Abstract
MAD family proteins are transcriptional repressors that antagonize the functions of MYC oncoproteins. In particular, MAD1 has been demonstrated to interfere with MYC-induced proliferation, transformation and apoptosis. The MAD1 gene is expressed in distinct patterns, mainly associated with differentiation and quiescence. We observed that MAD1 is directly activated by G-CSF in promyelocytic cell lines. To investigate the transcriptional regulation of the human MAD1 gene, we have cloned and characterized its promoter. A region of high homology between the MAD1 orthologs of human, mouse and rat contains the core promoter, marked by open chromatin, high GC content and the lack of a TATA box. Using deletion constructs we identified two CCAAT-boxes occupied by C/EBPα and β in the homology region that mediate responsiveness to G-CSF receptor signaling. The necessary signals include the activation of STAT3 and the RAS/RAF/ERK pathway. STAT3 does not bind directly to promoter DNA, but is recruited by C/EBPβ. In summary, our studies provide a first analysis of the MAD1 promoter and suggest STAT3 functions as a C/EBPβ cofactor in the regulation of the MAD1 gene. Our findings provide the base for the characterization of additional signal transduction pathways that control the expression of MAD1.
INTRODUCTION
The transcription factors of the MYC/MAX/MAD network have evolved as critical regulators of many aspects of cell physiology, including proliferation, differentiation and apoptosis. Furthermore, extensive work documents that MYC functions as an oncoprotein both in human tumors and animal models (1–4). The proteins of this network form dimeric complexes containing one MAX protein and one member of the MYC or MAD group of proteins or MAX itself. Dimer formation is a prerequisite for sequence-specific DNA binding, both being mediated by the bHLHZip domains, highly conserved regions common to all MYC/MAX/MAD network members (5). In general, DNA-bound MYC/MAX heterodimers activate gene transcription by recruiting a number of different cofactors, including different histone acetyl transferases, that regulate chromatin and polymerase function (1,6). MYC/MAX complexes bind to DNA sequences with the consensus 5′-CACGTG-3′, referred to as E-boxes. These elements are also recognized by MAD/MAX heterodimers and at least in vitro little difference in the binding specificity between MYC/MAX and MAD/MAX complexes could be observed (5). These findings suggest that the different dimers compete for binding to specific DNA sites. This model is particularly attractive since MAD proteins have been shown to recruit corepressor complexes with histone deacetylase activity (7). Thus, not only can MYC/MAX and MAD/MAX complexes compete for DNA binding but they also recruit opposing cofactor functions to target genes. In summary, these findings indicate that activating MYC/MAX and repressing MAD/MAX complexes are part of a switch mechanism that regulates the activities mentioned at the beginning.
The MAD group of proteins consists of six members, the four highly related small MAD1, MXI1 (MAD2), MAD3 and MAD4, and the considerably larger MNT and MGA (7). It has been suggested earlier on that these proteins might function as tumor suppressors since they can antagonize MYC function. Indeed, tissue culture transformation assays demonstrated that MAD proteins interfere with MYC-dependent transformation (8–16). Furthermore mxi1–/– mice exhibit an increased susceptibility to tumor formation (17). However, despite extensive effort, little evidence has been obtained to support the tumor suppressor notion for human tumors. This lack of evidence might be, at least in part, due to the rather broad expression pattern of all MAD genes and proteins, making it unlikely that enough MAD function can be eliminated during tumorigenesis (7). In addition to effects on transformation, MAD1 has been implicated in controlling aspects of proliferation, differentiation and apoptosis (9,18–23). The latter is at least in part due to the repression of the PTEN gene by MAD1 (24). This results in the activation of the AKT kinase and subsequently inhibition of apoptosis. In addition, ectopic expression of MAD1 in mice resulted in early postnatal lethality and dwarfism (25). Of particular importance are the studies in mice with a targeted disruption of mad1 (26). Although the overall phenotype was rather mild, in the absence of Mad1 granulopoiesis is distorted. While the number of mature granulocytes is unaltered, the precursor cells undergo extra rounds of cell division prior to terminal differentiation and the mature cells are highly susceptible to apoptosis (26). Consistent with this latter finding, granulocytes of mad1 transgenic animals are more resistant to apoptotic conditions (25). Together, these studies provide evidence for important regulatory functions of MAD proteins.
The available expression studies suggest that the small MAD genes are transcribed preferentially during proliferation arrest and differentiation (7). Specifically, MAD1 expression has been documented in differentiating hematopoietic cells as well as a number of additional cell types (16,19,21,26–33). Together, these findings suggest that the MAD1 gene must be the target of a number of distinct signaling pathways that control the expression of this gene in the different cell types. Previously, we have observed that MAD1 expression can be activated in human promyelocytic cell lines by different stimuli, including the phorbol ester TPA and retinoic acid, that induce differentiation (29,30). In addition, the cytokine granulocyte-colony stimulating factor (G-CSF) was capable of enhancing MAD1 expression (32). G-CSF is a key cytokine to regulate the proliferation and survival of myeloid progenitor cells and to stimulate their differentiation and maturation toward granulocytes (34–36). The G-CSF receptor (G-CSFR) activates multiple signaling pathways, similar to other cytokine receptors, that include the JAK/STAT and the RAS/RAF/MAPK pathways (37–41). G-CSF regulates a large set of genes that mediate its physiological effects, however, few direct targets have been identified (42–44). One immediate early G-CSF target is SOCS3, which upon activation interacts with the tyrosine phosphorylated G-CSFR and downregulates its signaling capacity (45–47).
In order to obtain insight into the regulation of MAD1 by G-CSF, we cloned the human MAD1 promoter region and identified the elements relevant for the G-CSF response. We observed that the JAK/STAT and the RAS/RAF/ERK pathways contribute to the activation of MAD1 through C/EBPβ. Our findings suggest that STAT3 is recruited to the promoter by binding to C/EBPβ.
MATERIALS AND METHODS
Cell culture and treatment conditions
HL60 (ATCC CCL-240) and U937 promyelocytes were cultured in RPMI 1640 medium with 10% FCS. HEK293 and RK13 were cultured in DMEM and MEM medium, respectively, with 10% FCS. The different cell lines were treated with 10 ng/ml G-CSF (Chugai Pharmaceutical Co., Ltd, Tokyo, Japan) for the indicated times.
Transient transfection and luciferase assay
RK-13 and HEK293 cells were transfected by the calcium phosphate coprecipitation method as described previously (48). For luciferase assays, 7–10 μg of plasmids were cotransfected. The cells were incubated for 24–28 h in complete medium and before harvesting treated with 10 ng/ml G-CSF for 6 h. The relative luciferase activities were normalized to the β-galactosidase activities.
Cloning and expression constructs
All promoter mutant constructs were generated using the QuickChange® site-directed mutagenesis protocol (Stratagene, Heidelberg, Germany) with complementary primers spanning the mutation site, for verification, all mutated plasmids were sequenced. The expression vectors pLNCX-GCSF/R pLNCX-GCSF/RΔ649, pLNCX-GCSF/RΔ670, pLNCX-GCSF/RΔ685 were obtained from K. Welte (Hannover Medical School, Germany). The expression constructs for the G-CSFR (pLNCX-GCSF/RmA, pLNCX-GCSF/RmB, pLNCX-GCSF/RmC, pLNCX-GCSF/RmD and pLNCX-GCSF/Rm0) were provided by I. Touw (Rotterdam University, The Netherlands) and have been described (38). The cDNA of C/EBPα and β were cloned into CMV-expression vector pCB6+ (49). Human C/EBPε was cloned into pcDNA3. The pCAGGS-neo-HA-STAT1, pCAGGS-neo-HA-STAT3, pCAGGS-neo-HA-STAT3(Y705F) and pECESTAT5α were provided by G. Müller-Newen (RWTH Aachen University, Germany).
Western blot and immunoprecipitation
Whole-cell lysates were prepared on ice in F-buffer [50 mM HEPES, pH7.4, 50 mM NaCl, 30 mM Na4P2O7, 100 µM Na3VO4, 10% glycerol, 0.2% (w/v) Triton X-100, 1 mM NaF] containing protease inhibitors. Cell extracts were resolved by SDS–PAGE and transferred onto nitrocellulose membranes. Blots were probed with the indicated antibodies in PBS-T [0.1% (v/v) Tween-20/PBS], washed and incubated with peroxidase-conjugated secondary antibodies. Detection was performed with the chemiluminescence kit ECL (Amersham, Piscataway, NJ, USA). For immunoprecipitation assays, whole-cell extracts were incubated with 15 µl protein A or G agarose beads, and 2 µg antibody rotating at 4°C for 2 h, washed with F-buffer and specific proteins were identified by western blot analysis.
Antibodies
Cytochrom C (Sc-7159, Santa Cruz), Polymerase II (Sc-900, Santa Cruz), AcH3 (#06-866,Upstate), C/EBPα (Sc-61, Santa Cruz), C/EBPβ (Sc-746 and Sc-150, Santa Cruz), C/EBPε (Sc-158, Santa Cruz), STAT3-P (#9131, Cell Signaling), STAT3 (Sc-482, Santa Cruz), actin (A2066, Sigma), HA(3F10) (Boehringer, Mannheim, Germany)
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed in U937 cells as described previously (50). The following primers were used: P1, 5′-AGTTGCGAATCCTGTCACCA-3′; P1’, 5′-TTCTCTTGACAGGCCAGCTT-3′; G1, 5′ATATTGTAGGTGACACAAACTGC-3′; G1’, 5′-ATCTCACTTGAAGCTTCCACAG-3′. These primer pairs amplified a 224 bp and a 257 bp product, respectively. PCR products were separated on a 2% agarose gel using a 100 bp DNA ladder as a size marker (Fermentas).
Total RNA preparation and quantitative RT–PCR
Total cellular RNA was prepared using the RNeasy Mini Kit (Qiagen) following the manufacturer's instructions, and residual genomic DNA was removed by DNase I treatment (Qiagen). Total RNA (1.5 µg) was reverse transcribed to cDNA with Omniscript Reverse Transcriptase (Omniscipt RT Kit, Qiagen) and analyzed by real time PCR using TaqMan Universal PCR reagents with ABI 7000 instrument (PE Applied Biosystems, Darmstadt, Germany). The relative quantification of MAD1 mRNA was analyzed by the comparative CT method, and normalized to that of β-GLUCURONIDASE.
MAD1 probe 6-Fam-TGGACAGCATCGGCTCCACC-Tamra
MAD1-f 5′-GAGAAGCTGGGCATTGAGAG-3′
MAD1-r 5′-ACGTCGATTTCTTCCCTGTC-3′
β-GLUCURONIDASE probe 6-Fam-TGAACAGTCACCGACGAGAGTGCTGG-Tamra
β-GLUCURONIDASE-f 5′-CTCATTTGGAATTTTGCCGATT-3′
β-GLUCURONIDASE-r 5′-CCGAGTGAAGATCCCCTTTTTA-3′
5′-RACE (rapid amplification of cDNA-ends)
U937 cells were grown to a density of 5 × 106 cells/ml and stimulated with 10 ng/ml human G-CSF for 2 h. mRNA was purified using the Oligotex mRNA-Kit (Qiagen) according to the manufacturer's instruction. For 5′-RACE we used the 5′/3′-RACE-Kit (Roche) following the manufacturer's protocol. The 2 µg of purified mRNA were incubated with AMV-Reverse-Transcriptase and specific primer Sp1 (5′-CGGAGTCGGAGCGCTCCG) to yield MAD1 cDNA. The single-stranded cDNA was purified with High Pure PCR Purification Kit (Roche). The PCR reactions were preformed with the purified cDNA as template and the oligo-dT-primer together with the second specific primer Sp2 (5′-AGCTGGTCGATTTGGTGAACG). The oligo-dT-anchor-primer with the third specific primer Sp3 (5′-CACAAGCAAGATGAGCCCG) were employed for the nested PCR reaction. Amplified DNA was extracted from agarose gel using the JetSorbKit (Genomed), cloned using the TA-cloning-kit (Invitrogen) and sequenced (MWG Biotech). Alternatively, the products of the reverse transcription with primer Sp1 were analyzed further by PCR with Sp2 as reverse primer and various forward primers from the nucleosome mapping (described below). Quantification of the products was done by real time PCR.
Nucleosomal mapping
U937 cells were grown to a density of 5 × 105 cells/ml. The cells were pelleted, washed two times in ice-cold PBS, resuspended in TBS at a concentration of 2 × 107 cells/ml and lyzed in TBS/1% Tween-20 (1:1) containing the proteinase-inhibitor PefaBloc. The nuclei were resuspended in 1 × TBS/25% sucrose (w/v) at a concentration of 4 × 106 nuclei/ml and purified by sucrose-gradient centrifugation (3000g, 4°C, 15 min). The washed nuclei were resuspended in digestion-buffer (0.32M sucrose, 50 mM Tris–HCl pH7.5, 4 mM MgCl2, 1 mM CaCl2, 5 mM Na-Butyrat pH8, PefaBloc). DNA-concentration was photometically determined (Eppendorf). Fifty units of Micrococcal Nuclease (MBI-Fermentas) were added per 0.5 mg DNA and incubated at 37°C for 5 min. The MNase-reaction was stopped by adding 0.5 M EDTA pH8 to a final concentration of 5 mM. The lysates were centrifuged at 16 000 g for 15 min. In parallel, input DNA was sonified (Branson Sonifier, Cell Disrubtor B15). Purification of DNA occurred by addition of proteinase K (500 µg/ml), RNAse (50 ng/ml) and SDS to a final concentration of 0.25% and incubation at 37°C for 6 h. Following phenol/chloroform extraction and isopropanol precipitation, purified DNA was resuspended in TE-buffer at pH7.5. PCR analysis was performed using 25 primer pairs as indicated in Figure 4. The PCR products had an average length of 100 bp and overlapped by about 15 to 20 bp. The PCR-program (30 cycles) was optimized and adjusted for each primer pair. PCR products were separated by agarose gel electrophoresis and quantified using a Chemi-Doc-Documentation-System (BioRad).
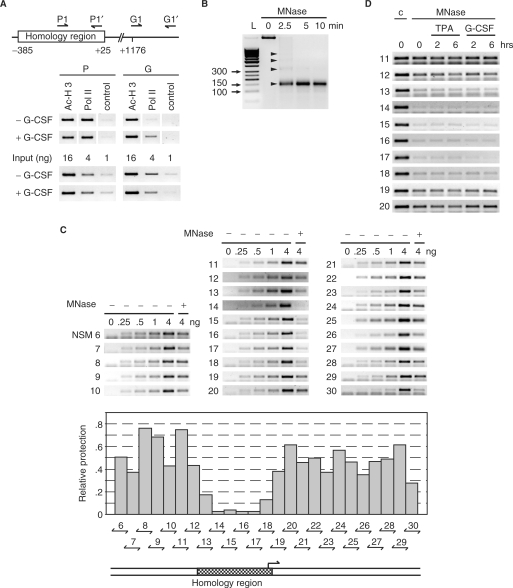
Figure 4.
The homology region of the MAD1 promoter possesses open chromatin. (A) Upper panel: schematic representation of the positions of the primers used for the ChIP experiments relative to the homology region. Lower panel: chromatin immunoprecipitations were performed with antibodies specific for the indicated proteins on lysates of U937 cells treated with or without G-CSF (10 ng/ml) for 30 min. For control serial dilutions of the input were analyzed. (B) U937 cells were lyzed in F-buffer, nuclei prepared and digested with micrococcal nuclease (MNase) for the indicated times. DNA was then extracted and analyzed by agarose gel electrophoresis. The arrowheads identify mono-, di-, tri-, and tetra-nucleosomes. L: molecular size markers. (C) MNase or untreated DNA from exponentially growing U937 cells was used in PCR analysis with the indicated primer pairs. Untreated DNA was titrated (0,.25,.5, 1, and 4 ng) for the control PCR reactions and compared to 4 ng of Mnase-treated DNA. The agarose gels with the PCR products are shown in the top panel. The middle panel shows a quantification of the signal obtained from the MNase-treated DNA compared to the control signals. The bottom panel shows the position of the homology region relative to the analyzed promoter region. (D) U937 cells were grown exponentially or treated for the indicated times with TPA or G-CSF as indicated. MNase or untreated DNA was amplified with primer pairs that span the homology region and neighboring genomic DNA.
The following primer sequences (forward, f; reverse, r) were used (see Figure 4 for location within the MAD1 promoter fragment):
6f: 5′-TACTGCAGCCCGATTCCAGC-3′;
6r: 5′-TTGAGAAGAAAAGCCGGCCG-3′
7f: 5′-TTCTTCTCAAGCCCCGCAGC-3′;
7r: 5′-TCCCGAAGAGCGGTGCAGG-3′
8f: 5′-AGCCTGCACCGCTCTTCGGG-3′;
8r: 5′-TGGTGGCCGCGGCCCTGG-3′
9f: 5′-TCCAGGGCCGCGGCCACC-3′;
9r: 5′-AATGGAAGGCTGGCACCTCG-3′
10f: 5′-AGGTGCCAGCCTTCCATTCG-3′;
10r: 5′-CCGGCCTTGCCTCGGACC-3′
11f: 5′-AGGTCCGAGGCAAGGCCG-3′;
11r: 5′-ATTGGCTGGGAGGGCCGG-3′
12f: 5′CGGCCCTCCCAGCCAATGC-3′;
12r: 5′-TTTCTCCAGGCCAGGCGAGC-3′
13f: 5′-TCGCCTGGCCTGGAGAAAGG-3′;
13r: 5′-AGAAGGCAAGCACCTCGCCG-3′
14f: 5′-GACCGGCGAGGTGCTTGC-3′;
14r: 5′-CCTTGTTTCCCATTGGTTCTATACC-3′
15f: 5′-GTTGTTGGTATAGAACCAATGGG-3′;
15r: 5′-GCTCCACCCCCTCTTTCC-3′
16f: 5′-TGGAAAGAGGGGGTGGAGC-3′;
16r: 5′-ACACTTGTGCGATTGGGAGAGG-3′
17f: 5′-AAGCCCTCTCCCAATCGCAC-3′;
17r: 5′-TCTCTTGACAGGCCAGCTTCG-3′
18f: 5′-AGCTGGCCTGTCAAGAGAAGG-3′;
18r: 5′-AGAGCAGCAGCCGCCACC-3′
19f: 5′-TGGCGGCTGCTGCTCTGC-3′;
19r: 5′-AACCACGCTCGACAAGAGAGG-3′;
20f: 5′-TCTCTTGTCGAGCGTGGTTGC-3′;
20r: 5′-AGCCCGCTATGGAGCCCGC-3′;
21f: 5′-AGCGGGCTCCATAGCGGGC-3′;
21r: 5′TGCACCGGGGGCCGGAGC-3′
22f: 5′-TCCCGTGGCTCCGGCCC-3′;
22r: 5′-CCGCCGCTCCAGATAGTCG-3′
23f: 5′-GACTATCTGGAGCGGCGGG-3′;
23r: 5′-TGAGGCCGGCCCCACAGG-3′;
24f: 5′-TGTGGGGCCGGCCTCAGG-3′;
24r: 5′-AGCCAGAGGGGTTGGGAGC-3′
25f: 5′-TCCCAACCCCTCTGGCTCTC-3′;
25r: 5′GCAGCCCCTTCTCCACCCC-3′;
26f: 5′-AATGAATGGGGTGGAGAAGGGG-3′;
26r: 5′-AGGCTATCGGAGGGGGC-3′;
27f: 5′GCCGGGGCCCCCTCCG-3′;
27r: 5′-AGACTGGGGCGCCGGGC-3′;
28f: 5′-CCCGGCGCCCCAGTCTCC-3′;
28r: 5′-ACCAGCTCAGGCGAAGGTGC-3′
29f: 5′-ACCTTCGCCTGAGCTGGTTC-3′;
29r: 5′-TGGCTAGGCTCACGCAGCC-3′;
30f: 5′-AGGCTGCGTGAGCCTAGCC-3′;
30b: 5′-AAATTGTGTGAATATCGTCAATTCGG-3′
RESULTS
Characterization of the MAD1 promoter
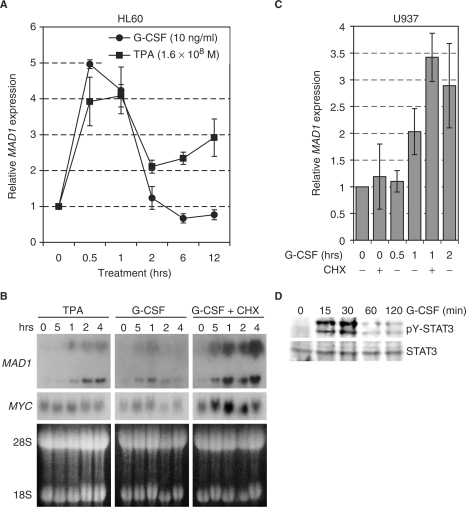
We observed previously that the MAD1 gene is activated in the human promyelocytic cell line HL60 upon treatment with G-CSF (32). We verified this activation in a time course experiment by performing qRT–PCR analysis. G-CSF induced MAD1 expression in HL60 cells to a similar extend and with similar kinetics as TPA (Figure 1A). Similarly, G-CSF induced MAD1 in the human promyelocytic line U937 as analyzed by northern blot (Figure 1B). The stimulation was insensitive to cycloheximide and was also observed by qRT–PCR (Figure 1B and C). These findings suggested that MAD1 expression is regulated by G-CSF independent of de novo protein synthesis. An important signal transducer of the G-CSFR is STAT3 that is activated in response to G-CSF. Indeed, in both U937 and HL60 cells phosphorylation of STAT3 at Tyr705 is enhanced by G-CSF (Figure 1D and data not shown).
Figure 1.
The MAD1 gene is regulated by G-CSF. (A) Exponentially growing HL60 promyelocytic cells were treated with the phorbol ester TPA (1.6 × 10–8 M) or recombinant G-CSF (10 ng/ml) for the indicated times. MAD1 mRNA was quantified by qRT–PCR with β-GUS as the internal standard. Mean values and standard deviations of three independent experiments are shown. (B) Exponentially growing U937 promyelocytic cells were treated as described in (A). Cycloheximide (25 µM) was added 15 min prior to treatment with G-CSF. Total RNA was extracted, 15 µg/lane separated on a formaldehyde–agarose gel and blotted. The hybridization was performed with probes specific for human MAD1 and MYC. The two distinct MAD1 mRNA species observed are roughly 3.8 and 6.0 kb long and are most likely the result of alternative splicing in exon 6, 3′ of the open reading fame, creating an additional non-coding exon 7. For loading control, the ethidium bromide-stained gel is shown with the 28S and 18S ribosomal RNAs indicated. (C) The experiments were performed as in (A) and (B). A qRT–PCR analysis of MAD1 mRNA is displayed. The mean values and standard deviations of three independent experiments performed in duplicates are shown. (D) U937 cells were stimulated with G-CSF (10 ng/ml) for the indicated times, total cell lysates generated in RIPA buffer and the samples immunoblotted for STAT3 (lower panel) and for Tyr705 phosphorylated STAT3 (upper panel).
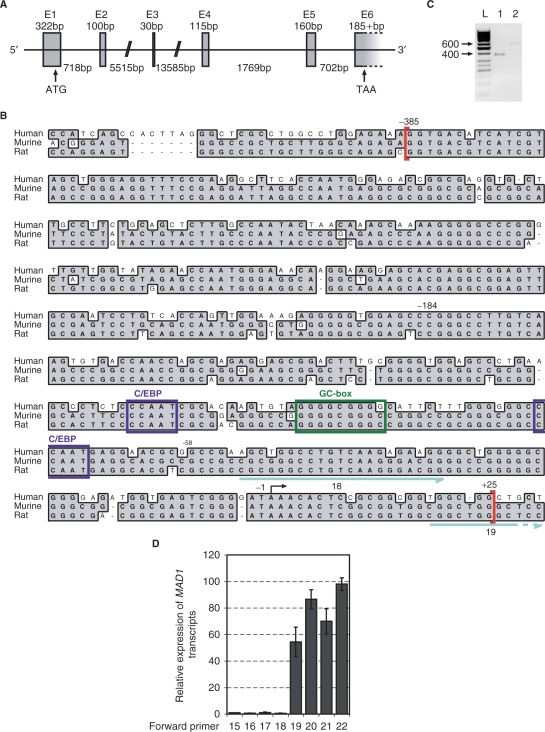
In order to study the regulation of MAD1 by G-CSF in more detail, we cloned the human MAD1 promoter. The MAD1 gene spans over 23 kbp and extends over 6 exons (alternatively 7 exons) on chromsome 2p13-p12 (Figure 2A, HTGS Acc. No. AC019206). Upstream of the most 5′ mRNA human MAD1 sequence, we identified a GC-rich region of about 400 bp that revealed a high degree of sequence homology to murine and rat mad1 (Figure 2B). In the following, we refer to this sequence as the homology region (Figure 3A). To determine whether the transcriptional start site(s) (TSS) is located within this region, we performed 5′-RACE experiments. Originally we cloned two products that indicated a major TSS at –250 and one at –46 relative to the translational initiation site (Figure 2C). The former is located within the homology region and was used subsequently for the numbering of the MAD1 promoter (Figure 2B). Additional 5′RACE analysis and comparison with the 5′-ends of MAD1 clones available in the data base suggested that multiple TSS are used, primarily downstream of the -250 site. This was further supported by the analysis of the primary 5′-RACE products. These were amplified with the Sp2 reverse primer and several forward primers using quantitative PCR (Figure 2D). A sharp increase in the amount of product was observed between primers 18 and 19, which flank the major TSS (Figure 2B and D). Primer 20, which is located further 3′, showed an increase compared to primer 19, while primer 22, which is close to the ATG of the TSS, revealed little additional product (Figure 2D). A primer located at the beginning of intron 1 was used for control but gave no signal (data not shown). Together, these findings are consistent with multiple TSS, comparable to what has been reported for other CpG-island promoters (the number of CpG dinucleotides is given in the legend to Figure 2) (51).
Figure 2.
Identification of the putative promoter region of MAD1. (A) The genomic organization of human MAD1 was deduced from comparing the cDNA to genomic clone HTGS Acc. No. AC019206. The ATG start codon is indicated in exon 1. E, exon sequences. (B) Upstream of the most 5′ cDNA sequences of human, rat and mouse MAD1, a genomic region of sequences homology is shown. This region encompasses nucleotides –385 to +25 relative to the start of transcription and is indicated with red brackets. The positions of nucleotides –184 and –58 are indicated, which delineate the region that is responsive to G-CSF/G-CSFR-mediated signals and contain two CCAAT- and a GC-box. The start site of transcription that was identified by 5′RACE is indicated by an arrow. The number of CpG dinucleotides within the homology region of the three genomic regions is: 23 out of 410 nt in human, 43 out of 405 in rat and 45 out of 405 in mouse. In addition, two of the primers (18 and 19, blue arrows) are indicated that were used to analyze 5'-RACE products (see panel D). (C) Total RNA was generated from G-CSF-treated U937 cells and reverse transcribed. 5′ RACE was performed and the main products were identified and sequenced. The results of two independent reactions are shown. Sequencing of the 600 bp product revealed the start site of transcription indicated in panel B. (D) The primary 5′-RACE products were amplified with the indicated forward primers and the Sp2 reverse primer by qPCR. The amount of products obtained with primer 15 was set as 1. Mean values and standard deviations of two independent experiments performed in duplicates are displayed.
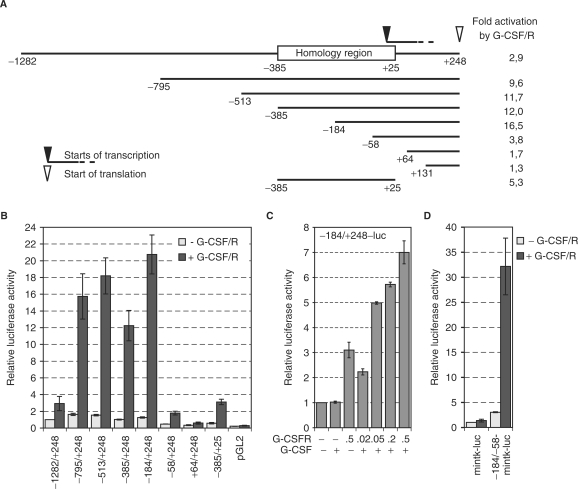
Figure 3.
The MAD1 promoter is activated by G-CSF. (A) Schematic representation of the MAD1 promoter fragments that were cloned into pGL2. The homology region that shows high sequence conservation between human, mouse and rat is indicated. Furthermore, the major start sites of transcription and the position of the ATG are shown. The fold activation in response to G-CSF/G-CSFR of each promoter construct is indicated. These numbers are derived from the data shown in (B). (B) RK13 cells were transiently transfected with plasmids expressing the G-CSFR (0.5 µg) and the indicated promoter-luciferase reporter gene construct (1 µg) and treated with G-CSF (G-CSF/R). Luciferase activities were standardized with co-expressed β-galactosidase. The mean values and standard deviations of three independent experiments performed in duplicates are displayed. (C) The experiments were performed as in panel B with the indicated reporter construct and increasing concentrations of the G-CSFR expressing plasmid. (D) The experimental design was as in panel B. The –184 to –58 fragment of the MAD1 promoter was cloned 5′ of the minimal thymidin kinase promoter (mintk)-luciferase reporter gene construct.
MAD1 reporter gene constructs are G-CSF/G-CSFR responsive
The functionality of the putative MAD1 promoter was addressed experimentally by cloning different portions into the pGL2 luciferase vector and analyzing the activities of these reporter gene constructs in RK13 cells (Figure 3A). We observed that constructs with at least the 3′ half of the homology region possessed promoter activity as compared to the pGL2 control (Figure 3B). Shorter constructs beginning at –58, +64 and +131 (the numbers define nucleotides relative to the 5′ start site of transcription) and the isolated homology region from –385 to +25 showed little basal activity (Figure 3B). Thus sequences within the homology and the 5′ untranslated region seem to be necessary for basal promoter activity (Figure 3B). These findings are in agreement with the mapping of the start of transcription. Next we determined the G-CSF-responsiveness of the different MAD1 promoter constructs. RK13 cells do not respond to human G-CSF and therefore human G-CSFR was co-expressed. This resulted in weak activation, probably due to dimerization and auto-activation (Figure 3C). The largest MAD1 promoter fragment (–1282 to + 248) was activated roughly 3-fold (Figure 3A and B). However, deletion of ∼500 bp (resulting in –795 to + 248) substantially enhanced the response to G-CSF/G-CSFR indicating that this distal promoter region contains negative regulatory elements. A high level of activation by G-CSF/G-CSFR was maintained with several MAD1 promoter fragments including the region from –184 to +248 (Figure 3). Deletion of a further 126 bp (in –58 to +248) substantially reduced and deletion beyond the start site of transcription abolished the G-CSF/G-CSFR response. A reporter gene construct with the isolated homology region of the MAD1 promoter showed an intermediate level of activity upon stimulation with G-CSF/G-CSFR (Figure 3A and B). The findings above suggested that the region from –184 to –58 within the homology domain contains the G-CSF/G-CSFR responsive elements. To verify this conclusion, the –184 to –58 fragments were cloned into a reporter construct with a minimal core promoter of the thymidine kinase gene (mintk-luc). This MAD1 promoter fragment was sufficient to confer G-CSF/G-CSFR responsiveness to the minimal tk promoter (–184/–58-mintk-luc) (Figure 3D). Together, these data define the MAD1 core promoter and a segment of the homology region that mediates the G-CSF/G-CSFR response.
The homology region shows distinct chromatin organization
To further evaluate the MAD1 promoter, we performed ChIP experiments. In the absence of G-CSF, the MAD1 promoter was occupied by RNA-polymerase II (Pol II) in U937 cells (Figure 4A). In contrast, no Pol II binding was seen within the gene body. This finding provided further evidence that the 3′ half of the homology region contains the core promoter. Upon G-CSF stimulation Pol II loading remained constant on the core promoter, but Pol II binding was now apparent further downstream (Figure 4A). This observation suggested that G-CSF induced MAD1 transcription by activating pre-assembled Pol II.
Since Pol II was occupying the MAD1 promoter prior to G-CSF stimulation, we analyzed the chromatin organization of this promoter. Nuclei of U937 cells were treated with micrococcal nuclease (MNase) to generate nucleosomal-sized DNA fragments (Figure 4B). This DNA was then analyzed by PCR using primers that amplified overlapping fragments of about 100 bp of the MAD1 promoter. The protection against MNase was poor over the core of the homology region spanning about 250 to 300 bp, while the neighboring portions of the promoter were well protected (Figure 4C). This suggested that the homology region contained either low number of nucleosomes or that the nucleosomes were poorly positioned. In ChIP experiments, acetylated histone H3 was associated with the homology region (Figure 4A). However, the poorly protected region is most likely too small to be separated clearly from neighboring promoter chromatin by ChIP. In summary, these findings demonstrate that the G-CSF-responsive region of the MAD1 promoter shows distinct chromatin organization. Next, we addressed whether G-CSF signaling affected chromatin organization at the MAD1 promoter. G-CSF did not appreciably affect the extent of histone H3 acetylation in U937 cells (Figure 4A). Furthermore, neither G-CSF nor TPA treatment changed the sensitivity of the MAD1 promoter to MNase (Figure 4D). From these experiments it appeared that the chromatin organization did not change grossly during stimulation of MAD1 expression.
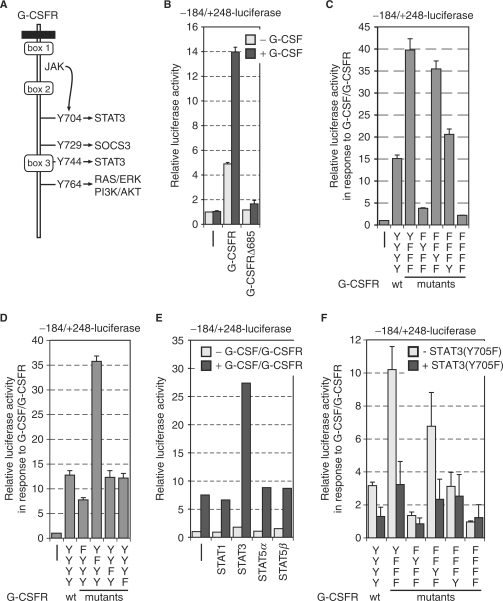
G-CSFR signal transduction pathways that mediate MAD1 activation
G-CSFR signaling is mediated by proteins recruited to the region proximal to the transmembrane domain and, upon phosphorylation, through four tyrosine residues (Y704, Y729, Y744 and Y764, summarized in Figure 5A) (35,36). To address the role of these elements for signaling to the MAD1 promoter, we used receptor deletion mutants. Loss of the distal region of the cytoplasmic portion of the G-CSFR in three different deletion mutants (52), including all four tyrosine residues, completely abolished stimulation of the –184 to +248 MAD1 promoter construct (Figure 5B). This suggested that the Tyr residues might be critical to mediate the signaling to the MAD1 promoter. Therefore, we used G-CSFR mutants, in which only a single tyrosine residue was maintained while the others were mutated to phenylalanine (41). We observed that when either only Y704 or only Y744 was present (lacking the SOCS3-binding sites that mediates repression), stimulation was more pronounced than with the wild-type (wt) G-CSFR (Figure 5C). These studies were complemented with G-CSFR mutants, in which individual tyrosines were mutated to phenylalanine (41). Indeed, the role of Y704, the major STAT3-binding site, was further substantiated. Mutation of this tyrosine in G-CSFR(Y704F) was sufficient to significantly reduce activation of the MAD1 promoter construct (Figure 5D). In contrast, mutation of Y744 had little consequence (Figure 5D), consistent with the finding that through this tyrosine STAT3 can be activated but to a lesser degree than through Y704 (41). These findings suggest that efficient activation of STAT3 is important to stimulate the MAD1 promoter.
Figure 5.
STAT3 is important to activate the MAD1 promoter. (A) Schematic representation of the cytoplasmic portion of the G-CSF receptor (G-CSFR). The conserved boxes 1–3 and the four tyrosine residues that are known to be phosphorylated in response to G-CSF are indicated. The signal transduction pathways that are connected to the individual tyrosine residues are given. (B) RK13 cells were cotransfected with the –184 to +248 MAD1 promoter luciferase reporter construct and expression plasmids for the indicated wild-type G-CSFR or deletion mutants. The cells were stimulated with G-CSF prior to harvesting and measuring luciferase and β-galactosidase activity. A typical experiment performed in triplicates is shown. (C and D) The experimental design was as in (B). G-CSFR mutants with the indicated changes of either individual tyrosine residues or combination of tyrosines were analyzed. (E) The transfections were performed as in panel B with the additional co-expression of different STAT factors as indicated. A typical experiment performed in duplicates is shown. (F) The experiment was done as in panel B. A dominant negative version of STAT3 [STAT3(Y705F)] was co-expressed as indicated.
A receptor mutant with only Y764 showed comparable activity to wt G-CSFR (Figure 5C). However, loss of this tyrosine in G-CSFR(Y764F) was not sufficient to reduce the stimulatory effect on the MAD1 promoter (Figure 5D), indicating that most likely alternative mechanisms exist to activate the MAPK and/or PI3K/AKT signal transduction pathways. In contrast to the other Tyr residues, when only Y729 was present substantially reduced MAD1 promoter stimulation was observed (Figure 5C), while G-CSFR(Y729F) was substantially more active (Figure 5D). Thus it appears that Y729, which recruits the suppressor of cytokine signaling (SOCS) 3, is mainly involved in signal repression, as suggested before, and demonstrated here by the strong effects on MAD1 promoter activity.
Y704 and Y744 are implicated in recruiting STAT3. This is supported by the analysis of the different G-CSFR mutants in combination with MAD1 promoter analysis. Indeed co-expression of STAT3, but not other STATs, was sufficient to enhance the G-CSF/G-CSFR response of the –184 to +248 MAD1 promoter reporter construct (Figure 5E). Furthermore, co-expression of a dominant negative mutant of STAT3 (STAT3(Y705F)) that cannot be phosphorylated on this tyrosine was sufficient to reduce the G-CSF/G-CSFR-dependent activation of the –184/+248-luc or of the –184/–58-mintk-luc reporter gene constructs (Figure 5F and data not shown). Similarly, the activities of G-CSFR mutants with only one Tyr residue, i.e. Y704 or Y744, were sensitive to STAT3(Y705F) (Figure 5F). The G-CSFR mutant with only Y764 was weakly sensitive to STAT3(Y705F), possibly a consequence of the reported tyrosine-independent activation of STAT3 by G-CSFR [41,53]. In summary, these observations support the notion that STAT3 is an important downstream mediator of the G-CSFR in the activation of the MAD1 promoter.
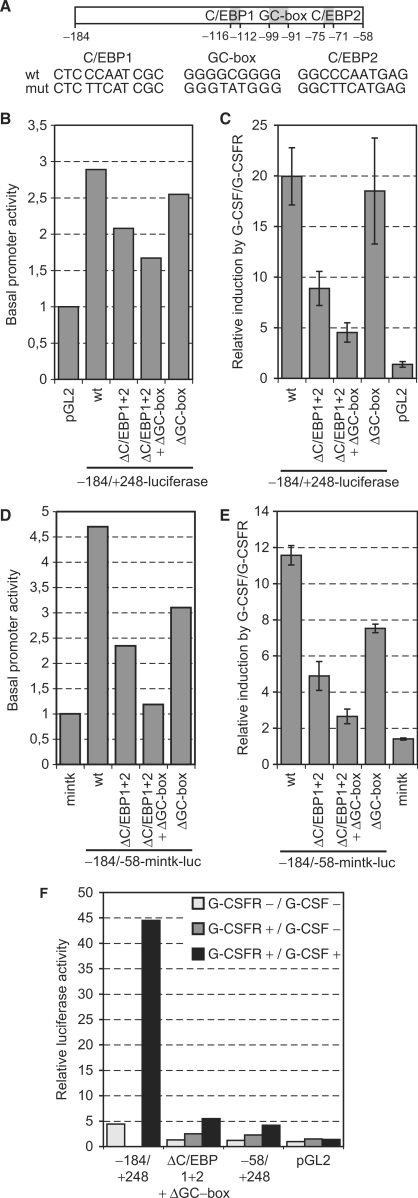
The C/EBP response elements are important to activate the MAD1 promoter in response to G-CSF/G-CSFR
Since Y704 of the G-CSFR was sufficient to mediate MAD1 promoter activity, STAT3 appeared to be important. However, inspection of the –184 to +248 MAD1 promoter sequence did not reveal any STAT consensus DNA-binding site. However, we noticed two CCAAT-boxes that can serve as binding sites for C/EBP factors (Figures 2B and 6A). These proteins are involved in regulating differentiation processes (54,55). In addition, a GC-box is present in the G-CSF/G-CSFR responsive region of the homology domain (Figures 2B and 6A). These elements are recognized by transcription factors of the SP family and are commonly found in CpG-island promoters, mediating basal activity in many instances (56,57). To address whether these sites might be important to mediate the G-CSF/G-CSFR effect, we mutated the CCAAT-boxes and the GC-box either individually or in combination (Figure 6A). These mutations reduced the basal promoter activity but affected the G-CSF responsiveness more severely (Figure 6B and C). While mutating the GC-box had little effect, the loss of the two CCAAT-boxes reduced substantially the response to G-CSF/G-CSFR of the –184 to +248 MAD1 promoter fragment (Figure 6C). When all three sites were mutated the promoter fragment was only poorly stimulated by G-CSF/G-CSFR. To determine whether these three sites were also relevant in combination with an unrelated core promoter, the mutants were introduced into the –184/–58-mintk-luc reporter. Similar to the findings with the MAD1 promoter, these mutations affected both basal as well as stimulated activity (Figure 6D and E). In particular, mutating the two CCAAT-boxes reduced the effect by G-CSF/G-CSFR (Figure 6E). In combination with the tk promoter, loss of the GC-box also affected G-CSF signaling. When all three sites were mutated little stimulation was observed (Figure 6E). To verify the findings described earlier, we compared the stimulation by G-CSF/G-CSFR of the –184 to +248 promoter fragment with the corresponding fragment mutated at the two CCAAT-boxes and at the GC-box. In addition, we included a MAD1 promoter fragment without the responsive region (–58 to +248) and the luciferase vector without a promoter. Mutation of the three sites resulted in a strong reduction in the responsiveness to G-CSF/G-CSFR with the remaining activity being comparable to the fragment without responsive region (Figure 6F). Both constructs still revealed a small but reproducible stimulation by G-CSF/G-CSFR (Figure 3). In summary, these findings suggested that the three conserved sites, the two CCAAT-boxes and to a lesser extent the GC-box, mediate a substantial part of the G-CSF/G-CSFR-response that controls the MAD1 promoter.
Figure 6.
C/EBP-binding sites and a GC-box mediate the G-CSF/G-CSFR signal. (A) Schematic representation of the –184 to –58 promoter region with the two CCAAT-boxes and the GC-box. The mutations that were generated in the three binding sites are shown. (B) RK13 cells were transfected with the indicated reporter gene constructs and the basal activity determined. The mean values of two experiments performed in duplicates are shown. (C) RK13 cells were cotransfected with the indicated reporter gene constructs and a plasmid encoding the G-CSFR. The stimulation by G-CSF/G-CSFR is shown on different –184 to +248 MAD1 promoter reporter gene constructs that are mutated in either the C/EBP-binding sites and/or in the GC-box. The mean values and standard deviations of three experiments performed in duplicates are displayed. (D) The experiments were performed as in panel B but with the –184 to –58 MAD1 promoter region cloned 5′ of the minimal tk promoter. (E) The experiments were as in panel C with the exception that the –184 to –58 region of the MAD1 promoter, and mutants thereof, were tested in combination with the mintk promoter. (F) The experimental design was as in (C). A typical experiment performed in duplicates is displayed.
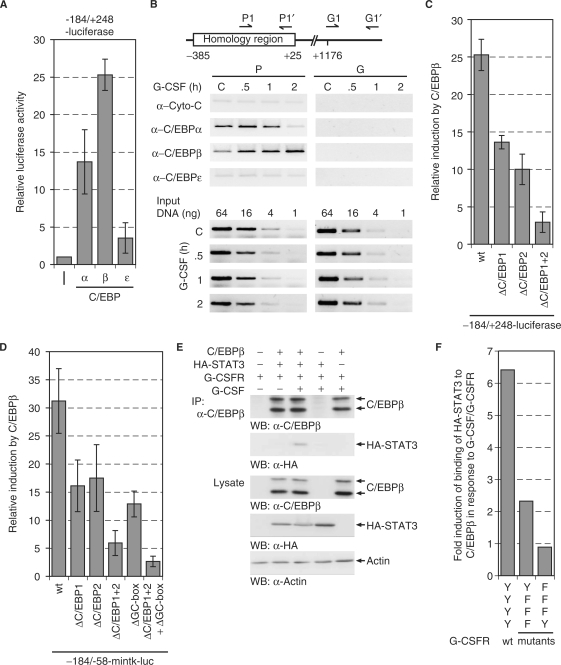
C/EBP transcription factors directly regulate the MAD1 promoter and interact with STAT3
To address the role of CCAAT-box binding factors, we analyzed the effect of different C/EBP proteins on MAD1 reporter genes. Of the three tested C/EBP proteins, C/EBPα and C/EBPβ activated the –184 to +248 MAD1 promoter fragment strongly, while C/EBPε was a poor stimulator (Figure 7A). This indicated that C/EBP factors might be involved in regulating the MAD1 promoter. Therefore, we tested whether these proteins could interact with the promoter in cells. U937 cells were stimulated with G-CSF, subsequently crosslinked and the binding of C/EBPα, β and all three are expressed in U937 cells (data not shown), to the MAD1 promoter was analyzed by ChIP. Near the core promoter, C/EBPα and C/EBPβ were constitutively bound while no interaction with C/EBPε could be seen. In response to G-CSF, a shift from preferentially C/EBPα to C/EBPβ was detected (Figure 7B). We further analyzed C/EBPβ and observed that stimulation of the MAD1 promoter was dependent on the two CCAAT-boxes with both elements contributing roughly equally to C/EBPβ-mediated activation (Figure 7C). Similarly in the context of the minimal tk promoter, both CCAAT-boxes were relevant and, in addition, also the GC-box (Figure 7D). Of note is that SP1 interacted with the MAD1 promoter by ChIP but the expression of wt or dominant negative SP1 had little effect on MAD1 reporter gene constructs (data not shown). Thus C/EBP transcription factors and their response elements are involved in regulating the MAD1 promoter.
Figure 7.
C/EBP proteins regulate the MAD1 promoter. (A) The –184 to + 248 MAD1 promoter reporter gene construct was cotransfected with plasmids expressing different C/EBP proteins. The mean values and standard deviations of three experiments performed in duplicates are shown. (B) U937 cells were stimulated for the indicated times with G-CSF. The cells were then crosslinked, lyzed and the indicated proteins immunoprecipitated. The associated DNA was analyzed by PCR using primers that correspond to the 3′ portion of the homology region or to a downstream fragment within the MAD1 gene body. For control serial dilutions of input DNA was analyzed by PCR. (C) Transient transfections were performed as described in (A). Reporter constructs with the indicated mutations were used as shown. (D) Transient transfections were performed as described (A) with –184 to –58-mintk promoter constructs and mutants thereof. (E) C/EBPβ, HA-STAT3 and G-CSFR were expressed in HEK293 cells. Prior to harvesting, the cells were treated with or without G-CSF as indicated. C/EBPβ was immunoprecipitated and the associated STAT3 detected by western blotting using an antibody specific for the HA-tag. The lysates show the expression controls. (F) The experimental approach was as in panel E. The relative STAT3 interaction with C/EBPβ upon stimulation with G-CSF and the indicated receptor mutants is displayed.
The results described earlier suggest that the G-CSF/G-CSFR response is mediated at least in part by STAT3. Since no obvious STAT3-binding sites could be detected, we evaluated whether C/EBPβ and STAT3 could interact. We expressed the two proteins in HEK293 cells in the presence of G-CSFR. The cells were then stimulated with G-CSF, low stringency lysates prepared and C/EBPβ immunoprecipitated. STAT3 was co-immunoprecipitated only when the cells were stimulated with G-CSF (Figure 7E). In addition, the signal dependency of the interaction of STAT3 with C/EBPβ was assessed by using G-CSFR mutants. With only Y704 present, which is responsible for STAT3 activation, the interaction of STAT3 with C/EBPβ was 3-fold weaker than with the wt receptor (Figure 7F). In contrast, a receptor with only Y764 did not stimulate the interaction between STAT3 and C/EBPβ. The interaction was weak indicating that it might be highly transient, providing an explanation for the lack of STAT3-binding sites in the MAD1 promoter. Thus, we propose that STAT3 is recruited by C/EBPβ to the MAD1 promoter.
DISCUSSION
We have identified the promoter of the MAD1 transcriptional repressor. Differential expression of MAD1 and the other small MAD genes and proteins has been well documented in different tissues and during development (7). However, relatively little is known about how these expression patterns are controlled. The analysis of the MAD1 promoter in U937 cells revealed an unexpected chromatin organization (Figure 4). The MNase analysis allowed us to identify a region of chromatin that was highly accessible to nuclease treatment. This region overlapped with the homology region, a promoter sequence that is highly conserved between human, rat and mouse and has the hallmarks of a CpG-island (56). What is common to all four MAD gene promoters, here shown for MAD1, is that they are TATA-less and contain GC-rich regions (58–60). SP transcription factors are frequently associated with GC-rich promoters, where they fulfill basal activities, but they can also be the target of signal transduction pathways (56,57). SP1 was described as a critical factor to control the expression of the MXI1 gene (58). SP1 was also found associated with the MAD1 promoter, as determined by ChIP experiments, and interacted with the GC-boxes in electrophoretic mobility shift experiments (data not shown). However, the manipulation of SP1 or SP3 activity did not have substantial effects on the MAD1 promoter reporter gene constructs used in this study (data not shown). These findings are consistent with a basal activity of the GC-boxes and associated SP factors (56,57). The promoter of the murine mad3 gene also contains potential binding sites for SP factors but no evidence for their involvement in mad3 expression has been found (59). Unlike the other MAD genes, mad3 is also expressed in some cycling cells, peaking in S-phase (21,61). E2F1, a member of the E2F family of cell cycle regulators (62), binds to the mad3 promoter and regulates its cell cycle-specific expression. The response element for E2F factors is also conserved in the human MAD3 gene, suggesting that its regulation is also controlled by E2F (59). The murine mad4 promoter is activated during differentiation. Here an initiator sequence element was found to be critical to repress mad4 in proliferating cells. This element is controlled by the MIZ1/MYC complex (60). MYC was previously identified as a repressing cofactor of MIZ1, providing a molecular explanation how MYC represses gene transcription (1). Together, these studies define various transcription factors and signaling pathways that control the expression of the four MAD genes. As expected from the overlapping but also distinct expression patterns observed, both common and distinct regulatory factors are associated with the different genes. The common factors could include SP proteins that might be relevant for the broad expression pattern of individual genes. In addition, specific factors might determine the expression in a cell-type and signal-specific fashion. E2Fs for mad3 and C/EBPs for MAD1 provide examples for the latter. Nevertheless, we assume that the GC-boxes are of functional relevance since their mutation reduces basal promoter activity and since they contribute to C/EBP-dependent gene activation (Figures 6 and 7).
Interestingly, Pol II was bound to the promoter constitutively, suggesting that in U937 the MAD1 promoter is in a pre-active state. In this state, the incoming G-CSF/G-CSFR signals rapidly increase promoter activity by stimulating STAT3 activity and most likely the RAS/RAF/ERK pathway that controls the activity of C/EBP transcription factors. The C/EBP proteins, similar to Pol II, were constitutively bound to the MAD1 promoter. Whether this is consistent with the presence of an enhanceosome (63), potentially with SP and C/EBP factors at its core, remains to be addressed.
The mutagenesis of the MAD1 promoter suggested strongly that the CCAAT-boxes are critical to mediate the G-CSF/G-CSFR response (Figures 6). C/EBP transcriptional regulators are involved in the regulation of proliferation and differentiation in many different cell types and function as executors of lineage commitment (54,55). In particular, C/EBP proteins have been demonstrated to be involved in the control of myeloid and granulocytic cell physiology. Four members of the family (α, β, δ and ε) are expressed in myeloid cells (54). C/EBPα is downregulated during terminal granulocytic differentiation (64), an observation that is in agreement with the loss of C/EBPα interaction to the MAD1 promoter in response to G-CSF (Figure 7). The importance of this C/EBP member is also documented by the observation that c/ebpα–/– mice fail to undergo myeloid differentiation and lack neutrophils (65). In addition, C/EBPε was suggested to be important for granulopoiesis since this factor is preferentially expressed during granulocytic differentiation (66,67). Although this factor is expressed in U937 cells (data not shown), we could not detect it on the MAD1 promoter and it was a poor activator of the MAD1 reporter genes (Figure 7). Similarly, C/EBPδ could not activate the MAD1 reporter genes (data not shown). The fourth C/EBP family member analyzed, C/EBPβ, was capable to activate the MAD1 reporter genes in a CCAAT-box-dependent manner and bound to the promoter in cells (Figure 7). C/EBPβ is not essential for myeloid development in the mouse (68). However, recent findings indicate that C/EBPβ-deficient neutrophils display enhanced apoptosis (69). A role of C/EBPβ in controlling MAD1 expression is in line with the anti-apoptotic function of this protein (20,24,26). Thus C/EBP factors, and in particular C/EBPβ, appear to be important to control different aspects of myeloid cell physiology. Since our ChIP experiments revealed that both C/EBPα and C/EBPβ are bound to the MAD1 promoter, it is possible that these proteins function as homo- or heterodimers in controlling promoter activity. We note however that the two CCAAT boxes represent only ‘half-sites’ and binding of C/EBPβ to these sites in electrophoretic mobility shift experiments is poor (data not shown). Thus it is possible that additional factors are required for efficient binding of C/EBP proteins to the proposed sites in the MAD1 promoter.
Nevertheless, the mutation of the CCAAT-boxes reduced substantially the responsiveness to G-CSF/G-CSFR; our findings suggest that C/EBP proteins are critical for the activation of the MAD1 promoter. Since STAT3 seems to be relevant as well, but no binding sites could be found in the responsive promoter fragment, and since we failed to detect STAT3 in ChIP experiments on the promoter, we suggest that STAT3 is recruited by C/EBP proteins to the promoter. Indeed, we observed co-immunoprecipitation of the two proteins when overexpressed and stimulated with G-CSF/G-CSFR (Figure 7). Probably owing to the weak interaction and possibly the transient nature of this complex, we were unable to detect this complex with endogenous proteins. This model is in agreement with a recent study that showed interaction of STAT3 with C/EBPα in response to G-CSF/G-CSFR (70). These authors find that a synthetic C/EBPα reporter is stimulated by STAT3. Thus, it appears that different C/EBP factors can interact with STAT factors and cooperate in gene activation. These findings suggest that STAT3 functions as cofactor in these circumstances. In the future the relevance of the activity of STAT3 in various settings, including tumor formation, will be important to investigate.
ACKNOWLEDGEMENTS
We thank G. Müller-Newen, I. Touw, and K. Welte for reagents, J. Baron and A. Greindl for help in the early phase of the project and L.-G. Larsson for many helpful discussions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 542 TB8) to B.L. Funding to pay the Open Access publication charges for this article was provided by the Deutsche Forschungsgemeinschaft (SFB542).
Conflict of interest statement. None declared.
REFERENCES
- 1.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell. Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 2.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 3.Luscher B. Function and regulation of the transcription factors of the Myc/Max/Mad network. Gene. 2001;277:1–14. doi: 10.1016/s0378-1119(01)00697-7. [DOI] [PubMed] [Google Scholar]
- 4.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 5.Luscher B, Larsson LG. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene. 1999;18:2955–2966. doi: 10.1038/sj.onc.1202750. [DOI] [PubMed] [Google Scholar]
- 6.Cole MD, Nikiforov MA. Transcriptional activation by the Myc oncoprotein. Curr. Top. Microbiol. Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- 7.Rottmann S, Luscher B. The mad side of the Max network: antagonizing the function of Myc and more. Curr. Top. Microbiol. Immunol. 2006;302:63–122. doi: 10.1007/3-540-32952-8_4. [DOI] [PubMed] [Google Scholar]
- 8.Cerni C, Bousset K, Seelos C, Burkhardt H, Henriksson M, Luscher B. Differential effects by Mad and Max on transformation by cellular and viral oncoproteins. Oncogene. 1995;11:587–596. [PubMed] [Google Scholar]
- 9.Cerni C, Skrzypek B, Popov N, Sasgary S, Schmidt G, Larsson LG, Luscher B, Henriksson M. Repression of in vivo growth of Myc/Ras transformed tumor cells by Mad1. Oncogene. 2002;21:447–459. doi: 10.1038/sj.onc.1205107. [DOI] [PubMed] [Google Scholar]
- 10.Hurlin PJ, Queva C, Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 11.Hurlin PJ, Queva C, Koskinen PJ, Steingrimsson E, Ayer DE, Copeland NG, Jenkins NA, Eisenman RN. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurlin PJ, Steingrimsson E, Copeland NG, Jenkins NA, Eisenman RN. Mga, a dual-specificity transcription factor that interacts with Max and contains a T-domain DNA-binding motif. EMBO J. 1999;18:7019–7028. doi: 10.1093/emboj/18.24.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koskinen PJ, Ayer DE, Eisenman RN. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- 14.Lahoz EG, Xu L, Schreiber-Agus N, DePinho RA. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc. Natl Acad. Sci. USA. 1994;91:5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi AI, DePinho RA. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 16.Vastrik I, Kaipainen A, Penttila TL, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J. Cell. Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiber-Agus N, Meng Y, Hoang T, Hou H, Jr, Chen K, Greenberg R, Cordon-Cardo C, Lee HW, DePinho RA. Role of Mxi1 in ageing organ systems and the regulation of normal and neoplastic growth. Nature. 1998;393:483–487. doi: 10.1038/31008. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Willingham T, Margraf LR, Schreiber-Agus N, DePinho RA, Nisen PD. Effects of the MYC oncogene antagonist, MAD, on proliferation, cell cycling and the malignant phenotype of human brain tumour cells. Nat. Med. 1995;1:638–643. doi: 10.1038/nm0795-638. [DOI] [PubMed] [Google Scholar]
- 19.Cultraro CM, Bino T, Segal S. Function of the c-Myc antagonist Mad1 during a molecular switch from proliferation to differentiation. Mol. Cell Biol. 1997;17:2353–2359. doi: 10.1128/mcb.17.5.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehring S, Rottmann S, Menkel AR, Mertsching J, Krippner-Heidenreich A, Luscher B. Inhibition of proliferation and apoptosis by the transcriptional repressor Mad1. Repression of Fas-induced caspase-8 activation. J. Biol. Chem. 2000;275:10413–10420. doi: 10.1074/jbc.275.14.10413. [DOI] [PubMed] [Google Scholar]
- 21.Pulverer B, Sommer A, McArthur GA, Eisenman RN, Luscher B. Analysis of Myc/Max/Mad network members in adipogenesis: inhibition of the proliferative burst and differentiation by ectopically expressed Mad1. J. Cell Physiol. 2000;183:399–410. doi: 10.1002/(SICI)1097-4652(200006)183:3<399::AID-JCP13>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Roussel MF, Ashmun RA, Sherr CJ, Eisenman RN, Ayer DE. Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol. Cell. Biol. 1996;16:2796–2801. doi: 10.1128/mcb.16.6.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sommer A, Hilfenhaus S, Menkel A, Kremmer E, Seiser C, Loidl P, Luscher B. Cell growth inhibition by the Mad/Max complex through recruitment of histone deacetylase activity. Curr. Biol. 1997;7:357–365. doi: 10.1016/s0960-9822(06)00183-7. [DOI] [PubMed] [Google Scholar]
- 24.Rottmann S, Speckgens S, Luscher-Firzlaff J, Luscher B. Inhibition of apoptosis by MAD1 is mediated by repression of the PTEN tumor suppressor gene. FASEB J. 2007 doi: 10.1096/fj.07-9627com. doi: 10.1096/fj.07-9627com. [DOI] [PubMed] [Google Scholar]
- 25.Queva C, McArthur GA, Ramos LS, Eisenman RN. Dwarfism and dysregulated proliferation in mice overexpressing the MYC antagonist MAD1. Cell Growth Differ. 1999;10:785–796. [PubMed] [Google Scholar]
- 26.Foley KP, McArthur GA, Queva C, Hurlin PJ, Soriano P, Eisenman RN. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayer DE, Eisenman RN. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 28.Ayer DE, Kretzner L, Eisenman RN. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 29.Larsson LG, Bahram F, Burkhardt H, Luscher B. Analysis of the DNA-binding activities of Myc/Max/Mad network complexes during induced differentiation of U-937 monoblasts and F9 teratocarcinoma cells. Oncogene. 1997;15:737–748. doi: 10.1038/sj.onc.1201390. [DOI] [PubMed] [Google Scholar]
- 30.Larsson LG, Pettersson M, Oberg F, Nilsson K, Luscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 31.Lymboussaki A, Kaipainen A, Hatva E, Vastrik I, Jeskanen L, Jalkanen M, Werner S, Stenback F, Alitalo R. Expression of Mad, an antagonist of Myc oncoprotein function, in differentiating keratinocytes during tumorigenesis of the skin. Br. J. Cancer. 1996;73:1347–1355. doi: 10.1038/bjc.1996.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sommer A, Bousset K, Kremmer E, Austen M, Luscher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- 33.Werner S, Beer HD, Mauch C, Luscher B. The Mad1 transcription factor is a novel target of activin and TGF-beta action in keratinocytes: possible role of Mad1 in wound repair and psoriasis. Oncogene. 2001;20:7494–7504. doi: 10.1038/sj.onc.1204937. [DOI] [PubMed] [Google Scholar]
- 34.Avalos BR. Molecular analysis of the granulocyte colony-stimulating factor receptor. Blood. 1996;88:761–777. [PubMed] [Google Scholar]
- 35.Touw IP, van de Geijn GJ. Granulocyte colony-stimulating factor and its receptor in normal myeloid cell development, leukemia and related blood cell disorders. Front. Biosci. 2007;12:800–815. doi: 10.2741/2103. [DOI] [PubMed] [Google Scholar]
- 36.van de Geijn GJ, Aarts LH, Erkeland SJ, Prasher JM, Touw IP. Granulocyte colony-stimulating factor and its receptor in normal hematopoietic cell development and myeloid disease. Rev. Physiol. Biochem. Pharmacol. 2003;149:53–71. doi: 10.1007/s10254-003-0014-0. [DOI] [PubMed] [Google Scholar]
- 37.de Koning JP, Soede-Bobok AA, Schelen AM, Smith L, van Leeuwen D, Santini V, Burgering BM, Bos JL, Lowenberg B, Touw IP. Proliferation signaling and activation of Shc, p21Ras, and Myc via tyrosine 764 of human granulocyte colony-stimulating factor receptor. Blood. 1998;91:1924–1933. [PubMed] [Google Scholar]
- 38.Hermans MH, van de Geijn GJ, Antonissen C, Gits J, van Leeuwen D, Ward AC, Touw IP. Signaling mechanisms coupled to tyrosines in the granulocyte colony-stimulating factor receptor orchestrate G-CSF-induced expansion of myeloid progenitor cells. Blood. 2003;101:2584–2590. doi: 10.1182/blood-2002-07-2062. [DOI] [PubMed] [Google Scholar]
- 39.Rausch O, Marshall CJ. Tyrosine 763 of the murine granulocyte colony-stimulating factor receptor mediates Ras-dependent activation of the JNK/SAPK mitogen-activated protein kinase pathway. Mol. Cell Biol. 1997;17:1170–1179. doi: 10.1128/mcb.17.3.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao H, Xu X, Jing N, Tweardy DJ. Unique structural determinants for Stat3 recruitment and activation by the granulocyte colony-stimulating factor receptor at phosphotyrosine ligands 704 and 744. J. Immunol. 2006;176:2933–2941. doi: 10.4049/jimmunol.176.5.2933. [DOI] [PubMed] [Google Scholar]
- 41.Ward AC, Hermans MH, Smith L, van Aesch YM, Schelen AM, Antonissen C, Touw IP. Tyrosine-dependent and -independent mechanisms of STAT3 activation by the human granulocyte colony-stimulating factor (G-CSF) receptor are differentially utilized depending on G-CSF concentration. Blood. 1999;93:113–124. [PubMed] [Google Scholar]
- 42.Iida S, Kohro T, Kodama T, Nagata S, Fukunaga R. Identification of CCR2, flotillin, and gp49B genes as new G-CSF targets during neutrophilic differentiation. J. Leukoc. Biol. 2005;78:481–490. doi: 10.1189/jlb.0904515. [DOI] [PubMed] [Google Scholar]
- 43.Liu XL, Yuan JY, Zhang JW, Zhang XH, Wang RX. Differential gene expression in human hematopoietic stem cells specified toward erythroid, megakaryocytic, and granulocytic lineage. J. Leukoc. Biol. 2007;82:986–1002. doi: 10.1189/jlb.0107014. [DOI] [PubMed] [Google Scholar]
- 44.Redell MS, Tsimelzon A, Hilsenbeck SG, Tweardy DJ. Conditional overexpression of Stat3{alpha} in differentiating myeloid cells results in neutrophil expansion and induces a distinct, antiapoptotic and pro-oncogenic gene expression pattern. J. Leukoc. Biol. 2007;82:975–985. doi: 10.1189/jlb.1206766. [DOI] [PubMed] [Google Scholar]
- 45.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 46.Irandoust MI, Aarts LH, Roovers O, Gits J, Erkeland SJ, Touw IP. Suppressor of cytokine signaling 3 controls lysosomal routing of G-CSF receptor. EMBO J. 2007;26:1782–1793. doi: 10.1038/sj.emboj.7601640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhuang D, Qiu Y, Haque SJ, Dong F. Tyrosine 729 of the G-CSF receptor controls the duration of receptor signaling: involvement of SOCS3 and SOCS1. J. Leukoc. Biol. 2005;78:1008–1015. doi: 10.1189/jlb.0105032. [DOI] [PubMed] [Google Scholar]
- 48.Luscher-Firzlaff JM, Westendorf JM, Zwicker J, Burkhardt H, Henriksson M, Muller R, Pirollet F, Luscher B. Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene. 1999;18:5620–5630. doi: 10.1038/sj.onc.1202967. [DOI] [PubMed] [Google Scholar]
- 49.Oelgeschlager M, Nuchprayoon I, Luscher B, Friedman AD. C/EBP, c-Myb, and PU.1 cooperate to regulate the neutrophil elastase promoter. Mol. Cell. Biol. 1996;16:4717–4725. doi: 10.1128/mcb.16.9.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Luscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 52.Herbst A, Koester M, Wirth D, Hauser H, Welte K. G-CSF receptor mutations in patients with severe congenital neutropenia do not abrogate Jak2 activation and stat1/stat3 translocation. Ann. NY. Acad. Sci. 1999;872:320–325. doi: 10.1111/j.1749-6632.1999.tb08476.x. discussion 325–327. [DOI] [PubMed] [Google Scholar]
- 53.Chakraborty A, Dyer KF, Cascio M, Mietzner TA, Tweardy DJ. Identification of a novel Stat3 recruitment and activation motif within the granulocyte colony-stimulating factor receptor. Blood. 1999;93:15–24. [PubMed] [Google Scholar]
- 54.Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene. 2002;21:3377–3390. doi: 10.1038/sj.onc.1205324. [DOI] [PubMed] [Google Scholar]
- 55.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell. Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 56.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol. Life Sci. 2003;60:1647–1658. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 58.Benson LQ, Coon MR, Krueger LM, Han GC, Sarnaik AA, Wechsler DS. Expression of MXI1, a Myc antagonist, is regulated by Sp1 and AP2. J. Biol. Chem. 1999;274:28794–28802. doi: 10.1074/jbc.274.40.28794. [DOI] [PubMed] [Google Scholar]
- 59.Fox EJ, Wright SC. The transcriptional repressor gene Mad3 is a novel target for regulation by E2F1. Biochem J. 2003;370:307–313. doi: 10.1042/BJ20021583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kime L, Wright SC. Mad4 is regulated by a transcriptional repressor complex that contains Miz-1 and c-Myc. Biochem. J. 2003;370:291–298. doi: 10.1042/BJ20021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fox EJ, Wright SC. S-phase-specific expression of the Mad3 gene in proliferating and differentiating cells. Biochem. J. 2001;359:361–367. doi: 10.1042/0264-6021:3590361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- 63.Merika M, Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 64.Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- 65.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl Acad. Sci. USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morosetti R, Park DJ, Chumakov AM, Grillier I, Shiohara M, Gombart AF, Nakamaki T, Weinberg K, Koeffler HP. A novel, myeloid transcription factor, C/EBP epsilon, is upregulated during granulocytic, but not monocytic, differentiation. Blood. 1997;90:2591–2600. [PubMed] [Google Scholar]
- 67.Yamanaka R, Kim GD, Radomska HS, Lekstrom-Himes J, Smith LT, Antonson P, Tenen DG, Xanthopoulos KG. CCAAT/enhancer binding protein epsilon is preferentially up-regulated during granulocytic differentiation and its functional versatility is determined by alternative use of promoters and differential splicing. Proc. Natl Acad. Sci. USA. 1997;94:6462–6467. doi: 10.1073/pnas.94.12.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akagi T, Saitoh T, O’Kelly J, Akira S, Gombart AF, Koeffler HP. Impaired response to GM-CSF and G-CSF, and enhanced apoptosis in C/EBP{beta}-deficient hematopoietic cells. Blood. 2007 doi: 10.1182/blood-2007-04-087213. doi: 10.1182/blood-2007-04-087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Numata A, Shimoda K, Kamezaki K, Haro T, Kakumitsu H, Shide K, Kato K, Miyamoto T, Yamashita Y, et al. Signal transducers and activators of transcription 3 augments the transcriptional activity of CCAAT/enhancer-binding protein alpha in granulocyte colony-stimulating factor signaling pathway. J. Biol. Chem. 2005;280:12621–12629. doi: 10.1074/jbc.M408442200. [DOI] [PubMed] [Google Scholar]