Abstract
Many sequence variations of the 8–17 RNA-cleaving deoxyribozyme have been isolated through in vitro selection. In an effort to understand how these sequence variations affect cleavage site selectivity, we systematically mutated the catalytic core of 8–17 and measured the cleavage activity of each mutant deoxyribozyme against all 16 possible chimeric (RNA/DNA) dinucleotide junctions. We observed sequence-function relationships that suggest how the following non-conserved positions in the catalytic core influence selectivity at the dinucleotide (5′ rN18-N1.1 3′) cleavage site: (i) positions 2.1 and 12 represent a primary determinant of the selectivity at the 3′ position (N1.1) of the cleavage site; (ii) positions 15 and 15.0 represent a primary determinant of the selectivity at the 5′ position (rN18) of the cleavage site and (iii) the sequence of the 3-bp intramolecular stem has relatively little influence on cleavage site selectivity. Furthermore, we report for the first time that 8–17 variants have the collective ability to cleave all dinucleotide junctions with rate enhancements of at least 1000-fold over background. Three optimal 8–17 variants, identified from ∼75 different sequences that were examined, can collectively cleave 10 of 16 junctions with useful rates of ≥0.1 min−1, and exhibit an overall hierarchy of reactivity towards groups of related junctions according to the order NG > NA > NC > NT.
INTRODUCTION
A small deoxyribozyme known as 8–17 catalyses the site-specific cleavage of RNA. This DNA enzyme has been the subject of much basic and applied research interest, due to several favourable characteristics including a simple secondary structure, relatively fast reaction rate and generalizable substrate recognition. The utility of the 8–17 deoxyribozyme has been demonstrated through a wide variety of applications including (i) the in vitro manipulation and analysis of RNA (1–3), (ii) fluorescent and colorimetric biosensors (4–6), (iii) DNA computational elements (7–9) and (iv) potential antiviral/gene control agents (10–12). Because of its simplicity, 8–17 has also been frequently employed as a model system in the demonstration of new concepts (13–21). Further investigation into 8–17's structure and function is expected to facilitate the development of new and/or improved applications.
Thus far, several studies have provided various insights into the kinetic and mechanistic details (22,23), metal-ion dependency (24,25), folding (26–29) and sequence requirements of the 8–17 deoxyribozyme (25,30). Aside from the potential practical benefits, basic research on 8–17 has also been motivated by its widespread recurrence. Since 1997, this catalytic motif has been isolated from five independent in vitro selection experiments (24,30–34), making it the most common solution to deoxyribozyme-mediated RNA-cleavage in sequence space. Its recurrence can be largely attributed to the fact that only four of the ∼15 nt that comprise its catalytic core are absolutely conserved, suggesting there may be thousands of active sequence variants. Understanding the functional significance of these many sequence variations represents a challenging, but important objective that has only recently begun to be addressed.
Peracchi and colleagues previously analysed the sequence requirements of the catalytic core to show how sequence variations influence the reaction rate (25). This study used a substrate that contains a 5′-AG-3′ dinucleotide junction as the target cleavage site, since 8–17 was originally selected to cleave this junction. However, Cruz et al. have recently demonstrated that sequence variations can also influence the selectivity of the cleavage site (34). In that study, 16 parallel in vitro selections were conducted, each using a substrate containing one of the 16 possible dinucleotide junctions as the target cleavage site. Ultimately, more than a 100 different 8–17 variants were isolated at various frequencies in nearly all of the selections. The cleavage versatility of five representative 8–17 variants was tested, and together these variants can cleave 14 out of 16 junctions with rate enhancements ranging from ∼103–107 fold over background. By comparison, the well-known 10–23 deoxyribozyme can reportedly cleave only four of the 16 junctions (33). These results prompted the hypothesis that 8–17 might actually have a latent capacity to cleave all 16 junctions proficiently, masked only by the right combination of nucleotides.
Herein, we wanted to investigate the functional limitations of 8–17 mediated catalysis. In particular, we sought to understand how the sequence of its catalytic core affects selectivity of the dinucleotide cleavage site. Towards this end, we conducted a systematic mutational analysis, and measured the effects against all 16 dinucleotide junctions. In addition to providing new insight into the structure and function of 8–17, we expect this study will also broaden its general utility.
MATERIALS AND METHODS
Oligonucleotides and reagents
Deoxyribozyme and substrate oligonucleotides were prepared by automated DNA synthesis using cyanoethylphosphoramidite chemistry (Integrated DNA Technologies; Mobix Central Facility, McMaster University). Oligonucleotides were purified by 10% preparative denaturing (8 M urea) polyacrylamide gel electrophoresis (PAGE) and their concentrations were determined by spectroscopic methods. [γ-32P]ATP was purchased from Amersham Pharmacia. T4 polynucleotide kinase (PNK) was purchased from MBI Fermentas. All chemical reagents were purchased from Sigma.
Kinetic methods
To assess the cleavage ability of more than 75 different mutants against 16 different substrates, simple assays based on single-timepoint measurements were preferred to complete kinetic characterizations. A similar strategy has been used previously to determine the sequence requirements in the catalytic core of the 10–23 deoxyribozyme (35). Each 8–17 mutant was separately incubated with each 5′-32P-labelled substrate in trans under single-turnover conditions (deoxyribozyme:substrate ∼800:1). Substrate and deoxyribozyme were heated together at 90°C for 30 s, and allowed to cool at room temperature for ∼10 min. A 2× reaction buffer was added to initiate the reaction giving a final concentration of 1.66 μM deoxyribozyme to 0.002 μM substrate. The reaction was terminated after a designated period of time by the addition of quenching buffer containing 60 mM EDTA, 7 M urea and loading dye solution.
All reactions were conducted at room temperature (∼23°C) for a designated period of time, using a reaction buffer (1 × concentration = 400 mM KCl, 100 mM NaCl, 7.5 mM MgCl2, 7.5 mM MnCl2 and 50 mM HEPES pH 7.0 at 23°C) previously demonstrated to support diverse 8–17 cleavage activity (34). Under these reaction conditions, the background rate of substrate cleavage was determined to be ∼10−7 min−1, based on both theoretical (36) and experimental determination (data not shown). We therefore chose reaction times ranging from 15 s to 72 h depending on the dinucleotide junction. These reaction times provided detectable but unsaturated cleavage yields by the wild type (WT) 8–17, such that mutants with lower or higher activity could be subsequently identified. The reaction rate was estimated using the equation kobs = −ln(1−fraction cleaved)/time. We assumed the endpoint of reactions to be ≥80% (although experimental observations often showed endpoints up to 95%). Therefore, any reactions that exceeded 75% cleavage product at an initial timepoint were subsequently assayed with a shorter timepoint(s) to ensure accuracy. When necessary, slower reactions were followed for at least 72 h (and up to 6 days when complete timecourses were conducted). If a distinct cleavage signal was not observed in this period of time, the rate was assumed to be the background rate of ∼10−7 min−1.
All reaction rates for 8–17NG, 8–17NA and 8–17NC were also independently determined with complete timecourses (data fit to a single-exponential equation), and the variation between the two methods was generally within 30%, suggesting that our simplified assay method provided a very good approximation of the rate. In general, rate constants calculated from either the simple assay or full timecourse were determined from at least two independent measurements that typically differed by <30%. Negative control reactions were conducted with each experiment, in which each substrate was incubated with reaction buffer in the absence of the deoxyribozyme for the same period of time. Any cleavage products detected in the negative controls were subtracted from the results of the deoxyribozyme reactions prior to analysis. This background cleavage was typically <1%.
RESULTS AND DISCUSSION
8–17 sequence and secondary structure
Figure 1 illustrates the sequence and secondary structure of an 8–17 motif, which is typically characterized by a 3-bp stem-triloop and a single-stranded turn region of 4–5 nt. Two binding arms engage the substrate through Watson–Crick base pairs on either side of the cleavage site. Deoxyribozyme 8–17 does not have any reported sequence requirements in the substrate, other than the identity of the dinucleotide cleavage junction. The catalytic core is composed of 14–15 nt, of which only 4 nt (i.e. A6, G7, C13 and G14) have been shown to be absolutely conserved and essential for activity (25,34). The sequence requirements may be even more flexible, as Cruz et al. have previously isolated 8–17 variants that contained one or two mismatches in the stem, insertions, deletions and combinations thereof (34). However, the activities of these sequence variants have not been confirmed. For simplicity, we use the term 8–17 generically to describe both the canonical motif first described in 1997 (33), and all related sequence variants.
Figure 1.
Wild type (WT) 8–17 sequence and secondary structure. This 8–17 sequence variant was used as a reference, against which the activity of each mutant was subsequently compared. Deoxyribozyme nucleotides in black. Substrate nucleotides in gray. Arrow denotes dinucleotide cleavage junction. rN18 = G, A, C or U ribonucleotide. N1.1 = G, A, C or T deoxyribonucleotide. Underlined nucleotides are absolutely conserved.
In an effort to understand how these sequence variations influence 8–17's cleavage site selectivity, we comprehensively mutated the catalytic core and analysed the effects against each of the 16 possible dinucleotide junctions. In addition to systematically replacing each position with the other three nucleotides, we also assayed the effects of various deletions, insertions and combination mutations. The 8–17 variant depicted in Figure 1 was chosen as the WT sequence, against which the activity of each mutant was subsequently compared. This particular 8–17 sequence was the fastest variant reported in a previous study by our laboratory for cleavage of 5′-GG junctions (37). Sixteen chimeric substrates (i.e. composed entirely of DNA except for a single-embedded scissile ribonucleotide) were designed with the sequence shown in Figure 1, differing only in the identity of the two nucleotides at the target cleavage site (denoted as positions rN18-N1.1 in Figure 1). Therefore, any respective difference in cleavage reactivity could be attributed exclusively to the identity of this dinucleotide junction. The nucleotide numbering system used in Figure 1 is analogous to the system originally proposed for the hammerhead ribozyme (38), and subsequently adopted for the 8–17 deoxyribozyme (25). For future reference, all dinucleotide junctions are written in the 5′–3′ direction, and N = G, A, C or T/U (deoxy)ribonucleotides.
One 8–17 sequence can cleave all 16 dinucleotide junctions
The canonical 8–17 deoxyribozyme was first reported to cleave AG junctions (33), and a sequence variant was subsequently shown to have the ability to cleave all four NG junctions (23). Most recently, our laboratory extended the list of known cleavage sites to include as many as 14 different dinucleotide junctions, based on a set of five different 8–17 sequence variants (34). Only the CT and UT junctions could not be cleaved by these 8–17 variants. However, for practical reasons these conclusions were based on a minimum observed reaction rate of ∼0.0001 min−1, which corresponds to a ∼1000-fold enhancement over the background rate of ∼10−7 min−1. In the present study, we reasoned it would be instructive to determine if any detectable level of catalysis were possible. Even a 10-fold rate enhancement could provide better insight into understanding the functional limitations and potential of 8–17.
The cleavage site versatility of the WT 8–17 sequence is illustrated in Figure 2. Remarkably, all 16 dinucleotide junctions were in fact susceptible to cleavage. Rate enhancements varied by several orders of magnitude, from as low as ∼40-fold (UT junction) to as high as ∼5 × 107 fold (GG junction) over background. As a group, the NG dinucleotide junctions were the most susceptible to cleavage, with rates ranging from ∼0.1 to 5 min−1. The remaining NA, NC and NT junctions were at least two orders of magnitude less reactive, with rates that did not exceed ∼0.001 min−1. Within each group of dinucleotide junctions defined by the same 3′-nt (i.e. the NG group, NA group, NC group and NT group), the reactivity consistently followed the order N = G > A > C > U.
Figure 2.
Cleavage versatility of the WT 8–17 sequence. (A) Autoradiogram demonstrating cleavage of all 16 dinucleotide junctions. 5′-32P-labelled substrate was incubated with (+) or without (−) WT for a designated period of time. Reaction buffer: 400 mM KCl, 100 mM NaCl, 7.5 mM MgCl2, 7.5 mM MnCl2 and 50 mM HEPES pH 7.0 at 23°C. Cleavage products were separated from uncleaved substrate by 10% denaturing PAGE. Dinucleotide junctions are organized into related groups, and written 5′-3′ as rN18N1.1. (B) Histogram of rate constants versus dinucleotide junction. kobs values represent the average of at least two independent trials, which typically differed by <30%. Background rate of cleavage under reaction conditions is ∼10−7 min−1.
Positions 2.1 and 12 influence selectivity at the 3′ nucleotide (N1.1) of the cleavage site
The relative activity of WT 8–17 variants mutated at positions 2.1 and 12 are summarized in Figure 3, along with all other positions excluding the 3-bp intramolecular stem. For clarity and comparison, the data has been organized into eight different groups of related dinucleotide junctions defined by either a common 3′-nt (i.e. the NG group, NA group, NC group and NT group) or common 5′-nt (i.e. the GN group, AN group, CN group and UN group). Mutations at position 2.1 had the same effect against junctions with the same 3′-nt, as reflected by the near superimposition of the relevant graphs (Figure 3, left panel of graphs). However, these mutations clearly exhibited different effects against junctions with a different 3′-nt (Figure 3, right panel of graphs), both in terms of the direction of the effect (i.e. increase or decrease in activity) and the magnitude. These observations suggest that position 2.1 influences the selectivity through the 3′ position N1.1 of the dinucleotide cleavage site. Nucleotide T2.1 has the greatest functional significance for the NG group of dinucleotide junctions. Substitution with one of the three other nucleotides impairs catalysis by ∼10–103 fold. Furthermore, deletion of T2.1 reduces the reaction rate by ∼104 fold, to a level approximately at par with the WT activity exhibited against the other junctions. In contrast, substitution or deletion of T2.1 has only a modest effect (i.e. decrease or increase of <10-fold) on the cleavage of NA, NC and NT junctions. These results indicate that mutation of position 2.1 is required, but not sufficient to confer a significant change in the cleavage site selectivity.
Figure 3.
Relative rate of various WT 8–17 mutants against 16 dinucleotide junctions. Designated nucleotide positions of the WT sequence were individually substituted with each of the other three nucleotides or deleted as indicated. krel is defined as kobs, mutant/kobs, WT. The kobs, WT values are provided in Figure 2B. Values represent the average of at least two independent trials, which typically differed by <30%. For clarity, dinucleotide junctions are organized into groups of related junctions. Deletions are denoted by a triangle (Δ). Position ‘15(Δ15.0)’ denotes a group of combination mutants in which position 15.0 was deleted, while position 15 was either substituted with one of the other three nucleotides or deleted. It should be noted that mutations at position 7 and 14 reduce activity to background levels for most junctions, and the magnitude of this decrease will differ between junctions depending on their respective WT cleavage rates.
Unlike position 2.1, substitutions at position 12 showed a similar pattern of effects against all dinucleotide junctions, initially suggesting that this position does not contribute to cleavage site selectivity. The magnitude of these effects was <10-fold. However, the deletion of T12 had a significant impact on the cleavage of NG junctions, reducing the reaction rate by ∼103–104 fold. Deletion of T12 caused <10-fold change in activity against all other junctions. Therefore, position 12 likely plays some role in cleavage site selectivity. Interestingly, position 12 has also previously been implicated as functionally important for the metal-ion selectivity of 8–17 (25).
Positions 15 and 15.0 influence selectivity at the 5′ nucleotide (rN18) of the cleavage site
Mutations at positions 15 and 15.0 exhibited the same effects against junctions with the same 5′-nt, as reflected by near superimposition of the graphs at these positions in the right panel of Figure 3. However, different effects were observed against junctions with a different 5′-nt, suggesting that position 15 and 15.0 influence selectivity through the 5′ position of the dinucleotide cleavage junction. Substitutions at position 15 and 15.0 generally exhibited moderate effects (<100-fold). Deletion of A15.0 caused a modest decrease in activity (<10-fold). Deletion of both A15 and A15.0 simultaneously, also caused a relatively modest decrease in activity for most junctions (<50-fold), but up to ∼1000-fold decrease for some of the CN junctions. Double mutations, in which position 15 was substituted with another nucleotide while position 15.0 was deleted, showed deleterious effects as high as ∼1000-fold for some junctions.
Though not officially part of the catalytic core of 8–17, we decided to evaluate the functional significance of position 16.1 given its proximity to the cleavage site. A previous study conducted on the 10–23 RNA-cleaving deoxyribozyme, which engages its substrate in an analogous manner, suggested that this position may benefit from the conformational freedom afforded by a mismatch (39). However, mutations to this position in 8–17 exhibited either neutral or deleterious effects (up to ∼100-fold) on the reaction rate.
Positions 6–8, 13 and 14 show no influence on cleavage site selectivity
Mutations at positions 6–8, 13 and 14 exhibited similar effects against all 16 dinucleotide junctions (Figure 3), which argues against their involvement in conferring selectivity. This result is logical, since these positions are very highly conserved (with the exception of position 8), and therefore probably form the basis of the catalytic strategies employed by 8–17.
Mutations at position 6 impaired catalysis by 10–104 fold, with the largest reduction observed with an A6T substitution. G7 was the most crucial nucleotide in the loop region. Substitution of G7 generally reduced the reaction rate to background levels. Position C8 was the most tolerable to mutation, with substitutions causing changes <50-fold. In agreement with prior observations (25), a C8T substitution was the most tolerable, and in some cases even led to a very modest increase in the reaction rate. As expected, C13 and G14 were crucial for activity. Substitution at these positions generally reduced the activity to background or near background levels for all junctions.
Mutations to the core stem show little influence on selectivity
The functional consequences of mutations to the 3-bp stem region were also addressed. Nucleotide positions 3, 4 and 5 were individually substituted with each of the other three nucleotides, and positions 11, 10 and 9 respectively, were co-mutated to maintain base pairing. The results of these substitutions as well as several combination mutants are illustrated in Figure 4. In general, substitutions to the stem region yielded a similar pattern of effects against all dinucleotide junctions, although the magnitude of the effect differed between junctions (typically <10-fold, but up to ∼100-fold in some cases), and tended to be more comparable for junctions with the same 3′-nt. The G3C4C5 and G3T4C5 stem sequence combinations provided some modest improvements in the rate from ∼2-fold for the NG, NA and NT junctions, to as high as ∼10-fold for the NC junctions. The largest deleterious effect was seen with the C5G substitution, which led to ∼100-fold decrease in activity against GC, AC, CC, GT and AT junctions. These differential effects tentatively suggest that the sequence of the stem may play some small role in the cleavage site selectivity of 8–17. It should also be noted that the stability of the stem was not directly correlated with the catalytic activity, as there were several examples of stem mutants exhibiting higher activity with fewer GC pairs.
Figure 4.

Relative rate of various WT 8–17 stem mutants against 16 dinucleotide junctions. Nucleotide positions 3, 4 and 5 representing half of the intramolecular stem of WT 8–17, were individually substituted with each of the other three nucleotides. Nucleotide positions 11, 10 and 9 were co-mutated with positions 3, 4 and 5 respectively, to maintain base pairing. krel is defined as kobs, mutant/kobs, WT. The kobs, WT values are provided in Figure 2B. Combination mutants are denoted by the sequence of positions 3, 4 and 5 in that order. For clarity, dinucleotide junctions are organized into groups of related junctions.
Single-nucleotide insertions/bulges show little influence on selectivity
Single-nucleotide insertions or bulges have been observed at various positions among in vitro selected 8–17 variants (34). An insertion of thymidine between positions 8 and 9 has been most frequently observed, followed by bulges (typically thymidine) between positions 9 and 12. The functional consequences of such mutations were investigated by systematically inserting thymidine residues between positions 3 and 13, with the results presented in Figure 5. Single-nucleotide T insertions generally exhibited the same pattern of effects among all dinucleotide junctions, suggesting little or no influence on the cleavage site selectivity. The reaction rate progressively decreased between positions 3–6 (by up to ∼104 fold), then progressively increased up to the WT activity levels by position 9 or 10, before once again declining by ∼10-fold at position 13.
Figure 5.
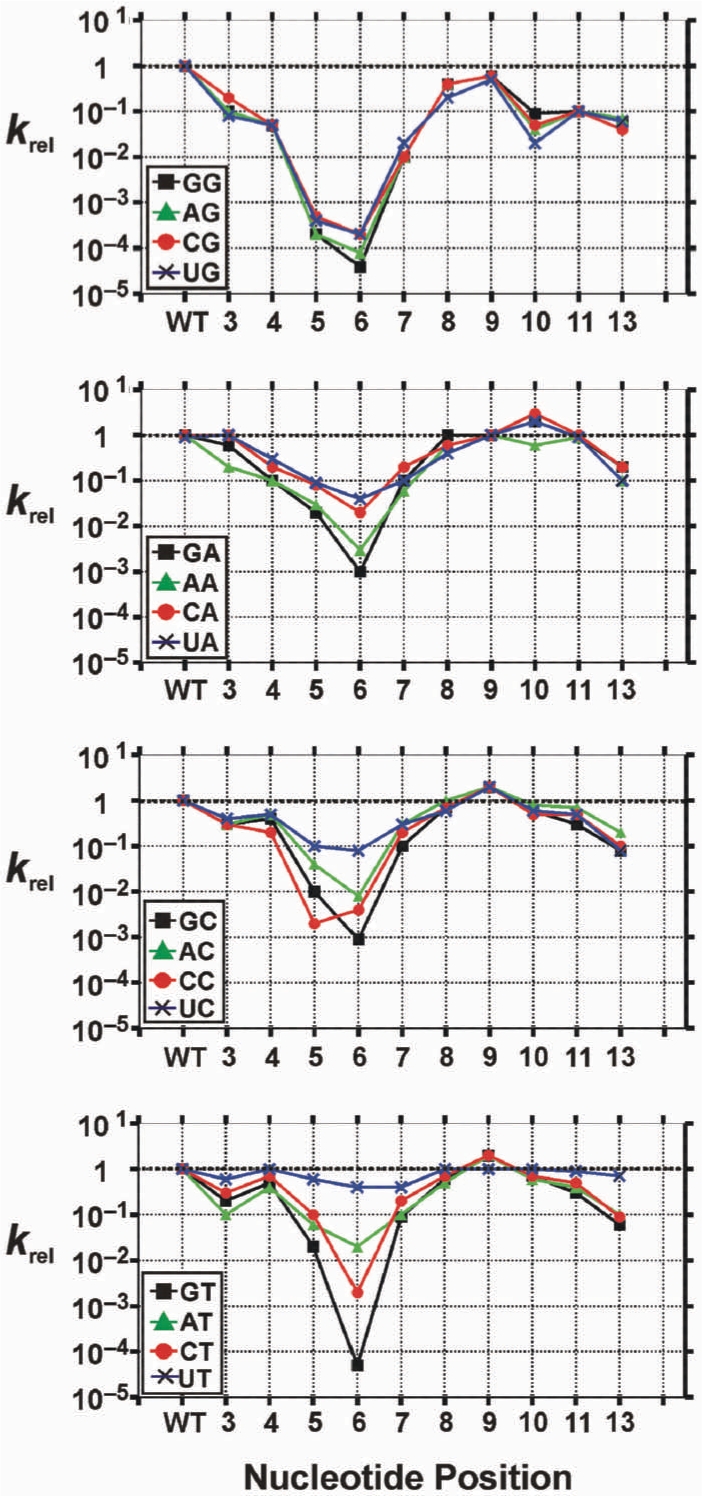
Relative rate of single-nt insertion mutants against 16 dinucleotide junctions. Thymidine nucleotide was systematically inserted along positions 3–13 of the WT 8–17 sequence. An insertion at position 3 should be considered as being located between positions 3 and 4, etc. krel is defined as kobs, mutant/kobs, WT. kobs, WT values are provided in Figure 2B. For clarity, dinucleotide junctions are organized into groups of related junctions. A T-bulge at position 12 is equivalent to a T-bulge at position 11, and therefore is not shown.
Cleavage site selectivity versus mutational distance
Although the WT sequence used in this study had the ability to cleave all 16 dinucleotide junctions, it showed a marked selectivity for the group of NG junctions. Cleavage of these four junctions proceeded ∼102–106 fold faster than the 12 remaining junctions. During the systematic mutation of the WT sequence, we did not observe any significant changes to this selectivity, despite testing more than 60 mutants within 1–2 mutations of the WT sequence. Nevertheless, previous data indicates that large changes in selectivity are accessible within a relatively short mutational distance. For instance, Cruz et al. described an 8–17 variant called E5112 (sequence 5′–3′: -2.1 GTC AGC8T8 GAC TCGAA) that differed by only two mutations from our G3T4C5 stem mutant (sequence 5′–3′: T2.1 GTC AGC8 GAC TCGAA), but showed at least 1000-fold selectivity for the group of NA junctions versus NG. This large change in selectivity is somewhat unexpected since one of the mutations (i.e. an insertion of T at position 8), is not expected to confer selectivity based on the results from our systematic analysis. The insertion of a T residue was shown to have similar effects against all dinucleotide junctions (Figure 5). This discrepancy illustrates an important point. The observed functional significance of a given mutation may change depending on the context of the surrounding sequence. Thus, the right combination of mutations could act cooperatively to elicit a large change in cleavage site selectivity.
One inherent limitation of this study is the potential bias associated with our choice of starting WT sequence. We therefore wondered if the conclusions from our mutational analysis would still be relevant, even as the mutational distance from the WT sequence increased. To provide some additional insight into this issue, we decided to look for faster NA, NC and NT-cleaving 8–17 variants. Since the WT sequence appeared to be already optimized for the cleavage of NG junctions (as reflected by the fact that nearly all point mutations were deleterious), we reasoned that more proficient cleavage of the other 12 junctions might require a larger mutational distance to escape the specialized fold of the WT sequence. This hypothesis is supported by the results of a study conducted by Bartel and colleagues, in which they demonstrated that the probability of deriving new function from an existing ribozyme increases with the mutational distance (40). In addition, Cruz et al. previously isolated many 8–17 sequences that contained various combinations of deletions, insertions and stem mismatches (34). The fact that these unusual sequence variants survived selection, suggests they may be functionally important. Fourteen such candidate sequences were chosen from pools of sequences that had been selected to cleave one of the less reactive NA, NC and/or NT dinucleotide junctions (34). Several of these sequences were very slow, or interestingly, exhibited higher activity against junctions for which they were not originally selected to cleave (data not shown). However, we did identify two sequences that provided ∼4–40-fold higher activities against NA and NC junctions than the best sequences reported by Cruz et al. under the same buffer conditions. These two sequences, denoted as 8–17NA and 8–17NC, respectively, differed from the WT sequence by nine mutations. Figure 6A shows the sequence of 8–17NA, 8–17NC and the sequence of one of the fastest NG-cleaving 8–17 variants (denoted as 8–17NG) that were identified during our systematic mutational analysis. 8–17NG also showed the highest activity against the group of NT junctions. However, it should be noted that the double mutant A15T/ΔA15.0, actually provided the largest rate enhancement for the UT junction (∼1000-fold over background). The rate constants for 8–17NG, 8–17NA and 8–17NC are presented in Figure 6B against each dinucleotide junction.
Figure 6.
Sequence and kinetic characterization of optimal 8–17 variants. (A) The sequences of three optimal 8–17 variants identified by systematic mutation (8–17NG) or random screening of previous in vitro selected deoxyribozymes (8–17NA and 8–17NC). Secondary structures are hypothetical. (B) Histogram of rate constants versus dinucleotide junctions. Dinucleotide junctions are organized into related groups, and written 5′-3′ as rN18N1.1. kobs values represent the average of at least two independent trials, which typically differed by <30%. Reaction endpoints ranged from 80–95%, with the following exceptions: 8–17NA versus AA = 58%, 8–17NC versus AA = 71%. Background rate of cleavage under reaction conditions is ∼10−7 min−1.
The selectivity of 8–17NA followed the approximate scheme NA > NC > NT ∼NG. The decrease in activity against NG junctions (compared to 8–17NG), is expected with the deletion of T2.1. Within each of these groups, the selectivity at the 5′-nt followed the order G > A > U > C, rather than the G > A > C > U order exhibited by 8–17NG. This change is consistent with the observed A15.0G substitution, since position 15.0 is expected to influence the selectivity at the 5′-nt of the cleavage site. Similarly, 8–17NC also showed reduced activity against NG junctions (selectivity = NC > NA > NT ∼NG), which can be reconciled by the deletion of T2.1 and/or T12. Positions 15 and 15.0 of 8–17NC were identical to 8–17NG, and therefore the same selectivity was observed at substrate position N18 in groups of junctions defined by the same 3′-nt (such as GG > AG > CG > UG, GT > AT > CT > UT and so on). The G3A4T5 stem utilized by 8–17NA is expected to modestly decrease the activity against NG and NT junctions, while having a relatively neutral effect for NA junctions and slightly beneficial effect for NC junctions. 8–17NC also utilized a G3A4T5 stem, but had a G–A mismatch between positions 3 and 11. The functional significance of this mismatch is still unclear, but it does reaffirm the notion that a stable stem is not an absolute prerequisite for robust activity (at least against NC junctions). 8–17NA and 8–17NC had single-nucleotide bulges/insertions at positions 10 and 8, respectively. These mutations are expected to cause only a modest decrease in the reactivity against NG junctions, and slightly increase the activity (by ∼2-fold) against NA, NC and NT junctions.
In general, the observed selectivity and sequence variations in 8–17NA and 8–17NC are consistent with the functional significance assigned to specific positions during our systematic analysis of the catalytic core. This provides some measure of confidence that the conclusions from this study are not artefacts of the chosen WT sequence. However, additional studies will be necessary to fully understand the interactions between different nucleotide positions and their functional consequences. High-resolution structural studies, including NMR spectroscopy and/or X-ray crystallography, would be particularly useful, and could benefit in turn from the many sequence variants characterized herein.
Prospects for faster 8–17 variants
Herein we have shown that the 8–17 deoxyribozyme has the ability to cleave all dinucleotide junctions, but exhibits a hierarchy of reactivity against groups of dinucleotide junctions according to the order NG > NA > NC > NT. This order was based on the fastest 8–17 variants (i.e. 8–17NG, 8–17NA and 8–17NC) that could be identified from a sample of ∼75 different sequence variants, representing some of the most commonly observed mutations. In theory, however, there are still thousands of additional sequence permutations that conform to the sequence and secondary structure requirements of the 8–17 motif. Thus, it remains to be determined if this hierarchy reflects the true functional limitations of 8–17, or alternatively, a possible sampling artefact. The in vitro selection results reported by Cruz et al. provides evidence against the latter possibility, because the observed hierarchy of reactivity closely parallels the relative frequency with which 8–17 variants were isolated against each of the various junctions (34). Nevertheless, these selection results are not sufficient to completely discount the prospect of faster sequence variants. This is because the 43-nt random domain used by Cruz et al. is too large to sample all possible sequence permutations. In addition, this study used a constant and relatively permissive reaction time during each round of selection, which, as our laboratory has subsequently shown, may contribute to the random loss of deoxyribozymes, including proficient catalysts (41). Therefore, faster 8–17 variants for cleavage of the NA, NC and NT junctions are still plausible.
Practical implications, limitations and future directions
The three optimal 8–17 sequence variants described herein can collectively cleave 10 junctions with useful rates of ≥0.1 min−1 (Figure 6). The number of junctions and/or the reaction rate can be further supplemented by other deoxyribozymes like the 10–23 (33) and bipartite (42). However, proficient deoxyribozymes for the cleavage of pyrimidine–pyrimidine junctions have yet to be reported. This study represents one step closer towards the goal of a complete set of practical deoxyribozymes for the site-specific cleavage of all 16 dinucleotide junctions. These enzymes would be of immediate benefit to researchers conducting structural/functional studies on RNA. Deoxyribozyme-mediated RNA cleavage offers several advantages over the alternative ribozyme (43) and RNase H (44) techniques including extra versatility, simplicity, stability and/or cost effectiveness.
This study also provides new insight into the distribution of RNA-cleaving deoxyribozymes in sequence space, and more importantly, how best to access them. For instance, several studies have suggested that longer random libraries (for in vitro selection) provide greater structural complexity (45–47), which in turn can be correlated with greater functional activity (48). We shared this view in the past, and therefore used the longest random-sequence domain (i.e. 80-nt) ever reported for the in vitro selection of RNA-cleaving deoxyribozymes (32). Contrary to expectations, however, the fastest deoxyribozymes that emerged were 8–17 variants. These results are complemented by the current study, in which we have demonstrated that all junctions can actually be cleaved by a deoxyribozyme as small and as simple as 8–17. These findings suggest that our goals may be better served from the complete sampling of sequence space afforded by a library containing a small random domain (N < 25), rather than from the increased structural complexity afforded by a library with a larger random domain. This hypothesis is currently under our investigation. Yarus and co-workers have previously investigated the relative merits of different random library sizes for the selection of isoleucine-binding RNA aptamers (49). Their results also suggest that the selection of certain functional motifs can benefit from smaller library sizes.
Although it is beyond the scope of the current study, further examination of the substrate and metal-ion dependency of these 8–17 deoxyribozymes may be the subject of a future report. In the meantime, it remains to be determined if the 8–17 variants described herein will be equally effective against all-RNA substrates, and whether the observed cleavage site versatility can be supported by different buffer conditions. However, it should be noted that 8–17 has been isolated from in vitro selection experiments using both chimeric (24,31,34) and all-RNA substrates (32,33). Furthermore, an 8–17 variant described by Lu and colleagues was only ∼4-fold less active against an all-RNA substrate versus the corresponding chimeric version (24). Previous studies have also indicated that 8–17 can utilize a wide variety of divalent metal ions as cofactors, with transition metal ions generally providing greater activity than the alkaline earth metal ions (22,24).
CONCLUSIONS
Deoxyribozyme 8–17 represents an ideal model system to address theoretical questions about the functional limitations of nucleic acid-mediated catalysis. This RNA-cleaving deoxyribozyme also represents considerable practical interest because of its utility in many diverse applications. Herein, we conducted a comprehensive mutational study to understand the functional significance of 8–17 sequence variations on cleavage site selectivity. We report for the first time that the 8–17 motif has the unique ability to cleave all 16 dinucleotide junctions, with rate enhancements of at least 1000-fold over background. To the best of our knowledge, no comparable deoxyribozyme or ribozyme has been reported. Sequence space is densely populated by 8–17 sequence variants that differ widely in both reaction rate and cleavage site preference. We have provided evidence suggesting how each nucleotide position in the catalytic core might contribute to the observed cleavage site selectivity of a given 8–17 sequence. In particular, core positions 2.1 and 12 represent a primary determinant of selectivity at the dinucleotide 3′ position, N1.1, while core positions 15 and 15.0 represent a primary determinant of selectivity at the dinucleotide 5′ position, rN18.
The optimal 8–17 variants identified in this study can cleave 10 out of 16 junctions with rates ≥0.1 min−1, and exhibit an overall hierarchy of reactivity towards groups of related junctions according to the order NG > NA > NC > NT. However, our results also suggest that the identification of faster sequence variants against less reactive junctions may still be possible through focused in vitro selection efforts. The remarkable structural and functional plasticity demonstrated by this very simple deoxyribozyme, also serves to challenge the general perception that complexity is a prerequisite for greater function.
ACKNOWLEDGEMENTS
This work was supported by a research grant from the Canadian Institutes for Health Research (grant number MOP37964). Y.L. is a Canada research chair. K.S. holds an NSERC CGS doctoral award. Funding to pay the Open Access publication charges for this article was provided by Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Buchhaupt M, Peifer C, Entian KD. Analysis of 2′-O-methylated nucleosides and pseudouridines in ribosomal RNAs using DNAzymes. Anal. Biochem. 2007;361:102–108. doi: 10.1016/j.ab.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Purtha WE, Coppins RL, Smalley MK, Silverman SK. General deoxyribozyme-catalyzed synthesis of native 3′-5′ RNA linkages. J. Am. Chem. Soc. 2005;127:13124–13125. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Silverman SK. Directing the outcome of deoxyribozyme selections to favor native 3′-5′ RNA ligation. Biochemistry. 2005;44:3017–3023. doi: 10.1021/bi0478291. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Lu Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 5.Chiuman W, Li Y. Efficient signaling platforms built from a small catalytic DNA and doubly labeled fluorogenic substrates. Nucleic Acids Res. 2007;35:401–405. doi: 10.1093/nar/gkl1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sando S, Sasaki T, Kanatani K, Aoyama Y. Amplified nucleic acid sensing using programmed self-cleaving DNAzyme. J. Am. Chem. Soc. 2003;125:15720–15721. doi: 10.1021/ja0386492. [DOI] [PubMed] [Google Scholar]
- 7.Stojanovic MN, Mitchell TE, Stefanovic D. Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 8.Stojanovic MN, Stefanovic D. Deoxyribozyme-based half-adder. J. Am. Chem. Soc. 2003;125:6673–6676. doi: 10.1021/ja0296632. [DOI] [PubMed] [Google Scholar]
- 9.Stojanovic MN, Semova S, Kolpashchikov D, Macdonald J, Morgan C, Stefanovic D. Deoxyribozyme-based ligase logic gates and their initial circuits. J. Am. Chem. Soc. 2005;127:6914–6915. doi: 10.1021/ja043003a. [DOI] [PubMed] [Google Scholar]
- 10.Vlassov AV, Ilves H, Johnston BH. Inhibition of hepatitis C IRES-mediated gene expression by 8–17 deoxyribozymes in human tissue culture cells. Dokl. Biochem. Biophys. 2006;410:257–259. doi: 10.1134/s1607672906050012. [DOI] [PubMed] [Google Scholar]
- 11.Chakraborti S, Banerjea AC. Inhibition of HIV-1 gene expression by novel DNA enzymes targeted to cleave HIV-1 TAR RNA: potential effectiveness against all HIV-1 isolates. Mol. Ther. 2003;7:817–826. doi: 10.1016/s1525-0016(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 12.Chakraborti S, Banerjea AC. Identification of cleavage sites in the HIV-1 TAR RNA by 10–23 and 8–17 catalytic motif containing DNA enzymes. Biomacromolecules. 2003;4:568–571. doi: 10.1021/bm025698i. [DOI] [PubMed] [Google Scholar]
- 13.Kankia BI. A real-time assay for monitoring nucleic acid cleavage by quadruplex formation. Nucleic Acids Res. 2006;34:e141. doi: 10.1093/nar/gkl594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Sen D. Light-regulated catalysis by an RNA-cleaving deoxyribozyme. J. Mol. Biol. 2004;341:887–892. doi: 10.1016/j.jmb.2004.06.060. [DOI] [PubMed] [Google Scholar]
- 15.Flynn-Charlebois A, Prior TK, Hoadley KA, Silverman SK. In vitro evolution of an RNA-cleaving DNA enzyme into an RNA ligase switches the selectivity from 3′-5′ to 2′-5′. J. Am. Chem. Soc. 2003;125:5346–5350. doi: 10.1021/ja0340331. [DOI] [PubMed] [Google Scholar]
- 16.Yim TJ, Liu J, Lu Y, Kane RS, Dordick JS. Highly active and stable DNAzyme-carbon nanotube hybrids. J. Am. Chem. Soc. 2005;127:12200–12201. doi: 10.1021/ja0541581. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari D, Peracchi A. A continuous kinetic assay for RNA-cleaving deoxyribozymes, exploiting ethidium bromide as an extrinsic fluorescent probe. Nucleic Acids Res. 2002;30:e112. doi: 10.1093/nar/gnf111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wernette DP, Swearingen CB, Cropek DM, Lu Y, Sweedler JV, Bohn PW. Incorporation of a DNAzyme into Au-coated nanocapillary array membranes with an internal standard for Pb(ii) sensing. Analyst. 2006;131:41–47. doi: 10.1039/b510071b. [DOI] [PubMed] [Google Scholar]
- 19.Pei R, Taylor SK, Stefanovic D, Rudchenko S, Mitchell TE, Stojanovic MN. Behavior of polycatalytic assemblies in a substrate-displaying matrix. J. Am. Chem. Soc. 2006;128:12693–12699. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- 20.Wang DY, Lai BH, Feldman AR, Sen D. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 2002;30:1735–1742. doi: 10.1093/nar/30.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang DY, Lai BH, Sen D. A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J. Mol. Biol. 2002;318:33–43. doi: 10.1016/S0022-2836(02)00046-3. [DOI] [PubMed] [Google Scholar]
- 22.Bonaccio M, Credali A, Peracchi A. Kinetic and thermodynamic characterization of the RNA-cleaving 8–17 deoxyribozyme. Nucleic Acids Res. 2004;32:916–925. doi: 10.1093/nar/gkh250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown AK, Li J, Pavot CM, Lu Y. A lead-dependent DNAzyme with a two-step mechanism. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peracchi A, Bonaccio M, Clerici M. A mutational analysis of the 8–17 deoxyribozyme core. J. Mol. Biol. 2005;352:783–794. doi: 10.1016/j.jmb.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 26.Kim HK, Liu J, Li J, Nagraj N, Li M, Pavot CM, Lu Y. Metal-dependent global folding and activity of the 8–17 DNAzyme studied by fluorescence resonance energy transfer. J. Am. Chem. Soc. 2007;129:6896–6902. doi: 10.1021/ja0712625. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Lu Y. FRET study of a trifluorophore-labeled DNAzyme. J. Am. Chem. Soc. 2002;124:15208–15216. doi: 10.1021/ja027647z. [DOI] [PubMed] [Google Scholar]
- 28.Leung EK, Sen D. Electron hole flow patterns through the RNA-cleaving 8–17 deoxyribozyme yield unusual information about its structure and folding. Chem. Biol. 2007;14:41–51. doi: 10.1016/j.chembiol.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Kim HK, Rasnik I, Liu J, Ha T, Lu Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007;3:763–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peracchi A. Preferential activation of the 8–17 deoxyribozyme by Ca(2+) ions. Evidence for the identity of 8–17 with the catalytic domain of the Mg5 deoxyribozyme. J. Biol. Chem. 2000;275:11693–11697. doi: 10.1074/jbc.275.16.11693. [DOI] [PubMed] [Google Scholar]
- 31.Faulhammer D, Famulok M. The Ca2+ ion as a cofactor for a novel RNA-cleaving deoxyribozyme. Angew. Chem. Int. Ed. Engl. 1996;35:2809–2813. [Google Scholar]
- 32.Schlosser K, Li Y. Tracing sequence diversity change of RNA-cleaving deoxyribozymes under increasing selection pressure during in vitro selection. Biochemistry. 2004;43:9695–9707. doi: 10.1021/bi049757j. [DOI] [PubMed] [Google Scholar]
- 33.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cruz RP, Withers JB, Li Y. Dinucleotide junction cleavage versatility of 8–17 deoxyribozyme. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Zaborowska Z, Furste JP, Erdmann VA, Kurreck J. Sequence requirements in the catalytic core of the “10–23” DNA enzyme. J. Biol. Chem. 2002;277:40617–40622. doi: 10.1074/jbc.M207094200. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Breaker RR. Kinetics for specific base catalysis of RNA degradation by transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121:5364–5372. [Google Scholar]
- 37.Schlosser K, Lam JC, Li Y. Characterization of long RNA-cleaving deoxyribozymes with short catalytic cores: the effect of excess sequence elements on the outcome of in vitro selection. Nucleic Acids Res. 2006;34:2445–2454. doi: 10.1093/nar/gkl276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hertel KJ, Pardi A, Uhlenbeck OC, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, et al. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cairns MJ, King A, Sun LQ. Optimisation of the 10–23 DNAzyme-substrate pairing interactions enhanced RNA cleavage activity at purine-cytosine target sites. Nucleic Acids Res. 2003;31:2883–2889. doi: 10.1093/nar/gkg378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtis EA, Bartel DP. New catalytic structures from an existing ribozyme. Nat. Struct. Mol. Biol. 2005;12:994–1000. doi: 10.1038/nsmb1003. [DOI] [PubMed] [Google Scholar]
- 41.Schlosser K, Li Y. Diverse evolutionary trajectories characterize a community of RNA-cleaving deoxyribozymes: a case study into the population dynamics of in vitro selection. J. Mol. Evol. 2005;61:192–206. doi: 10.1007/s00239-004-0346-7. [DOI] [PubMed] [Google Scholar]
- 42.Feldman AR, Sen D. A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J. Mol. Biol. 2001;313:283–294. doi: 10.1006/jmbi.2001.5058. [DOI] [PubMed] [Google Scholar]
- 43.Ferre-D’Amare AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapham J, Crothers DM. RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA. 1996;2:289–296. [PMC free article] [PubMed] [Google Scholar]
- 45.Knight R, Yarus M. Finding specific RNA motifs: function in a zeptomole world? RNA. 2003;9:218–230. doi: 10.1261/rna.2138803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knight R, De Sterck H, Markel R, Smit S, Oshmyansky A, Yarus M. Abundance of correctly folded RNA motifs in sequence space, calculated on computational grids. Nucleic Acids Res. 2005;33:5924–5935. doi: 10.1093/nar/gki886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sabeti PC, Unrau PJ, Bartel DP. Accessing rare activities from random RNA sequences: the importance of the length of molecules in the starting pool. Chem. Biol. 1997;4:767–774. doi: 10.1016/s1074-5521(97)90315-x. [DOI] [PubMed] [Google Scholar]
- 48.Carothers JM, Oestreich SC, Davis JH, Szostak JW. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 2004;126:5130–5137. doi: 10.1021/ja031504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Legiewicz M, Lozupone C, Knight R, Yarus M. Size, constant sequences, and optimal selection. RNA. 2005;11:1701–1709. doi: 10.1261/rna.2161305. [DOI] [PMC free article] [PubMed] [Google Scholar]






