Abstract
The Saccharomyces cerevisiae protein Cdc13 tightly and specifically binds the conserved G-rich single-stranded overhang at telomeres and plays an essential role in telomere end-protection and length regulation. The 200 residue DNA-binding domain of Cdc13 (Cdc13-DBD) binds an 11mer single-stranded representative of the yeast telomeric sequence [Tel11, d(GTGTGGGTGTG)] with a 3 pM affinity and specificity for three bases (underlined) at the 5′ end. The structure of the Cdc13-DBD bound to Tel11 revealed a large, predominantly aromatic protein interface with several unusual features. The DNA adopts an irregular, extended structure, and the binding interface includes a long (∼30 amino acids) structured loop between strands β2-β3 (L2–3) of an OB-fold. To investigate the mechanism of ssDNA binding, we studied the free and bound states of Cdc13-DBD using NMR spectroscopy. Chemical shift changes indicate that the basic topology of the domain, including L2–3, is essentially intact in the free state. Changes in slow and intermediate time scale dynamics, however, occur in L2–3, while conformational changes distant from the DNA interface suggest an induced fit mechanism for binding in the ‘hot spot’ for binding affinity and specificity. These data point to an overall binding mechanism well adapted to the heterogeneous nature of yeast telomeres.
INTRODUCTION
Single-stranded DNA (ssDNA) is present in small quantities in the cell, and its presence is limited predominantly to highly regulated and controlled processes, such as transcription, DNA replication and maintenance of specialized nucleoprotein structures (telomeres). If left unattended, ssDNA triggers a damage response, impacting cellular proliferation and survival (1). Specialized proteins bind ssDNA for subsequent actions such as repair, degradation or, in the case of telomeres, to sequester it from other ssDNA-binding proteins. Recognition and binding of ssDNA creates unique challenges due to the inherent flexibility of ssDNA as compared to double-stranded DNA (dsDNA) or ordered RNAs. Many ssDNA-binding proteins, including the telomere end-protection (TEP) family of proteins and the autoantibody DNA-1, specifically recognize their ligands through stacking interactions between aromatic amino acids and the bases and specific hydrogen-bonding interactions (2,3). Intra-molecular interactions within the ssDNA can also play a role, for example, in the telomere end-binding protein protection of telomeres 1 (Pot1) from humans and fission yeast and the relaxase TraI (4–6). Shape complementarity on the surface of the protein contributes to recognition by TraI (7). Characterization of the mechanism of ssDNA binding has been hampered by the limited number of studies of the unbound state of these proteins, especially in OB-fold family of ssDNA-binding proteins (8).
The oligonucleotide/oligosaccharide-binding fold (OB-fold) is the most common topology involved in the recognition of ssDNA (8). Proteins in this family are involved in diverse functions in the cell including replication, repair and recombination of DNA and in chromosomal end-protection (8). Nearly all OB-fold proteins bind their ligands along a similar interface and with the same nucleic acid polarity despite very low sequence conservation (8). Interestingly, the specific determinants of recognition vary greatly even within a closely related sub-family, such as the telomere-end protection proteins (2,9). Within this family, the number of OB-folds, the amino acid composition at the interface, the region of the ligand required for recognition and the amount of solvent accessible area vary widely (2). Structural information is available for the bound states of the telomere end-binding proteins from Oxytricha nova (TEBPαβ), fission yeast (SpPot1), human (HsPot1) and budding yeast (Cdc13) (4,5,10–13). However, comparative studies of the free and ssDNA-bound states are necessary to understand the structural changes that occur upon binding ssDNA.
We have probed the mechanism of ssDNA recognition exhibited by the DNA-binding domain of Cdc13 (Cdc13-DBD), the essential telomere end-binding protein from Saccharomyces cerevisiae. Cdc13 binds the conserved G-rich single strand overhang found at telomere ends, and provides a platform on which higher order complexes are assembled (14). Cdc13 regulates key telomere functions including end-capping and telomerase activity (15–18). The structure of the Cdc13-DBD was solved in complex with an 11-base ssDNA oligonucleotide Tel11 [d(GTGTGGGTGTG)] representative of yeast telomere sequence (11,12). Tel11 represents the minimal DNA required for high affinity binding (Kd = 3 pM), with truncation from either the 5′ or 3′-end resulting in loss of affinity (J. N. Roberts, DSW, unpublished). The Cdc13-DBD adopts an OB-fold with Tel11 bound in a completely extended configuration and the bases interacting with a large surface of the protein.
One striking feature of the Cdc13-DBD/Tel11 complex is an unusually long well-ordered loop of ∼30 amino acids between β-strands 2 and 3, L2–3. Comparative analysis with other OB-folds reveals that the canonical ligand-binding site (between loops 1–2 and 4–5, which bind the 5′-end of the ligand) is augmented in the Cdc13-DBD complex with L2–3. This loop is integral to DNA binding, and occupies one-quarter of the protein/DNA interface. Mutational analysis of L2–3 shows a number of residues that are thermodynamically important for high affinity binding, but these sites do not appear to be the primary mediators of sequence-specific recognition (9,19). This loop confers the requirement for a large DNA ligand, and presumably contributes to the unusually high affinity Cdc13-DBD has for ssDNA. While uncommon, a long L2–3 that contributes to nucleic acid recognition is also found in other OB-fold containing proteins, including Escherichia coli Rho, T. thermophilus ribosomal protein S17 and E. coli SSB (8).
To better understand the mechanism of ssDNA recognition, specifically the reorganization that occurs in the protein upon binding, we have compared the free and ssDNA-bound states of the Cdc13-DBD. As changes in both static structural features and dynamic behavior of proteins are linked to biological function (20), we have probed the roles of both structural rearrangements and changes in dynamic behavior upon binding. Our analysis shows that the free state has the same global fold as the bound state and does not experience a dramatic folding event upon ssDNA binding (20,21). Rather, the binding of ssDNA induces localized conformational changes in a previously unappreciated region of the protein distant from the binding site, suggesting a tightening of this region as it orders around the DNA. In the absence of DNA, L2–3 is constrained at the tip but has increased flexibility at either end. This constraint could serve to pre-position the loop until subsequent DNA binding locks it into place, thus providing some malleability for recognition of the heterogeneous yeast telomere ligand.
MATERIALS AND METHODS
All chemicals and reagents were obtained from Fisher Scientific (Pittsburg, PA) unless otherwise indicated.
Expression and purification procedures
Cdc13-DBD
The C-terminally His-tagged DNA-binding region of Cdc13, residues 497–694 was expressed in E. coli as previously described (9,22). Incorporation of 15N and 13C isotopes for NMR analysis was accomplished by growing in media containing (15NH4)2SO4 and 13C glucose as the sole nitrogen and carbon sources, respectively.
Cells were resuspended in 50 mM potassium phosphate, pH 7.0, 300 mM NaCl, 150 mM Na2SO4, Complete EDTA-free protease inhibitor (Roche Applied Science, Germany) and 5 mM β-mercaptoethanol (βME) and lysed via sonication. Following centrifugation, residually bound DNA was removed from the protein by the addition of 0.05% polyethylene imine (PEI) stirring at 4°C for an hour. The sample was centrifuged and the supernatant was retained for a second round of PEI-precipitation at 1 M NaCl. The supernatant was dialyzed overnight against 50 mM potassium phosphate, pH 7.0, 50 mM NaCl, 5 mM βME. The protein was purified by cation exchange using two tandem 5 ml Hi-TRAP SP columns on an ACTA FPLC (GE Healthcare), with a 0–1 M NaCl gradient. Protein-containing fractions were pooled and further purified using a Ni-chelating sepharose column (GE Healthcare). The chelating column was washed with increasing concentrations of imidazole (10, 20, 50 and 100 mM) and the protein eluted with 250 and 500 mM imidazole in 50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 100 mM Na2SO4.
Tel11
Oligonucleotides with the sequence d(GTGTGGGTGTG) were obtained from IDT (Coralville, IA) at 1–10 μM scale. DNA was resuspended in 1 ml of deionized water per micromole of DNA, purified on a C18 reversed-phase column (Vydac) and eluted with a gradient of 5–40% 10 mM triethanolamineacetate, pH 6.5/80% acetonitrile in water. Fractions from HPLC purification were lyophilized and resuspended with deionized water three times before final resuspension in 200 μl of deionized water. Final concentrations were determined by absorbance at 260 nm using the extinction coefficient (106 900 M−1 cm−1).
Preparation of NMR samples
Purified Cdc13-DBD protein was exchanged into the NMR sample buffer (50 mM imidazole-d4, 300 mM NaCl, 100 mM Na2SO4, 10% 2H2O, 0.02% NaN3, ∼2 mM DTT-d10 at pH 7) through several rounds of concentration, dilution with the NMR buffer and then re-concentration. The final concentration of the free-state samples was determined by absorbance at 280 nm using an extinction coefficient of 10 382 M−1 cm−1 and ranged from 0.3 to 0.7 mM. Samples could not be concentrated further due to solubility issues. While these concentration levels are too low for structure determination by NMR, they are sufficient for making backbone resonance assignments and measuring relaxation parameters.
Preparation of NMR samples for the bound state followed the same procedure as for the free state; the protein was exchanged into the NMR buffer (50 mM imidazole-d4, 300 mM NaCl, 100 mM Na2SO4, 10% 2H2O, 0.02% NaN3, ∼2 mM DTT-d10 at pH 7) through several rounds of dilution/concentration. On the last round of concentration, the DNA was added to a final ratio of 1:1 with protein. Since the bound state is more stable than the free state, the bound sample was concentrated further to ∼0.7 to 1.0 mM.
NMR analysis
Assignments and chemical shift analysis
Spectra for assignments of 13C 15N-labeled Cdc13-DBD free and bound to Tel11 were obtained on a Varian 600 Inova MHz spectrometer equipped with a cold probe and on a Varian Inova 800 MHz spectrometer equipped with an RT probe. The bound state of the protein was reassigned in regions that were altered by sample buffer differences and the presence of the affinity tag, and the free state was assigned de novo. Standard two- and three-dimensional sensitivity enhanced, gradient selected non-TROSY experiments were acquired using Varian BioPack pulse sequences with minor modifications; HSQC, HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH and HNCO (23). Spectra were processed using the nmrPipe package (24) and assignments were made using ANSIG (25). Chemical-shift differences are reported using [(ΔHppm)2 + (0.17*ΔNppm)2]1/2, where ΔH is the difference in chemical shift from free to bound of the amide proton, ΔN is the difference in the nitrogen chemical shift.
Secondary structure elements were determined using the Cα chemical shift index in which the random coil Cα value was subtracted from the actual Cα chemical shift for each amino acid (26–28). Residues with positive differences have helical propensity and negative values suggest β-sheets. In order to assess similarities between free and bound predicted secondary structure, the ΔCαbound-random values for the bound state were subtracted from the free state ΔCαfree-random values.
Hydrogen exchange
NMR samples used in hydrogen-exchange experiments were transferred to 100% 2H2O buffer (50 mM imidazole-d4, 300 mM NaCl, 100 mM Na2SO4, 0.02% NaN3, ∼2 mM DTT-d10 at pH 7) using 3 × 200 μl G-25 Sepharose spin columns. A series of 2 h 15N HSQC spectra at 800 MHz were obtained immediately after buffer exchange and every 2 h until 6 h post-exchange, at which point spectra were obtained every 4 h until 50 h post-exchange. Peak volumes at each time point were determined using the NLinS portion of the nmrPipe software (24). The first spectrum was used as the reference in the normalization of subsequent spectra. The volumes were then fitted to a single exponential decay to determine a rate of exchange. The rate difference between free and bound was determined in order to directly compare the two states. For the purposes of calculating a rate differences, residues in which the corresponding peak had fully exchanged by 2 h were assigned a minimal estimated rate of 2 h−1 (the fastest observed rate in this analysis was 1.9 h−1) and residues in which the peak intensity was unchanged over the course of the experiment were assigned a rate of 0 h−1.
Dynamics data
The 15N relaxation times T1, T2 and values for the {1H}15N NOE were measured at 600 MHz using two-dimensional experiments obtained from the Kay lab (29). The T1 and T2 relaxation delays were interleaved in a ‘pseudo-random’ order to minimize systematic errors (30) with delays of 11, 44.1, 88.1, 154.2, 220, 440.1, 770.9, 770.9, 1101.3, 1431.7, 1982.4 ms for T1 and 0, 9.4, 18.7, 28.0, 37.4, 37.4, 56.1, 74.8, 93.5, 112.2, 130.9 ms for T2. Values for T1 and T2 were determined by plotting peak volumes versus delay time and fitting the curve to a single exponential decay using xcrvfit (31). The error derived from the fit of the curve was used to assess the quality of the data, similar T1 and T2 values were obtained using data collected on more dilute samples. The values for the {1H}15N NOE were determined by the average of two experiments on different samples of the ratio of peak volumes with and without 1H saturation. Errors were estimated based on the ratio of peak height to noise in the spectrum.
The Rex values resulting from milli- to microsecond motions in the protein were determined via a constant relaxation time CPMG experiment collected at 800 MHz (32,33). The peak intensities for each spectrum in which the spacing between pulses (τCPMG) was varied were determined as for the 15N fast internal dynamics measurements. For each residue, the normalized peak intensities were plotted versus the delay time and fit to a two-state model for conformational exchange on the ‘fast’ timescale using the program Curvefit (Free Software Foundation, Inc., Boston, MA) with the following equation: R2eff(τcp) = R2 + Rex*(1–2τx tanh(1/(2τx))) (see Supplementary Figure 2 for sample data). Where R2 is the transverse relaxation rate, Rex = pa pb δω2/kex; pa and pb are the site populations, δω is the chemical shift difference between sites (1/s), kex is the exchange rate constant and τ = 1/kex (34). Errors were derived from the fit.
RESULTS
The solution structure of the Cdc13-DBD bound to its cognate ligand, Tel11 [d(GTGTGGGTGTG)] showed that the ssDNA binds in an extended, irregular conformation across a large protein interface that includes a long structured loop between strands β2 and β3 (L2–3) (11,12). This binding event induces a fixed ssDNA conformation in which numerous aromatic residues in the Cdc13-DBD make extensive stacking interactions with the bound DNA. To better understand the mechanism of ssDNA binding by the Cdc13-DBD, we have compared the structural and dynamical features of the free and bound states of the protein. The free state of the protein has been difficult to study in isolation, presumably due to the highly aromatic interface exposed to solution in the absence of DNA. Due to the instability of the free protein, a C-terminal His-tag was added to the Cdc13-DBD to enhance solubility. The addition of this modification had no impact on binding affinity (9,19), however, it did allow us to identify conditions where the protein was stable enough to obtain data for backbone assignments. These conditions were not sufficient to obtain side chain assignments and the extensive NOE data required for de novo structure determination. Therefore, conformational changes in the protein that accompany binding were evaluated by chemical shift mapping of fully assigned free and bound states and through differences in H/D exchange rates in the free and bound Cdc13-DBD. Additionally, the DNA-dependent changes in the backbone dynamics of Cdc13-DBD on fast (ps–ns) and on slower timescales (μs–ms) were determined using 15N relaxation experiments.
Backbone assignments of the free and bound states of the Cdc13-DBD
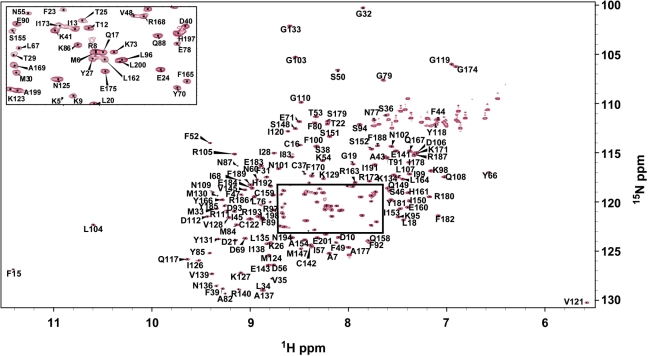
The Cdc13-DBD/Tel11 complex is in slow exchange, thus the free-state assignments could not be derived from ssDNA titrations using assignments from the bound state. Assignments for backbone atoms (1HN, 15N, 13Cα, 13Cβ, CO) were made using standard triple-resonance experiments for Cdc13-DBD free in solution and bound to the cognate ssDNA ligand, Tel11 [d(GTGTGGGTGTG)] (Figure 1, and Supplementary Table 1). A total of 169 amide backbone assignments (out of 195 possible) were determined for the free state (87%) and 175 for the bound state (90%). In the free state, there are only four peaks of normal intensity in the 15N HSQC spectrum that were not assignable, suggesting that the majority of the 25 unassigned residues are exchange broadened. The missing residues in the free state are found at the extreme N-terminus (residues M1–S4), in L2–3 and in β3 (residues D51, V58, Q59, Y61-R65, E72, L74, E75, K81), and in loop α2–β4 (residues E113–S116). In the bound state, 19 residues could not be assigned, with seven unassigned peaks in the 15N HSQC spectrum. The pattern of missing assignments at the N- and C-termini is similar to that of the free state, with seven assignments missing from the N-terminus and two from the C-terminus. Five residues (N132, S157, S176, G195, S196) are unassigned in both free and bound states. In contrast to the free state, the bound state does not have any missing assignments in L2–3, instead residues preceding and in α3 (H147, S155-E160, L162, L164) could not be assigned.
Figure 1.
1H-15N HSQC spectrum of free Cdc13-DBD with assignments labeled. Boxed region labelled in inset.
The chemical shifts of the bound state are similar in most regions to those reported previously for the non-His-tagged complex (35). The affinity tag and altered buffer conditions cause minor chemical shift changes, predominantly in the C-terminal helices, presumably due to local effects of the His tag on the C-terminus. Assignments could not be obtained for virtually identical regions of the protein.
Chemical shift changes indicate local (but not global) conformational changes accompany binding
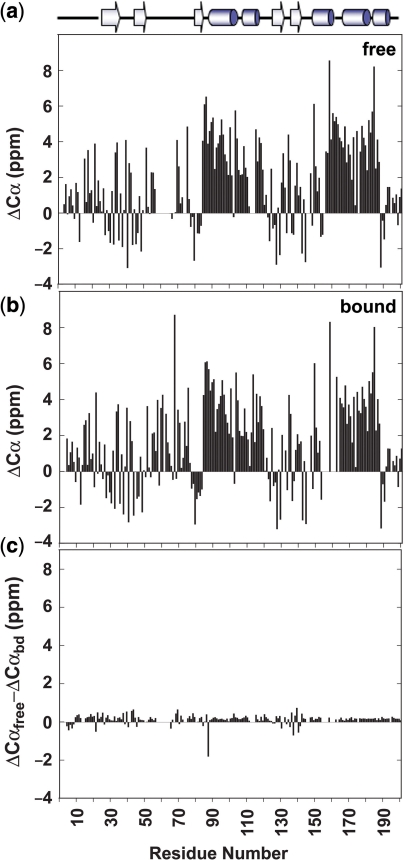
Binding DNA has no dramatic effect on the secondary structure of the Cdc13-DBD as assessed by the chemical shift index for Cα atoms. The difference in actual Cα chemical shift from the random coil value predicts the type of secondary structure for each residue with positive values indicative of helices and negative values of β-sheets (26–28). The pattern of secondary structure elements of the free state is likely to be the same as for the bound state since they have nearly identical values for the Cα chemical shift differences from random coil values (Figure 2a and b). One notable exception is residue Q88 at the start of helix 1, which is less helical in the free state as shown by the large negative value for ΔCαfree-ΔCαbound (Figure 2c).
Figure 2.
Graphs showing the difference in Cα chemical shift from random coil values for the free state (a) and the bound state (b). Positive values indicate helical propensity and negative values show regions in β-strands. The difference of free to bound differences to random coil values shows virtually no change in chemical shift index value (c).
Further evidence for the similarity in global topology is the observation that over half of the residues (85 out of 158) have <0.07 ppm change in composite chemical shift from the free to bound states (in black in Figure 3a). This similarity indicates that the majority of the core residues and the N- and C-termini are unaffected by DNA binding, which is consistent with the observation of identical patterns of proteolytic protection of the free and bound states (E. M. Anderson and DSW, unpublished).
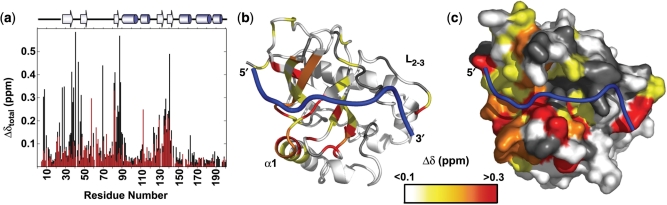
Figure 3.
Graph of combined chemical shift difference from free to bound showing the improved analysis of using actual chemical shift values (black) over minimal analysis (red) (a). Combined chemical shift difference values were determined using the following equation Δδ = [(ΔH)2 + (0.17*(ΔN))2]1/2 (11). Chemical shift differences mapped onto the structure of Cdc13-DBD/Tel11 [PDB accession number 1S40 (12)] (b and c). The differences are shown in a color ramp from red to yellow with the largest changes in red (>0.3 ppm), residues with very small changes are shown in white, and residues with no data are in grey [figure was prepared using PyMol (46)].
In the absence of assignments for the free state, an estimate of residues involved in either binding or conformational change was determined by minimal chemical shift analysis (11). In this analysis, a peak in the unassigned free-state spectrum is assumed to arise from the nearest known assigned bound state peak, providing the most conservative estimate of chemical shift change. Because this method underestimates the true chemical shift change, we assigned the free protein de novo. Comparison of the true chemical shift changes (in black in Figure 3a) with the minimal chemical shift changes (in red in Figure 3a, performed in the absence of free assignments) illustrates how minimal chemical shift changes underestimate environmental changes. Generally, the regions that experience changes are similar regardless of the method used to calculate chemical shift differences, however the magnitude and the number of residues experiencing a significant change are much greater when free state assignments are used in the analysis. Additionally, new regions with mechanistic significance emerged from analysis of the exact chemical shifts. For example, minimal perturbation analysis did not find the changes in residues D21-V48 or in α1.
As expected, the majority of residues located at the interface with DNA have significant chemical shift changes (Figure 3b and c). Analysis of the Cdc13-DBD/Tel11 structure shows that 39 amino acids have atoms within 5 Å of the DNA, of which nine are prolines or are unassigned (2). Twenty-one of these interface residues have >0.1 ppm chemical shift change (70%). Only one of the remaining nine contact residues with a <0.1 ppm chemical shift difference has the amide nitrogen atom within 3 Å to the DNA, residue Y66. This lack of a shift upon binding could reflect a similar overall environment for the amide upon DNA binding or may indicate that the amide is in fact further from the DNA. All of the residues in the thermodynamic ‘hotspot’ of binding affinity have large chemical shift changes (19).
Three areas of the protein exhibit unexpected chemical shift behavior. The first region, L2–3, is remarkable for the lack of change upon binding. This loop, consisting of residues S50-E78, is thermodynamically important for binding the 3′-end of Tel11 and in maintaining the extended state of the DNA (19). The majority of non-contact residues in the loop (D50-K54, D56, I57, L67-D69, L76-G79) have chemical shift changes <0.1 ppm, suggesting that these residues do not significantly alter their chemical environment upon binding DNA. It is likely that these residues are in the same conformation in the free and bound states and may indicate that the structure of the loop is preformed in the free state prior to binding DNA.
The second region of unusual chemical shifts is in the N-terminal half of α1. A series of residues with moderate to large chemical shifts extends from residue G79 in the center of β3 through residue S94 in the center of α1. Residues within the loop between the secondary structure elements contact DNA and are important for binding affinity, however, the extension of chemical shift change from the interface deep into α1 was surprising and suggests a conformational change transferred from the actual binding residues through that helix. This could be mediated by nearby residue E141, which also has a large chemical shift change.
The third region of chemical shift change is located near the 5′-end binding site of the DNA. This region of the protein is critical for conferring DNA-binding affinity and specificity (9,19). Residue T29, which is >5 Å from the DNA, has a large 0.35 ppm composite change. T29 is near a known critical recognition residue Y27, which contacts G1, but the bound structure provides no clear reason for the shift in T29. T29 and Y27 side chains do not interact and none of the surrounding residues have altered chemical shifts. Interestingly, the first two assignable residues of the bound protein, A7 and R8, both have large chemical shift changes upon addition of DNA. These data, combined with the observation that D10 contributes to DNA-binding affinity (V. Lundblad and DSW, unpublished data), suggest that the N-terminal region of the protein undergoes a conformational change upon binding.
Hydrogen exchange indicates protection at the protein/DNA interface and highlights regions of conformational change
We probed the DNA-dependent changes in local and global conformation and dynamics by determining hydrogen exchange rates for each amide in both the free and bound states of the Cdc13-DBD. A total of 121 sites were resolved enough in both states for analysis (Supplementary Table 2). In both the free and bound states, 42 of the 121 residues were fully exchanged by the first time point at 2 h post-transfer into 2H2O buffer. In addition, the free state had an additional nine residues exchanged by 2 h. There are 31 slow exchanging residues in common between the free and bound states and an additional 16 slowly exchanging residues in the bound state. The majority of residues (79%) were essentially unchanged (<±0.075 h−1 difference) in the presence of DNA (see top curves and bottom curves in Figure 4a). However, in 21% of residues the observed rates varied widely between free and bound (see middle curves in Figure 4a).
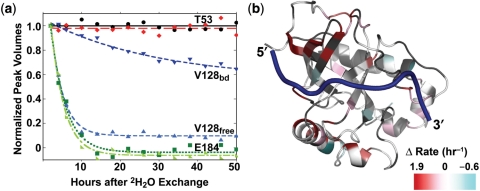
Figure 4.
Exponential decay curves of selected hydrogen exchange data showing that some residues have similar rates in free and bound whether slow (T53) or fast (E184) (a). Residue V128 is an example of a residue with very different rates between free and bound. Mapping the difference in the free and bound rates shows that residues in red near the 5′-end of the DNA and in α1 have faster hydrogen exchange rates in the free state (b).
In general, the backbone amides of the Cdc13-DBD are more protected from solvent in the DNA-bound state. As observed in many proteins, the slowest rates are in the solvent-protected core of the protein in both free and bound protein, with surface residues exchanging readily (Supplementary Table 2). The free protein has fewer residues exhibiting a large degree of protection, suggesting reduced stability in the free state (21). Mapping the difference in the rates reveals several regions that have altered hydrogen exchange properties in the presence of DNA (Figure 4b). As expected, residues along the DNA-binding interface, especially near the 5′-end and L2–3 residues, are more protected in the bound state (residues in red in Figure 4b; K26, Y27, I28, F44, Y66, M130, Y131, K134, N136, R140, E141). A second region with changes in the patterns of protection coincides with a region identified in the chemical shift analysis, the beginning of α1 and part of α2. Finally, we identified a region in α3 in the C-terminal helical extension in which the bound state of the protein exhibits a faster rate of exchange than the free state. This is consistent with this region displaying dynamic behavior in the bound state, as evidenced by a lack of observable NH peaks likely due to exchange broadening.
Limited changes occur in the fast timescale dynamic properties of the free and bound Cdc13-DBD
Internal dynamics on the ps–ns timescale were determined for the free and bound states using standard 2D 15N HSQC-based relaxation experiments (36). Both free and bound states experience dynamic behavior at the extreme termini as evidenced by pronounced deviations from the average values for T1, T2 and {1H}15N NOE (Supplementary Figure 1 and Table 2). In addition, several loop regions also appear to be more dynamic based on their decreased T1 and NOE values. There do not appear to be regions of the protein that behave dramatically differently in the free state with respect to the bound. However, if the residues with low {1H}15N NOE values (<0.7) are mapped onto the structure (Figure 5a), we observe that the free state has increased flexibility in residues near the 5′-end of the DNA. Interestingly, the L2–3 loop does not have significantly decreased NOE values in either the bound or free states, providing further evidence that the loop is ordered with respect to the whole protein.
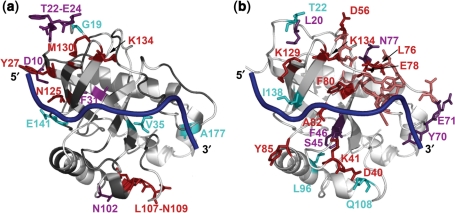
Figure 5.
Residues with average {1H}15N NOE values lower than 0.7 are mapped onto the Cdc13-DBD/Tel11 structure (a). Residues with low NOE values in both free and bound are shown in purple, residues with only low values in free are in red, only in bound are in cyan and residues with no data are in grey. Rex values are mapped on the Cdc13-DBD/Tel11 structure (b). More residues have slow and intermediate dynamics in the free state (residues in red have measurable Rex values and in salmon are the exchange broadened unassigned residues in L2–3) versus the bound (cyan). Residues exhibiting relaxation dispersion in both the free and bound states are shown in purple.
Conformational exchange data suggest altered slow timescale dynamics
Motions on the μs–ms timescale are associated with conformational changes and correlate with functional regions of proteins (20,37,38). The relaxation rate (Rex) due to conformational exchange was determined using a Carr–Purcell–Meiboom–Gill (CPMG) R2 relaxation dispersion experiment (32,33). The decay in the effective R2 relaxation rate as a function of applied field strength was fitted to the fast exchange regime to obtain values for Rex, R2eff and τ as described in the ‘Materials and Methods’ section.
Conformational exchange was observed for several sites in both the free and bound states (Supplementary Table 2). The free state had more residues (14) that experienced conformational exchange under these conditions than the bound state (11) (Figure 5b). Six of these residues are common to both states (L20, S46, F47, Y70, E71 and N77). The free state experienced additional conformational exchange along the DNA-binding interface and at the C-terminal end of L2–3.
DISCUSSION
The yeast telomeric ssDNA is unstructured in solution but adopts an extended conformation upon binding the Cdc13-DBD (12). We investigated the changes in the structure and dynamic behavior of the Cdc13-DBD upon binding DNA in order to increase our understanding of ssDNA recognition, providing the first insight into DNA-dependent changes within this important family of proteins. We found that, although the nature of the contacting amino acids is the same, the mechanism of binding differs at the 5′ and 3′-ends of the cognate ligand. These differences appear to correlate with regions most important for specificity and affinity, and provide a basis for understanding specificity in the recognition of DNA.
Conformational and dynamical changes upon binding suggest a globally preformed binding site with local folding in the region of the binding site most important for affinity and specificity
Chemical shift, H/D exchange and dynamics data all indicate that the overall topology of the Cdc13-DBD free state is quite similar to the bound state. As expected, the majority of chemical shift changes are localized to the DNA-binding interface, and DNA binding suppresses dynamics at the interface.
Two regions of the protein appear to undergo conformational structuring upon binding, α1 and residues at the 5′-end of the DNA. In the absence of DNA, these regions are less protected from hydrogen exchange and have increased fast timescale motions. The first region, α1, is at the base of the barrel and does not directly contact the DNA, however, it lies directly under T4. This helix exhibits unexpectedly large chemical shift changes upon binding DNA, indicating a change in the local environment due to a conformational change. Taken together with the decrease in dynamic behavior upon DNA binding and an increase in helical character, it appears that the N-terminal region of α1 is somewhat unwound in the free state. Since the helix supports the loop regions that interact directly with the DNA, we suspect that the reorientation of the loops leads to stabilization of the helix. The conformational and dynamic changes in this helix may regulate protein–protein interactions in the larger telomere end-binding complex. The second site of local folding or tightening of the structure upon binding occurs near the 5′-end of the binding site and includes residues in the N-terminus that are distant from the DNA. This potential local folding event explains why a residue in the N-terminus, D10, is important for binding and for cell viability (V. Lundblad and DSW, unpublished data) despite a lack of direct contact with the DNA.
These sites coincide precisely with the region of the protein and DNA most important for affinity and specificity, suggesting an induced fit mechanism for binding this region. The critical determinants for recognition of the DNA are located at the 5′ region of the DNA and in protein residues nearby (9,19). However, it is not clear how the specificity determining bases are recognized since there is no evidence of traditional Watson–Crick-like interactions. Instead the three critical bases (GXGT) appear to be recognized via both hydrogen bonding interactions and shape complementarity (9). Specificity for these sites within the cognate ligand could be attained through an induced fit model whereby residues important for hydrogen bonding are positioned correctly and the protein collapses around the DNA in order to create the appropriate shape for the base being recognized.
This result is similar but not as dramatic as the induced folding of a much smaller OB-fold protein, the cold shock protein from Bacillus subtilis, Bs-CspB (39). Binding of dT7 to Bs-CspB dramatically stabilizes the protein as assessed by a pronounced reduction in the number of residues experiencing conformational exchange, a significant increase in protection from hydrogen exchange, and increased {1H}15N NOE values (39). The global changes observed could be due to the extremely small size of this protein, the OB-fold contains just the barrel region and no surrounding helices, or to its role as an mRNA chaperone at low temperatures.
In contrast to the behavior of α1 and the 5′-binding site, the binding of DNA increased dynamical behavior (as observed by increased H/D exchange and the presence of intermediate exchange in the bound state) on the C-terminal helix, which is one the opposite face of the protein from the DNA-binding site. Increases in dynamics may represent thermodynamic compensation for ordering at the interface and in the loop (20). Alternatively, this may indicate allosteric regulation of the protein where binding of the DNA relays information to other sites in the protein as in the entropic switch described for acireductone dioxygenase (ARD) (40). DNA-binding-induced conformational changes may allow this region of the Cdc13-DBD to interact with the putative OB-fold located in the C-terminal domain of Cdc13 or with protein partners required for end-protection and telomerase activation (41).
Binding reduces slow and intermediate time scale dynamics in the L2–3 loop
The L2–3 is well ordered in the complex structure (11,12) and appears to adopt a similar orientation in the free structure. Several residues in the middle of the loop (Y66-D69, K73) and non-contacting residues (D52-K54, D56, I57, L76-G79) have no discernible chemical shift differences between the free and bound states, indicating that the loop is essentially preformed in the free state. The loop makes extensive packing interactions with the β4–β5 loop, the C-terminal long helix (α3) and with several strands in the barrel (12). In addition, a hydrogen bond between the carbonyl of N55 and the side chain amide of N77 ties the loop to itself halfway between the connection with the barrel and the region that interacts with the DNA. The majority of residues (S50-T53, N55, D56, Y61-D64, Y66-Y70, N72-L74, N77) in the loop are well-conserved (>66%) throughout Cdc13 proteins from closely related yeast species, suggesting that this structural element has evolved to pack against the barrel (12). The preordering of the loop may provide a means to keep the entropic cost of binding low enough to be compensated by enthalpic contributions and contribute to the unusually high affinity of the Cdc13-DBD for its cognate substrate.
While not completely unfolded in the free state, the loop does exhibit increased dynamic behavior in the free state on the microsecond to millisecond timescale. The first suggestion of increased intermediate to slow dynamics was the difficulty assigning residues in L2–3, nearly half of the resonances are absent in our spectra due to exchange broadening. Relaxation dispersion experiments confirm this observation, with four residues undergoing motion in the free but not the bound state. Thus, slower timescale dynamics in the loop are suppressed upon binding, a phenomenon observed in other protein/ligand interactions [for a review (20)]. The dynamic nature of the loop may be essential for binding affinity as was observed in the Pin1-WW domain, in which binding affinity was correlated with the intrinsic dynamic nature of the recognition loop (42). Similarly, dynamics in the loop may provide a means of adapting to the heterogeneous nature of yeast telomeric DNA.
The β2–β3 loop provides an extended nucleic acid binding site for several OB-fold proteins. However, L2–3 in the Cdc13-DBD is unique in its association with the barrel. The E. coli Rho and SSB proteins and the ribosomal protein S17 from T. thermophilus all have long L2–3 but in each of these cases the loop extends from the barrel to interact with nucleic acid (43–45). Dynamic loops do play a large role in binding for many OB-fold proteins, for example large DNA-dependent conformational changes are observed in L1–2 and L4–5 in the human RPA70 protein and in E. coli Rho, L1–2 becomes structured upon binding DNA (8). Unlike the loops in these proteins, the Cdc13-DBD L2–3 does not experience a large conformational change and is tethered in the middle preventing the loop from becoming completely unstructured in the absence of DNA.
This analysis on the DNA-dependent changes in conformation and dynamics in Cdc13-DBD suggests that the mechanisms of binding are different for the 5′ specificity-determining region and the region that binds the 3′-end of the DNA, L2–3. The probable co-folding event near the 5′ region of the DNA may contribute to specificity for these bases and the slower dynamics in L2–3 may assist in binding to the heterogeneous telomeric DNA found in budding yeast.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Funding was provided by National Institutes of Health (GM-059414 to DSW); the National Science Foundation (MCB-0617956 to DSW); the Keck Foundation and the Arnold and Mabel Beckman Foundation. We thank Dr Andrew Fowler for assistance with the NMR relaxation experiments and Dr Johnny Croy for helpful comments on the manuscript. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 2002;16:1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croy JE, Wuttke DS. Themes in ssDNA recognition by telomere-end protection proteins. Trends Biochem. Sci. 2006;31:516–525. doi: 10.1016/j.tibs.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Schuermann JP, Prewitt SP, Davies C, Deutscher SL, Tanner JJ. Evidence for structural plasticity of heavy chain complementarity-determining region 3 in antibody-ssDNA recognition. J. Mol. Biol. 2005;347:965–978. doi: 10.1016/j.jmb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Lei M, Podell E, Cech TR. Structure of human Pot1 bound to telomeric DNA single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 5.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the crystal structure of the Pot1 (Protection of Telomeres)-ssDNA complex. Nature. 2003;426:198–204. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 6.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Datta S, Larkin C, Shildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eldridge AM, Halsey WA, Wuttke DS. Identification of the determinants for the specific recognition of single-strand telomeric DNA by Cdc13. Biochemistry. 2005;46:871–879. doi: 10.1021/bi0512703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 11.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 12.Mitton-Fry RM, Anderson EM, Glustrom LW, Theobald DL, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 13.Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein alpha subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- 14.Lustig AJ. Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Biol. 2001;8:297–299. doi: 10.1038/86157. [DOI] [PubMed] [Google Scholar]
- 15.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans SK, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J. Cell. Sci. 2000;113:3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 17.Meier B, Driller L, Jaklin S, Feldmann HM. New function of CDC13 in positive telomere length regulation. Mol. Cell. Biol. 2001;21:4233–4245. doi: 10.1128/MCB.21.13.4233-4245.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- 19.Anderson EM, Halsey WA, Wuttke DS. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- 20.Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem. Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- 21.Englander SW, Sosnick TR, Englander JJ, Mayne L. Mechanisms and uses of hydrogen exchange. Curr. Opin. Struct. Biol. 1996;6:18–23. doi: 10.1016/s0959-440x(96)80090-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of S. cerevisiae Cdc13. Nucleic Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett T, Farmer BT, Mueller L. Unambiguous resonance assignments in 13C, 15N-labelled nucleic acids by 3D triple resonance NMR. J. Am. Chem. Soc. 1993;115:11040–11041. [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Kraulis PJ. ANSIG: a program for the assignment of protein 1H 2D NMR spectra by interactive computer graphics. J. Magn. Reson. 1989;84:627–633. [Google Scholar]
- 26.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 27.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 28.Wishart DS, Sykes BD. The 13C chemical shift index: a simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 29.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 30.Pawley NH, Clark MD, Michalczyk R. Rectifying system-specific errors in NMR relaxation measurements. J. Magn. Reson. 2006;178:77–87. doi: 10.1016/j.jmr.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Boyko R, Sykes BD. (University of Alberta); 1994. xcrvfit: a graphical X-windows program for binding curve studies and NMR spectroscopic analysis, developed by Boyko, R. and Sykes, B.D. [Google Scholar]
- 32.Loria JP, Rance M, Palmer AG., 3rd A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J. Biomol. NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- 33.Tollinger M, Skrynnikov NR, Mulder FA, Forman-Kay JD, Kay LE. Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 34.Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 35.Mitton-Fry RM, Wuttke DS. 1H, 13C and 15N resonance assignments of the DNA-binding domain of the essential protein Cdc13 complexed with single-stranded telomeric DNA. J. Biomol. NMR. 2002;22:379–380. doi: 10.1023/a:1014932727059. [DOI] [PubMed] [Google Scholar]
- 36.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 37.Mulder FA, Hon B, Muhandiram DR, Dahlquist FW, Kay LE. Flexibility and ligand exchange in a buried cavity mutant of T4 lysozyme studied by multinuclear NMR. Biochemistry. 2000;39:12614–12622. doi: 10.1021/bi001351t. [DOI] [PubMed] [Google Scholar]
- 38.Kern D, Eisenmesser EZ, Wolf-Watz M. Enzyme dynamics during catalysis measured by NMR spectroscopy. Methods Enzymol. 2005;394:507–524. doi: 10.1016/S0076-6879(05)94021-4. [DOI] [PubMed] [Google Scholar]
- 39.Zeeb M, Max KE, Weininger U, Low C, Sticht H, Balbach J. Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution. Nucleic Acids Res. 2006;34:4561–4571. doi: 10.1093/nar/gkl376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju T, Goldsmith RB, Chai SC, Maroney MJ, Pochapsky SS, Pochapsky TC. One protein, two enzymes revisited: a structural entropy switch interconverts the two isoforms of acireductone dioxygenase. J. Mol. Biol. 2006;363:823–834. doi: 10.1016/j.jmb.2006.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theobald DL, Wuttke DS. Prediction of multiple tandem OB-folds in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 42.Peng T, Zintsmaster JS, Namanja AT, Peng JW. Sequence-specific dynamics modulate recognition specificity in WW domains. Nat. Struct. Mol. Biol. 2007;14:325–331. doi: 10.1038/nsmb1207. [DOI] [PubMed] [Google Scholar]
- 43.Bogden CE, Fass D, Bergman N, Nichols MD, Berger JM. The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell. 1999;3:487–493. doi: 10.1016/s1097-2765(00)80476-1. [DOI] [PubMed] [Google Scholar]
- 44.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 45.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMol Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.