Abstract
Over the past 5 years, the availability of dozens of whole genomic sequences from a wide variety of eukaryotic lineages has revealed a very large amount of information about the dynamics of intron loss and gain through eukaryotic history, as well as the evolution of intron sequences. Implicit in these advances is a great deal of information about the structure and evolution of surrounding sequences. Here, we review the wealth of ways in which structures of spliceosomal introns as well as their conservation and change through evolution may be harnessed for evolutionary and genomic analysis. First, we discuss uses of intron length distributions and positions in sequence assembly and annotation, and for improving alignment of homologous regions. Second, we review uses of introns in evolutionary studies, including the utility of introns as indicators of rates of sequence evolution, for inferences about molecular evolution, as signatures of orthology and paralogy, and for estimating rates of nucleotide substitution. We conclude with a discussion of phylogenetic methods utilizing intron sequences and positions.
INTRODUCTION
Patterns and evolution of intron–exon structures
Spliceosomal introns are sequences that interrupt eukaryotic genes and are removed from RNA transcripts by the spliceosome, a complex cellular RNA–protein machine incorporating five RNAs and hundreds of proteins (1). Our understanding of the evolution of spliceosomal introns has increased exponentially over the past few years due to the release of many genome sequences from most major eukaryotic lineages, both about intron loss and gain dynamics (2–8), as well as the evolution of intron sequences and splicing (9–13). [Several reviews have recently tackled the question of the evolution of introns in eukaryotes (14–19)]. During the first 25 years after their discovery in 1977 (20), much of the study of spliceosomal introns focused on a debate about the timing of origin of the first introns (21), whether before the divergence of eukaryotes from prokaryotes (which lack spliceosomal introns) (22–25) or within the evolutionary history of eukaryotes (26–31). Although this debate continues, the momentum has clearly tipped towards the perspective that introns appeared once in early (or pre-) eukaryotic evolution by the proliferation and transformation of type II self-splicing introns (26,27,32,33), possibly transferred from the mitochondrion.
Over the past 5 years, the focus has shifted away from the question of the ultimate origin of introns to attempts to track the history of intron loss/gain and intron sequence evolution during eukaryotic history. We can now be confident that large numbers of introns were present by early eukaryotic history (14,34–40) and that many or even most modern introns date to the times of early eukaryotic ancestors. Over at least recent eukaryotic evolution (say, the last ∼100 My), intron gain has been a very rare event, with most lineages experiencing rates of gain corresponding to <0.0002 gains per gene per million years (7,8,41–46). Rates of intron loss have been more variable: in some lineages, rates of loss are perhaps 10% per 100 My, whereas other lineages have experienced almost no intron loss over tens or hundreds of millions of years (2,6,7,42,44–48). Figure 1 shows the example of metazoans, where the majority of intron positions have been retained between vertebrates and basal animals.
Figure 1.
Intron positions are often conserved over long evolutionary times. Protein-level alignments of the translation initiation factor 4A gene (TIF4A) from a variety of metazoan species are shown. Intron positions are indicated by digits corresponding to the phase of the intron relative to the surrounding codons (phases 0, 1 and 2 introns fall before the first, second and third bases of a codon, respectively). Most intron positions are conserved at the exact homologous position and phase over all of animal history within these species, all the way from the placozoan Trichoplax adhaerens to chordates. Abbreviations: Hsa, Homo sapiens; Mmu, Mus musculus; Cin, Ciona intestinalis; Bfl, Branchiostoma floridae; Cap, Capitella sp; Lgi, Lottia gigantea; Nve, Nematostella vectensis; Tad, Trichoplax adhaerens.
Moreover, intron positions have been shown to be very constant over time—i.e. that intron ‘sliding’, in which an intron would migrate a few base pairs along a gene, is a very rare occurrence (4,49,50). Several studies have also documented the mechanisms of intron loss: patterns of intron loss including 3′-biased intron loss (7,51–53), exact removal of intron sequences (2), apparently coincident loss of adjacent introns (7,47,54) and possibly germline-biased intron loss (unpublished data), all seem to indicate that intron loss proceeds via reverse transcription of RNA intermediates (55,56). In addition, comparative analyses have uncovered an apparent (though incomplete) correspondence between rates of intron loss and rates of sequence evolution—degree of loss of ancestral introns appears directly correlated with degree of sequence change [(57) and unpublished data].
Introns as the repository of information about gene structure
The focus of this article is not on these patterns of evolution themselves, but on their implications for analysis of genome structures and eukaryotic evolution in general. Although introns have largely been regarded as a hindrance for genome analysis given the difficulties associated with gene annotation in the presence of introns, intron positions and sequences are potentially very useful in addressing a wide variety of important genomic and evolutionary problems. In particular, intron loss/gain has been shown to be a very slow process in many lineages relative to other genetic characters (sequence evolution of proteins, genes and non-coding DNA, insertion and deletion of transposable elements and even genome rearrangement), thus intron positions contain (and retain) a large amount of information about genome structure and deep evolutionary history.
Over the past few years, a variety of researchers from disparate fields have developed methods that harness this information for purposes ranging from reconstructing evolutionary phylogenies to improving gene prediction, from alignment of homologous protein sequences to assignment of orthology in large protein families. Many of these methods are already quite powerful, though often known primarily to those working on introns themselves, rather than those working on the problems addressed by the methods. Other methods are still largely undeveloped, and represent promising future lines of work. Here, we review these approaches, and delineate possible uses in genomic and evolutionary study.
USES OF SPLICEOSOMAL INTRONS FOR GENOME SEQUENCING, ANNOTATION AND ALIGNMENT
Intron length distributions and genome assembly and annotation
A potential utility of introns for large-scale sequencing efforts involves the distribution of intron lengths. Since introns are removed from protein-coding transcripts, intron lengths are not expected to respect coding frame: across the genome, we expect roughly equal proportions of introns that are multiple of three bases (‘3n’ introns), one more than a multiple of three bases (3n + 1) and two more (3n + 2). However, one of us and David Penny (58) recently reported a survey of predicted genes from genome annotations across 29 different species in which we found common deviations from this expectation. In some cases, the number of 3n introns was much larger than the numbers of 3n + 1 or 3n + 2 introns (Figure 2A), in other cases less. Further investigation indicated that many such cases seemed to be due to genome-wide problems in annotation. Such an internal check for genome annotations could constitute an important step in improving genome annotations before their public release. [While this paper was in press, a new report by Jaillon et al. (Nature 2008 451:359–62) showed a real biological deficit of 3n introns owing to selection for nonsense mediated decay. This result cannot however explain predicted proteomes with other types of skewed intron length distribution].
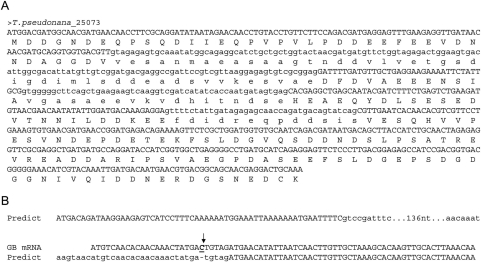
Figure 2.
Intron length distributions and gene prediction. Since introns are removed from transcripts, they are not expected to respect coding frame, predicting roughly equal numbers of introns with lengths of a multiple of three nucleotides (3n), 3n + 1 and 3n + 2. (A) The majority of predicted introns in the genome of the diatom Thalassiosira pseudonana preserve reading frame (i.e. they are 3n nucleotides and lack in frame stop codons), suggesting that many predicted introns are instead coding sequence. All three introns of this predicted gene (JGI ID25063) preserve reading frame (56). (B) Predicted introns in the E. histolytica genomes are disproportionately 3n + 2. This excess appears to reflect single bases left out of the assembly (i.e. indels), with introns predicted in order to restore reading frame. Here, a predicted intronic sequence from the genome annotation is present in an mRNA from GenBank, and is missing a single cytosine nucleotide (7).
Our previous study (8) also showed a case in which analysis of gene and intron annotations was able to identify a previously unnoticed large number of indels in a genome assembly (example in Figure 2B). In the case of Entamoeba histolytica, the publicly available annotation showed a pronounced excess of 3n + 2 introns. Genome-wide computational and manual inspection of predicted introns indicated that the majority of these excess 3n + 2 introns were associated with a single missing base in the assembled genome sequence—many of these cases appear to be actual coding sequence which had been disrupted by the lack of a base, leading to false prediction of an intron in order to keep the predicted gene sequence in frame. Thus, in some cases, scrutiny of genome-wide intron length distributions from preliminary gene predictions could indicate otherwise undetected errors in genome assembly.
Intron position conservation and improved gene annotation
Intron positions are very often conserved over very long evolutionary distances (Figure 1). In some lineages, this reaches extremes. In Theileria apicomplexans, 99.7% of intron positions are conserved between T. parva and T. annulata, diverged roughly 82 Mya (2). In mammals, 99.9% of intron positions are conserved between human and dog, diverged around 100 Mya (42). Lineages with significant numbers of introns are particularly difficult to annotate in the absence of exhaustive transcript sequence information. Here, comparison with species which are known to have very similar intron–exon structures in orthologous regions could vastly improve uncertain annotations (59). When predicted intron positions are mapped onto protein-level alignments of predicted orthologs, the results are often very clear—protein-level sequence similarity will cease abruptly at the boundary of a species-specific intron position [(2,60,61) and unpublished observations]. While this could in fact reflect biological reality, it seems very likely that this often reflects misprediction of the intron in one species—either there is an intron present in both species, which has gone unpredicted in one, or there is no intron at that position in either species, and truly exonic sequence in one species has been predicted as an intron.
Figure 3 shows a pair of orthologs from Plasmodium falciparum and P. yoelii. Clear protein sequence similarity continues through conserved intron positions, and then ends abruptly at a P. yoelii-specific predicted intron. Alignment of the regions at the DNA level clearly shows that the sequence of the P. yoelii-specific predicted intron is highly similar to the P. falciparum sequence. This pattern strongly suggests that the predicted intron sequence is instead coding sequence homologous to the P. falciparum coding sequence.
Figure 3.
Reconciling orthologous intron–exon structures to improve gene predictions. (A) Clear protein similarity between the MAL7P1.99 and PY05856 genes of P. falciparum and P. yoelii continues through two conserved intron positions, and then ends abruptly at a predicted P. yoelii-specific intron. (B) Alignment of the genes at the DNA level shows strong sequence similarity between the predicted P. yoelii intron and the predicted P. falciparum coding sequence, strongly suggesting that the predicted P. yoelli intron instead is coding.
Reconciliation between protein models in species pairs or clusters could greatly improve gene predictions, particularly in species where highly skewed sequence composition or frequent repetitive sequence renders accurate gene prediction most difficult. For example, Coghlan and Durbin (62) recently presented a new method to combine predictions from different gene finders used for one species by comparing intron/exon structures of the different predictions and between gene models of closely related species, building gene structures based on the most conserved exons. They applied this methodology to the nematodes Caenorhabditis briggsae and C. remanei, obtaining increases of >10% in exon-level specificity and almost 3% in sensitivity, compared to the best outputs from previous gene finders.
Improved protein sequence alignments
Since introns often maintain their positions over very long evolutionary timescales, intron positions can often retain information about gene homology after protein sequences have experienced enough change as to render alignment difficult. As such, intron positions can be used as check points in protein alignments, improving their quality. Recently, Csuros and coauthors (63) developed a method to use intron positions to improve protein-level alignments in regions of questionable alignment (Figure 4). They introduced alignment penalties and rewards for intron positions into the alignment matrixes, considering intron position matches/mismatches scoring alternative alignments. The use of these algorithms significantly improved the quality of some gapped alignments.
Figure 4.
Using intron positions to improve protein sequence alignments. Protein sequence alignment in regions with significant change is often ambiguous (left). Alignment of intron positions indicates the likely true alignment (from 61).
INTRONS AS TOOLS TO STUDY MOLECULAR EVOLUTION
Intron density as a surrogate for rates of sequence evolution
A central goal of full genome sequencing is understanding the evolutionary history of ourselves and other organisms. Identification of slow evolving lineages (the so-called ‘living fossils’) is thus of central interest, both for what they can reveal about organismal complexity and genome structure evolution. In this context, intron density may serve as an important indicator for branch length. Since rates of intron gain across a wide variety of lineages have been very low (2,6,7,42,44–47), intron numbers in modern species often largely reflect the extent of intron loss since intron-rich ancestors. Intron number is therefore inversely correlated to ‘branch length’, in terms of intron loss.
Interestingly, and somewhat surprisingly given the very different sets of mutation and presumed evolutionary forces controlling intron loss and sequence evolution, a correspondence between degree of intron loss and degree of sequence change has been found in some eukaryotic lineages. The most straightforward case involves a genomic survey across metazoan genomes (57). The genomes which had experienced the least intron loss since the metazoan ancestor (vertebrates and the marine annelid Platynereis) also had experienced less sequence change since the ancestor than other studied lineages (from among insects, urochordates and nematodes). A second example concerns the multitude of nearly intronless protists that also exhibit very high degrees of sequence change (29,64). If in fact a relationship between intron number and branch lengths holds generally, intron density as estimated from small-scale genomic sequencing efforts could be useful in identifying short-branch taxa.
Inferences about molecular evolution
Patterns of intron distribution and evolution can also provide insights into other aspects of molecular evolution. First, intron presence/absence is useful in inferring the mechanism of gene duplication, since intron absence is a hallmark of gene duplication by retroposition (65). Thousands of intron-less copies of the so-called ‘processed pseudogenes’ are present in many eukaryotic genomes (66,67), originated by retrotranscription of processed mRNAs and subsequent insertion into the genome (65). This mechanism can be easily distinguished from segmental genome duplication by the absence of introns (65,68). Second, the correspondence of intron positions with the boundaries of domains whose reshuffling contributed to the origin of new proteins in metazoan lineages allows the reconstruction of the mechanism of origin of multi-domain protein-encoding genes (69–73).
Given the apparent dependency of intron loss on reverse transcriptase, rates of intron loss could also provide information about the presence and activity of retroelements through evolutionary history. For instance, we recently showed that rates of intron loss have varied by orders of magnitude in the history of apicomplexan evolution (74). Given the apparent dependence of intron loss on retroelement activity (52), the lack of known active retroelements in modern Plasmodium and Theileria species is consistent with the lack of recent intron loss (74). If so, the much more extensive loss in both the Plasmodium and Theileria ancestors since the genera's divergence suggests retroelement activity. Thus the pattern of intron loss through time may provide information about the activity of retroelements over evolutionary depths where the actual retroelement insertion history has been erased by subsequent mutation.
Intron positions as signatures of orthology and paralogy
Since the rate of intron loss in modern organisms is often very low, the pattern of intron positions can be used as an indication for orthology among paralogous groups (75–78). These studies usually complement others such as classical phylogenetic analysis of gene families or synteny comparisons, and may be especially useful in the annotation of newly sequenced genomes in assigning orthology among large gene families with very similar domains (such as kinases, TGF-βs, immunoglobulins, etc.) that are hard to distinguish by traditional phylogenetic methods. Some evidence suggests that patterns of intron gain and loss might be different among paralogous groups (79) [although see (80)]. If so, orthology inferences could be hampered by the higher rate of intron change. On the other hand, increased rates of loss and gain could increase intron positions’ usefulness in identifying orthology even over short evolutionary times (with little sequence differentiation).
Estimation of neutral rates of nucleotide substitution
Intron sequences themselves appear to tolerate sequence changes quite easily. Putatively neutrally evolving (portions of) intron sequences are thus a key tool in estimating neutral rates of mutation. Hoffman and Birney (81) recently published a new method to estimate neutral rates of nucleotide substitution based on the study of the substitutions occurring on the alignable introns sequences. They compared their method to a previous method also based on intron sequences (82) and more classic methods based on substitutions in synonymous coding sites, finding a strong correlation between estimates from the two types of methods on different species comparisons. Interestingly, synonymous sites have been shown to be under purifying selection [reviewed in (83,84)], or even under positive selection (85–87). However, introns are also known to contain different types of functional elements (88–90) and thus selection of regions of estimation of neutral rates requires caution.
INTRONS IN PHYLOGENETIC ANALYSIS
Intron sequences and positions contain a record of the evolutionary history of a species or group of species, which presumably contains valuable phylogenetic information. Two phylogenetic strategies have utilized introns as phylogenetic markers at two very different evolutionary depths. Intron sequences, which are relatively fast evolving, have been commonly used to resolve relationships between closely related species. At the other end, evolution of gene structures by intron loss and gain, which can be very slow in some lineages, has been used in order to resolve deeper nodes over which our confidence in traditional sequence methods can be reduced by the large amount of change over the studied species.
Intron sequences as phylogenetic markers
The use of intron sequences for resolution of relationships between closely related species was established more than 15 years ago (91,92), and it is so common as to barely require comment (93–98). The appeal of intronic sequences for phylogenetics of recent divergences owes to their plausibly being both more rapidly and more neutrally evolving than protein coding and other clearly functional sequences (Figure 5A). As such, relatively simple phylogenetic methods are thought to be of use in utilizing intronic sequences over depths where multiple mutation is unlikely, and over which protein sequences may have experienced too little change to yield sufficient signal. Moreover, obtaining informative intron sequence data sets from non-model organisms is relatively easy and fast using PCR-based methods (91), and the global lack of functional constraints and high potential phylogenetic information content make introns a good complement to mtDNA-based phylogenies for poorly studied groups (99–101).
Figure 5.
Introns as tools for phylogenetics. (A) Intron sequences themselves are often used to resolve phylogenetics relationships between closely related species. Here, four phylogenetically informative sites in an alignment of sequences of orthologous introns from five Drosophila species (the 1st intron from the CG10050 locus) are consistent with the accepted consensus relationship of the species (left). (B) Intron loss/gain and position conservation as phylogenetic characters. Intron position conservation across subsets of species suggests phylogenetic groupings. Under many circumstances, the high degree of intron loss/gain change requires correction for the possibility of multiple changes at a site (3). (C) Nearby pairs of intron positions as phylogenetic characters. Krauss et al. (115) suggest a novel class of intron change in order to diminish the possibility of homoplasy. Since short exons are rare across various genomes, introns at nearby positions in orthologous genes are unlikely to have coexisted, suggesting multiple rare changes (an intron loss followed by a nearby gain) between putatively ancestral and derived gene structures. Here, two pairs of nearby intron positions in the CDC5-like gene of metazoans support grouping the three vertebrates (top three lines) with Ciona intestinalis (Cin_CDC5L). (D) Intron boundary sliding as a possible phylogenetic character. Rarely, an intron boundary may shift, leading to conversion of exonic sequence to intron (or vice versa). In this hypothetical case, mutation of the 5′ splice boundary (gt→ct) leads to use of an upstream previously exonic GT site. To our knowledge, no study has yet employed such changes for phylogenetic analysis. Abbreviations: Hsa, Homo sapiens; Mmu, Mus musculus; Tru, Takifugu rubripes; Cin, Ciona intestinalis; Bfl, Branchiostoma floridae; Spu, Strongylocentrotus purpuratus; Nve, Nematostella vectensis; Tad, Trichoplax adhaerens.
Intron positions as phylogenetic markers
As mentioned above, intron presence/absence is a relatively very slowly evolving character in most lineages studied to date. Whereas the average number of changes per nucleotide site in putatively unconstrained nucleotide sequence between mouse and human is estimated to be Ks = 0.6 (102), and the degree of protein sequence change around 21.5% (102), a survey of more than 150 000 intron positions between the species found only 120 changes (0.08%), a degree of change three orders of magnitude lower (42). Stajich and Dietrich (47) found <1% intron loss/gain change across a clade of four Cryptococcus species, compared to 35% change in nucleotide sites across the same species (47). Median dS is estimated to be around 0.49 for the Plasmodium parasites P. falciparum and P. yoelii (103), but fewer than 1.5% of intron sites have experienced a loss/gain event (45).
Relative to sequence-based studies, phylogenetics using intron presence/absence is truly in its infancy. Among the very few published studies, there are essentially three strategies. First, some authors have used one or a few intron loss/gain patterns to group species into a clade (78,104,105) (Figure 5B), since as rare genome change (RGC) losses/gains could be theoretically highly parsimonious (106). As RGCs, introns have the advantage of having a very wide taxonomic resolution, potentially low homoplasy and applicability to a broad range of eukaryotic groups (106). Here, however, significant caution is necessary. First, while such ‘magic bullet’ approaches may work well for groups with very few intron changes, the possibility of homoplasy (in particular, of multiple loss of the same intron) is likely in cases where there is more change, for instance, nematodes and dipterans (107); second, individual cases attest to the recurrent loss of the same intron, even while flanking introns remain intact (107,108). Until the degree of such variation across sites is better known, individual cases of intron loss/gain as phylogenetic markers should in our opinion be viewed as non-conclusive in many cases.
A second strategy involves using explicit phylogenetic models to analyze large collections of intron presence/absence data across species. One of us and Walter Gilbert (109) used intron presence/absence data for 684 sets of eukaryotic orthologs across eight animal, fungus, plant, and apicomplexan species (4) to develop a method for resolving deep nodes in metazoan phylogeny. Nguyen and coauthors (38) then developed a more general phylogenetic method for analysis of the same kind of data. These data were used to address the relationship between arthropods, nematodes and deuterostomes, with both analyses placing deuterostomes as the outgroup.
Again, there is reason for caution here. Both analyses assumed constancy of intron loss rates across intron sites, an assumption which is unlikely to hold generally, and which might bias methods towards long branch attraction and other recurrent problems in phylogenetics, especially important in this particular phylogeny (110–114). Moreover, the small absolute number of characters obtainable from even such an exhaustive comparative genomic effort may make it difficult to obtain sufficient knowledge of the shape of the distribution of rates across sites, which may make these shortcomings difficult to correct for. On the other hand, the availability of representatives of the groups in question that have experienced less intron loss will presumably allow for more confident reconstruction.
The third and perhaps most promising (as well as intriguing) strategy was developed by Krauss and coauthors (115), and aims to overcome these problems by restricting the analysis to cases in which ancestral and derived states can be more confidently inferred. Very short exons (e.g. shorter than 50 bp) are very rare across most characterized species, likely due to problems associated with accurate splicing of such regions. Therefore, introns found at nearby positions in orthologous coding regions are unlikely to have coexisted. Assuming that multiple insertions into the same site are very rare, one can then confidently infer that there is an edge on the phylogenetic tree (along which both intron loss and gain has occurred) separating those species that share one of the positions from those that share the other (Figure 5C). In cases in which a known outgroup shares one of the two positions, one can furthermore infer the directionality of this change, and therefore place all derived species into a clade. However, it remains to be seen whether (and in what cases) sufficient numbers of such intron pairs will be obtainable, thus it is not yet clear to what extent this model will be generalizable.
A fourth possibility that has not to our knowledge been explored would utilize shifts of intron boundaries (i.e. shortening or lengthening of adjacent coding sequence) (Figure 5D). Analysis of large numbers of alignments in various lineages shows that such changes are very rare indeed, and that such changes might represent useful genomic changes. A major concern in developing methods using these changes will be exclusion of incorrect genome annotation as an explanation.
An important caveat to the seeming usefulness of intron loss/gain across species is the possibility of highly skewed distributions of rates across sites (alluded to above). In a few cases, careful study of closely related species with known phylogenies has indicated that a single intron position has been subject to striking recurrent intron loss while other intron positions have remained intact (107,108). In such cases, the observed phylogenetic position will give support to inaccurate phylogenetic groups. It is not currently known how general this pattern of recurrent loss of the same introns is, and so it is not yet clear how much of a problem this may constitute in large-scale or genome-level comparisons. Strong confidence in use of intron losses/gains as phylogenetic characters awaits a better understanding of the causes and generality of large differences in loss rates across sites.
CONCLUSION
Problems associated with signal-to-noise ratios are ubiquitous in bioinformatic, genomic and evolutionary analyses. As slowly evolving characters, intron positions provide useful and otherwise scarce information. We have reviewed well-developed, early-stage and potential uses of introns as tools for addressing a wide range of problems. We look forward to development of further methods.
ACKNOWLEDGEMENTS
M.I. was funded by the Spanish Ministerio of Educación y Ciencia, through the FPI grant (BFU2005-00252), and S.W.R. by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding to pay the Open Access publication charges for this article has been provided by the National Institutes of Health. We thank Eugene Koonin and Jordi Garcia-Fernandez and their groups for intellectual support and stimulation, for financial support, and for fostering environments of open intellectual exploration in their respective groups. SWR thanks Rogério da Silva and all of Marcelo Ferreira's group for heroics of hospitality and friendship during the course of this project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 2.Roy SW, Penny D. Large-scale intron conservation and order-of-magnitude variation in intron loss/gain rates in apicomplexan evolution. Genome Res. 2006;16:1270–1275. doi: 10.1101/gr.5410606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy SW, Penny D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 2007;24:1447–1457. doi: 10.1093/molbev/msm048. [DOI] [PubMed] [Google Scholar]
- 4.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- 5.Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen C, Friedman B, Birren B, Burge C, Galagan J. Patterns of intron gain and loss in fungi. PLoS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy SW, Gilbert W. The pattern of intron loss. Proc. Natl Acad. Sci. USA. 2005;102:713–718. doi: 10.1073/pnas.0408274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy SW, Irimia M, Penny D. Very little intron gain in Entamoeba histolytica genes laterally transferred from prokaryotes. Mol. Biol. Evol. 2006;23:1824–1827. doi: 10.1093/molbev/msl061. [DOI] [PubMed] [Google Scholar]
- 9.Kupfer DM, Drabenstot SD, Buchanan KL, Lai H, Zhu H, Dyer DW, Roe DA, Murphy JW. Introns and splicing elements of five diverse fungi. Eukaryotic Cell. 2004;3:1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bon E, Casaregola S, Blandin G, Llorente B, Neuveglise C, Munsterkotter M, Guldener U, Mewes H-W, Helden JV, Dujon B, et al. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irimia M, Penny D, Roy SW. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Irimia M, Rukov JL, Penny D, Roy SW. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol. Biol. 2007;7:188. doi: 10.1186/1471-2148-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz S, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogozin I, Sverdlov A, Babenko V, Koonin E. Analysis of evolution of exon-intron structure of eukaryotic genes. Brief Bioinform. 2005;6:118–134. doi: 10.1093/bib/6.2.118. [DOI] [PubMed] [Google Scholar]
- 15.Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Trelles F, Tarrio R, Ayala FJ. Origins and evolution of spliceosomal introns. Annu Rev. Genet. 2006;40:47–76. doi: 10.1146/annurev.genet.40.110405.090625. [DOI] [PubMed] [Google Scholar]
- 17.Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118:123–131. [PubMed] [Google Scholar]
- 18.Lynch M, Richardson AO. The evolution of spliceosomal introns. Curr. Opin. Genet. Dev. 2002;12:701–710. doi: 10.1016/s0959-437x(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 19.Zhaxybayeva O, Gogarten JP. Spliceosomal introns: New insights into their evolution. Curr. Biol. 2003;13:R764–R766. doi: 10.1016/j.cub.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Berget SM, Sharp PA. A spliced sequence at the 5′-terminus of adenovirus late mRNA. Brookhaven Symp. Biol. 1977;12–20:332–344. [PubMed] [Google Scholar]
- 21.Doolittle WF. Genes in pieces: were they ever together? Nature. 1978;272:581–582. [Google Scholar]
- 22.Gilbert W. The exon theory of genes. Cold Spring Harb. Sym. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 23.de Souza SJ, Long M, Schoenbach L, Roy SW, Gilbert W. Intron positions correlate with module boundaries in ancient proteins. Proc. Natl Acad. Sci. USA. 1996;93:14632–14636. doi: 10.1073/pnas.93.25.14632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long M, de Souza SJ, Gilbert W. Evolution of the intron-exon structure of eukaryotic genes. Curr. Opin. Genet. Dev. 1995;5:774–778. doi: 10.1016/0959-437x(95)80010-3. [DOI] [PubMed] [Google Scholar]
- 25.Darnell J., Jr Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science. 1978;202:1257–1260. doi: 10.1126/science.364651. [DOI] [PubMed] [Google Scholar]
- 26.Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985;315:283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- 27.Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. 1991;7:145–148. [PubMed] [Google Scholar]
- 28.Stoltzfus A. Origin of introns-early or late? Nature. 1994;369:526–527. doi: 10.1038/369526b0. [DOI] [PubMed] [Google Scholar]
- 29.Logsdon J. The recent origins of spliceosomal introns revisited. Curr. Opin. Genet. Dev. 1998;8:637–648. doi: 10.1016/s0959-437x(98)80031-2. [DOI] [PubMed] [Google Scholar]
- 30.Logsdon J, Jr, Tyshenko M, Dixon C, D-Jafari J, Walker V, Palmer J. Seven newly discovered intron positions in the triose-phosphate isomerase gene: evidence for the introns-late theory. Proc. Natl Acad. Sci. USA. 1995;92:8507–8511. doi: 10.1073/pnas.92.18.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dibb NJ, Newman AJ. Evidence that introns arose at proto-splice sites. EMBO J. 1989;8:2015–2021. doi: 10.1002/j.1460-2075.1989.tb03609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cech TR. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell. 1986;44:207–210. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 33.Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 34.Fedorov A, Merican A, Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc. Natl Acad. Sci. USA. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Archibald J, O’Kelly C, Doolittle W. The chaperonin genes of jakobid and jakobid-like flagellates: implications for eukaryotic evolution. Mol. Biol. Evol. 2002;19:422–431. doi: 10.1093/oxfordjournals.molbev.a004097. [DOI] [PubMed] [Google Scholar]
- 36.Roy SW, Gilbert W. Complex early genes. Proc. Natl Acad. Sci. USA. 2005;102:1986–1991. doi: 10.1073/pnas.0408355101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sverdlov A, Rogozin I, Babenko V, Koonin E. Conservation versus parallel gains in intron evolution. Nucleic Acids Res. 2005;33:1741–1748. doi: 10.1093/nar/gki316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen H, Yoshihama M, Kenmochi N. New maximum likelihood estimators for eukaryotic intron evolution. PLoS Comput. Biol. 2005;1:e79. doi: 10.1371/journal.pcbi.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshihama M, Nakao A, Nguyen HD, Kenmochi N. Analysis of ribosomal protein gene structures: implications for intron evolution. PLoS Genet. 2006;2:e25. doi: 10.1371/journal.pgen.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slamovits CH, Keeling PJ. A high density of ancient spliceosomal introns in oxymonad excavates. BMC Evol. Biol. 2006;6:34. doi: 10.1186/1471-2148-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Csurös M. Third RECOMB Satellite Workshop on Comparative Genomics. 2005. pp. 47–60. Springer LNCS 3678. [Google Scholar]
- 42.Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedorov A, Roy S, Fedorova L, Gilbert W. Mystery of intron gain. Genome Res. 2003;13:2236–2241. doi: 10.1101/gr.1029803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy SW, Gilbert W. Rates of intron loss and gain: implications for early eukaryotic evolution. Proc. Natl Acad. Sci. USA. 2005;102:5773–5778. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy SW, Hartl DL. Very little intron loss/gain in Plasmodium: intron loss/gain mutation rates and intron number. Genome Res. 2006;16:750–756. doi: 10.1101/gr.4845406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy SW, Penny D. Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 2007;24:171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- 47.Stajich JE, Dietrich FS. Evidence of mRNA-mediated intron loss in the human-pathogenic fungus Cryptococcus neoformans. Eukaryotic Cell. 2006;5:789–793. doi: 10.1128/EC.5.5.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carmel L, Rogozin IB, Wolf YI, Koonin EV. Evolutionarily conserved genes preferentially accumulate introns. Genome Res. 2007;17:1045–1050. doi: 10.1101/gr.5978207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogozin IB, Lyons-Weiler J, Koonin EV. Intron sliding in conserved gene families. Trends Genet. 2000;16:430–432. doi: 10.1016/s0168-9525(00)02096-5. [DOI] [PubMed] [Google Scholar]
- 50.Sakharkar MK, Tan TW, de Souza SJ. Generation of a database containing discordant intron positions in eukaryotic genes (MIDB) Bioinformatics. 2001;17:671–675. doi: 10.1093/bioinformatics/17.8.671. [DOI] [PubMed] [Google Scholar]
- 51.Sverdlov A, Babenko V, Rogozin I, Koonin E. Preferential loss and gain of introns in 3′ portions of genes suggests a reverse-transcription mechanism of intron insertion. Gene. 2004;338:85–91. doi: 10.1016/j.gene.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 52.Mourier T, Jeffares DC. Eukaryotic intron loss. Science. 2003;300:1393. doi: 10.1126/science.1080559. [DOI] [PubMed] [Google Scholar]
- 53.Lin K, Zhang D-Y. The excess of 5′ introns in eukaryotic genomes. Nucleic Acids Res. 2005;33:6522–6527. doi: 10.1093/nar/gki970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Niu D-K, Hou W-R, Li S-W. mRNA-Mediated intron losses: evidence from extraordinarily large exons. Mol. Biol. Evol. 2005;22:1475–1481. doi: 10.1093/molbev/msi138. [DOI] [PubMed] [Google Scholar]
- 55.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 56.Fink G. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- 57.Raible F, Tessmar-Raible K, Osoegawa K, Wincker P, Jubin C, Balavoine G, Ferrier D, Benes V, de Jong P, Weissenbach J, et al. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310:1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- 58.Roy SW, Penny D. Intron length distributions and gene prediction. Nucleic Acids Res. 2007;35:4737–4742. doi: 10.1093/nar/gkm281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siegel N, Hoegg S, Salzburger W, Braasch I, Meyer A. Comparative genomics of ParaHox clusters of teleost fishes: gene cluster breakup and the retention of gene sets following whole genome duplications. BMC Genomics. 2007;8:312. doi: 10.1186/1471-2164-8-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Louis A, Ollivier E, Aude J-C, Risler J-L. Massive sequence comparisons as a help in annotating genomic sequences. Genome Res. 2001;11:1296–1303. doi: 10.1101/gr.177601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy S, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc. Natl Acad. Sci. USA. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coghlan A, Durbin R. Genomix: a method for combining gene-finders’ predictions, which uses evolutionary conservation of sequence and intron-exon structure. Bioinformatics. 2007;23:1468–1475. doi: 10.1093/bioinformatics/btm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Csuros M, Holey JA, Rogozin IB. In search of lost introns. Bioinformatics. 2007;23:i87–i96. doi: 10.1093/bioinformatics/btm190. [DOI] [PubMed] [Google Scholar]
- 64.Dacks JB, Marinets A, Ford Doolittle W, Cavalier-Smith T, Logsdon JM., Jr Analyses of RNA Polymerase II genes from free-living protists: phylogeny, long branch attraction, and the eukaryotic big bang. Mol. Biol. Evol. 2002;19:830–840. doi: 10.1093/oxfordjournals.molbev.a004140. [DOI] [PubMed] [Google Scholar]
- 65.Vanin EF. Processed pseudogenes: characteristics and evolution. Annual Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
- 66.Harrison PM, Gerstein M. Studying genomes through the aeons: protein families, pseudogenes and proteome evolution. J. Mol. Biol. 2002;318:1155–1174. doi: 10.1016/s0022-2836(02)00109-2. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Z, Carriero N, Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 68.D’Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief. Funct. Genom. Proteom. 2004;3:157–167. doi: 10.1093/bfgp/3.2.157. [DOI] [PubMed] [Google Scholar]
- 69.Benito-Gutierrez E, Nake C, Llovera M, Comella JX, Garcia-Fernandez J. The single AmphiTrk receptor highlights increased complexity of neurotrophin signalling in vertebrates and suggests an early role in developing sensory neuroepidermal cells. Development. 2005;132:2191–2202. doi: 10.1242/dev.01803. [DOI] [PubMed] [Google Scholar]
- 70.Kaessmann H, Zollner S, Nekrutenko A, Li W-H. Signatures of domain shuffling in the human genome. Genome Res. 2002;12:1642–1650. doi: 10.1101/gr.520702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vibranovski M, Sakabe N, Oliveira R, Souza S. Signs of ancient and modern exon-shuffling are correlated to the distribution of ancient and modern domains along proteins. J. Mol. Evol. 2005;61:341–350. doi: 10.1007/s00239-004-0318-y. [DOI] [PubMed] [Google Scholar]
- 72.Patthy L. Genome evolution and the evolution of exon-shuffling — a review. Gene. 1999;238:103–114. doi: 10.1016/s0378-1119(99)00228-0. [DOI] [PubMed] [Google Scholar]
- 73.Patthy L. Modular assembly of genes and the evolution of new functions. Genetica. 2003;118:217–231. [PubMed] [Google Scholar]
- 74.Roy SW, Penny D. Widespread intron loss suggests retrotransposon activity in ancient apicomplexans. Mol. Biol. Evol. 2007;24:1926–1933. doi: 10.1093/molbev/msm102. [DOI] [PubMed] [Google Scholar]
- 75.Ferrier DEK, Minguillon C, Holland PWH, Garcia-Fernandez J. The amphioxus Hox cluster: deuterostome posterior flexibility and Hox14. Evol. Dev. 2000;2:284–293. doi: 10.1046/j.1525-142x.2000.00070.x. [DOI] [PubMed] [Google Scholar]
- 76.Endo Y, Liu Y, Kanno K, Takahashi M, Matsushita M, Fujita T. Identification of the mouse H-ficolin gene as a pseudogene and orthology between mouse ficolins A/B and human L-/M-ficolins. Genomics. 2004;84:737–744. doi: 10.1016/j.ygeno.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 77.Franck E, Madsen O, van Rheede T, Ricard GN, Huynen MA, de Jong WW. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004;59:792–805. doi: 10.1007/s00239-004-0013-z. [DOI] [PubMed] [Google Scholar]
- 78.Jordal BH. Elongation factor 1 alpha resolves the monophyly of the haplodiploid ambrosia beetles Xyleborini (Coleoptera: Curculionidae) Insect Mol. Biol. 2002;11:453–465. doi: 10.1046/j.1365-2583.2002.00354.x. [DOI] [PubMed] [Google Scholar]
- 79.Babenko V, Rogozin I, Mekhedov S, Koonin E. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 2004;32:3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roy SW, Penny D. On the incidence of intron loss and gain in paralogous gene families. Mol. Biol. Evol. 2007;24:1579–1581. doi: 10.1093/molbev/msm082. [DOI] [PubMed] [Google Scholar]
- 81.Hoffman MM, Birney E. Estimating the neutral rate of nucleotide substitution using introns. Mol. Biol. Evol. 2007;24:522–531. doi: 10.1093/molbev/msl179. [DOI] [PubMed] [Google Scholar]
- 82.Castresana J. Estimation of genetic distances from human and mouse introns. Genome Biol. 2002;3:research0028.0021–research0028.0027. doi: 10.1186/gb-2002-3-6-research0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 84.Xing Y, Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl Acad. Sci. USA. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Resch AM, Carmel L, Marino-Ramirez L, Ogurtsov AY, Shabalina SA, Rogozin IB, Koonin EV. Widespread positive selection in synonymous sites of mammalian genes. Mol. Biol. Evol. 2007;24:1821–1831. doi: 10.1093/molbev/msm100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nielsen R, Bauer DuMont VL, Hubisz MJ, Aquadro CF. Maximum likelihood estimation of ancestral codon usage bias parameters in Drosophila. Mol. Biol. Evol. 2007;24:228–235. doi: 10.1093/molbev/msl146. [DOI] [PubMed] [Google Scholar]
- 87.Neafsey D, Galagan J. Positive selection for unpreferred codon usage in eukaryotic genomes. BMC Evol. Biol. 2007;7:119. doi: 10.1186/1471-2148-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeo GW, Nostrand ELV, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Epstein DJ, McMahon AP, Joyner AL. Regionalization of sonic hedgehog transcription along the anteroposterior axis of the mouse central nervous system is regulated by Hnf3-dependent and -independent mechanisms. Development. 1999;126:281–292. doi: 10.1242/dev.126.2.281. [DOI] [PubMed] [Google Scholar]
- 90.Kabat JL, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler AM. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comp. Biol. 2006;2:e86. doi: 10.1371/journal.pcbi.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lessa E. Rapid surveying of DNA sequence variation in natural populations. Mol. Biol. Evol. 1992;9:323–330. doi: 10.1093/oxfordjournals.molbev.a040723. [DOI] [PubMed] [Google Scholar]
- 92.Slade RW, Moritz C, Heideman A, Hale PT. Rapid assessment of single-copy nuclear DNA variation in diverse species. Mol. Ecol. 1993;2:359–373. doi: 10.1111/j.1365-294x.1993.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 93.Pecon-Slattery J, Pearks Wilkerson AJ, Murphy WJ, O’Brien SJ. Phylogenetic assessment of introns and SINEs within the Y chromosome using the cat family Felidae as a species tree. Mol. Biol. Evol. 2004;21:2299–2309. doi: 10.1093/molbev/msh241. [DOI] [PubMed] [Google Scholar]
- 94.Eick GN, Jacobs DS, Matthee CA. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera) Mol. Biol. Evol. 2005;22:1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- 95.Willows-Munro S, Robinson TJ, Matthee CA. Utility of nuclear DNA intron markers at lower taxonomic levels: phylogenetic resolution among nine Tragelaphus spp. Mol. Phylogenet. Evol. 2005;35:624–636. doi: 10.1016/j.ympev.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 96.Creer S, Pook CE, Malhotra A, Thorpe RS. Optimal intron analyses in the Trimeresurus radiation of asian pitvipers. Syst. Biol. 2006;55:57–72. doi: 10.1080/10635150500431213. [DOI] [PubMed] [Google Scholar]
- 97.Matthee CA, Eick G, Willows-Munro S, Montgelard C, Pardini AT, Robinson TJ. Indel evolution of mammalian introns and the utility of non-coding nuclear markers in eutherian phylogenetics. Mol. Phylogenet. Evol. 2007;42:827–837. doi: 10.1016/j.ympev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 98.Matocq MD, Shurtliff QR, Feldman CR. Phylogenetics of the woodrat genus Neotoma (Rodentia: Muridae): implications for the evolution of phenotypic variation in male external genitalia. Mol. Phylogenet. Evol. 2007;42:637–652. doi: 10.1016/j.ympev.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 99.Slade R, Moritz C, Heideman A. Multiple nuclear-gene phylogenies: application to pinnipeds and comparison with a mitochondrial DNA gene phylogeny. Mol. Biol. Evol. 1994;11:341–356. doi: 10.1093/oxfordjournals.molbev.a040117. [DOI] [PubMed] [Google Scholar]
- 100.Prychitko TM, Moore WS. Comparative evolution of the mitochondrial cytochrome b gene and nuclear {beta}-fibrinogen intron 7 in woodpeckers. Mol. Biol. Evol. 2000;17:1101–1111. doi: 10.1093/oxfordjournals.molbev.a026391. [DOI] [PubMed] [Google Scholar]
- 101.Prychitko TM, Moore WS. Alignment and phylogenetic analysis of {beta}-fibrinogen intron 7 sequences among avian orders reveal conserved regions within the intron. Mol. Biol. Evol. 2003;20:762–771. doi: 10.1093/molbev/msg080. [DOI] [PubMed] [Google Scholar]
- 102.Waterston R, Lindblad-Toh K, Birney E, Rogers J, Abril J, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 103.Hall N, Karras M, Raine JD, Carlton JM, Kooij TWA, Berriman M, Florens L, Janssen CS, Pain A, Christophides GK, et al. A Comprehensive survey of the plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 104.Venkatesh B, Ning Y, Brenner S. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc. Natl Acad. Sci. USA. 1999;96:10267–10271. doi: 10.1073/pnas.96.18.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rokas A, Kathirithamby J, Holland PWH. Intron insertion as a phylogenetic character: the engrailed homeobox of Strepsiptera does not indicate affinity with Diptera. Insect Mol. Biol. 1999;8:527–530. doi: 10.1046/j.1365-2583.1999.00149.x. [DOI] [PubMed] [Google Scholar]
- 106.Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 107.Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DHA. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl Acad. Sci. USA. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krzywinski J, Besansky NJ. Frequent intron loss in the white gene: a cautionary tale for phylogeneticists. Mol. Biol. Evol. 2002;19:362–366. doi: 10.1093/oxfordjournals.molbev.a004091. [DOI] [PubMed] [Google Scholar]
- 109.Roy SW, Gilbert W. Resolution of a deep animal divergence by the pattern of intron conservation. Proc. Natl Acad. Sci. USA. 2005;102:4403–4408. doi: 10.1073/pnas.0409891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aguinaldo AMA, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 111.Dopazo H, Dopazo J. Genome-scale evidence of the nematode-arthropod clade. Genome Biol. 2005;6:R41. doi: 10.1186/gb-2005-6-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Philippe H, Lartillot N, Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of ecdysozoa, lophotrochozoa, and protostomia. Mol. Biol. Evol. 2005;22:1246–1253. doi: 10.1093/molbev/msi111. [DOI] [PubMed] [Google Scholar]
- 113.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 114.Irimia M, Maeso I, Penny D, Garcia-Fernandez J, Roy SW. Rare coding sequence changes are consistent with ecdysozoa, not coelomata. Mol. Biol. Evol. 2007;24:1604–1607. doi: 10.1093/molbev/msm105. [DOI] [PubMed] [Google Scholar]
- 115.Krauss V, Pecyna M, Kurz K, Sass H. Phylogenetic mapping of intron positions: a case study of translation initiation factor eIF2-gamma. Mol. Biol. Evol. 2005;22:74–84. doi: 10.1093/molbev/msh255. [DOI] [PubMed] [Google Scholar]