Abstract
MicroRNAs (miRNA) are endogenous tissue-specific short RNAs that regulate gene expression. Discriminating each let-7 family member expression is especially important due to let-7's abundance and connection with development and cancer. However, short lengths (22 nt) and similarities between multiple sequences have prevented identification of individual members. Here, we present ProDeG, a computational algorithm which designs imperfectly matched sequences (previously yielding only noise levels in microarray experiments) for genome-wide microarray “signal” probes to discriminate single nucleotide differences and to improve probe qualities. Our probes for the entire let-7 family are both homogeneous and specific, verified using microarray signals from fluorescent dye-tagged oligonucleotides corresponding to the let-7 family, demonstrating the power of our algorithm. In addition, false let-7c signals from conventional perfectly-matched probes were identified in lymphoblastoid cell-line samples through comparison with our probe-set signals, raising concerns about false let-7 family signals in conventional microarray platform.
INTRODUCTION
MicroRNA (miRNA) is an endogenous non-coding short RNA which regulates gene expression (1–3). Processing of the hairpin-shaped precursor miRNA (∼75 nt) by the Dicer enzyme results in mature miRNA about ∼22 nt long (4,5). There are 470 human miRNAs registered in the miRBase database version 9.1 (6), many of which are conserved across several species and highly similar to other miRNAs in the genome. There is a great demand for accurate expression profiling of these miRNAs to better understand their tissue specificities (7–10) and their role in development (11–14) and disease (15–19).
Techniques for determining miRNA expression include northern blot analyses (20), quantitative RT-PCR (15), and microarrays (21). Among these, the oligonucleotide microarray platform offers a simple and high-throughput experimental procedure for genome-wide miRNA profiling. Barad et al. have shown in expression profiling experiments that mature miRNA sequences, not their precursors, are responsible for fluorescence signals (8). By positioning short probes away from a solid support via an unrelated linker sequence, they have demonstrated efficient miRNA hybridization to the probes.
However, miRNA arrays pose several challenges. One is the ability of design strategies to distinguish many highly similar sequences that differ by only a few nucleotides. Another is the mere ∼22 nt length of miRNA, which allows no choice for a probe sequence other than the miRNA itself. Given the diverse range of miRNA melting temperatures (Tm), it is almost impossible to find one experimental condition to satisfy all genomic miRNA hybridizations simultaneously. Currently there exist two major strategies for balancing Tm: (i) by incorporating chemically modified nucleotides with higher affinity (22) and (ii) altering probe sizes (23). However, discriminating highly similar sequences, thus featuring similar Tm, remains a challenge. Such sequences will hybridize similarly to the probes and the signal will not be specific anymore. Guo et al. have shown experimentally that the introduction of an artificial nucleotide (lacking hybridization ability) into the probes enhanced specificity and allowed discrimination of single nucleotide polymorphisms (24). However, the small data set and use of an artificial nucleotide limit genome-wide application, as no microarray could utilize this feature.
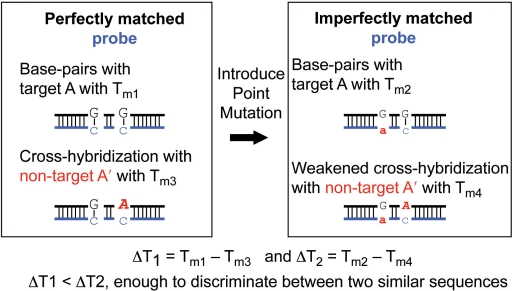
In contrast, based on extensive computational experiments, we suggested utilizing mismatches with naturally occurring nucleotides in designing mRNA probes (25). Conventionally, mismatched sequences have been used in assessing noise levels rather than signals because hybridization can disappear with single or double nucleotide mismatches. The problem arises that background signals produced by these mismatched probes can be as strong as those of the matched probes. In a previous study, we identified mismatched sequences and positions which induced minimal or maximal changes in oligonucleotide hybridization compared to perfectly matched sequences. In addition, we found Tm variance with two-point mismatches to be greater than twice that with one-point mismatches (25). By carefully introducing mismatches into a probe sequence, we can increase differences in stabilities of hybridization between target and non-target sequences sufficient to achieve discrimination, as shown in Figure 1.
Figure 1.
Schematic example of including an imperfectly matched probe to increase specificity. Probe strand is shown in blue and in lower case characters. Target A and non-target A′ differ by one base at the position shown as sequence A. After incorporating a base change (sequence a), the difference in Tm between probe-target and probe-non-target pairs (right, ΔT2 = Tm2 − Tm4) is sufficient to discriminate similar sequences as compared to the difference in Tm before point mutation (left, ΔT1 = Tm1 − Tm3).
Here, we present highly specific microarray probes with a narrower calculated Tm range, produced using Probe Design Guru, or ProDeG (pronounced ‘prodigy’), the first algorithm to implement our previous findings in a genome-wide microarray probe design. We applied ProDeG to miRNA sequences as a first step in validating our probes based on the importance and feasibility. This technique allows the reduction of probe-target hybridization melting temperatures (Tm) when they significantly exceed the Tm of most other probe-target pairs. Introducing mismatches in the probe sequence can also serve to eliminate secondary structures of probes. Since the probes do not include any modified oligonucleotides or change of lengths, our methods are easy to incorporate into any microarray platform.
Applying this method to human mature miRNAs from miRBase version 9.1, we found specific probes for all the let-7 family members. Two types of probes were designed: probes which mimicked the conventional cDNA microarray experiments (cDNA samples) and probes which followed current miRNA profiling (RNA samples). The let-7 family is especially important for its abundant expression and connection with developmental processes and cancer (26,27). We experimentally verified probe specificity and homogeneity for this family, which has not been achieved before, with cDNA spiked-in samples. We also verified probe specificity using RNA spiked-in samples and further identified non-specific signals of the let-7 family that arise from conventional perfectly matched probes by comparing them with signals from our probe-set for lymphoblastoid cell-line samples. This discrimination was confirmed with quantitative RT-PCR, verifying the efficacy of our probes.
MATERIALS AND METHODS
ProDeG algorithm
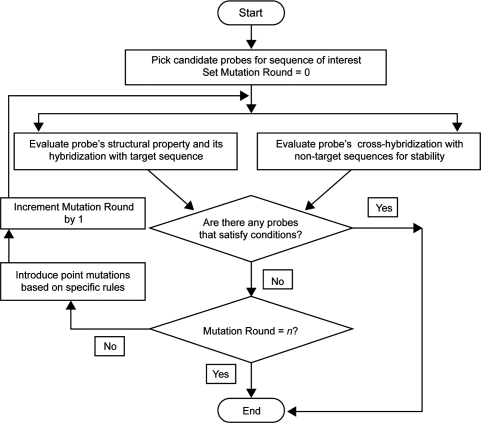
ProDeG follows a series of steps in scrutinizing each of the probes before reporting them as specific to a targeted miRNA. The flow chart in Figure 2 details all the steps in processing before final reporting on probes. Initially, the sole candidate probe is the mature miRNA sequence. Following this, probes are evaluated in two broad stages, first addressing probe quality in respect to a target and secondly checking non-targets. In the first stage, probes are assessed for their structural properties and for their hybridization with the target sequence. Undesirable stable hairpin formations in probes and uniform Tm are evaluated. Melting temperature as a measure of hybridization stability is calculated using the nearest neighbor thermodynamics model (28) with licensed software Oligonucleotide Modeling Platform (OMP; http://www.dnasoftware.com). Observing that OMP calculations correlated with experimental Tm better than our in-house program containing publicly available parameters, we then calculated Tm variance dependency on mismatch positions (25). Interestingly, Pizhitkov et al. recently reported a similar mismatch position dependency (29) based on microarray signal intensity data, leading us to utilize OMP in the miRNA probe design.
Figure 2.
The ProDeG flowchart.
In the second stage, we use BLAST to search for similarities among all the other human miRNA sequences, making sure that the DUST program is turned off using the –F option so all sequence stretches are considered. Candidates predicted to cross-hybridize with matches ≥14 nucleotides are retained for further processing. Predicted cross-hybridizations between probes and non-target sequences may, in fact, not occur due to unstable interactions. Such interactions then undergo a round of thermodynamic stability evaluations using OMP. Probes without any stable cross-hybridization are then reported as specific.
Next, imperfectly matched probes are used to identify a target sequence when candidates fail to satisfy the conditions set forth in the two prior stages of evaluation. If stable cross-hybridizations are present, we change bases in order to alter binding stabilities enough to distinguish between target and non-target sequences (Figure 1). If target-candidate hybridization Tm is above a set temperature (75°C in the current case) or the candidates have strong secondary structure, imperfectly matched candidates are generated to destabilize secondary structure and also reduce excessively high Tm of hybridization between a candidate and its target. To assess the probe characteristics after base changes have been introduced, each of these modified probes is made a new candidate for which evaluations are repeated from the start. When imperfectly matched probes satisfy all the set criteria, they can be reported as specific to the target.
If a single round of changes in the probe sequence fails to weaken secondary structure formation, reduce high Tm of hybridization with the target, or eliminate hybridization with non-target sequences, we subject the probe to a defined maximum number n of rounds of base changes (currently, n = 2) and evaluate its hybridization properties. In spite of having two sets of introduced mutations, miRNA probes still showing some cross-hybridizations are reported as such.
Mature human miRNAs have a very wide Tm range of about 36°C, the lowest and highest melting temperatures being 56°C and 92°C for miR-620 and miR-663, respectively (1 M salt concentration). Since there are numerous miRNAs with melting temperatures between 65°C and 75°C, we set the ceiling for the Tm range at 75°C. The discriminating ΔT for our program is based on data from experiments conducted by Thomson et al. which showed that sequences with one mismatch are distinguishable (21). By calculating Tm for miR-124a and the reverse complements of the perfectly and imperfectly matched probe sequences used in their experiments (data not shown), we concluded that 5°C is sufficient. Even though absolute Tm is a function of parameters in the nearest neighbor model, Tm difference is not. Since our criteria for lowest hybridization Tm between probe and target is 56°C, we set the maximum hybridization Tm between probe and non-target to be 51°C. Detailed calculation parameters are given in Table 1.
Table 1.
Parameters used in ProDeG microarray probes for mature human miRNA
| Parameters | |
|---|---|
| Assay Temperaturea | 53°C |
| Maximum hybridization Tm | 75°C |
| Maximum monomer folding Tma (secondary structure measurement) | 65°C |
| Minimum hybridization Tm between probe and target | 56°C |
| Maximum hybridization Tm between probe and non-target | 51°C |
| Na+ concentration | 1 M |
| K+ concentration | 0 M |
| Probe concentration | 100 nM |
| Target concentration | 100 nM |
| BLAST word size | 7 |
aSpecific parameters in the OMP software.
ProDeG is a bundle of programs for the UNIX programming environment written in PERL and C++, process flow being controlled by a PERL script which calls all other programs within it. ProDeG uses two external programs, BLAST and OMP. BLAST is available for download for several platforms; OMP is a licensed application available on several platforms as well and may be purchased from the vendor. By calculating Tm using the nearest neighbor model and published parameters (28), licensed OMP may feasibly be replaced.
Microarray platform
Microarray services were provided by LC Sciences Inc. (Houston, TX), which made the detection probes by in situ synthesis using photogenerated reagent chemistry on a microfluidic chip. We augmented their microarray layout with custom probes to experimentally validate our probe design strategy. The block of whole probe sets is repeated six times in a microarray. Custom probes include DNA sequences to the let-7 family in Table 2 (as a control) and the ProDeG let-7 family probes in Table 3 for the cDNA spiked-in experiments. Custom probes also include the reverse complementary sequences of the let-7 family (as a control; Supplementary Table 1), as well as ProDeG-designed probes for RNA samples (Table 4) of both spiked-in and total RNA from the lymphoblastoid cell lines.
Table 2.
Mature human let-7 family sequences in DNA and their hybridization Tm with perfectly complementary pairs
| Name (control) | Sequencea | Tm (°C)b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| let-7a | let-7b | let-7c | let-7d | let-7e | let-7f | let-7g | let-7i | miR-98 | ||
| let-7a | TGAGGTAGTAGGTTGTATAGTT | 64 | 58 | 59 | 58 | 57 | 59 | 52 | 51 | 51 |
| let-7b | TGAGGTAGTAGGTTGTG TG GTT | 58 | 70 | 65 | 51 | 51 | 54 | |||
| let-7c | TGAGGTAGTAGGTTGTATGGTT | 59 | 64 | 67 | 51 | 51 | 52 | 52 | 51 | 51 |
| let-7d | AGAGGTAGTAGGTTGCATAGT | 55 | 51 | 51 | 66 | |||||
| let-7e | TGAGGTAGGAGGTTGTATAGT | 62 | 55 | 57 | 56 | 66 | 55 | |||
| let-7f | TGAGGTAGTAGATTGTATAGTT | 57 | 62 | |||||||
| let-7g | TGAGGTAGTAGTTTGTACAGT | 64 | 55 | |||||||
| let-7i | TGAGGTAGTAGTTTGTGCTGT | 55 | 68 | |||||||
| miR-98 | TGAGGTAGTAAGTTGTATTGTT | 63 | ||||||||
aMismatch sequences compared to let-7a are shown in bold italics.
bHybridization Tm to the target is in bold; 51°C in italics, for reference, is not expected to produce signals in our design criteria.
Table 3.
ProDeG-designed probe sequences for cDNA of mature human let-7 family and their hybridization Tm with targets and non-targets
| Name (probe) | Sequencea | Tm (°C)b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| let-7a | let-7b | let-7c | let-7d | let-7e | let-7f | let-7g | let-7i | miR-98 | ||
| let-7a | TGAGaTAGTAGGTTGTATAGTT | 57 | 51 | |||||||
| let-7b | TGtGGTAGTAGGcTGTGTGGTT | 57 | ||||||||
| let-7c | TGtGGcAGTAGGTTGTATGGTT | 57 | ||||||||
| let-7d | AGAGGTAGTAaGTTGCATAGT | 58 | ||||||||
| let-7e | TGAcGTAGGAGGTTGTATAGT | 51 | 57 | |||||||
| let-7f | TGcGGTAGTAGATTGTATAGTT | 51 | 57 | |||||||
| let-7g | TGAGGTAaTAGTTTGTACAGT | 56 | ||||||||
| let-7i | TGAGGTAGTAcTTTGTGCTGT | 58 | ||||||||
| miR-98 | TGAGGTAGTAAGTTGTATTGTT | 63 | ||||||||
aMismatch sequences compared to the original are shown in bold lower case.
bHybridization Tm to the target is in bold; 51°C in italics, for reference, is not expected to produce signals in our design criteria.
Table 4.
ProDeG-designed probe sequences for mature human let-7 family (RNA as sample) and their respective hybridization Tm with targets and non-targets
| Name (probe) | Sequencea | Tm (°C)b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| let-7a | let-7b | let-7c | let-7d | let-7e | let-7f | let-7g | let-7i | miR-98 | ||
| let-7a | AACTATACAACCTACTAtCTCA | 59 | 52 | 56 | 52 | 57 | ||||
| let-7b | AACCACACAACtTACTACCaCA | 60 | 51 | |||||||
| let-7c | AACCATACAACCTAtTACCTtA | 55 | 58 | |||||||
| let-7d | AcCTATGCcACCTACTACCTCT | 59 | ||||||||
| let-7e | AtCTATAaAACCTCCTACCTCA | 58 | ||||||||
| let-7f | tACTATACAgTCTACTACCTCA | 53 | 57 | |||||||
| let-7g | AACTGTACtAACTACTACCTCA | 51 | 60 | 52 | ||||||
| let-7i | AACAGCACcAACTACTACCTCA | 63 | ||||||||
| miR-98 | AACAATtCAACTTACTACCTCA | 58 | ||||||||
aMismatch sequences compared to the original are shown in bold lower case.
bHybridization Tm to the target is in bold; 51°C in italics, for reference, is not expected to produce signals in our design criteria.
let-7 family spiked-in experiments
DNA and RNA oligonucleotides with fluorescence dye attached to their 5′-end were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). DNA sequences are reverse complementary to the mature let-7 member sequences, while RNA sequences are the same as the mature let-7 family. For pairing, let-7a, let-7c and let-7f were labeled with Cy-5 and let-7b, let-7d, let-7e with Cy-3. LC Sciences performed custom microarray fabrication, hybridization and signal reading. All hybridization was performed for 1 h in the presence of hybridization buffer (25% formamide, 6 × SSPE, pH 6.8) on a µParaflo microfluidic chip using a micro-circulation pump (Atactic Technologies, Inc.; Houston, TX). The signal intensities of each pair (let-7a/7d, let-7b/7c and let-7e/7f) were recorded at seven temperature conditions (25°C–55°C) for both cDNA and RNA cases. Because the microarray platform is microfluidic, the hybridization solution contains formamide, which reduces hybridization temperature (30) to minimize bubble formation in the chamber. Internal controls were used to compare multiple experiments. Hybridization images were collected using a laser scanner (GenePix 4000B, Molecular Devices, Inc; Sunnyvale, CA) and digitized using Array-Pro image analysis software (Media Cybernetics, Inc). Data were analyzed by first subtracting the background and then normalizing the signals using a LOWESS filter (Locally-weighted Regression) to compensate for the intensity difference between Cy5 and Cy3.
Lymphoblastoid cell line preparation
Lymphoblastoid cell lines were prepared from blood draws of six human subjects using established methods (31). Briefly, peripheral blood mononuclear cells were isolated from whole blood with Histopaque reagent (Sigma). For each blood sample, 10 ml of Histopaque was added to a 50 ml sterile conical tube. In another 50 ml conical tube, 10 ml of well-inverted blood was mixed with 10 ml of RPMI 1640 medium (Invitrogen). We then gently layered the blood and RPMI mixture on top of the Histopaque, and centrifuged at 1500–1700 r.p.m. for 30 min.
In a bar-coded T25 flask, we added 0.15 ml of phytohemagglutinin reagent and 6 ml of 30% FBS complete medium. When blood centrifugation was complete, we aspirated off the top layer and transferred the white cloudy middle layer into a new 50 ml conical tube to wash the PBM cells with RPMI 1640 medium. We then re-suspended the cell pellet in 2 ml of RMPI 1640 medium. In the T25 flask prepared as described above, we added 2 ml of filtered EBV and the suspended pellet. We then filled the flask with 30% FBS complete medium up to 10 ml of total volume. The cells were placed in a CO2 incubator for 6–8 weeks. At the half-way point (∼3 weeks), we fed the cells with 10% FBS complete medium. When the culture grew to a confluency of 106 cells/ml, we collected the cells and made stocks with freezing medium, storing the cell stocks in freezers at −140°C.
Hybridization experiments using total RNA of lymphoblastoid cell lines
Total RNA from each human lymphoblastoid cell line was isolated with Trizol reagent (Invitrogen) according to the manufacturer's protocol (Invitrogen Cat No. 15596). Following the recommendation of LC Sciences, Inc., we used 1.5 ml of isopropyl alcohol per 1 ml of Trizol Reagent for the initial homogenization. We incubated samples at −20°C overnight and centrifuged them at no more than 12 000 × g for 10 min at 4°C. These modifications were necessary for the recovery of small RNAs from our cell line samples (based on preliminary study), which would be lost otherwise.
Microarray assay was performed using a service provider (LC Sciences). The assay started from 2–5 μg total RNA sample, which was size fractionated using a YM-100 Microcon centrifugal filter (from Millipore) and the small RNAs (<300 nt) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye Cy-3 staining. Hybridization took place at 34°C. Wash temperatures for control and ProDeG probes were 53 and 47°C, respectively.
Quantitative RT-PCR
We purchased commercially available real-time reverse transcription quantitative PCR (qRT_PCR) reagent kits from Applied Biosystems, Inc (ABI, C.A.), including TaqMan® assays for human let-7 family miRNAs (7a, 7b, 7c, 7d, 7e, 7f), a control assay (RNU6B), TaqMan® MicroRNA Reverse Transcription Kit (Part # 4366596) and TaqMan® Universal PCR Master Mix (Part # 4324018). All TaqMan assays were performed using a two-step procedure following the supplier's manual (Pat # 4364031 Rev. B, ABI). First, we performed single-stranded cDNA synthesis from total RNA using the TaqMan® MicroRNA Reverse Transcription Kit. Briefly, for each 7 μl of reverse transcription (RT) master mixture, we combined 0.15 μl of 100 mM dNTPs, 1 μl of MultiScribe™ Reverse Transcriptase, 1.5 μl of 10X RT buffer, 0.19 μl of RNase inhibitor, and 4.16 μl of nuclease-free water. The 7 μl of RT master mixture was then combined in a fresh tube with 5 μl total RNA (10 ng) and 3 μl of RT primer (specific to each TaqMan assay), and gently mixed. The RT reactions were then performed on an iCycler thermal cycler (Bio-Rad) programmed to incubate the reactions at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. The synthesized cDNA were diluted ten times with H2O and stored in a −20°C freezer for qRT-PCR analysis.
Second, we carried out TaqMan qRT-PCR assays according to the assay developer's recommended conditions (ABI, CA) on a 7900HT Fast Real-Time PCR System (ABI, CA). Each qRT-PCR reaction was performed in 20 μl volume, containing 10 μl of TaqMan® Universal PCR Master Mix, 1 μl of 20X TaqMan MicroRNA Assay mix, and 9 μl of the 10x diluted single-stranded cDNA product. Three replicates were used for each sample. We employed SDS2.2.1 software (ABI, CA) for quantification analysis in conjunction with the 2−ΔΔCt method (33), using the RNU6B as the reference control for normalization.
RESULTS
Variance of Tm by introducing mismatches and the let-7 family
One of the most abundant and well-studied miRNAs is the let-7 family, associated with most cancers (26,27). The let-7 family of sequences and their corresponding DNA hybridization Tm with perfectly complementary pairs are shown in Table 2. Each family member differs by only one or two nucleotides. Predicted cross-hybridizations with Tm ≥ 52°C are also presented in Table 2. With perfectly matched probes, there is no way to prevent cross-hybridizations (23). Utilizing our finding that Tm variance with two-point mutations is greater than twice that with one-point mutations (25), discrimination is now possible. This synergetic effect is not limited to nearest neighbor two-point mutation sites. Rather, most positions of an oligonucleotide show this, unless they are close to the chain end. The discrimination of let-7e and let-7a exemplifies the process diagram in Figure 1. One nucleotide among these differs near the middle of the sequences. Tm of a perfectly-matched let-7e probe–target is 66°C; Tm of a perfectly-matched let-7e probe–non-target (let-7a) is 62°C (Table 2), so that ΔT1 = Tm1 − Tm3 = 4°C. When we change the 10th position sequence A of the let-7e probe to T, Tm for target and non-target becomes 60 and 54°C, respectively (ΔT2 = Tm2 − Tm4 = 6°C). After incorporation of a base change, the Tm difference between target and non-target is increased (ΔT2 > ΔT1). This technique, moreover, allows probe-target hybridization Tm's to be reduced when they significantly exceed the Tm of most other probe-target pairs. Introducing mismatches in the probe sequence can also serve to eliminate secondary structures.
ProDeG probes for cDNA of human miRNAs
Taking advantage of the fact that Tm variance with two-point mutations is greater than twice that with one-point mutations, ProDeG processed mature human miRNAs to design microarray probes with the parameters in Table 1 and predicted probes for all 470 of them. Calculations treat samples as reverse complementary DNA sequences to mature miRNA and DNA probes as equivalent to mature miRNA, in accordance with cDNA microarray experiments. These cDNA probes will validate that microarray signals produced by ProDeG from highly similar sequences are discriminated. Moreover, as miRNA amplification methods become more advanced, probes for miRNA cDNA may prove valuable. ProDeG probes for the let-7 family are shown in Table 3 along with predicted cross-hybridizations where Tm ≥ 52°C. Following several mutation steps, all cross-hybridizations predicted in Table 2 have been eliminated. In addition, all the probes shown in Table 3 have uniform melting temperatures (mostly 57 and 58°C). Note that miR-98 did not undergo the mutation steps because our Tm ceiling criterion was set at 75°C. All calculated miRNA probe sequences are searchable and downloadable from the web (http://oligo.ctaalliance.org/miRNA).
Characteristics of ProDeG probes for cDNA of human miRNAs
Among the probes for the 470 mature miRNA sequences, those for 432 miRNAs are target specific, including imperfectly matched probes for 224 miRNAs, 160 of them due to eliminating cross-hybridizations of perfectly matched probes. Secondary structures were eliminated in probes for 27 miRNAs. High Tm was eliminated in probes for 76 miRNAs. We were able to overcome these obstacles (cross-hybridization, secondary structures, and high Tm) using imperfectly matched probe sequences. Designed probes for 38 mature miRNAs presented cross-hybridization with non-target miRNAs (mostly with one other); the detailed sequences and Tm are in Supplementary Table 3. 20 out of 38 miRNAs were 100% identical to at least one other miRNA except for bases at either end of the sequences, 5 of the 20 being complete subsets of the other miRNAs. 10 other miRNAs contained one mismatch with other mature miRNA sequences at the second or third position from the 3′-end. The remaining eight miRNAs have one middle A which differs from G in another miRNA sample, leading to T (probe)-A (target sample) and T (probe)-G (non-target sample) discrimination tasks.
Verification of ProDeG cDNA probe specificity using let-7 spike-in experiments
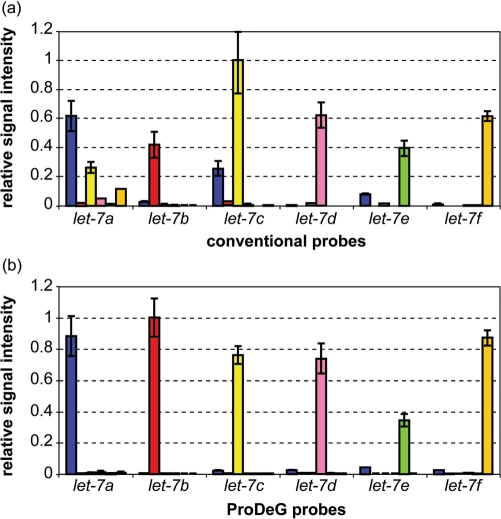
Spiked-in experiments were performed to verify designed probe specificity within the let-7 family. Since there is no significant cross-hybridization for let-7g, 7i or miR-98, we used designed probes for let-7a to 7f (Table 3). Based on the Tm calculations (Tables 2), we paired let-7a/7d, let-7b/7c and let-7e/7f for two-color hybridization experiments. Average fluorescent signals from six adjacent spots of perfectly matched probes (controls) and of our probes are shown in Figure 3a and b. Each control or probe signal value is chosen for its optimal discriminating temperature [35 and 30°C, respectively, with formamide addition (30)] from 55 to 25°C data and normalized with the highest signal value from the respective control set or probe set. Two clear advantages over the controls become apparent. First, probe-target signal intensities align except in the case of let-7e probes, yielding much more homogeneous fluorescence signals, as predicted. Second, cross-hybridizing signals appearing in the control sets are mostly removed. In addition, the highest signal intensity value from the control set is nearly 4 times greater than that of the probe set. The minimal cross-hybridization signals in Figure 3b are practically non-existent. The question arises whether the signals from our probes are strong enough for use in an application.
Figure 3.
Relative signal intensities of the let-7 family with spiked-in cDNA sequences are shown using perfectly matched conventional (control) sequences as probes at 35°C (a) and ProDeG designed probes at 30°C (b). Each x axis category indicates probes used while the corresponding series shows the relative probe intensities normalized with highest intensity value. Spiked-in sample notations are as follows: blue bars, let-7a; red bars, let-7b; yellow bars, let-7c; pink bars, let-7d; lime bars, let-7e and orange bars, let-7f.
ProDeG probes for RNA samples of human miRNAs and let-7 spike-in experiments
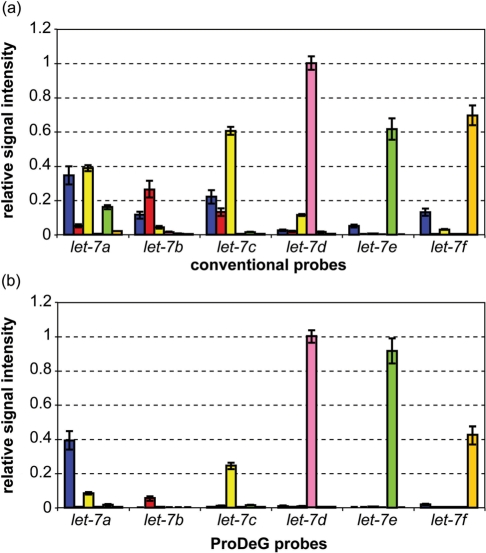
Since most miRNA profiles use fractionated small RNAs from total RNA, we designed probes for RNA samples using hybridization parameters of DNA-RNA pairs. Again, all 470 probes for RNA samples were predicted. Table 4 shows let-7 family probes and their predicted cross-hybridizations using the same criteria of Tm ≥ 52°C. Probes for RNA samples are predicted to present some cross-hybridization on let-7a probe with let-7c and let-7e samples and on let-7c probe with let-7b. Tm's for targets are less uniform and a bit higher than cDNA sample cases.
When we performed RNA spike-in experiments with these probes over 7 temperature points from 25–55°C, we found that the hybridizations were more stable than those in the case of DNA–DNA. Since some signals of the mismatched probes were much stronger than with cDNA, we prepared the normalized signal graph at 40°C for both control and ProDeG probes in Figure 4. The Tm calculations are basically held in the signal intensities except for the let-7b probe (Figure 4b). If we set aside the let-7b probe signal, the specificity of ProDeG probes were dramatically superior to the control probes (Figure 4a), with only mild cross-hybridization of let-7c on let-7a probes. Please note that the overall cross-hybridization of control probes was also much more prevalent compared to the case of cDNA. The normalized intensity of control probes is about three times higher than that of ProDeG probes.
Figure 4.
Relative signal intensities of the let-7 family with spiked-in RNA sequences are shown using perfectly matched conventional (control) sequences as probes (a) and ProDeG designed probes (b) at 40°C. Each x axis category indicates probes used while the corresponding series shows the relative probe intensities normalized with highest intensity value. Spiked-in sample notations are as follows: blue bars, let-7a; red bars, let-7b; yellow bars, let-7c; pink bars, let-7d; lime bars, let-7e and orange bars, let-7f.
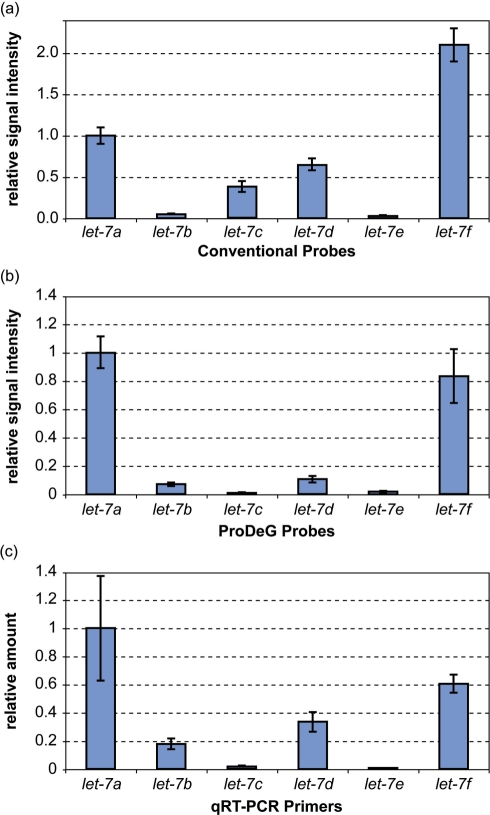
Expression signals of ProDeG let-7 probes from human lymphoblastoid cell lines
We prepared total RNA of lymphoblastoid cell lines from a human subject to obtain miRNA profiles. In addition to LC Sciences probes, we incorporated custom probes containing controls (perfectly matched sequences) and ProDeG probes to compare signals among them. Since the hybridization temperature was 34°C, optimized for the company's probes, gentle wash condition (47°C) was performed to detect ProDeG signals compensating weaker signals in addition to the normal wash condition (53°C). Each microarray contained probe blocks repeated six times. The relative signal intensities compared to the let-7a signal are shown in Figure 5a and b for control and ProDeG probes, respectively. Interestingly, let-7b signal from ProDeG probes was detectable, in spite of the unusually low let-7b spike-in signal in Figure 4b. Rather, the let-7b signal in control probes was minimal. On the other hand, the let-7c signals from the control probe were significant, while those of the ProDeG probe were non-existent.
Figure 5.
Total RNA sample data from a lymphoblastoid cell-line. All data are normalized against let-7a data. Relative signal intensity of the let-7 family conventional (a) and ProDeG (b) probes are shown after Cy-3 labeled total RNA from a lymphoblastoid cell-line was hybridized at 34°C. The relative amount of each let-7 family miRNA was quantified using TaqMan® qRT-PCR assay (c).
In order to verify the presence of each let-7 family, we performed qRT-PCR on the same total RNA. The relative amount compared to let-7a quantity is shown in Figure 5c. The relative amount pattern strikingly resembles the ProDeG probe signal intensity: practically non-existent let-7c and let-7e, while let-7a amount is the largest followed by let-7f amount. We therefore conclude that the let-7c signals from the conventional perfectly-matched probe were actually false signals from other let-7 family members (probably from cross-hybridization with let-7a based on Figure 4a). ProDeG probes are highly reproducible using qRT-PCR and proved to be specific in our study.
DISCUSSION
Which miRNAs need to be discriminated? Even though we definitely removed most cross-hybridizations, at least in computational terms, several remain (Supplementary Table 2). Eliminating these involves discriminating one nucleotide difference near or at the end of the miRNA and discriminating T-A and T-G pairs. We reported that mutation in the first or last three bases of a sequence produces minimal Tm changes. Moreover, the interaction energy between T-A and T-G are similar, indicating limited discrimination by mismatched probes.
This limitation would be overcome when discriminating one nucleotide difference near or at the end of the miRNA by simulating an internal mismatch which may be obtained by padding two or three nucleotides during sample preparation. This concept has already been implemented by other researchers (23). ProDeG can then be applied to mismatch probe design. However, among the miRNAs listed in Supplementary Table 2, some are only predicted, without experimental confirmation. Also, a nuclease might have cut one or more sequences in the process of miRNA maturation. We do not feel compelled to go further in discriminating end sequence differences.
Discrimination of T-A and T-G pairs can be addressed using reverse complementary sequences as probes and mature miRNAs as samples. T (probe)-A (target sample) and T (probe)-G (non-target sample) pairs in the original set become A (probe)-T (target sample) and A (probe)-C (non-target sample) pairs in this reverse set. There should be no miRNAs in common in the G-U wobble category of predicted cross-hybridizations. Therefore, two sets of experiments, one using probes with mature miRNAs and the other using their reverse complements, will ultimately discriminate T-A and T-G pairs.
ProDeG probes for cDNA samples are of significant value both in terms of Tm calculations (Table 3) and spike-in experiments (Figure 3b). One intrinsic concern, however, is that signal intensities from the ProDeG probes are relatively weak compared to the perfectly matched probes, thus raising a question regarding signal sensitivity in real applications. The next step is to optimize hybridization conditions and to find a balance between specificity and sensitivity. However, once techniques to obtain cDNAs of small RNAs are further developed and PCR amplification is routinely achievable, increased specificity to a target sequence using the ProDeG algorithm will be of some value.
RNA samples produced stronger signals and more cross-hybridization (Supplementary Table 1, Table 4 and Figure 4) than cDNA samples. Since signals from the ProDeG probes were strong enough, we could use the same hybridization temperature for both control and ProDeG probes in total RNA profiling experiments. During the revision process, the Sanger Institute miRBase updated its miRNA sequence database to version 10. Since cDNA samples established the correspondence between microarray signals and our calculations, cDNA data are meaningful by themselves. However, profiling total RNA involves endogenous miRNA, which needs to be updated based on the new information. In terms of the let-7 family, however, only one nucleotide was added at the 3′-end position for let-7d, e, g and i, whose influence is probably not significant. We added a corresponding sequence A to each let-7 ProDeG probe, as calculated with version 9.1 and used for probes for RNA samples.
Comparing Tm calculation (Table 4) and spike-in experimental data (Figure 4b), either the let-7b Tm calculation was wrong or let-7b RNA synthesis was not desirable or both. Since there was no let-7e cross-hybridization to the let-7a ProDeG probe (different from the Table 4 prediction), thermodynamic parameters of RNA–DNA pairs might be less accurate than those of DNA–DNA pairs. Improved thermodynamic parameters will increase the quality of designed probes. On the other hand, considering the let-7b signal detection using total RNA samples (Figure 5b), there might not be a high-purity yield of let-7b RNA, as the company warned, due to the difficulty of incorporating Cy-5 into RNA oligonucleotides. Despite these limitations, to our surprise, the relative signal intensity of total RNA using ProDeG probes matched the qRT-PCR data excellently, demonstrating the utility of ProDeG probes.
The presence of let-7c signal in the control emphasizes the false positive signal in miRNA microarray data which is prone to generate incorrect inferences in terms of miRNA expression. Another miRNA, miR-99a, is transcribed right next to the let-7c transcription site in the same intron of Chromosome 21 open reading frame 34. The expressions of these two miRNAs were reported to be correlated (32). In our total RNA sample, the miR-99a signal was absent from the microarray data. However, significant false signaling of let-7c in the control probes (Figure 5a) would not yield such a correlation. With our probes, we can report both let-7c and miR-99a are probably absent from the transcription stage.
The ProDeG strategy is simple, powerful, cost-efficient and fully compatible with current profiling techniques, moreover considering only naturally occurring nucleotide hybridization. The use of mismatched sequences with natural nucleotides (less toxic than artificial ones) to enhance target specificity (minimal off-target effects) will allow safer in vivo applications. Like other hybridization calculations, ours lacks surface effects, which may have led to a lower than predicted let-7e signal in Figure 3b (note that let-7d and let-7e are one nucleotide shorter than other members according to v9.1 of miRBase). To our surprise, however, the overall calculation predicted microarray intensity very well. All experimental data point to the validity of our computational algorithm.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Dr Xiaochuan Zhou from LC Sciences for valuable discussion and experiments beyond initial arrangements, Doyoung Chung from the University of Michigan Bioinformatics Graduate Program for assisting with transcription site analysis, Zachary C. Wright for help with database and web page preparation, and Linda Gates for assistance in cell culture. Michigan Economic Development Corporation—Life Sciences Corridor Fund (N002831); National Institutes of Health (U54-DA021519) (to M.M. and B.A.); National Institute of Mental Health (MH064596) and Stanley Medical Research Institute Grant (to H.C.). Funding to pay Open Access publication charges for this article was provided by U54-DA021519.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol. Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 12.Bushati N, Cohen SM. microRNA Functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 13.Moss EG. Heterochronic genes and the nature of developmental time. Curr. Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Jay C, Nemunaitis J, Chen P, Fulgham P, Tong AW. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 18.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in Disease and Potential Therapeutic Applications. Mol. Ther. 2007;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 19.van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 22.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. RNA. 2007;13:151–159. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z, Liu Q, Smith LM. Enhanced discrimination of single nucleotide polymorphisms by artificial mismatch hybridization. Nat. Biotechnol. 1997;15:331–335. doi: 10.1038/nbt0497-331. [DOI] [PubMed] [Google Scholar]
- 25.Lee I, Dombkowski AA, Athey BD. Guidelines for incorporating non-perfectly matched oligonucleotides into target-specific hybridization probes for a DNA microarray. Nucleic Acids Res. 2004;32:681–690. doi: 10.1093/nar/gkh196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 27.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 28.SantaLucia J, Jr, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 29.Pozhitkov A, Noble PA, Domazet-Loso T, Nolte AW, Sonnenberg R, Staehler P, Beier M, Tautz D. Tests of rRNA hybridization to microarrays suggest that hybridization characteristics of oligonucleotide probes for species discrimination cannot be predicted. Nucleic Acids Res. 2006;34:e66. doi: 10.1093/nar/gkl133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutton JR. Renaturation kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic Acids Res. 1977;4:3537–3555. doi: 10.1093/nar/4.10.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum. Genet. 1986;73:320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- 32.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.