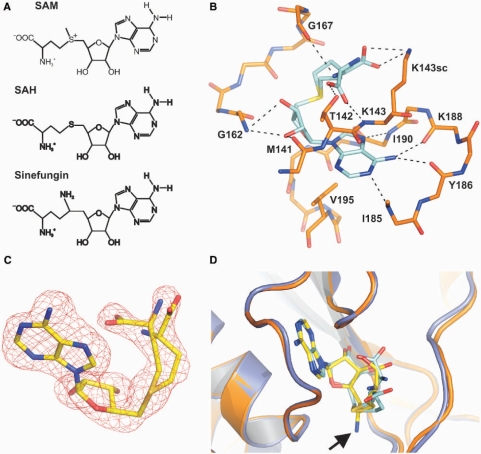
Two conformations of equal populations of the SAH homocysteine chain are observed in each monomer in the asymmetric unit. This results in two orientations for the amino- and carboxylate-groups of the homocysteine chain of SAH. Chain A and chain B in the structure of the free Nep1 have an RMSD of ∼0.21 Å for the backbone heavy atoms. Structural differences between monomers in the asymmetric unit are localized to two surface exposed loops (residues Pro 16–Val 28 and Lys 168–Glu 174) and do not include the SAH-binding site. The RMSD for the backbone heavy atoms between the Nep1 dimer bound to SAH and the dimer of free Nep1 is ∼0.18 Å. The 3D structure of the Nep1-dimer bound to SAH is shown in Figure 1A. An example for the quality of the obtained electron density maps is shown in Figure 1B. The coordinates and structure factors for MjNep1 in its apo-form as well as bound to SAH and sinefungin have been deposited in the Protein Data Bank under accession numbers 3BBE, 3BBD and 3BBH, respectively.
Figure 1.
Overall structure of the M. jannaschii Nep1-protein and quality of the data. (A) View of the Nep1-dimer in the asymmetric unit. Subunit A is shown in orange and subunit B is shown in blue. The bound co-factor S-adenosylhomocysteine (SAH) is shown as a stick model. (B) Typical example for the quality of the experimental 2Fo-Fc σA-weighted electron density map at the 1.5 σ-level for the Nep1 bound to SAH. Shown are residues G53-I57.
Structure of the Nep1 monomer
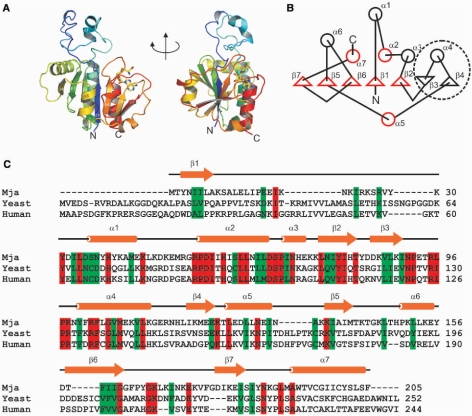
The Nep1 monomer is comprised of a single compactly folded α/β-sandwich domain and a long protruding globular loop (residues 13–53) that lacks regular structural elements with the exception of a distorted α-helix (α1) (Figure 2A). The core of the domain is formed by a seven-stranded twisted β-sheet with six strands in a parallel and one strand in an anti-parallel orientation. Five α-helices (α2, α3, α4, α6, α7) pack against one side of this β-sheet and one α-helix (α5) against the other. The β-sheet has a β7↑β5↑β6↑β1↑β2↑β3↓β4↑-topology (Figure 2B). The topology of the five parallel β-strands β7↑β5↑β6↑β1↑β2↑ corresponds to the topology of the five-stranded β-sheet (β5↑β3↑β4↑β1↑β2↑ that has been found as the minimal conserved core in methyltransferases containing the so-called SPOUT-domain (related to the SpoU- and TrmD-methyltransferases) (26,27,41).
Figure 2.
Fold of the Nep1-monomer. (A) Two views of the Nep1-monomer with the color gradient starting in blue at the N-terminus and changing to red at the C-terminus. The N- and C-terminus and the secondary structure elements are labelled. (B) Cartoon of the secondary structure of Nep1. α-helices are presented by circles and β-sheets are presented by triangles. The five-stranded parallel β-sheet typical for the fold of the SPOUT-class of methyl transferases corresponds to β-sheets β1, β2 and β5, β6 and β7. The structural elements that are part of the common core of SPOUT-class methyltransferase domains are highlighted in red. The β–α–β insertion element is encircled with a broken line. (C) ClustalW-alignment of the sequences of Nep1 from M. jannaschii (Mja), S. cerevisiae (yeast) and H. sapiens (human). Identical residues are highlighted in red, conservative replacements are indicated in green. A cartoon of the secondary structure is shown above the sequence alignment.
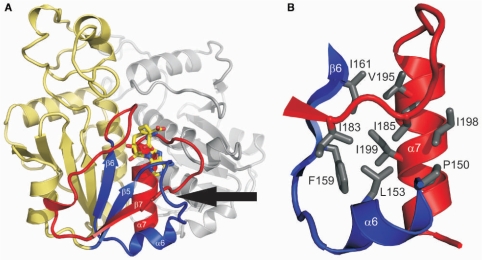
The C-terminal half of Nep1, including β-strands β5-β7 and α-helices α6 and α7, forms a deep trefoil knot where the last 22 residues (residues 184–205) of the protein including the α-helix α7 (residues 192–204) are tucked away beneath the loop connecting β5 and β6 (residues 146–157) which includes the α-helix α6 (residues 150–155, Figure 3A). This trefoil knot, which is involved in both the formation of the co-factor-binding site and part of the dimer interface (see below), is the other defining feature of the SPOUT-domain fold (41–43). The knot region is stabilized by an extended network of hydrophobic interactions that form a genuine hydrophobic core (Figure 3B). The first layer of this core is created by packing β-strand β6 (Phe 159, Ile 161) against the hydrophobic face of α-helix α7 (Val 195, Ile 198, Ile 199). In turn, Pro 150 and Leu 153 of α6 are packed against Ile 198 and Ile 199 of α-helix α7. The side chain of Ile 185, the first residue following the strand crossing, is wedged between the side chains of Ile 198 and Val 195 of α7 and also contacts Pro 150 and Leu 153 of α6. In addition, the side chain of Ile 183, the residue preceding the strand crossing, is packed against the side chain of Ile 185. A number of residues involved in this network of hydrophobic interactions are conserved residues or substituted by other hydrophobic residues in Nep1-sequences from other species. A hydrogen bond between the side chain of Ser 184, the residue at the chain crossing, and the backbone carbonyl oxygen of Tyr 186 provides another stabilizing interaction.
Figure 3.
The trefoil knot in the C-terminal part of Nep1. (A) Topology of the knot and its relative orientation to the ligand binding site and the dimer interface. The β-sheets β5 and β6 and their connecting loop including α-helix α6 are shown in blue. These structural elements form the ‘knotting loop’. Amino acids C-terminal of β6 are shown in red. This includes the terminal 22 amino residues and α7 following β7 that are threaded through the ‘knotting loop’. The point of the actual strand crossing is indicated by an arrow. Bound SAH is shown as a stick model. The second monomer of the Nep1 dimer is colored gray. (B) Hydrophobic core of the trefoil knot. The side chains of amino acids that contribute to the formation of the hydrophobic core are colored in gray and shown as stick models. The colour-coding for secondary structure elements is similar to Figure 2A.
There are two extensions in Nep1 relative to the common core motif of SPOUT-class methyltransferases. First, a globular loop (residues 13–53) lacking regular structure is inserted between β1 and α2 of the core motif. This loop is rich in basic residues such as Lys and Arg (11 of 41 residues). The second insertion is a β–α–β element that contains the anti-parallel β-strand β3 and β-strand β4 linked by α-helix α4. This element is inserted between β2 and α5. N-or C-terminal extensions or insertions in the core fold of the SPOUT-methyltransferases are commonly observed for many proteins of this class and vary from single α-helices or β-strands to complete domains (27). However, an insertion of a β–α–β element with an anti-parallel strand that extends the five-stranded β-sheet core has not been described so far for this class of proteins. Interestingly, the β–α–β insertion element contains a number of residues which are highly conserved in Nep1 proteins from different species (Figure 2C) and might be involved in RNA binding (see below). It therefore appears to be a signature motif for the Nep1-family of proteins.
The X-ray structure of Nep1 differs from a previous theoretical prediction (27) with regard to the domain arrangement and the location of functionally important extension elements. Instead of an N-terminal βααα-extension domain that precedes the C-terminal SPOUT-domain as predicted by bioinformatical methods (27), we find that the very N-terminus of the protein provides the β1-strand of the parallel β-sheet of the SPOUT-domain and two independent insertion elements are introduced following the canonical β-strands β1 and β2, respectively.
Structure of the dimer
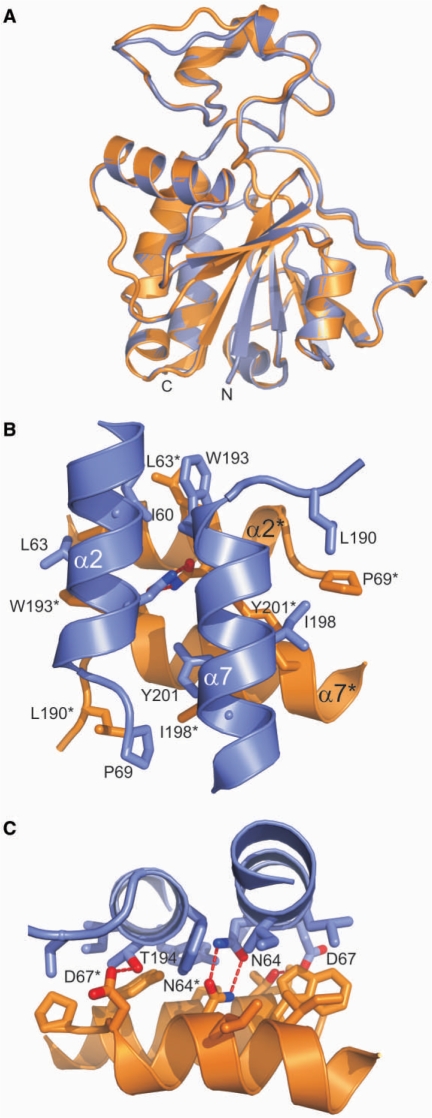
Nep1 crystallizes as a dimer in the asymmetric unit. The two monomers are structurally very similar with an RMSD for all backbone heavy atoms of 0.17 Å for the SAH/Nep1–complex (Figure 4A). The conformational differences are restricted to two solvent-exposed surface loops (residues Pro 16–Val 28 and Lys 168–Glu 174). Dimerization results in a large decrease of solvent accessible surface area of ∼1500 Å2 per monomer. The size of this buried surface area is typical for biologically relevant stable protein–protein interactions (44).
Figure 4.
Monomer superpositions, the dimer interface and the intermolecular four-helix bundle. (A) The superposition of the two monomers of the NepI dimer demonstrates that both chains adopt very similar conformations. Only very limited conformational differences are detectable in two surface exposed loops (residues Pro 16–Val 28 and Lys 168–Glu 174). (B) Orientation of the two pairs of parallel helices relative two each other. The helices α2 and α7 of chain A are colored blue whereas those of chain B are colored orange. The side chains of those residues involved in hydrophobic packing interactions are shown in a stick representation. Residues from different monomers are distinguished by an asterisk following the amino acid label. (C) Hydrogen bonding interactions across the dimer interface.
At the heart of the dimer interface is a four-helix bundle formed by the two parallel α-helices α2 (residues 56–67) and α7 (residues 192–204) of each monomer (Figure 4B). In contrast to standard α-helical bundles, the two parallel pairs of α-helices are oriented almost perpendicular to each other. The intermolecular interface of the helical bundle reveals two clusters of hydrophobic interactions involving the side chains of Ile 60, Leu 63 (α2) and Trp 193, Ile 198 and Tyr 201 (α7) from both monomers. In one cluster, the side chain of Leu 63 of one monomer is packed against the side chains of Ile 60 and Trp 193 of the associated monomer. In the other cluster, Leu 198 (α7) of one monomer packs against the aromatic ring of Tyr 201 (α7) in the associated monomer. In addition, two hydrogen bonds are observed between the Asn 64 (α2) side chains from both monomers and between the carboxylate group of the Asp 67 (α2) side chain of one monomer and the hydroxyl group of Thr 194 (α7) as well as the backbone amide group of Met 191 of the associated monomer (Figure 4C).
The dimer interface is extended by two additional contact areas. One involves the loop preceding α-helix α7 (residues 187–190) of one monomer and the loop following α2 (residues 68 and 69) in the other monomer with a prominent hydrophobic interaction between Leu 190 and Pro 69. The other contact area is formed by the N-terminal tip of α-helix α4 including its preceding residues (residues 98–102) and parts of the irregular insertion loop (residues 37 and 38) that interact with the equivalent regions of the associated monomer. Possible intermolecular hydrogen bonds are observed between the side chains of Asn 37 and Arg 102 as well as between Tyr 38 and Asn 99.
Remarkably, only a few of the residues involved in the interactions described earlier are conserved (Asn 37, Leu 63, Asp 67, Pro 69, Arg 102 and Leu 190).
We used analytical ultracentrifugation to address the question if the Nep1 dimer is also present in solution. Sedimentation equilibrium analysis at three different starting concentrations (85–200 μM) as described in Material and Methods section unequivocally demonstrated that the dimer with a molecular mass of ∼48 kDa is the predominant species in solution (Figure S1). No evidence for the presence of the ∼24 kDa monomer in solution was found.
Structurally similar proteins
To identify structurally similar proteins we performed a search using the DALI-server. The four best matches consisted of three bacterial RNA-methyltransferases (pdb codes 1x7o, 1zjr, 1uaj) and one uncharacterized archaebacterial protein (2o3a) from a structural genomics program. All four proteins display the common core fold for a SPOUT-domain methyltransferase, a β-sheet with at least five parallel strands and a β5↑β3↑β4↑β1↑β2↑-topology and a deep trefoil knot in its C-terminal half.
The protein with the highest structural similarity was found to be the antibiotic resistance mediating AviRb protein from Streptomyces viridochromogenes (1x7o), which methylates the 2′-hydroxyl group of U2479 in the bacterial 23S rRNA (45). The DALI-Z-score for AviRb is 7.8 and the RMSD between Nep1 and AviRb is 2.2 Å for 112 equivalent Cα-carbon positions. AviRb consists of an N-terminal RNA-binding domain with homology to ribosomal protein L30e and a C-terminal methyltransferase domain that belongs to the SPOUT-class. In contrast to Nep1, it contains an all-parallel six-stranded β-sheet with a β10↑β8↑β9↑β5↑β6↑β7↑-topology. In this case, an additional parallel β-strand (β7) and an additional α-helix are inserted into the core fold of the SPOUT-domain. Similar to Nep1, AviRb forms a dimer where the dimer interface involves a four-helix bundle. However, in contrast to Nep1, the angle between the two parallel helix pairs from each monomer is ∼40° and the most prominent contacts are made only between the two helices that would be equivalent to helix α7 of Nep1.
A comparable degree of similarity (Z-score 7.5, RMSD 2.6 Å for 116 equivalent Cα-positions) to Nep1 is found for TrmH from Aquifex aeolicus (1zjr), a methyltransferase that modifies the 2′-hydroxyl group of guanine G18 in bacterial tRNAs (46). TrmH is a single domain protein with a six-stranded all-parallel β-sheet with a β6↑β4↑β5↑β1↑β2↑β3↑-topology and N- and C-terminal α-helical extensions possibly involved in RNA binding. It also forms a dimer mediated by two α-helices corresponding to α7 with an approximately perpendicular orientation very similar to Nep1.
Interestingly, the two proteins mentioned earlier belong to the SpoU-subgroup of the SPOUT-domain methyltransferases and both modify a ribose 2′-OH position. The similarity of Nep1 to members of the TrmD-subgroup of the SPOUT-domain methyltransferases is less pronounced. This is indicated by a significantly lower Z-score (6.2) and higher RMSD (3.7 Å for 112 equivalent Cα-positions) to TrmD from Haemophilus influenzae (1uaj) (41). TrmD is a guanine N1-methyltransferase that modifies G37 in a subset of bacterial tRNAs. The structure consists of an N-terminal methyltransferase domain with a central five-stranded all-parallel β-sheet (β5↑β3↑β4↑β1↑β2↑) and a C-terminal domain with homology to the Trp-repressor possibly involved in RNA binding. An additional β-hairpin is inserted between β2 and β3. In contrast to β3 and β4 in the Nep1 structure, this β-hairpin is not connected to the central β-sheet. TmrD also forms a dimer mediated by a four-helix bundle, but in contrast to Nep1 the two pairs of helices are oriented in an antiparallel fashion.
The SAM-binding site
We crystallized Nep1 in its free form and bound to the SAH which corresponds to one of the products of the methyltransferase reaction (Figure 5A). In addition, we solved the structure of Nep1 bound to the general methyltransferase inhibitor sinefungin (Figure 5A). SAH and sinefungin bound in a very similar manner to equivalent binding sites in both monomers.
Figure 5.
The SAM-binding site. (A) Chemical structures of SAM, SAH and sinefungin. (B) Intermolecular interactions between the bound SAH and NepI. Amino acid residues participating in hydrogen bonding or hydrophobic interactions are labeled. (C) Simulated annealing omit map (Fo–Fc) contoured at the 3.0 sigma level for SAH bound to chain A of the Nep1-dimer revealing two possible conformations for the homocysteine side chain of the cofactor. (D) Comparison of the conformation of the Nep1-bound SAH and Nep1-bound sinefungin. Carbon atoms of SAH with its two possible conformations for the homocysteine side chain are colored in cyan whereas the carbon atoms of the bound sinefungin are colored yellow. The position of the additional NH2-group of sinefungin is indicated by an arrow. Nep1 secondary structure elements are shown as cartoon. Nep1 bound to SAH is colored blue whereas Nep1 bound to sinefungin is colored orange.
The two SAH-binding sites are ∼28 Å away from each other as measured by distance between the two sulfur atoms of the two bound SAH molecules and thus appear to be independent of each other (Figure 1A). However, the binding site in one monomer (chain B) was only 70% occupied due to an alternate conformation of SeMet 141 that partially occupies the binding site. We therefore use only chain A where the binding site is fully occupied by SAH to discuss the protein ligand interactions (Figure 5B). The ligand-binding site is located in the C-terminal half of the protein and involves the structural elements also required for the formation of the trefoil knot. Intermolecular contacts to the ligand involve β6 and residues in the adjacent loop to β7, residues in the loop between β5 and α6, and α7 and its preceding residues. The majority of contacts involve the adenine base and the ribose moiety of the ligand, whereas only limited contacts are observed with the homocysteine chain.
The adenine moiety is recognized by two hydrogen bonds between the backbone carbonyl groups of Tyr 186 and Lys 188 and the adenine N6-NH2-group. Additional hydrogen bonds are possible between the backbone amide group of Leu 190 and the N7 nitrogen of adenine as well as between the backbone amide of Ile 185 and the N1 nitrogen of adenine. Furthermore, a hydrophobic interaction is observed between the Val 195 side chain and the aromatic ring of the adenine. The backbone amide of Gly 162 is within hydrogen-bonding distance both to the 2′ and the 3′-hydroxyl group of the SAH ribose moiety. The 2′-hydroxyl group of SAH is also hydrogen bonded to the backbone carbonyl group of Met 141.
Two different conformations with equal populations are found for the homocysteine chain of the cofactor indicating that its orientation is not restricted (Figure 5C). In one case, the carboxylate group of the amino acid chain forms a salt bridge with the side chain of Lys 143. In the second case, the carboxylate group is hydrogen bonded to the main chain amide group of Lys 143 and to the side chain hydroxyl group of Thr 142. Surprisingly, the ligand protein hydrogen bond interactions are almost exclusively mediated by backbone amide or carbonyl groups. The prevalence of backbone-mediated interactions with the ligand correlates with the lack of sequence conservation for amino acids in the SAM-binding pockets between Nep1 and other members of the SPOUT-class of methyltransferases. On the other hand, the topology and the location of the co-factor-binding site are exactly conserved between Nep1 and other members of the SPOUT-family (27,41,45).
The adenine is in the anti-conformation relative to the ribose (the torsion angle O4′-C1′-N9-C4 is –156°). The ribose itself adopts the C3′-endo conformation found for the bound co-factor in other SPOUT-class methyltransferases (41,45) but different from the C2′-endo ribose conformation found for the bound co-factor in the Rossman-fold methyltransferases (47). The co-factor adopts a bent conformation with an O4′-C4′-C5′-S-torsion angle of −54° that brings the homocysteine chain in close proximity to the adenine base in both of its two observed conformations. This bent conformation of the co-factor with similar torsion angles is also found in other SPOUT-class methyltransferases (41,45) whereas the cofactor adopts an extended conformation in the classical Rossman-fold methyltransferases (47).
Sinefungin binds exactly in the same location as SAH and the interactions between the adenosine part of the ligand and the protein appear to be very similar to those found in the SAH/Nep1-complex (Figure 5D). Sinefungin also adopts a bent conformation that brings the amino acid side chain of the ligand into the vicinity of its adenine ring (Figure 5D). As observed for SAH its ribose is in the 3′-endo conformation. The interactions between the amino acid part of sinefungin and Nep1 resemble those described for the second conformer of SAH bound to Nep1 where the carboxylate group of sinefungin is hydrogen bonded to the backbone amide of Lys 143 and the sidechain hydroxyl group of Thr 142. The additional NH2-group (Figure 5A and D) of sinefungin will be mostly uncharged under our crystallization conditions. It is involved in two additional hydrogen bonds with the main chain carbonyl oxygen of Phe 164 and Pro 165.
The addition of SAM to the medium restores growth at elevated temperatures in yeast with temperature sensitive mutants (Scnep1-1ts) of the yeast Nep1 protein (25). Structure-based sequence alignments (Figure 2C) of the Nep1 proteins from M. jannaschii and S. cerevisiae, however, indicate that none of the mutated amino acid positions in the temperature sensitive yeast protein variants (V30, N69, T92, D222, yeast numbering) is in the vicinity of the SAM-binding site or close to the dimer interface. Therefore, the rescue effect of SAM appears not to be attributable to overcoming a decreased SAM-affinity in the mutant proteins by the increased intracellular SAM-concentrations. Rather, SAM binding might lead to an indirect stabilization of the temperature-sensitive mutant proteins.
Ligand binding does not lead to any global structural changes in the structure of Nep1. The superposition of the Nep1 dimer in its free form and bound to SAH indicates that there are no changes in the relative orientation of the two monomers with respect to each other and no rearrangements in the dimer interface. The RMSD for all backbone heavy atoms between the two dimer structures is only 0.18 Å. The superposition of the equivalent chains of the two dimers give equally low RMSD values. Only very minor adjustments are observed for the backbone and side chain conformations of residues in the SAH-binding pockets. Apparently, no ‘induced-fit’ is necessary to accommodate the cofactor and therefore, Nep1 has a preformed co-factor-binding site.
Possible RNA-binding site and active centre
MjNep1 is a highly positively charged protein with a pI of 9.4 and 37 basic residues (18%, 29 Lys and 8 Arg) in its sequence. However, many of these residues, especially the lysine residues are both not conserved and directly preceded or followed by Glu or Asp residues. Therefore, many lysine residues appear to be involved not in RNA binding but instead in the formation of surface salt bridges that have been found to stabilize proteins from thermophilic organisms (48) such as M. jannaschii.
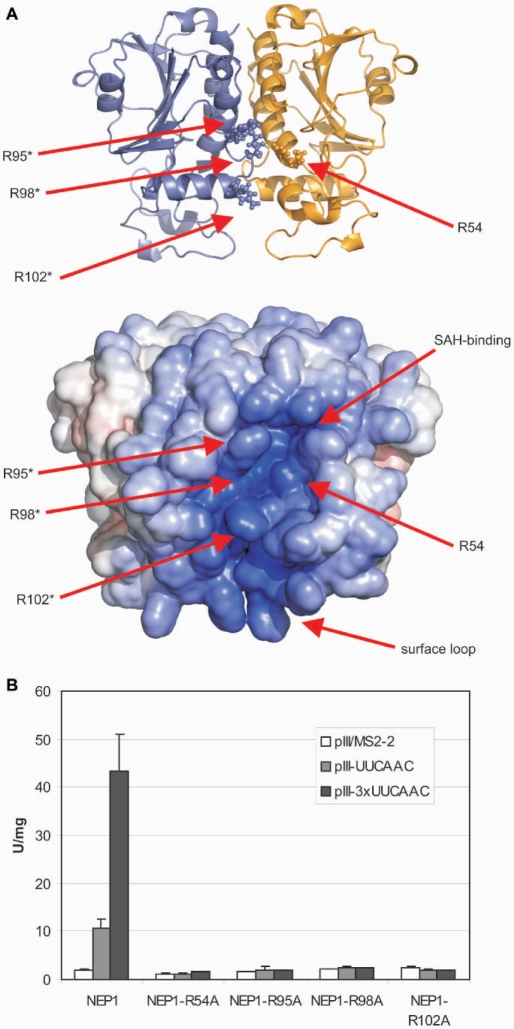
Nevertheless, when the electrostatic surface potential is plotted on the structure of the Nep1 dimer two positively charged areas stand out. One is centered on Arg 54 of one monomer and Arg 95*, Arg 98* and Arg 102* of the associated monomer (Figure 6A). Arg 54 is part of an Arg-Pro-Asp-Ile sequence, which is totally conserved among Nep1 proteins. Its side chain is precisely oriented by forming hydrogen bonds to the side chain of Asp 56, which is part of the same conserved sequence. Arg 95, Arg 98 and Arg 102 are located in α-helix α4 and its preceding loop which is part of the β–α–β insertion element unique to the Nep1-family. They are also conserved or in the case of Arg 102 substituted by lysine residues in Nep1 proteins from other species (data not shown). The level of sequence conservation observed for these residues strongly suggests a functional importance for these amino acids in RNA binding. The other positively charged area involves residues of the irregularly structured surface loop (residues 13–53) which is close in space to the cluster of conserved arginines. Whereas the sequence of this loop is poorly conserved, its overall basic character is preserved in Nep1-proteins from other species. Together, these two areas form a continuous positively charged crevice along the dimer interface that appears to be poised for RNA-binding (Figure 6A).
Figure 6.
Electrostatic surface potential representation of the NepI dimer and possible RNA-binding amino acid residues. (A) Cartoon of the Nep1 dimer structure with the side chains of the four conserved arginines shown in a ball and stick-representation (top) and electrostatic surface potential map (bottom) in the same orientation. The approximate locations of the side chains of Arg 54, Arg 95, Arg 98 and Arg 102 and of the SAH-binding site are indicated by arrows. Amino acid residues from different monomers are differentiated by an asterisk. (B) Yeast three-hybrid experiments with the yeast Nep1-protein mutants. The arginine side chains equivalent to Arg 54 (Arg 88), Arg 95 (Arg 129), Arg 98 (Arg 132) and Arg 102 (Arg 136) of the M. jannaschii protein were mutated to alanine in yeast Nep1. The yeast numbering is given in brackets. Whereas the wild-type yeast protein binds to RNAs containing either one (pIII/UUCAAC) or three copies (pIII/3×UUCAAC) of the RNA-motif 5′-UUCAAC-3′ as indicated by significant β-galactosidase activity (given in Miller units, U/mg) but not to a control (pIII/MS2-2), the RNA-binding activity of all four mutants is strongly reduced.
To address the role of the conserved arginine residues Arg 54, Arg 95, Arg 98 and Arg 102 for RNA binding, we mutated the equivalent residues in the yeast Nep1 protein Arg 88, Arg 129, Arg 132 and Arg 136, respectively, to alanine. The resulting four mutant proteins where then tested in the yeast three-hybrid system for binding to RNAs that contained either one or three copies of the 5′-UUCAAC-3′ motif. Whereas wild-type yeast Nep1 bound strongly to these RNAs as also demonstrated previously (26), all four mutant proteins lost the ability to bind these RNAs as indicated by the absence of β-galactosidase activity in these mutants (Figure 6B).
The utilization of the irregularly structured surface loop and the β–α–β element for RNA binding would be in agreement with the observation that in many other SPOUT-methyltransferases insertion or extension elements are exploited as auxiliary RNA-binding elements (27,41,45).
Interestingly, there are neither conserved basic or acidic residues nor glutamine or asparagine residues in the immediate vicinity of the S-adenosylhomocysteine sulfur atom, which would carry the reactive methyl group in SAM. The closest lysine side chain (Lys 143) is 7.5 Å away but shows no indication of conservation in other Nep1 sequences. The closest conserved residue that could be involved in the reaction chemistry is Asp 67 in α2 of the second monomer, which is 8.1 Å away from the SAH sulfur atom. This aspartate residue is present in most Nep1 sequences and sometimes substituted by glutamate or threonine. The distance between the SAH sulfur atom and the completely conserved arginine 54 sidechain is 9.5 Å.
However, the absence of a clearly recognizable catalytic general base is not unprecedented in methyltransferases. In a structurally unrelated N7-guanine methyltransferase that is responsible for the formation of the cap structure of eukaryotic messenger RNAs, catalysis occurs through the appropriate positioning of the guanine N7 nitrogen in proximity to the reactive methyl group of the co-factor (49). Similarly, in the structurally related AviRb no catalytic base could be unambiguously identified despite additional efforts in site-directed mutagenesis (45). In this case, a lysine that forms a hydrogen bond to the reactive ribose 2′-OH group is thought to be responsible for the correct positioning of the substrate, whereas the catalytic base is hypothesized to be water. An orbital steering mechanism has been suggested for a similarly positioned Lys in the structurally unrelated 2′-O-methyltransferase VP39 (50).
On the other hand, an aspartate is identified as the catalytic base in the structurally related guanine N1-methyltransferase TrmD, which deprotonates the guanine N1 imino group prior to the methyltransferase step (41,51). Interestingly, this catalytic aspartate is also derived from the second chain in the TrmD dimer. However, in the case of TrmD the aspartate side chain and the sulfur atom of the bound SAM are only 4.8 Å apart (41,51).
A catalytic mechanism that involves an arginine as the catalytic base has been suggested for the ribose-2′-O-methyltransferase TrmH, which is also a member of the SPOUT-class of methyltransferases (52,53). Here, the pKa of the arginine side chain is lowered by simultaneous hydrogen bonding to a conserved serine residue and a backbone phosphate group of the RNA-substrate so that it can activate the reactive ribose 2′-OH group by abstracting the proton prior to the methyl group transfer. Remarkably, in Nep1 the side chain of the totally conserved Arg 54 is strongly hydrogen bonded to the side chain of the totally conserved Asp 56 resembling the hydrogen bonding schema found for the catalytic Arg in TrmD.
Thus, in the absence of knowledge about the actual RNA-substrate of Nep1 it is difficult to decide if Asp 67 and Arg 54 play catalytic roles or are important for substrate positioning. The rather long distances to the sulfur atom of the bound SAH favour the latter possibility. However, given the fact that the RNA-binding interface includes residues from both monomers of the Nep1 dimer, binding of the RNA substrate might lead to a slight reorganization of the dimer interface that might bring at least Asp 67 into a position closer to the reactive site.
CONCLUSIONS
The X-ray structure of archaebacterial Nep1 from M. jannaschii reveals a three-layered α/β-sandwich fold (Figure 2A) with a seven-stranded β-sheet at its core and a deep trefoil knot in the C-terminal half of the protein. The β-sheet has a β7↑β5↑β6↑β1↑β2↑β3↓β4↑-topology. The order and connectivity of five of these seven β-strands (β7↑β5↑β6↑β1↑β2↑) is the same as in the central β-sheet in the core of all RNA-methyltransferases that belong to the SPOUT-class of methyltransferases (27,28,41,52). All SPOUT-class methyltransferases structurally characterized so far contain this deep trefoil knot in their C-terminal regions. Therefore, the fold of Nep1 strongly suggests that this protein is a genuine RNA-methyltransferase. As observed in all other structurally characterized SPOUT-class methyltransferases, Nep1 forms a dimer mediated by packing interactions between α-helices. Many SPOUT-class methyltransferases have unique insertions or extensions in their core fold that are used to adapt them to their specific functions and often play a role in RNA-binding (27). Nep1 displays two specific insertions, a large basic loop mostly devoid of regular structural elements inserted between β1 and α2 and a β–α–β-element inserted between β2 and β5. The latter interacts with the core five-stranded β-sheet and extends it into a seven-stranded β-sheet containing one antiparallel strand. While many members of the SpoU-related subgroup of the SPOUT-family contain a sixth parallel β-strand, an extension by two β-strands including an antiparallel strand has so far not been described for this protein family.
The suggestion that Nep1 is a genuine RNA-methyltransferase is based not only on the fold similarity. The structure of the SAH/Nep1 complex demonstrates that Nep1 is also capable of binding close homologs of the methyl group donor SAM. Remarkably, SAH is bound in a pocket topologically equivalent to those in other SPOUT-class methyltransferases despite the lack of obvious sequence similarities in this region. This lack of sequence conservation for residues forming the binding pocket is explained by the prevalence of backbone-mediated protein ligand interactions. The similarity of the co-factor-binding pocket between Nep1 and the other SPOUT-class members is reflected in the bent conformation of the bound SAH typical for this class (41,45,51,52). There the amino acid side chain of the cofactor is close to the purine ring in contrast to the extended conformation of the cofactor observed in other classes of methyltransferases. Interestingly, Nep1 also binds in a similar manner to the general methyltransferase inhibitor and antibiotic sinefungin, which also adopts a bent conformation. To the best of our knowledge, this is the first structure of a SPOUT-class methyltransferase bound to this antibiotic.
Whereas the fold homology and the SAH binding together with previous genetic data suggest that Nep1 is an 18S rRNA methyltransferase, the actual RNA-substrate of the methyl transfer carried out by Nep1 remains unidentified. The functionally characterized SPOUT-class methyltransferases either modify ribose-2′-groups, guanine N1 or uridine N3-nitrogens. The 5′-C/UUUCAAC-3′ RNA-binding motif identified for yeast Nep1 occurs three times in yeast 18S rRNA. In one case it is located in the vicinity of 2 nt of the 18S rRNA known to undergo modification: G1572 which is 2′-O-methylated and G1575 which is methylated in the N7-position of the guanine base. In the second example, it includes a hypermodified methylated pseudouridine nucleotide (m1acp3-Ψ1191).
Structurally, Nep1 appears to be most similar to SPOUT-class members that modify ribose 2′-positions. However, the modification at G1572 is known to be mediated by the C/D-box RNA snr57 and the general Nop1-methyltransferase in yeast (54). On the other hand, neither a guanine N7 nor a pseudouridine methylation activity has been reported thus far for a member of the SPOUT-class methyltransferases.
A Nep1 homologue is found in the archaebacterium Sulfolobus solfataricus, where the N7-methylation of G1575 is reported to be absent (55). If, however, Nep1 would also function as an rRNA-chaperone or S19 assembly factor as suggested by the genetic data the absence of the N7-modification in S. solfataricus would not necessarily rule out a N7-methylation activity for Nep1 in other organisms. An N7-methylation activity for Nep1 could in principle account for the long distances between the reactive center and basic or acidic side chains observed in the SAH/Nep1 complex as well as the absence of side chains with clear hydrogen bonding potential in the vicinity of the SAH sulfur atom that could be used to orient e.g. a ribose 2′-OH group. Instead, the conserved R54 and D67 would be used to orient a large purine ring system by hydrogen bonding. However, this remains speculative as long as conformational changes and relative reorientations of monomers upon RNA-binding cannot be ruled out.
Methylation of a pseudouridine, however, might be mechanistically similar to the N3 methylation of uridine nucleotides, a known enzymatic activity for SPOUT-class methyltransferases such as RsmE from E. coli (56). However, structurally characterized RsmE-related uridine N3-methyltransferases (57) differ from Nep1 with respect to the extension of the core SPOUT-class fold and the presence of an additional RNA-binding domain. The conserved five-stranded parallel β-sheet of the SPOUT-class methyltransferases is extended by a sixth parallel β-strand in these proteins in contrast to the β–α–β element found in Nep1. Furthermore, they contain an N-terminal PUA-domain involved in RNA-binding and their putative active side residues are apparently located in different structural elements. Thus, the structural similarity of the known N3 uridine methyltransferases with Nep1 is not sufficient to draw conclusions about a possible pseudouridine methylation function of Nep1.
Finally, it is a possibility that Nep1 is not enzymatically active as an RNA-methyltransferase and only functions as an RNA-chaperone or an S19-assembly factor. However, in this case, it would be hard to understand why the protein would have retained the ability to bind SAM throughout evolution. SAM is a valuable cofactor in many cellular processes and SAM binding to Nep1 without its subsequent use would contribute to draining the cellular SAM pools. A regulatory role of SAM binding for Nep1 function is also unlikely since ligand binding does not induce structural changes in the protein.
Further experiments to address the RNA-substrate specificity and the enzymatic activity of Nep1 are currently in progress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to C.A. Kim, M. Görlach, H. Schwalbe and A.P. Hinck for extensive and helpful discussions and their critical reading of the manuscript. We would like to thank K. W. Hakala and S. T. Weintraub for carrying out the mass-spectrometry analysis and the staff at the Advanced Light Source (ALS) beam line 8.2.2 for support. We also thank J.C. Nix for collecting and processing data at ALS beam line 4.2.2. S. Ullevig was involved in optimizing the Nep1-sinefungin crystals. This work was supported by start-up funds from the Department of Biochemistry, UTHSCSA, to J. W., the Robert A. Welch Foundation (grant AQ-1399 to P.J.H.), the Deutsche Forschungsgemeinschaft (DFG) through the SFB 579 ‘RNA–ligand interactions’ (K.-D.E. and P.K.) and the Excellence Center: Macromolecular Complexes (K.-D.E.). Support for the X-ray-Crystallography Core Laboratory of the Department of Biochemistry by the Executive Research Council of the University of Texas Health Science Center at San Antonio is also gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by a University Research Council grant (J.W.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Hage AE, Tollervey D. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- 3.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 4.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 5.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 6.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:793–795. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 7.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 8.Selmer M, Dunham CM, Murphy FVIV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 9.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae -tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 10.Melese T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr. Opin. Cell. Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 11.Scheer U, Hock R. Structure and function of the nucleolus. Curr. Opin. Cell. Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- 12.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 13.Kowalek JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J. Biol. Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- 14.Green R, Noller HF. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996;2:1011–1021. [PMC free article] [PubMed] [Google Scholar]
- 15.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 16.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 17.Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 18.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 19.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 2000;19:317–323. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 1998;12:527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine D, Delcour J, Glasser AL, Desgrès J, Vandenhaute J. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 1994;241:492–497. doi: 10.1006/jmbi.1994.1525. [DOI] [PubMed] [Google Scholar]
- 23.Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Liu PC, Thiele DJ. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol. Biol. Cell. 2001;12:3644–3657. doi: 10.1091/mbc.12.11.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eschrich D, Buchhaupt M, Kotter P, Entian KD. Nep1p (Emg1p), a novel protein conserved in eukaryotes and archaea, is involved in ribosome biogenesis. Curr. Genet. 2002;40:326–338. doi: 10.1007/s00294-001-0269-4. [DOI] [PubMed] [Google Scholar]
- 26.Buchhaupt M, Meyer B, Kotter P, Entian KD. Genetic evidence for 18S rRNA binding and an Rps19p assembly function of yeast nucleolar protein Nep1p. Mol. Genet. Genomics. 2006;276:273–284. doi: 10.1007/s00438-006-0132-x. [DOI] [PubMed] [Google Scholar]
- 27.Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007;8:73. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002;4:71–75. [PubMed] [Google Scholar]
- 29.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–24. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Cryst. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 32.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2006;364:215–30. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 33.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan K. An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 35.Lamzin VS, Perrakis A, Wilson KS. The ARP/wARP suite for automated construction and refinement of protein models. In: Rossmann MG, Arnold E, editors. International Tables for Crystallography. Volume F: Crystallography of biological macromolecules. The Netherlands: Kluwer Academic Publishers Dordrecht; 2001. pp. 720–722. [Google Scholar]
- 36.Emsley P, Cowtan P. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Grosse-Kunstleve RW, Hung LW, Loerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 39.Demeler B. UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, Harding SE, Rowe AJ, editors. Modern Analytical Ultracentrifugation: Techniques and Methods. UK: Royal Society of Chemistry; 2005. pp. 210–229. [Google Scholar]
- 40.Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid. Polym. Sci. 2007 doi: 10.1007/s00396-007-1699-4. [Google Scholar]
- 41.Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 2003;22:2593–2603. doi: 10.1093/emboj/cdg269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michel G, Sauve V, Laroque R, Li Y, Matte A, Cygler M. The structure of the RlmB 23S rRNA methyltransferase reveals a new methyltransferase fold with a unique knot. Structure. 2002;10:1303–1315. doi: 10.1016/s0969-2126(02)00852-3. [DOI] [PubMed] [Google Scholar]
- 43.Nureki O, Shirouzu M, Hashimoto K, Ishitani R, Terada T, Tamakoshi M, Oshima T, Chijimatsu M, Takio K, Vassylyev DG, et al. An enzyme with a deep trefoil knot for the active-site architecture. Acta Cryst. D. 2002;58:1129–1137. doi: 10.1107/s0907444902006601. [DOI] [PubMed] [Google Scholar]
- 44.Nooren IMA, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosbacher TG, Bechthold A, Schulz GE. Structure and function of the antibiotic resistance mediating methyltransferase AviRb from Streptomyces viridochromogenes. J. Mol. Biol. 2005;345:535–545. doi: 10.1016/j.jmb.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 46.Pleshe E, Truesdell J, Batey RT. Structure of a class II TrmH tRNA-modifying enzyme from Aquifex aeolicus. Acta Cryst. F. 2005;61:722–728. doi: 10.1107/S1744309105022980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin JL, McMillan FM. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 2002;12:783–793. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 48.Schuler B, Kremer W, Kalbitzer HR, Jaenicke R. Role of entropy in protein thermostability: folding kinetics of a hyperthermophilic cold shock protein at high temperatures using 19F NMR. Biochemistry. 2002;41:11670–11680. doi: 10.1021/bi026293l. [DOI] [PubMed] [Google Scholar]
- 49.Fabrega C, Hausman S, Shen V, Shuman S, Lima CD. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell. 2004;13:77–89. doi: 10.1016/s1097-2765(03)00522-7. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Xia Y, Gao X, Gershon PD. Mechanism of RNA 2′-O-methylation: evidence that the catalytic lysine acts to steer rather than deprotonate the target nucleophile. Biochemistry. 2004;43:5680–5687. doi: 10.1021/bi0359980. [DOI] [PubMed] [Google Scholar]
- 51.Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 2003;333:931–949. doi: 10.1016/j.jmb.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 52.Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, Hori H, Yokoyama S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure. 2004;12:593–602. doi: 10.1016/j.str.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe K, Nureki O, Fukai S, Ishii R, Okamoto H, Yokoyama S, Endo Y, Hori H. Roles of conserved amino acid sequence motifs in the SpoU (TrmH) RNA methyltransferase family. J. Biol. Chem. 2005;280:10368–10377. doi: 10.1074/jbc.M411209200. [DOI] [PubMed] [Google Scholar]
- 54.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 55.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 1998;180:2883–2888. doi: 10.1128/jb.180.11.2883-2888.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forouhar F, Shen J, Xiao R, Acton TB, Montelione GT, Tong L. Functional assignment based on structural analysis: crystal structure of the yggJ protein (HI0303) of Haemophilus influenzae reveals an RNA methyltransferase with a deep trefoil knot. Proteins. 2003;53:329–332. doi: 10.1002/prot.10510. [DOI] [PubMed] [Google Scholar]