Abstract
Nuclear factor kappaB (NF-κB) plays an important role in the transcriptional regulation of genes involved in immunity and cell survival. We show here in vitro and in vivo acetylation of RelA/p65 by p300 on lysine 314 and 315, two novel acetylation sites. Additionally, we confirmed the acetylation on lysine 310 shown previously. Genetic complementation of RelA/p65−/− cells with wild type and non-acetylatable mutants of RelA/p65 (K314R and K315R) revealed that neither shuttling, DNA binding nor the induction of anti-apoptotic genes by tumor necrosis factor α was affected by acetylation on these residues. Microarray analysis of these cells treated with TNFα identified specific sets of genes differently regulated by wild type or acetylation-deficient mutants of RelA/p65. Specific genes were either stimulated or repressed by the acetylation-deficient mutants when compared to RelA/p65 wild type. These results support the hypothesis that site-specific p300-mediated acetylation of RelA/p65 regulates the specificity of NF-κB dependent gene expression.
INTRODUCTION
The inducible transcription factor family nuclear factor κB (NF-κB) consists of dimeric proteins involved in many diverse processes such as immune and stress responses and the opposing processes of proliferation and apoptosis (1–3). NF-κB is induced in almost all cell types by different extracellular stimuli causing the activation of an enormous array of target genes (4). Thus, it is not surprising that the specificity of NF-κB responses is very important for the fate of a cell. It has been shown that abnormal NF-κB activity, which is not always associated with genetic alterations, plays a role in different inflammatory diseases and cancer (5–7).
NF-κB specificity is regulated at different levels in the cell (8). One level of regulation is the selective activation of distinct NF-κB complexes after induction by diverse stimuli. In mammals there exist five family members, c-Rel, RelB, p65 (RelA), p105/p50 (NF-κB1) and p100/p52 (NF-κB2) that can form a range of homo- and heterodimers (9). After regulated IκB (inhibitor of NF-κB)-dependent NF-κB translocation to the nucleus, these dimers bind with variable affinities to consensus NF-κB-binding sites in the promoter and enhancer regions of their target genes, often cooperatively with other transcription factors [e.g. IFNβ promoter (10)]. This integrates other signal transduction pathways with the NF-κB pathway giving additional levels of specificity and regulation to the transcriptional control of responsive genes. The interaction with cell-type-specific co-factor proteins has been shown to influence the transcriptional potential of NF-κB (11). One of the co-factors of NF-κB is the co-activator p300 and its homolog CBP (CREB-binding protein). They have been shown to interact with the RelA/p65 and the p50 subunit serving as molecular bridges between NF-κB and the transcription machinery (8,10,12–14). They contain intrinsic histone acetyltransferase activity catalyzing the acetylation of lysine residues in histones and non-histone proteins (15,16). A growing number of transcription factors are acetylated and regulated by p300/CBP including p53 (17), GATA-1 (18), E2F-1 (19,20) and YY1 (21). Post-translational acetylation influences different properties of these transcription factors such as DNA binding, protein–protein interactions, protein stability and transcriptional potential (22).
NF-κB is subject to a variety of post-translational modifications [e.g. phosphorylation (23), ubiquitination (24) or prolyl-isomerisation (25)] that modulate its activity. Phosphorylation of the RelA/p65 subunit by the PKAc, MSK1 and PKCζ kinases enhances its interaction with the co-activator p300/CBP and stimulates the NF-κB transcriptional activity (26–28). In contrast, ubiquitination of RelA/p65 on the promoter specifically terminates the NF-κB response (24).
It has recently been shown that RelA/p65 and p50 are reversibly acetylated by p300 and PCAF (29–31). Chen et al. identified lysine residues (K) 218, 221 and 310 of RelA/p65 as acceptor sites for p300 acetylation. They reported that lysine 221 acetylation enhanced DNA-binding activity of NF-κB in vitro and abolished the interaction with IκBα leading to a prolonged NF-κB response in the nucleus. The acetylation at lysine residue 310 was required for full transcriptional activity of RelA/p65 (32). Kiernan et al. identified lysine 122 and 123 in RelA/p65 as acetylation sites modified by both p300 and P/CAF. In contrast to K218, K221 and K310, acetylation of K122 and K123 decreased the DNA binding of RelA/p65 facilitating the removal of RelA/p65 from the DNA and the export from the nucleus by IκBα resulting in a faster termination of the NF-κB response (30). Furthermore, a recent report presented the TGF-β1 mediated acetylation of RelA/p65 at lysine 221 in vitro and in vivo enhancing the induced activation of NF-κB by bacteria (33).
Together, these data question the precise functional relevance of RelA/p65 post-translational acetylation in NF-κB-dependent gene regulation in vivo. Thus, our study aimed to identify the role of RelA/p65 acetylation in vivo. We found that p300 efficiently acetylated RelA/p65 in vitro and in cells at lysine 314 and 315—two novel acetyl acceptor sites. Additionally, our results confirmed the acetylation of RelA/p65 at the previously reported site of lysine 310 in vitro and in vivo. We generated acetylation-deficient lysine to arginine substitution mutants of RelA/p65 and stably complemented murine RelA/p65–/– cells with these mutants. The nuclear-cytosolic shuttling and the DNA binding of the acetylation-deficient mutants were similar to that of wild type RelA/p65. Furthermore, induction of anti-apoptotic genes by TNFα was not affected by the non-acetylatable mutants of RelA/p65. However, whole genome microarray analysis after TNFα stimulation indicated that the expression of specific genes was either positively or negatively affected by the K/R mutations. Our results imply that although general transcriptional activity of RelA/p65 was not affected by acetylation at lysine 310, 314 and 315, the expression of specific sets of genes was modulated by lysine-specific acetylation of RelA/p65. Thus, site-specific acetylation could serve as molecular mechanism to promote specificity of NF-κB-dependent gene expression.
MATERIAL AND METHODS
Plasmids
hGCN5L, mP/CAF and hTip60 were cloned into pFastBacHTb vector in frame with an N-terminal 6 ×His-tag. pph-CMV-Km-RelA/p65 wild type was previously described in (13). pph-CMV-Km-RelA/p65K310R (K310R), pph-CMV-Km-RelA/p65K314/K315R (K314/315R) and pph-CMV-Km-RelA/p65K310R/K314/K315R (KTR) were generated by site-directed mutagenesis according to the QuickChange protocol (Stratagene) using the following oligonucleotides:
K310R: 5′CGTAAAAGGACATACGAGACCTTCAGGAGCATCATGAAGAAGAGTCC3′,
5′GGACTCTTCTTCATGATGCTCCTGAAGGTCTCGTATGTCCTTTTACG3′,
K314/315R: 5′CCTTCAGGAGCATCATGCGGAGGAGTCCTTTCAGCGGACCC3′,
5′GGGTCCGCTGAAAGGACTCCTCCGCATGATGCTCCTGAAGG3′ (bold letters represent K/R mutation). pphCMV-Km-RelA/p65K122/123R, pphCMV-Km-RelA/p65K218/221R, pphCMV-Km-RelA/p65K218/221/310R were generated using the QuickChange site-directed mutagenesis protocol with pph-CMV-Km-RelA/p65 wild type as template vector. The specific primer sequences can be received upon request. The combinatorial mutant pphCMV-Km-RelA/p65K218/221/310/314/315R (KQR) was generated with the same protocol using pph-CMV-Km-RelA/p65K310R/K314/K315R as template vector. All introduced mutations were confirmed by sequencing.
Reagents and antibodies
Mouse TNFα, Trichostatin A (TSA), Nicotinamide (NAM), acetyl-Coenzyme A, calf thymus core histones (H7755) and Trichloroacetic acid (TCA) were purchased from Sigma. Sodium fluoride (NaF) and beta-glycerophosphate were purchased from Flucka. 14C-labeled acetyl Coenzyme A (MC269) was obtained from Moravek Biochemicals. Most of antibodies were from Santa Cruz Biotechnology: anti-RelA/p65 (C-20, sc-372), anti-α-tubulin (TU-02, sc-8035), control mouse IgG (sc-2025) and anti-PCNA (PC10, sc-56). The anti-p300 monoclonal antibody was purchased from BD Pharmingen (554215). The anti-p50 antibody was a generous gift from N. Rice (National Cancer Institute, Frederick, MD). Anti-myc 9E10 antibody was either purified from hybridoma cells according to standard protocol or purchased from Roche Applied Science. The polyclonal anti-acetylated-Lysine antibody was from Cell Signaling. A specific antibody against acetylated lysine 310 of RelA/p65 was generated in collaboration with Abcam. The anti-Ccl-7 antibody was purchased from Abcam (ab9911).
Tissue culture, cell transfections
Complemented RelA/p65–/– NIH 3T3 mouse embryonic fibroblasts (MEFs) and HEK 293T cells were maintained in DMEM supplemented with 10% FCS, 100 units/ml penicillin/streptomycin and non-essential amino acids (GIBCO). Cells were transfected using the calcium phosphate precipitation method.
Generation and purification of baculovirus expressed proteins
All recombinant proteins were expressed in Sf21 cells using the Bac-To-Bac (GIBCO) or BacPAK (Clontech) system. Recombinant His-tagged proteins were purified over Ni2+-beads (ProBond, Invitrogen).
In vitro acetylation assay
One microgram of recombinant human wild type or mutant RelA/p65 was incubated with 0.5–1 μg recombinant p300 or CBP or equimolar amounts of hGCN5L, mP/CAF or hTip60 in HAT buffer (50 mM Tris–HCl pH 8.0, 100 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin, 1 mM sodium butyrate) supplemented with 1.5 nmol 14C-acetyl CoA for 45 min at 30°C. Reactions were stopped by adding 10 × Laemmli-buffer and proteins resolved on SDS–PAGE with subsequent visualization by Coomassie brilliant blue or SyproRuby staining. The gel was immersed in 1 M sodium salicylate for 20 min at RT. After drying, the gel was exposed to X-ray films (Contatyp) at –80°C.
MS/MS
In vitro acetylated RelA/p65 was resolved on SDS–PAGE, fixed and stained with Coomassie brilliant blue. The corresponding protein band was then excised and washed twice with 50% acetonitrile. After tryptic digestion the protein sequence analysis was performed at the Harvard Mass Spectrometry and Proteomics Resource Laboratory by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry (μLC/MS/MS) on a ThermoFisher LCQ DECA XP quadrupole ion trap mass spectrometer.
GST-pull down experiments
GST, GST-RelA/p65wt and GST-RelA/p65KTR proteins were immobilized on glutathione beads (Amersham Pharmacia) and incubated with purified his-p300 in binding buffer (20 mM Hepes pH 7.5, 60 mM NaCl, 10% glycerol, 1.5 mM MgCl2, 1 mM DTT, 1 mM PMSF and 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin) for 2 h at 4°C rolling. Glutathione beads were washed with binding buffer. Proteins were boiled, resolved on SDS–PAGE and subjected to western blot analysis using anti-his antibody (Qiagen).
Acetylation assay in cells
Myc-tagged RelA/p65 wild type, RelA/p65 acetylation-deficient mutants or control empty vector were co-expressed with p300 in HEK 293T cells. After 15 h of transfection, cells were treated with HDAC inhibitors (HDACi: 2 μM TSA, 5 mM NAM) alone or in combination with TNFα (30 ng/ml) for 30 or 45 min, respectively. Whole cell extracts were prepared (50 mM Hepes pH 7.9, 420 mM NaCl, 0.5% NP-40, 1 mM PMSF, 0.5 mM DTT, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin). One milligram of whole cell extract and 2 μg of anti-myc antibody were used for subsequent immunoprecipitation. The immunocomplexes were analyzed by standard western blot analysis using anti-acetylated Lysine antibody. Membranes were reprobed with anti-myc antibody.
Lentiviral complementation of RelA/p65–/– MEFs
Virus production and transduction of RelA/p65–/– MEFs were performed as described in (34). Briefly, HEK 293T cells were transfected with 3.5 μg of the envelope plasmid, 6.5 μg of packaging plasmid, and 10 μg of pTV-myc-RelA/p65 wild type, RelA/p65 K/R mutants or the control pTV vector. After 24 h the viral supernatant was harvested and used to infect RelA/p65–/– MEFs. Thirty-six hours post infection cells were split into selective medium containing 2.5 μg/ml Blasticidin (Sigma). Expression of recombinant proteins in the complemented cells was screened by western blot analysis. Pools of cells were used for further analysis.
Electrophoretic mobility shift assay (EMSA)
Binding reactions were carried out in a total volume of 20 μl containing 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM DTT, 2 mM PMSF, 0.25 μg poly dI-dC, 7% (v/v) Ficoll/glycerol, 0.25 pmol HIV-LTR oligonucleotide containing 2 κB binding sites labeled 5′ with ATPγ32P and 7 μg of nuclear extract. The reaction was incubated for 20 min on ice and then resolved using a 5% polyacrylamide gel in 0.5 × TBE. The gel was run at 150 V for 3 h, dried and subjected to autoradiography. When supershifts were performed the binding reaction was pre-incubated with the indicated antibody for 20 min on ice before the labeled oligonucleotide was added.
Immunohistochemistry
Cells were plated at the density of 45 000 cells per chamber on poly-l-lysine (Sigma)-coated chamber slides (LAB-TEK) and incubated overnight at 37°C and 5% CO2. Next day the cells were treated with 30 ng/ml of TNFα for the indicated time. The cells were fixed in 4% paraformaldehyde and then permeabilized with 0.2% Triton-X-100/PBS. After blocking for 1 h in 2% BSA/0.1% Triton-X-100/PBS slides were incubated with anti-RelA/p65 C-20 antibody (1:300 dilution) followed by anti-rabbit Cy3 antibody (1:250 dilution, Jackson Immunology). The samples were washed and Vectashield mounting solution (Vector laboratories) was applied to prevent bleaching. Cells were visualized using an Olympus T50 microscope.
Whole cell extract preparation and immunoprecipitation for RelA/p65-p300 interaction
The complemented cell lines were treated with TNFα (30 ng/ml) for 30 min. Whole cell extracts were prepared (25 mM Hepes pH 7.9, 300 mM KAc, 1% NP-40, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin, 50 mM NaF, 20 mM beta-glycerophosphate). One milligram of whole cell extract was incubated with 2 μg of anti-p300 antibody or control IgG antibody for 2.5 h at 4°C. The immunocomplexes were analyzed by standard western blot analysis using anti-RelA/p65, anti-p300 and anti-tubulin antibodies.
Nuclear extract preparation and immunoprecipitation for in vivo acetylation of K310
Nuclear extracts were prepared as previously described in (35). Two hundred micrograms of TNFα (30 ng/ml) and HDAC inhibitor [TSA (2 μM), NAM (5 mM)] treated nuclear extracts of complemented cells were incubated with 2 μg of anti-RelA/p65 C-20 antibody for 2.5 h at 4°C rolling in binding buffer (20 mM Hepes pH 7.9, 80 mM NaCl, 2.5 mM MgCl2, 0.05% NP-40, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin). After incubation with protein G-sepharose beads (Amersham Pharmacia) for another hour, the immunocomplexes were extensively washed in washing buffer (20 mM Hepes pH 7.9, 100 mM NaCl, 2.5 mM MgCl2, 0.05% NP-40, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin). Proteins were boiled, resolved on SDS–PAGE and analyzed by western blot using the anti-RelA/p65ac310 antibody. Membranes were reprobed with anti-RelA/p65 antibody.
Cell survival assay
The complemented cells (pTV empty vector control, RelA/p65wt, RelA/p65K310R, RelA/p65K314/315R and RelA/p65KTR) were plated in 12-well dishes at a density of 90 000 cells/ml. After 24 h of incubation, cells were starved overnight with 0% FCS containing medium and then treated with TNFα (30 ng/ml) or left untreated for additional 10 h. The surviving cells were counted using Trypan blue. The ratios of TNFα-treated and untreated cells were calculated for each cell line. Each condition was carried out in duplicate. A representative assay of three independent experiments is shown.
RNA preparation
Total RNA was isolated three independent times from TNFα-treated lysates from the different complemented RelA/p65–/– MEFs with the ‘Total RNA isolation kit’ (Agilent Technologies). RNA quality was assessed with the RNA 6000 Nano kit using the Bioanalyzer 2001 (Agilent Technologies). Purified RNAs were converted into double-stranded cDNA and transcribed into Cy3-/Cy5 (PerkinElmer/NEN Life Science)—labeled cRNA using the ‘Low RNA Input Linear Amp Kit’ (Agilent Technologies). cRNA from wild type RelA/p65 cells was Cy5-labeled while the RelA/p65K/R mutant cRNAs were Cy3-labeled. The purification of the labeled cRNAs was performed with the RNeasy kit (Qiagen). Dye incorporation was measured on the ND-1000 Spectrophotometer (NanoDrop Technologies)
Gene expression profiling
Gene expression profiling was performed in the Functional Genomics Center Zurich using the two-colour Agilent Microarray system (Agilent Technologies). 1 μg of fragmented Cy5-labeled wild type and 1 μg of Cy3-labeled mutant cRNA were each co-hybridized on the Whole Mouse genome 60mer-oligo array (G4122A, Agilent Technologies) according to manufacturers protocol. The microarray analysis was performed in triplicates. Slides were scanned using the Agilent DNA microarray scanner and the scans were quantified with the Agilent Feature Extraction software.
Data analysis
Data analysis was performed with GeneSpring software (Silicon Genetics). For the statistical comparisons of the RelA/p65 mutants and RelA/p65 wild type we only considered genes that were present in either the mutant or the wild type. We declared a gene as present in a comparison, if the hybridisation intensity was in all mutant replicates or in all wild type replicates above 200. Student's t-test was used to compute the significance of differential expression and 0.01 was used as significance threshold. For each list of significant genes, the Benjamini–Hochberg false discovery rate was computed and is reported in the results section. Furthermore, genes were filtered according to their fold change and only genes exceeding a fold change of 1.5 up or down are reported.
Quantitative real time RT-PCR
Total RNA from untreated or 45 min TNFα-treated cell lines (pTV, RelA/p65wt, RelA/p65K310R, RelA/p65K314/315R and RelA/p65KTR) was reverse transcribed using the high capacity cDNA Archive kit according to manufacturers protocol (ABI). Real-time PCR was performed using mouse-specific TaqMan probes (Gene expression assays, ABI) for Ccl-7, Ifi-44, Ccl-20 and Gpb-2. TaqMan probes for 18SrRNA and Rps6 were used to normalize for differences in RNA input. Rotor-Gene3000A (Corbett) was used to perform the real-time PCR reactions and the REST program was applied for analysis (36). The figures show the averaged results of three independent experiments.
TCA protein precipitation and Ccl-7 protein detection
Complemented cells (RelA/p65wt, RelA/p65K310R, RelA/p65K314/315R and RelA/p65KTR) were starved with 0% FCS containing medium for 1.5 h and then treated with TNFα (30 ng/ml) or left untreated. After 4 h of TNFα treatment, the medium was collected and 250 ng of recombinant PCNA was added as a control for the TCA protein precipitation. The medium was incubated overnight at 4°C with one-fourth volume of TCA (100% w/v) and centrifuged. Pellets were washed twice with acetone and lysed with lysis buffer (50 mM Hepes pH 7.5, 420 mM NaCl, 2 mM EDTA, 0.5% NP-40, 15% glycerol, 1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml bestatin, 1 μg/ml leupeptin). Forty micrograms of proteins were analyzed by standard western blot analysis using anti-Ccl-7 and anti-PCNA antibodies.
RESULTS
RelA/p65 is acetylated in vitro by p300 or CBP
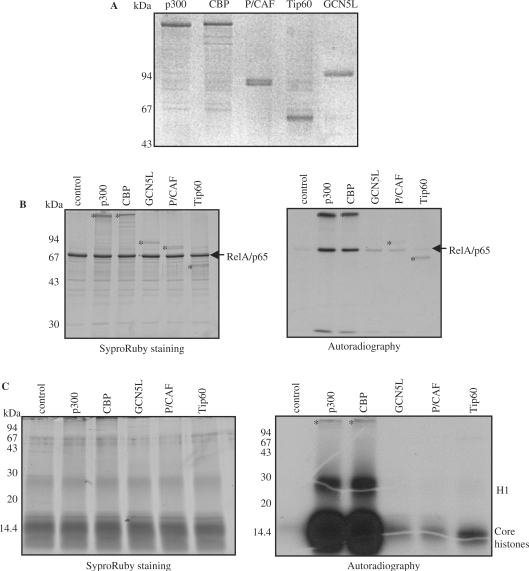
Since RelA/p65 was reported to interact directly with several histone acetyltransferases (HAT), we investigated whether RelA/p65 could serve as a substrate in an in vitro acetylation assay. We compared the ability of different HATs to acetylate RelA/p65 in vitro. Full-length RelA/p65 was incubated with recombinant p300, CBP, GCN5L, P/CAF or Tip60 (all proteins expressed and purified from insect cells, Figure 1A) in the presence of radioactively labeled acetyl-coenzyme A (acetyl-CoA) as a donor of the acetyl group (Figure 1B). All tested HATs, except for Tip60, acetylated RelA/p65 in vitro (Figure 1B). Calf thymus core histones were used as a positive control of acetylation (Figure 1C). p300 and CBP were the most potent HATs for RelA/p65 in our system. This prompted us to focus on p300 in this study.
Figure 1.
RelA/p65 is acetylated by p300 and CBP in vitro. (A) Recombinant histone acetyltransferases (HATs) expressed and purified from insect cells were analyzed by SDS–PAGE and Coomassie staining. Molecular weight markers are shown on the left. (B) In vitro acetylation assay using full-length RelA/p65 and indicated HATs. RelA/p65 was incubated in the presence of radioactively labeled [14C]-acetyl-CoA with 500 ng of p300 or CBP or the equimolar amount of GCN5L, P/CAF or Tip60. Proteins were resolved on SDS–PAGE, stained with SyproRuby (left) and exposed to X-ray films (right). RelA/p65 acetylation signals are indicated with an arrow. HATs or HAT autoacetylation signals are indicated with an asterisk (*). (C) In vitro acetylation of histones. In vitro acetylation assay was performed as described in (B).
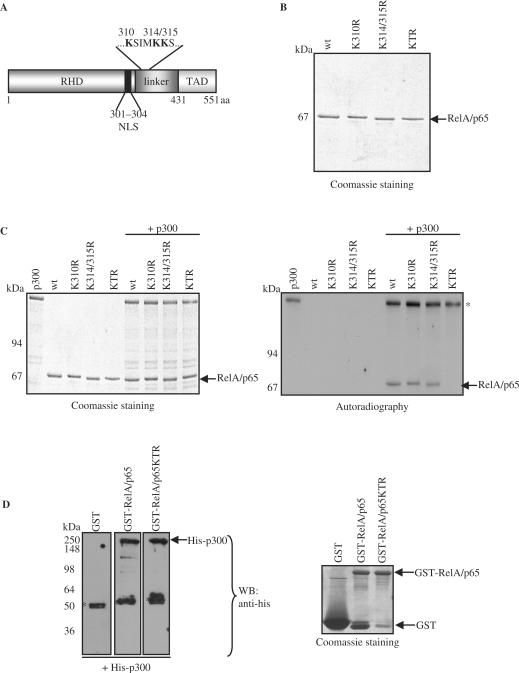
RelA/p65 is acetylated in vitro by p300 or CBP at lysine 310, 314 and 315
To identify the acetylation residues, in vitro acetylated RelA/p65 by p300 was digested with trypsin and the resulting peptides were analyzed by LC/MS/MS. 81.8% of the K310 comprising peptides contained an acetylated K310. 61.9% of K314 and 56.52% of K315 containing peptides showed acetylated K314 and acetylated K315, respectively. These data indicate that lysine 310, 314 and 315 were acetylated by p300. The identified lysine residues are located close to the C-terminus of the Rel homology domain (RHD) (Figure 2A). To confirm these findings, the corresponding lysines were replaced with arginine residues by site-directed mutagenesis. Substitution of lysine to arginine maintains the positive charge of the residue and may cause only minimal changes in the local environment of the protein. Wild type or mutated RelA/p65 harboring K310R, K314/315R or all three K310/314/315R (KTR) substitutions were expressed and purified from insect cells (Figure 2B). Subsequently, all proteins were subjected to in vitro acetylation by p300 or CBP (Figure 2C and Supplementary Figure 1). Acetylation of RelA/p65 mutated at single K310 or K314/315 was only slightly reduced compared to wild type, while mutation of all three lysine residues abolished acetylation of RelA/p65 (Figure 2C). When the purified protein substrates were tested only in the presence of acetyl-CoA, no acetylation was observed confirming that the acetylation was mediated by p300 and not by a co-purified acetyltransferase. These results clearly indicate that K310, K314 and K315 of RelA/p65 are the main acetylation sites of p300 in vitro. To confirm that the RelA/p65KTR mutant was still able to interact with p300, GST-pull down experiments were performed using GST-RelA/p65 wild type or GST-RelA/p65KTR as bait proteins in the presence of purified p300. Subsequent western blot analysis revealed that p300 was able to equally directly interact with both RelA/p65 wild type and the RelA/p65KTR mutant (Figure 2D). These results indicate that abolished acetylation of the RelA/p65KTR mutant was due to the lack of specific sites and not due to the inability of this mutant to interact with p300.
Figure 2.
RelA/p65 is acetylated by p300 at lysine 310, 314 and 315 in vitro. (A) Protein domain diagram of RelA/p65 depicts the location of the acetylated lysine residues in the ‘linker’ region of RelA/p65 (NLS, nuclear localization signal; RHD, Rel homology domain; TAD, transactivation domain). (B) Coomassie staining of recombinant arginine to lysine substitution mutants of RelA/p65 expressed and purified from insect cells: wt, RelA/p65 wild type; K310R, RelA/p65K310R; K314/315R, RelA/p65K314/315R and KTR, RelA/p65K310/314/315R. (C) In vitro acetylation of RelA/p65K/R mutants by p300. Proteins were analyzed as in Figure 1B. The autoacetylation signals of p300 are indicated with an asterisk (*). (D) GST-pull down experiments using GST, GST-RelA/p65wt and GST-RelA/p65KTR as bait proteins were performed in the presence of purified his-p300. Co-precipitated proteins were analyzed by western blot using anti-his antibody (left panel: arrow indicates his-p300, asterix indicate unspecific binding). Coomassie staining of the bait proteins for equal loading is shown on the right (arrows indicate GST, GST-RelA/p65wt and GST-RelA/p65KTR).
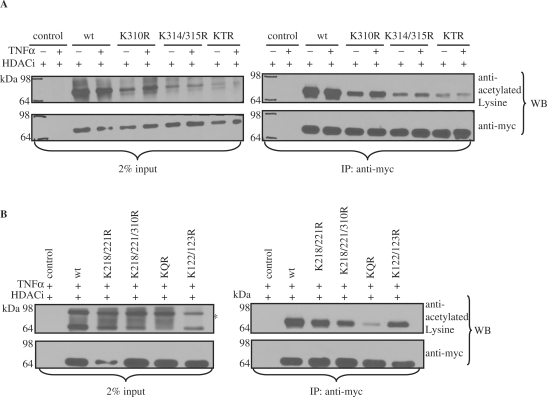
Lysine 314 and 315 of RelA/p65 are acetylated by p300 in cells
To examine if lysine residues 314 and 315 of RelA/p65 are acetylated in cells, myc-tagged wild type RelA/p65, RelA/p65 acetylation-deficient mutants (RelA/p65K310R, RelA/p65K314/315R, RelA/p65KTR) or myc-tagged empty control vector were ectopically expressed in HEK 293T cells together with the histone acetyltransferase p300. Previous experiments had revealed that overexpressed RelA/p65 wild type can only be detected as acetylated in the presence of ectopically expressed p300 [(29) and data not shown]. TNFα was applied to induce NF-κB, while HDAC inhibitors (HDACi: TSA and NAM) were used to inhibit histone deacetylases. Subsequent immunoprecipitation analysis with anti-myc antibody and western blot analysis using anti-acetylated Lysine antibody revealed acetylation of RelA/p65 wild type (Figure 3A). The acetylation of wild type RelA/p65 was already close to saturation before TNFα stimulation most probably due to the activation of the NF-κB pathway through overexpression of RelA/p65. However, an increase of acetylation after TNFα stimulation is detected in the RelA/p65K310R mutant, strongly suggesting that lysine 314 and 315 are acetylated upon TNFα induction. A significant decrease in acetylation was detected in the RelA/p65 acetylation-deficient double mutant (K314/315R). A small residual amount of acetylation was still detected when the triple mutant (KTR) of RelA/p65 was expressed in HEK 293T cells indicating that also other lysine residues are acetylated by p300 (Figure 3A).
Figure 3.
RelA/p65 is acetylated in cells on lysine 314 and 315 by p300. (A) Wild type myc-RelA/p65 and myc-RelA/p65K/R mutants were immunoprecipitated with anti-myc antibody from HDACi (TSA/NAM) and –/+ TNFα treated HEK 293T cells transfected with either empty control vector or different CMV-myc-RelA/p65 vectors (wt, K310R, K314/315R or KTR) along with CMV-p300. Western blot analysis of the immunocomplexes was performed after SDS–PAGE using anti-acetylated Lysine antibody (right panel). 2% input lanes are shown in the left panel. The membranes were reprobed with anti-myc antibody to assess equal input and immunoprecipitation. (B) HEK 293T cells were transfected with either control vector, myc-RelA/p65 wild type, myc-RelA/p65K122/123R, myc-RelA/p65K218/221R, myc-RelA/p65K218/221/310R or myc-RelA/p65KQR along with CMV-p300 and treated with HDACi (TSA and NAM) and TNFα. Wild type myc-RelA/p65 and myc-RelA/p65K/R mutants were immunoprecipitated with anti-myc antibody and the resulting immunocomplexes were resolved on SDS–PAGE. Western blot analysis was performed with anti-acetylated Lysine antibody. 2% input lanes are shown on the left. Membranes were reprobed with anti-myc antibody. Unspecific signals are indicted with an asterisk (*).
To investigate if one of the previously reported sites [K122, K123 (30) and K218, K221 and K310 (32)] is acetylated in our system a variety of acetylation-deficient point mutants of RelA/p65 were generated: RelA/p65K122/123R, RelA/p65K218/221R, RelA/p65K218/221/310R and a combinatorial mutant RelA/p65K218/221/310/314/315R (RelA/p65KQR) harboring the acetylation acceptor sites identified in this study and the sites identified by Chen et al. after TNFα stimulation (32). The different mutants of RelA/p65 were expressed in HEK 293T cells along with p300 in the presence of TNFα and HDAC inhibitors. Anti-myc immunoprecipitation and anti-acetylated Lysine western blot analysis revealed a slight decrease in acetylation of the RelA/p65K122/123R and RelA/p65K218/221R mutant when compared to wild type RelA/p65 (Figure 3B). However, RelA/p65K218/221/310R showed a higher reduction in acetylation while the RelA/p65KQR mutant showed a decrease in acetylation of 90% (when compared to the wild type) indicating that K314 and K315 greatly contribute to the detected p300-dependent acetylation level in RelA/p65 after TNFα treatment. It also points out that K218 and K221 might probably be the other lysine residues acetylated in cells by p300 (Figure 3A). Together, these data indicate that RelA/p65 is acetylated in cells mainly on lysine 314 and 315 by p300.
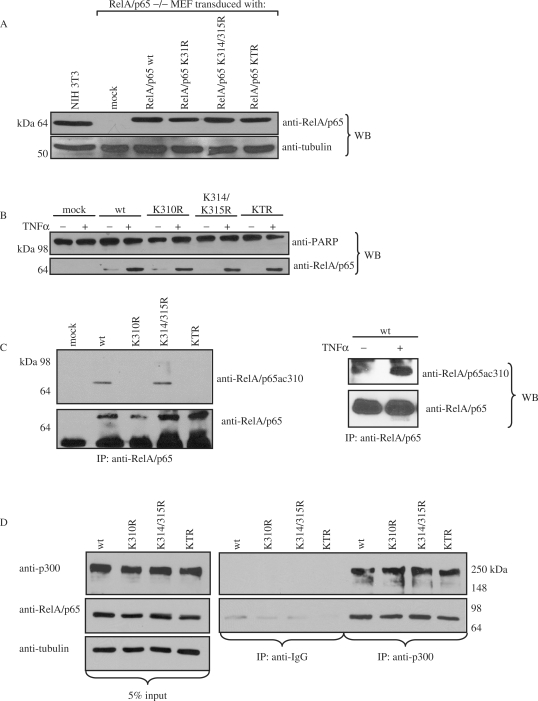
Endogenous RelA/p65 is acetylated in vivo in TNFα stimulated cells
To investigate the role of RelA/p65 acetylation of K310, K314 and K315 in vivo, RelA/p65–/– NIH 3T3 mouse embryonic fibroblasts (MEF) were genetically complemented using lentiviruses encoding myc-RelA/p65 wild type, myc-RelA/p65K310R, myc-RelA/p65K314/315R or myc-RelA/p65KTR. After appropriate selection cells were kept in pools and the expression of recombinant proteins was analyzed by western blotting using an anti-RelA/p65 antibody (Figure 4A). The cells transduced with control virus encoding the resistance gene [mock infected (pTV)] and non-transduced wild type NIH 3T3 cells expressing endogenous RelA/p65 protein were included as controls. The expression levels of the recombinant wild type and mutated RelA/p65 proteins were comparable to that observed for endogenous RelA/p65 in NIH 3T3 cells. Furthermore, cell growth analysis revealed that the proliferation rate was comparable between the tested cell pools under normal growth conditions (data not shown).
Figure 4.
Endogenous RelA/p65 is acetylated at lysine 310 in vivo in response to TNFα. (A) Western blot analysis with anti-RelA/p65 antibody of whole cell extracts from RelA/p65–/– MEFs complemented with RelA/p65 wild type (wt) and the different substitution mutants of RelA/p65 (K310R, K314/315R and KTR). Mock transduced (pTV) and NIH 3T3 cells were used as controls. The membrane was reprobed with anti-tubulin antibody as loading control. (B) Nuclear extracts of the complemented cell lines untreated or treated with TNFα (30 ng/ml) for 30 min were subjected to western blot analysis using anti-RelA/p65 antibody. The anti-PARP western blot was performed to check extract fractionation. (C) In the left panel, nuclear extracts of the complemented cell lines treated with TNFα and HDACi (TSA/NAM) were subjected to immunoprecipitation analysis using anti-RelA/p65 antibodies. Western blot analysis with the anti-RelA/p65ac310 antibody was performed. The membranes were reprobed with anti-RelA/p65 antibody. In the right panel, RelA/p65 was immunoprecipitated from whole cell extracts of the complemented cell lines treated –/+ TNFα. The immunocomplexes were analyzed by western blot using anti-RelA/p65ac310 antibody and the membrane was reprobed with anti-RelA/p65 antibody. (D) Endogenous interaction analysis of RelA/p65 with p300 in TNFα-treated complemented cell lines. Endogenous p300 was immunoprecipitated from whole cell extract of complemented cell lines after TNFα stimulation for 30 min using anti-p300 antibody. Immunocomplexes were resolved on SDS-PAGE and subsequently analyzed by western blot using anti-RelA/p65, anti-p300 and anti-tubulin antibodies. Left panel: 5% inputs are shown, middle panel: IgG control immunoprecipitation; right panel: anti-p300 immunocomplexes.
We and others showed that RelA/p65 is acetylated in cells in a TNFα-dependent manner [Figure 3A and (29)]. In order to investigate whether endogenous RelA/p65 is acetylated at the identified lysine residues in vivo, antibodies directed against RelA/p65 peptides acetylated at residues 310, 314 or 315 were generated. Nuclear extracts were prepared after treating the complemented cells with TNFα and HDAC inhibitors (TSA and NAM). Upon TNFα treatment RelA/p65 nuclear import was induced in RelA/p65wt and RelA/p65 mutant complemented cells (Figure 4B). Immunoprecipitation with an anti-RelA/p65 antibody and subsequent western blot analysis using the specific acetyl K310 antibody revealed that RelA/p65 was indeed acetylated at this site upon TNFα treatment (Figure 4C, left panel). TNFα induced acetylation of RelA/p65 was further confirmed by immunoprecipitating RelA/p65 from untreated and TNFα stimulated cells (Figure 4C, right panel). Acetylated RelA/p65 could only be detected in the extracts stimulated by TNFα. Acetylation of RelA/p65 could not be detected in the input lanes due to low detection sensitivity of the anti-acetyl 310 antibody. Antibodies raised against acetylated 314 and acetylated 315 were found to be unspecific in this analysis (data not shown).
To investigate if the abolished acetylation of RelA/p65K310R and RelA/p65KTR was due to their inability to interact with endogenous histone acetyltransferase p300, immunoprecipitation analyses with the different mutants and p300 were performed. Whole cell extracts were prepared from wild type and mutant RelA/p65 complemented cell lines after TNFα stimulation to induce NF-κB. Immunoprecipitation with either anti-p300 or control IgG antibody and subsequent western blot analysis using anti-RelA/p65 antibody revealed that endogenous RelA/p65K10R, RelA/p65K314/315R and RelA/p65KTR are able to interact with endogenous p300 upon TNFα stimulation to the same extend as wild type RelA/p65 (Figure 4D).
K314/315R substitution mutation does not affect the shuttling, the DNA-binding ability or the protection against TNFα-induced cell death of RelA/p65
The complemented cells were treated with TNFα and subcellular localization of the recombinant RelA/p65 proteins was analyzed by immunofluorescence analysis at different time points (Figure 5A). Nuclear translocation of the recombinant RelA/p65 wild type was detected after 20 min of TNFα treatment and relocation to the cytoplasm after 60 min. A similar shuttling kinetics was observed for endogenous RelA/p65 protein in NIH 3T3 cells (data not shown). Analysis of the cells complemented either with K310R, K314/315R or KTR revealed no significant differences in the shuttling kinetics between mutated RelA/p65 and RelA/p65 wild type proteins (Figure 5A). This suggested that acetylation of RelA/p65 at the three tested lysine residues was not essential for the regulation of the RelA/p65 nuclear-cytoplasmic redistribution. This observation is supported by our findings that IκBα was able to bind to RelA/p65 wild type and the acetylation-deficient mutants with similar affinity (data not shown).
Figure 5.
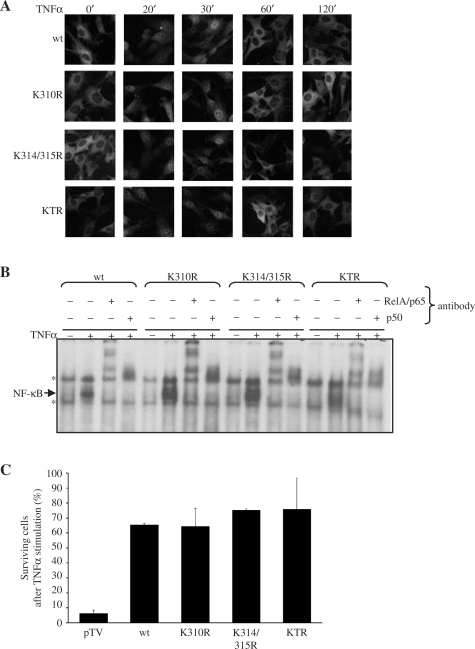
Mutation of the acetylation sites in RelA/p65 does not affect its nuclear translocation and DNA binding. (A) The complemented cells were treated with TNFα and fixed with paraformaldehyde followed by immunostaining using anti-RelA/p65 antibodies. Subcellular localization of RelA/p65 protein was analyzed by immunofluorescence microscopy. (B) Electro mobility shift assays were performed with nuclear extracts from the untreated or TNFα treated cells using 32P-labeled oligonucleotide DNA containing 2κB sites. The bound complexes were characterized by anti-RelA/p65 or anti-p50 supershift assays. Specific NF-κB complexes are indicated with an arrow, unspecific bands are marked with an asterisk (*). (C) Cell viability assay after TNFα treatment. The different complemented cell lines (pTV, RelA/p65wt, K310R, K314/315R and KTR) were starved for 14 h with 0% FCS medium and then left untreated or treated with TNFα (30 ng/ml) for 10 h. The number of surviving cells (%) after TNFα treatment was calculated as described in ‘Material and methods’ section.
Next we investigated the influence of p300-mediated RelA/p65 acetylation on the ability of RelA/p65 to bind DNA. Nuclear extracts of the complemented cells treated with TNFα were tested in an electro mobility shift assay using an oligonucleotide containing two κB elements of the HIV-LTR promoter. The experiments revealed that mutations of K310, K314 and K315 did not significantly influence the TNFα-induced DNA binding of RelA/p65 (Figure 5B). Binding was substantially reduced by competition with non-labeled oligonucleotides containing a wild type κB site, but not a mutated κB site, indicating that the binding was specific (data not shown). The presence of RelA/p65 and p50 in the complex was confirmed by supershift experiments using specific anti-RelA/p65 and anti-p50 antibodies, respectively (Figure 5B). Together, these findings show that the acetylation-deficient mutants are able to translocate to the nucleus and bind to DNA to the same extent as RelA/p65 wild type. These results are in accordance with previously published data where K310 mutation to arginine did not affect subcellular localization, DNA binding or interaction with IκBα (32).
The deletion of the RelA/p65 gene in mouse cells is known to render them vulnerable to TNFα-induced cell death, stressing the important role of RelA/p65 in cell survival (37). To investigate whether TNFα-induced acetylation of RelA/p65 would affect the protective function of RelA/p65, cell viability assays were performed. The control cell line pTV (RelA/p65–/– cells complemented with empty vector), RelA/p65 wild type and the three different acetylation-deficient RelA/p65 mutant cell lines were either left untreated or were stimulated with TNFα for 10 h. The number of surviving cells after treatment was determined using Trypan blue and compared to the number of untreated cells (Figure 5C). As expected, the pTV cells were not protected from TNFα-induced cell death due to their lack of RelA/p65. However, the non-acetylatable mutants of RelA/p65 (K310R, K314/315R and KTR) protected the cells from cell death to the same extend as RelA/p65 wild type, indicating that the acetylation of RelA/p65 at the three mutated lysines does not affect substantially the tested pathway or that a possible effect could be compensated by the expression of RelA/p65-independent genes.
Determination of genes regulated by p300-mediated acetylation of RelA/p65 on lysine 314 and 315
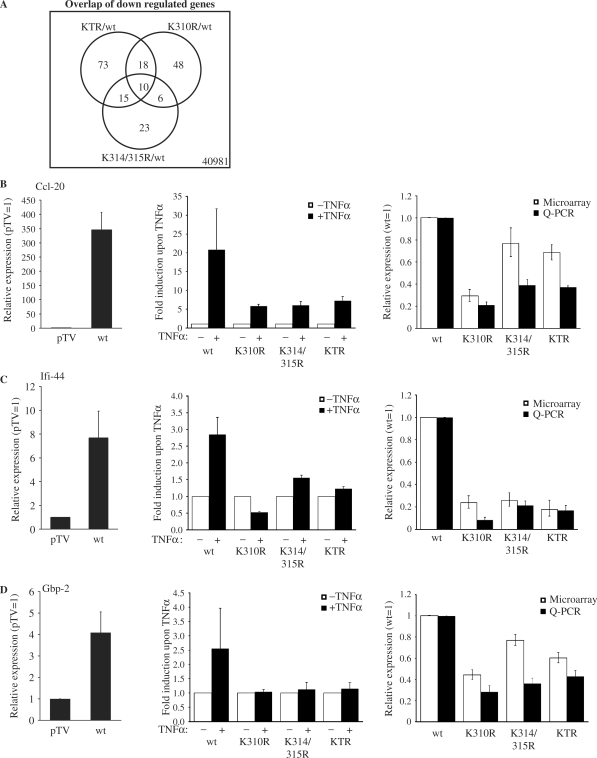
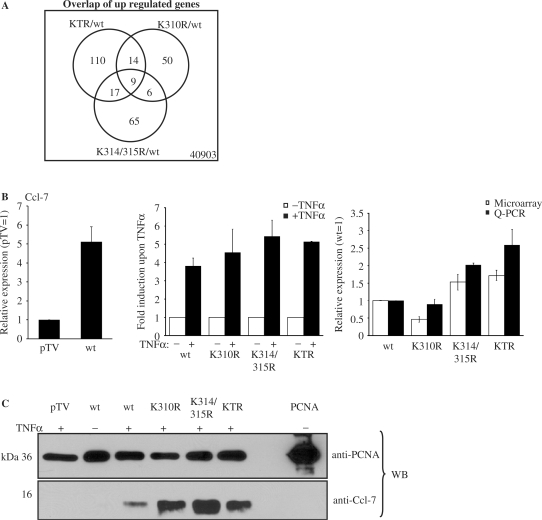
To investigate the functional relevance of lysine 314 and 315 acetylation, microarray analyses were performed using the Agilent Whole Mouse Genome Array. The complemented cell lines (RelA/p65 wild type, RelA/p65K310R, RelA/p65K314/315R and RelA/p65KTR) were treated for 45 min with TNFα to induce NF-κB and total RNA was isolated in three independent replicates from these cells. RT-PCR for known TNFα-dependent NF-κB target genes was performed to check whether the TNFα induction was successful. The IκBα mRNA was induced to the same extend in the wild type and acetylation-deficient mutant cell lines (data not shown). RNA was amplified and labeled. The RNA from the different mutants (K310R, K314/315R and KTR) was then each co-hybridized with wild type RNA (RelA/p65 wild type cells) to the two-color whole mouse genome array. Statistical analysis of the TNFα-induced expression profiles aimed at identifying differentially regulated genes when comparing the different mutant cell lines with the wild type cell line. After performing this comparison, we identified following numbers of potentially regulated genes for K310R RelA/p65 versus wild type RelA/p65: 1149 significantly regulated genes with a P value ≤0.01 and a signal intensity > 200 at a false discovery rate of 18.61%. Thereof 79 genes were upregulated more than 1.5 times and 82 downregulated more then 1.5 times at a P value ≤ 0.01 (Supplementary Table S1). In the K314/315R RelA/p65 versus wild type RelA/p65 comparison we found 735 potentially regulated genes at P ≤ 0.01 (signal intensity >200 and false discovery rate 28.43%). Ninety-seven genes were found to be upregulated with a fold change >1.5 and 54 downregulated with a fold change of <0.67 (P ≤ 0.01) (Supplementary Table S2). When we finally analyzed the KTR mutant we found 1233 genes significantly regulated with a signal >200 and P ≤ 0.01 at a false discovery rate of 17% (i.e. 216 false positives). Of these 1233 regulated genes 150 were upregulated with a fold change >1.5 and 116 downregulated with a fold change <0.67 (P ≤ 0.01) when compared to wild type RelA/p65 (Supplementary Table S3). To investigate if common regulated genes were present in the different cell lines the overlap of the significantly downregulated and upregulated genes was generated at a P value ≤0.01 (Figures 6A and 7A). The overlap of genes between the different RelA/p65 mutants was small, possibly indicating distinct sets of genes being regulated by the different mutants.
Figure 6.
Acetylation of RelA/p65 at lysine 310, 314 and 315 activates gene specific transcription. (A) Venn diagram showing the numbers of common down regulated genes between the significantly regulated genes in the different RelA/p65 acetylation-deficient cell lines compared to the wild type cell line found in the microarray analysis. (B–D) Quantitative real time RT-PCR validation for selected downregulated genes, Ccl-20 (B), Ifi-44 (C) and Gbp-2 (D). Left panels show the comparison of relative expression in the presence of TNFα (pTV was set as 1). Middle panels show the TNFα fold induction of the different cell lines upon TNFα stimulation. Right panels show the comparison of relative expression of microarray (white bars) and real time PCR (black bars) experiments in the presence of TNFα (RelA/p65 wild type was set 1 in both methods). For real-time PCR 18SrRNA or Rps6 were used to correct for differences in cDNA inputs. The mean values from three independent, normalized measurements are shown with standard deviations.
Figure 7.
Acetylation of lysine 314 and 315 downregulates gene and protein expression. (A) Venn diagram of differentially upregulated genes found in the microarray analysis when the different RelA/p65 acetylation-deficient cell lines were compared to the wild type cell line. (B) Quantitative real-time RT-PCR validation for Ccl-7. The expression in the wild type cell line relative to pTV cell line in the presence of TNFα is shown in the left panel. The TNFα induction of this gene in the different cell lines is shown in the middle panel. The comparison between the microarray (white bars) and the real-time PCR (black bars) experiment is shown on the right side. The mean values from three independent, normalized measurements are shown with standard deviations. (C) Differential expression of Ccl-7 protein in acetylation-deficient cell lines of RelA/p65. Proteins present in the medium of complemented cells treated with TNFα for 4 h or left untreated were precipitated using TCA. Total 250 ng of recombinant PCNA was added as control to the collected medium before the precipitation was carried out. Western blot analysis was performed using anti-Ccl-7 and anti-PCNA antibodies. Total 250 ng of recombinant PCNA were loaded on the last lane of the SDS–PAGE to control the protein precipitation.
To validate the results derived from the microarray studies, and to confirm the p65 dependency and TNFα induction of the analyzed genes, we performed quantitative RT-PCR for some selected genes. The following genes were chosen from the lists of overlapping differentially regulated genes at P value ≤0.01: Ccl-20, Ccl-7, Ifi-44 and Gbp-2. To perform quantitative real-time RT-PCR, RelA/p65–/– cells complemented either with the empty vector pTV, RelA/p65 wild type, RelA/p65K310R, RelA/p65K314/315R or RelA/p65KTR mutant were left untreated or were stimulated with TNFα for 45 min. Total RNA was isolated and reverse transcribed with random hexamer primers to obtain cDNA. Gene expression of Ccl-20, Ifi-44 and Gbp-2 was highly dependent on the acetylation of RelA/p65 at the identified sites, since only low levels of expression of these genes could be detected in cells expressing any of the mutants (Figure 6B–D, right panel, black bars). On the other hand, Ccl-7 expression was increased in cells expressing the K314/315R (2-fold) and the KTR mutant (2.59-fold) (Figure 7B, right panel). In the present analysis, the expression profiles obtained by microarray analysis were confirmed by quantitative RT-PCR analysis for all tested genes. However, the two methods showed higher correlation in the upregulated gene (Ccl-7; Figure 7B) than in the downregulated genes (Ccl-20, Ifi-44 and Gbp-2; Figure 6B–D).
In order to examine if these selected genes require RelA/p65 to be induced upon TNFα treatment, we compared their expression levels in the wild type cell line and in the control cell line pTV upon TNFα treatment. The four genes were upregulated in wild type cells, indicating that they were RelA/p65-dependent (left panels in Figures 6B–D and 7B). To determine the induction of these selected genes after TNFα stimulation in the different complemented cell lines, expression levels upon TNFα treatment were compared with unstimulated samples. All four genes were induced upon TNFα stimulation in wild type cells (middle panels from Figures 6B–D and 7B). However, among the downregulated genes, only Ccl-20 was stimulated after TNFα treatment in all acetylation-deficient mutants, though to a reduced amount than in the wild type cell line (Figure 6B–D). This suggests that gene expression of Ccl-20, Ifi-44 and Gbp-2 can only be properly induced by TNFα when RelA/p65 is acetylatable by harboring lysines at the position 310, 314 or 315. The Ccl-7 gene was induced by TNFα in all cell lines (Figure 7B, middle panel).
To demonstrate that transcriptional changes in gene expression correlate with changes in corresponding protein expression, western blot analysis was performed for Ccl-7 in the complemented cell lines. Upregulation of Ccl-7 protein was detected in the RelA/p65 acetylation-deficient mutant cell lines compared to wild type after 4 h of TNFα stimulation (Figure 7C).
Taken together, the results obtained in the microarray analysis could be confirmed by the complementary real time RT-PCR method for four selected genes. Interestingly, Ccl-7 protein expression analysis in the distinct acetylation-deficient cell lines indicated that the differential gene regulation detected in the microarray analysis is indeed translated into protein expression. Additionally, this analysis provides strong evidence that acetylation of RelA/p65 at lysines 310, 314 and 315 positively or negatively regulates the transcription of specific genes.
DISCUSSION
Increasing experimental evidence has indicated that NF-κB-dependent gene expression is regulated by different post-translational modifications including acetylation (38,39). The acetylation-dependent NF-κB regulation was shown to occur via many different mechanisms (40). For example acetylation of histones is known to regulate NF-κB-dependent gene accessibility for the transcriptional machinery (38). The direct acetylation of NF-κB was reported to regulate the transcriptional potential of NF-κB, the duration of the NF-κB response, its DNA-binding activity as well as protein–protein interactions with several transcription cofactors (32,41,42).
In this study we have addressed the role of p300-mediated acetylation of RelA/p65 in the regulation of gene expression in vivo. We identified three lysines (K310, K314 and K315) in RelA/p65 as targets for the acetylation by p300 in vitro and in cells upon TNFα stimulation (Figures 2A and 3A). Interestingly, lysine 310 was previously shown to be acetylated by p300 (32). However, we additionally found two novel lysine residues, lysine 314 and 315, acetylated in RelA/p65 not reported before. It is important to mention that among the previously reported acetylation sites by Kiernan et al. and Chen et al. (30,32), we could only confirm the acetylation of K310 in our experimental system (Figure 3B). This could be due to the different experimental procedures; stimuli (TNFα versus PMA) and cell lines (HEK 293T cells versus Jurkat T cells) used in the studies and could point towards a very specific stimulus and cell-type-dependent regulation of RelA/p65 through acetylation. However, our results suggest additional yet to be identified p300 specific acetylation sites in RelA/p65 other than lysine 218, 221, 310, 314 and 315 (Figure 3C). The relevance of the in vitro acetylation sites identified in our study was confirmed by the ability of endogenous RelA/p65 to be acetylated in vivo upon TNFα stimulation using an acetylation site-specific antibody (Figure 4C).
To investigate the role of RelA/p65 acetylation in cells we genetically complemented RelA/p65–/– fibroblasts with a control vector (pTV) or cDNAs encoding for RelA/p65 wild type, K310R, K314/315R and KTR mutant (Figure 4A). Analysis of the complemented cells revealed that the mutation of the acetylation sites did not affect the kinetics of the cytoplasmic-nuclear redistribution of RelA/p65 upon TNFα stimulation (Figure 5A). This implied that the upstream signalling events are not regulated by the acetylation of RelA/p65 at the identified sites. Furthermore, cell survival assays were performed to examine whether the acetylation of RelA/p65 is involved in the protection from TNFα-induced cell death. No difference was seen in cell survival when RelA/p65–/– cells complemented with wild type RelA/p65 were compared to cells complemented with the acetylation-deficient mutants, indicating that the acetylation of RelA/p65 on lysine 314 and 315 is not involved in the TNFα-dependent cell death pathway (Figure 5C).
In order to elucidate the influence of RelA/p65 acetylation on gene expression in vivo we performed genome-wide microarray analyses using total RNA isolated from the complemented cells after TNFα stimulation. We identified subsets of genes, which were specifically modulated depending on the ability of RelA/p65 to be acetylated at lysine 310, 314 or 315 (Supplementary Tables 1–3). The number of common significantly down or upregulated genes found in the current analysis of the different cell lines is small, indicating that acetylation of RelA/p65 at lysine 310, 314 and 315 is possibly modulating the expression of only a subset of genes (Figures 6A and 7A). Four genes from the distinct subsets of differentially expressed genes were selected for validation: Ifi-44, Ccl-20 (Mip-3α), Gbp-2 and Ccl-7 (MCP-3); thereof Ccl-20 is a known NF-κB target gene (43). Ifi-44, Ccl-20 and Gbp-2 gene expression showed to be dependent on acetylation of RelA/p65 at lysine 310, 314 and 315. Expression of Ccl-7 was found to be repressed by acetylated RelA/p65. This was supported by the protein analysis performed for Ccl-7, which revealed induction of Ccl-7 protein expression in the acetylation-deficient mutant cell lines (Figure 7C). The increase in Ccl-7 protein only slightly correlated with the increase of mRNA levels detected in the different mutants at 45 min of TNFα stimulation (compare Figure 7B, right panel). This could be explained by the differences in stimulation duration for mRNA (45 min) and protein (4 h) detection and by additional regulatory mechanisms (such as secretion), which might influence the quantity of proteins.
When the induction of gene expression upon TNFα stimulation was investigated, differences were detected between the four genes. From the selected genes, the chemokines Ccl-20 and Ccl-7 were the only genes induced by TNFα in the mutant cell lines. Even though the upstream events of TNFα-induced NF-κB signalling were shown to be normal in our study, Ifi-44 and Gbp-2 gene expression was not induced by TNFα in the K/R mutant cell lines suggesting that regulation by K310, 314 and 315 acetylation is essential for the first group of genes (Ifi-44, Gbp-2) and required for the latter group (Ccl-20 and Ccl-7).
The results of the microarray analysis and the quantitative real-time PCR revealed that the acetylation of RelA/p65 does not always correlate with activation of gene expression as has been suggested before (32). Rather, we provide evidence that the p300-mediated acetylation of RelA/p65 contributes to both gene-specific activation and repression of transcription. It is unknown how RelA/p65 acetylation signals to activate certain genes while simultaneously suppressing the expression of others. One hypothesis would be that the regulation of certain genes by p300-mediated acetylation of RelA/p65 promotes the recruitment of additional factors or stabilizes the formation of specific pre-initiation complexes at promoter sites. Like this, acetylated RelA/p65 could serve as a factor that regulates the recruitment of these proteins. It has been previously reported that many proteins can specifically recognize and bind acetylated lysine residues in histones and transcription factors through their bromo domains (44–46). Among them are well-characterized histone acetyltransferases (e.g. p300, P/CAF and GCN5) (47), subunits of chromatin remodelling complexes (e.g. ATPases of SWI2/SNF2 and proteins of the WAL/BAZ family of ISWI-associated proteins) (48) and a general cofactor of the basal transcription machinery, TAF1 (49). The nature of the recruited co-activator/co-repressor would determine the response of the specific genes. Another hypothesis would be that the presence of other cis-regulatory elements in a targeted gene and regulatory proteins recruited to these elements might be critical for the modulation of the acetylation dependent NF-κB response. Acetylation is known to be a reversible protein modification. In this regard HDAC-1, HDAC-2 and HDAC-3 were reported to repress NF-κB-dependent transcription upon treatment with inflammatory stimuli (32,38,40). These histone deacetylases were also shown to directly interact with several proteins involved in the NF-κB signalling pathway, including NF-κB itself (29,38,50). Deacetylation of RelA/p65 by HDAC-3 may provide a counterpart mechanism for p300 function. The exact molecular mechanism by which acetylation/deacetylation of RelA/p65 regulates the transcription activity of RelA/p65 in the context of chromatin remains to be investigated.
Together, acetylation of RelA/p65 presents an attractive regulatory mechanism for the control of NF-κB-dependent gene expression. The combination of acetylation with other post-translational modifications will broaden even more the potential of this regulation. Combinations of different modifications have already been proposed to serve as a ‘code’ for the interacting domains of different proteins (51,52). Furthermore, acetylation can be regulated by other post-translational modifications. For example phosphorylation of RelA/p65 at serine 276 and 536 was shown to enhance the acetylation of lysine 310 (53). It cannot be excluded that other post-translational modifications of RelA/p65 can also regulate the acetylation of this transcription factor. Our findings could help to explain the diversity of NF-κB-dependent gene expression upon different stimuli. We hypothesize that unique combinations of post-translational markers and the presence of cell-type-specific cofactors determine the specific NF-κB-mediated response. Recruitment analyses of RelA/p65 to the promoter region of the regulated genes using chromatin immunoprecipitation will help to understand how post-translational modifications of this transcription factor influence gene expression of its target genes.
In conclusion, our results support the hypothesis that p300-mediated acetylation of distinct sites in the RelA/p65 protein is important to modulate the expression of defined genes, thereby contributing to the specificity of the NF-κB response.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Funding for this study was provided by Krebsliga Zürich; the Swiss National Science Foundation program (31-109315.05). T.V. and M.O.H. are supported by the Kanton of Zurich. We thank P.A. Cole (Johns Hopkins School of Medicine, Baltimore, MD), S. Dent (University of Texas MD Anderson Cancer Center), D. Trouche (LBME, University of Toulouse), Lee W. Kraus (Cornell University, Ithaca, NY), D. Thanos (BSRC Al. Fleming, Athens, Greece), E. Ferrari and U. Huebscher (IVBMB, University of Zurich) for providing useful tools. We are grateful to the Functional Genomics Center Zurich (FGCZ) for technical support (thanks especially to Andrea Patrignani for his assistance in data generation). We are also grateful to all the members of the Institute of Veterinary Biochemistry and Molecular Biology (University of Zurich, Switzerland) for helpful advice and discussions. Funding to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation program 31-109315.05.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Shishodia S, Aggarwal BB. Nuclear factor-kappaB activation: a question of life or death. J. Biochem. Mol. Biol. 2002;35:28–40. doi: 10.5483/bmbrep.2002.35.1.028. [DOI] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 4.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 5.Perkins ND. The Rel/NF-kappa B family: friend and foe. Trends Biochem. Sci. 2000;25:434–440. doi: 10.1016/s0968-0004(00)01617-0. [DOI] [PubMed] [Google Scholar]
- 6.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 7.Courtois G, Gilmore TD. Mutations in the NF-kappaB signaling pathway: implications for human disease. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 8.Perkins ND. Achieving transcriptional specificity with NF-kappa B. Int. J. Biochem. Cell Biol. 1997;29:1433–1448. doi: 10.1016/s1357-2725(97)00088-5. [DOI] [PubMed] [Google Scholar]
- 9.Karin M. The NF-kappa B activation pathway: its regulation and role in inflammation and cell survival. Cancer J. Sci. Am. 1998;4(Suppl. 1):S92–S99. [PubMed] [Google Scholar]
- 10.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 11.Silkov A, Wolstein O, Shachar I, Dikstein R. Enhanced apoptosis of B and T lymphocytes in TAFII105 dominant-negative transgenic mice is linked to nuclear factor-kappa B. J. Biol. Chem. 2002;277:17821–17829. doi: 10.1074/jbc.M200696200. [DOI] [PubMed] [Google Scholar]
- 12.Gerritsen ME, Williams AJ, Neish AS, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl Acad. Sci. USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J. Biol. Chem. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 14.Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 15.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 16.Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 17.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 18.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. Embo J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J. Biol. Chem. 2000;275:10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 21.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Vermeulen L, De Wilde G, Notebaert S, Vanden Berghe W, Haegeman G. Regulation of the transcriptional activity of the nuclear factor-kappaB p65 subunit. Biochem. Pharmacol. 2002;64:963–970. doi: 10.1016/s0006-2952(02)01161-9. [DOI] [PubMed] [Google Scholar]
- 24.Saccani S, Marazzi I, Beg AA, Natoli G. Degradation of promoter-bound p65/RelA is essential for the prompt termination of the nuclear factor kappaB response. J. Exp. Med. 2004;200:107–113. doi: 10.1084/jem.20040196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 26.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1) Embo J. 2003;22:1313–1324. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duran A, Diaz-Meco MT, Moscat J. Essential role of RelA Ser311 phosphorylation by zetaPKC in NF-kappaB transcriptional activation. Embo J. 2003;22:3910–3918. doi: 10.1093/emboj/cdg370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 30.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 31.Furia B, Deng L, Wu K, Baylor S, Kehn K, Li H, Donnelly R, Coleman T, Kashanchi F. Enhancement of nuclear factor-kappa B acetylation by coactivator p300 and HIV-1 Tat proteins. J. Biol. Chem. 2002;277:4973–4980. doi: 10.1074/jbc.M107848200. [DOI] [PubMed] [Google Scholar]
- 32.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. Embo J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishinaga H, Jono H, Lim JH, Kweon SM, Xu H, Ha UH, Xu H, Koga T, Yan C, Feng XH, et al. TGF-beta induces p65 acetylation to enhance bacteria-induced NF-kappaB activation. Embo J. 2007;26:1150–1162. doi: 10.1038/sj.emboj.7601546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Andaloussi N, Valovka T, Toueille M, Steinacher R, Focke F, Gehrig P, Covic M, Hassa PO, Schar P, Hubscher U, et al. Arginine methylation regulates DNA polymerase beta. Mol. Cell. 2006;22:51–62. doi: 10.1016/j.molcel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 38.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 39.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 40.Schmitz ML, Mattioli I, Buss H, Kracht M. NF-kappaB: a multifaceted transcription factor regulated at several levels. Chembiochem. 2004;5:1348–1358. doi: 10.1002/cbic.200400144. [DOI] [PubMed] [Google Scholar]
- 41.Chen LF, Greene WC. Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation. J. Mol. Med. 2003;81:549–557. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 42.Deng WG, Zhu Y, Wu KK. Up-regulation of p300 binding and p50 acetylation in tumor necrosis factor-alpha-induced cyclooxygenase-2 promoter activation. J. Biol. Chem. 2003;278:4770–4777. doi: 10.1074/jbc.M209286200. [DOI] [PubMed] [Google Scholar]
- 43.Tomimori K, Uema E, Teruya H, Ishikawa C, Okudaira T, Senba M, Yamamoto K, Matsuyama T, Kinjo F, Fujita J, et al. Helicobacter pylori induces CCL20 expression. Infect. Immun. 2007;75:5223–5232. doi: 10.1128/IAI.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 45.Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 2001;21:5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 47.Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]
- 48.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 49.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 50.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 52.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 53.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.