Abstract
Progesterone triggers the resumption of meiosis in the amphibian oocyte through a signaling system at the plasma membrane. Analysis of [3H]ouabain and [3H]progesterone binding to the plasma membrane of the Rana pipiens oocyte indicates that progesterone competes with ouabain for a low affinity ouabain binding site on a 112 kDa α1-subunit of the membrane Na/K-ATPase. Published amino acid sequences from both low and high affinity ouabain binding α1-subunits are compared, together with published site-directed mutagenesis studies of ouabain binding. We propose that the progesterone binding site is located in the external loop (23 amino acids) between the M1-M2 transmembrane helices. Analysis of loop topology and the countercurrent hydrophobicity/polarity gradients within the M1-M2 loop further suggest that the polar β and hydrophobic α surfaces of the planar progesterone molecule interact with opposite sides of the amino acid loop. The 19-angular methyl group of progesterone is essential for activity; it could bind to the C-terminal region of the M1-M2 loop. Maximum biological activity requires formation of hydrogen-bond networks between the 3-keto group of progesterone and Arg118, Asp129 and possibly Glu122-124 in the C-terminal region of the loop. The 20-keto group hydrogen may in turn hydrogen bond to Cys111 near the M1 helix. Peptide flexibility undergoes a maximal transition near the midway point in the M1-M2 loop, suggesting that folding occurs within the loop, which further stabilizes progesterone binding.
Keywords: Progesterone, oocyte, ouabain, plasma membrane, receptor, topology, α1-subunit Na/K-ATPase
1. Introduction
Steroids affect cell metabolism through two different pathways. Cytosol steroid receptors which bind to DNA and alter gene expression are characterized by a latent period between steroid uptake and cell response (reviewed in [1]). The second pathway acts via receptors at the plasma membrane, is associated with a much more rapid response, and does not directly affect transcription (reviewed in [2-4]). Progesterone-induced meiosis in the Rana pipiens oocyte was one of the first demonstrations of a steroid response system originating in the plasma membrane [5,6]. The meiotic divisions in amphibian oocytes are initiated by progesterone, secreted by ovarian follicle cells in response to pituitary gonadotropins. As demonstrated with enucleated oocytes, progesterone induces changes in the oocyte membrane and cytosol which, in intact oocytes, lead to completion of the first meiotic division and arrest at metaphase II [7].
The present study discusses evidence for the interaction of progesterone with a specific site on an integral plasma membrane protein, the ouabain-binding α1-subunit of the Na/KATPase. We find that progesterone competes with ouabain for a low affinity, but not a high affinity, binding site on the oocyte plasma membrane [8]. This suggests that the low affinity ouabain binding site may correspond to the progesterone receptor on the oocyte surface. Evidence from other laboratories indicates that the low affinity and a high affinity sites are on different α1-subunits [9]. Since the low and high affinity ouabain binding α1-subunits differ by only a few amino acids, largely concentrated in the first external loop between the first and second transmembrane helices, it should be possible to identify the amino acids associated with progesterone binding. A comparison of topology and site-directed mutagenesis data of ouabain binding in the low and high affinity forms are used to propose a model for the interaction of progesterone with a specific site on the oocyte plasma membrane.
Two other steroid binding sites have been proposed for the vertebrate oocyte plasma membrane. A G-protein-linked 40 kDa peptide with seven transmembrane domains has been isolated from fish ovarian segments and shown to bind steroids (reviewed in [10]). Alternatively, it has been proposed that the classical progesterone (cytosol) receptor mediates the non-genomic action that triggers meiosis in X. laevis oocytes [11]. Thus, including the steroid binding Na/K-ATPase α1-subunit described here, there are at least three candidate membrane receptor peptides associated with non-genomic steroid events in the amphibian/fish oocyte.
2. Experimental
The present study uses: 1) our published data on progesterone-ouabain competition for a binding site on the external surface of isolated plasma membranes from Rana pipiens oocytes [8, 12], 2) site-directed mutagenesis studies of ouabain binding to sheep and human Na/K-ATPase α1-subunits by other investigators (e.g. [13]), and 3) the known amino acid sequence of α1-subunit isoforms from protein data banks. Previously unpublished data describes the meiotic inducing activity and electron spin resonance (ESR) data for 3-DOXYL-17β-OH-5α-androstane (Table 1).
Table 1.
Comparison of [3H]Progesterone and [3H]Ouabain Binding to Isolated Plasma-vitelline Membranes from Rana pipiens Oocytes
| Oocyte Plasma-vitelline Membrane | |||||
|---|---|---|---|---|---|
| Steroid | [Progesterone]o, μM | Kd | fmols/oocyte | M.W. | Ref |
| Progesterone | …. | 5.1±1.1 × 10−7 M | 66±19 | 110-114 kDa | 15 |
| Ouabain | 0 | 3.2±0.5 × 10−8 M | 15±4 | 8,12 | |
| 0 | 2.0±0.4 × 10−6 M | 80±9 | 112 kDa1 | 8,12 | |
| 1.62 | 1.8±0.26 × 10−8 M | 28±5 | 8 | ||
M.W. of α-subunit of Na/K-ATPase [27].
Oocytes were pretreated with 1.6 μM progesterone for 1 h.
2.1 Use of Rana pipiens to study the meiotic divisions
Rana pipiens (Leopard frog) has been widely used in developmental biology. Its oocytes are particularly advantageous for the study of steroid action at the plasma membrane. Mature females contain up to several thousand fully grown, yolk-filled oocytes, arrested in prophase of the first meiotic division. In contrast to Xenopus laevis (African clawed frog), Rana ovulates only once a year (late spring for females from the Northeastern United States), following winter hibernation. However, oocytes can be removed from the ovary of the hibernating R. Pipiens female from October thru May, and, immersed in Ringer's solution containing progesterone, will complete the first meiotic division and arrest at metaphase II in vitro (reviewed in [14]). The chromosomes are tightly condensed and remain condensed throughout the meiotic divisions. Since all fully grown oocytes are in exactly the same stage, all synchronously complete the first meiotic division and advance to metaphase of the second meiotic division. The disappearance or “breakdown” of the nuclear membrane is generally used as the criterion for initiation of the first meiotic division. This can be monitored by dissection of heat-fixed oocytes. (Completion of the second meiotic division is triggered by sperm entry). R. pipiens nuclear membrane breakdown occurs 8-10 h after exposure to progesterone at 20° C, compared to about 3 h in fully grown Stage VI X. laevis oocytes. (The oocyte nucleus has been referred to as the germinal vesicle and germinal vesicle breakdown as GVBD.)
2.2 Isolation of Intact Plasma Membrane from Amphibian Oocytes
The large size of the R. pipiens oocyte (1.8-2.2 mm diameter) and nucleus (0.4 mm diameter) are particulary useful for biochemical, biophysical and microinjection studies. Intact or enucleated oocytes can be pulse-labeled with radiolabeled compounds or studied noninvasively using nuclear magnetic resonance techniques (reviewed in [14]). Prophase oocytes free of adherent follicle cells (referred to as “denuded”) can be obtained after brief exposure to EDTA-containing Ca2+-free Ringer's solution (Masui et al. (cit. [15]). (This avoids modification of plasma membrane proteins by prolonged treatment with collagenase, often employed to free X. laevis oocytes from follicle and thecal cells.)
The ligand-binding experiments described here were carried out with plasma-vitelline membranes from fully grown, denuded R. pipiens oocytes. Membranes were isolated, one at a time, from denuded oocytes immersed in isotonic sucrose containing 1.0 mM CaCl2. Each oocyte was manually penetrated with a fine-tipped pipette, and the yolk contents carefully rinsed out with the sucrose solution, leaving a ghostlike translucent sphere [15,16]. The presence of the vitelline membrane, an acellular fibrous mesh-like glycoprotein structure overlying the plasma membrane, supports the plasma membrane and prevents fragmentation. Care was taken to avoid contact of the membrane preparation with the air-water interphase (if this occurs there is a rapid dispersal of membrane fragments as an oily layer on the surface of the aqueous phase).
2.3 Peptide Topology Analysis
The UniprotKB/Swiss-Prot/EMBL data base (www.expasy.org/uniprot) was the source for sequence data for α-subunit isoforms of the Na/K-ATPase from a variety of species. The sequence data for rat, sheep, frog and human Na/K-ATPase α1-subunit (see Accession numbers indicated in the appropriate text) were used to plot the conformational differences between the low vs. high affinity ouabain binding forms that are shown in the figures below. SIB BLAST searches were performed using the BLAST network service: NCBI BLAST program reference [PMID:9254694]. No X-ray crystallographic or nuclear magnetic resonance protein structure studies have been published for the α1-subunits of the Na/K-ATPase from any species.
The Residue-based Diagram editor (RbDe) Web site maintained by the Department of Physiology and Biophysics, Weill Medical College of Cornell University, New York, NY was employed to illustrate the topology of the Na/K-ATPase α-subunits of various species [17]. Chem 3D Ultra v. 10 (Cambridgesoft, Cambridge Scientific Computing, Cambridge, MA) was used to visualize the structure of external loops and helicies of the α1-subunit of Na/K-ATPase shown in Figures 3 and 8. Hydrophobic and polarity gradients within the loops were measured as described by Kyte and Doolittle [18] and of Grantham [19], respectively. Hydrophobic cluster analysis [20] on the amino acid sequence of the first external loop (M1-M2) was carried out using the internet site bioserv.rpbs.jussieu.fr. The modeling software used in this study are either programed for single letter or three letter amino acid codes. Therefore, the amino acid codes used in each computer generated figure are also used in the corresponding discussion.
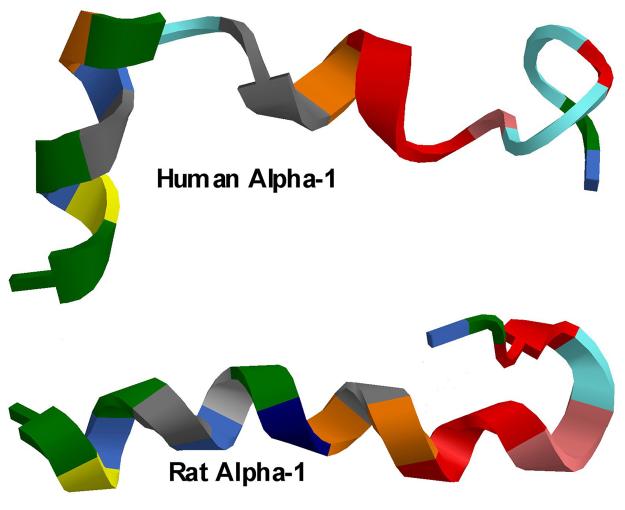
Figure 3.
A comparison of 3D projection of the 23 amino acid sequence in the first external loop of the α1 subunit of the Na/K-ATPase of the high affinity ouabain binding isoform (human, upper cartoon) and low affinity ouabain binding isoform (Rat, lower cartoon). Plots were generated using Chem 3D Ultra v. 10.0 (Cambridgesoft.com). Colors indicate individual amino acids.
Figure 8.

An illustration of the possible progesterone binding site within the first external loop of the low affinity ouabain binding isoform of the α1-subunit of the Na/K-ATPase. The graphic represents a view looking down on the cell surface, with an end view of the first (M1) and second (M2) transmembrane helices and the interconnecting loop of 23 amino acids. The peptide bonds between the helices and the external loop amino acids are discontinuous to allow for 90 degree rotation of the loop sequence. The arrows indicate the direction of insertion of the planar steroid into the loop. The amino acids in the external loop are color coded and the amino acids thought to be critical for ouabain binding are indicated in single letter codes (C111, R118, S122, P128, D129, D131). The planar progesterone (shown on its side with the α hydrophobic face upward and to the left) interacts with hydrophobic clusters in the N-terminal region. The 19-methyl group (facing down and to the right) interacts with residues at the beginning of the transmembrane helix (M2). As proposed, a hydrogen-bonding network forms between the 3-keto group and the conserved residues Gln120, Asn131 and Tyr118 in the first external loop of the α-1 subunit of the rat Na/KATPase.
3.0 Comparison of Meiotic Inducing Activity of Progesterone and Cardiotonic Steroids on the Amphibian Oocyte
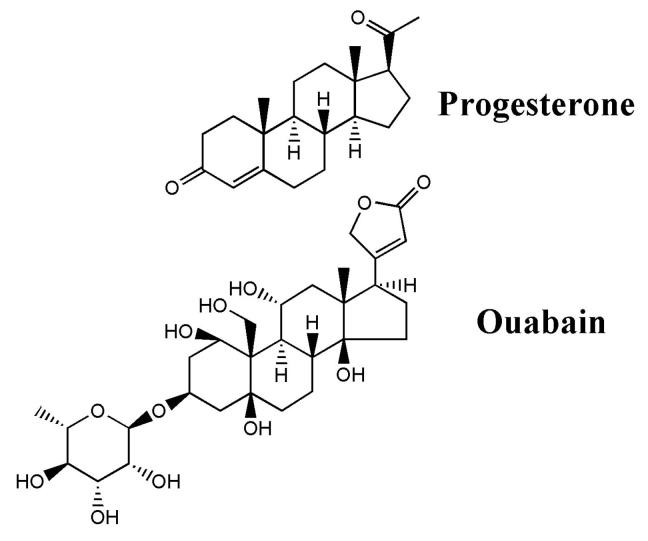
Figure 1 compares the structure of Δ4-pregnane 3,20-dione (progesterone) with that of the steroid, ouabain (G-strophanthin). Ouabain is a digitalis derivative, frequently used in the laboratory as an inhibitor of the Na/K-ATPase (Na+-pump). It differs from progesterone in that it contains rhamnose in the 3 position, an unsaturated lactone ring in the 20 position, and hydroxyl groups on carbons 1, 5, 11, 14 and 21 of the steroid nucleus. Ouabain-like immunoreactivity has been found in vertebrate tissues and in plasma; the highest levels being found in the adrenal cortex, hypophysis and hypothalamus (reviewed in [21]). Its concentration in plasma varies from pmolar to nmolar responding to physiological stress, causing it to be considered for hormone status. Endogenous mammalian ouabain appears to be identical to plant-derived ouabain [21]. Pregnenolone and progesterone have both been shown to be precursors of endogenous ouabain in vertebrates [22-24]. Smaller amounts of related compounds (bufadieholides, marinobufogenin,19-norbufalin) have also been identified in vertebrates (cit [21]).
Figure 1.
Structures of Δ4-pregnane-3,20-dione (progesterone) and the cardiotonic steroid ouabain. Compounds with an unsaturated five-membered lactone ring are cardenolides and those with an unsaturated six-membered lactone ring are bufadienolides. In the formulae, bonds or atoms or groups above the plane of the ring system are depicted as solid wedges; those below the plane are depicted as open, dashed wedges.
The meiotic activity of two cardiotonic steroids (ouabain and digitoxigenin) is compared with that of progesterone in Table 1. Vitto and Wallace found that while ouabain alone will not induce meiosis in X. laevis oocytes, it will induce nuclear membrane breakdown in Xenopus oocytes after a brief exposure to progesterone [25]. Cartaud et al. [26] subsequently confirmed the ouabain facilitation effect and found that digitoxigenin, a digitalis steroid that differs from ouabain in that it lacks hydroxyl groups in the 1, 5 and 11 positions, also induced nuclear membrane breakdown in X. Laevis oocytes with a half-maximal response of 3 x 10-5 M. These findings suggest that initial progesterone binding permits the plasma membrane receptor to subsequently bind ouabain.
4.0 Comparison of Progesterone and Ouabain binding to the Rana pipiens Oocyte Plasma Membrane
4.1 Progesterone Binding to the Isolated Plasma-Vitelline Membrane
The surface of the prophase R. pipiens oocyte is covered with numerous microvilli that increase the effective surface area of the prophase oocyte 10-12 fold [27]. The isolated plasma-vitelline membrane complex does not metabolize progesterone [15], which facilitates accurate determination of [3H]progesterone binding. Table 2 summarizes the progesterone binding characteristics of the oocyte plasma-vitelline membrane. We find that progesterone binds to a single site (Kd = 5 ×10−7 M) on the plasma membrane of the R. pipiens oocyte which is specific, saturable and which has a slow off-rate [15]. Photoaffinity labeling with the progestin R5020 demonstrates a major 110-114 kDa component in these membranes [16]. Similarly, Western blots of plasma-vitelline membranes, using SDS/PAGE, contain a single 110-114 kDa band which binds an antibody to the steroid-binding C-terminal domain of the human/rat progesterone cytosol receptor [8]. In contrast, a polyclonal antibody to an N-terminal region (sequence 375-564) of the human progesterone receptor immunoreacted with oocyte cytosol but not with oocyte plasma membrane proteins [8]. Thus, the oocyte plasma membrane receptor seems to be homologous to carboxyterminal steroid binding region of the human/rat intracellular progesterone receptor but differs in the N-terminal region of the oocyte and rat/human cytosol progesterone receptor.
Table 2.
 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Steroid | C-3 | Δ4 DB | C-1 | C-10 | C-5 | C-13 | C-11 | C-14 | C-17 | Response1 | Ref |
| Progesterone | keto | Yes | H | CH3 | H | CH3 | H | H | 20-keto | Induces | 44 |
| Ouabain | Sugar | No | OH | CHOH | OH | CH3 | OH | OH | Lactone | Potentiates | 21 |
| Digitoxigenin | OH | No | H | CH3 | H | CH3 | H | OH | Lactone | Induces | 22 |
| 3-DOXYL-17βol-5α-androstane | DOXYL | Yes | H | CH3 | H | CH3 | H | H | OH | Induces | …. |
| 5α-androstane-3,17-diol | OH | Yes | H | CH3 | H | CH3 | H | H | OH | Induces | 44 |
| 19-Nor-androstane-3,17-diol2 | OH | Yes | H | H | H | CH3 | H | H | OH | Inactive | 44 |
Nuclear membrane breakdown in amphibian oocytes in vitro (24h at 20-22° C).
19-Nor-testosterone is also inactive (ref. 44).
The progestin R5020 also binds to a single class of sites on both 80 and 110 kDa components in 105,000g cytosol prepared from R. pipiens oocytes denuded of follicle cells [28]. These binding sites have a Kd (3 × 10−8 M) that is an order of magnitude higher than that of the plasma membrane receptor [15]. Therefore, based on Kd and Western blot analysis, the 110-114 kDa progesterone-binding component present in the oocyte cytosol appears to be distinct from that found in the plasma membrane. A progesterone receptor recently cloned from X. laevis oocytes [29] may represent the oocyte cytosol receptor [28], rather than a plasma membrane receptor.
4.2 Ouabain Binding to the R. pipiens Plasma-Vitelline Membrane
The outer surface of the amphibian oocyte plasma membrane is known to have binding sites for both progesterone [15] and ouabain [12]. Table 2 summarizes the dissociation constants, binding capacity and estimated molecular weights of the ouabain binding component in plasma-vitelline membranes from prophase-arrested (unstimulated) R. pipiens oocytes. Ouabain binding to the oocyte plasma membrane was measured after incubation of prophase-arrested oocytes (denuded of follicle cells) in [3H]ouabain, followed by the rapid manual isolation of plasma membranes [12]. Scatchard analysis of [3H]ouabain binding to the isolated plasma membrane indicates the presence of at least two components: a K+-sensitive, high affinity site (Kd = 3 × 10−8 M) linked to inhibition of the pump, and a low affinity site (Kd = 2 × 10−6 M) which does not appear to be associated with transport inhibition [12]. The binding capacity for the low-affinity ouabain site and the progesterone site are comparable. However, progesterone has a Kd one order of magnitude greater than that for low affinity ouabain binding site (Table 2).
Pretreatment of prophase Rana oocytes (denuded of follicle cells) with 10 μM exogenous progesterone for 60 min eliminates the low affinity, but not the high affinity ouabain binding site (Table 1). This finding, plus the fact that the estimated molecular weight of the oocyte plasma membrane progesterone binding component and that of the steroid-binding α1 subunit of the Na/K-ATPase are comparable (112 kDa, reviewed in [30]), and that ouabain and its analogues either facilitate or induce meiosis (Table 1), suggests that progesterone binds to the steroid-binding α-subunit of the Na/K-ATPase.
The total surface area of the prophase-arrested Rana oocyte can be estimated to be about 0.6 cm2 per oocyte, based on capacitance measurements [27]. Scatchard analysis of low affinity ouabain binding and progesterone binding to the prophase-arrested oocyte indicates that 4,280±720 and 3,960±910 (mean±SD, N = 3) molecules of ouabain and progesterone, respectively, are bound per μm2. This indicates that progesterone and ouabain bind in a 1:1 ratio, consistent with a common binding site.
5.0 The Plasma Membrane Na/K-ATPase: Cardiotonic Steroid Binding to the α-Subunits of the Na/K-ATPase
5.1 Properties of the α-subunits of the Na/K-ATPase
Four genes encoding the Na/K-ATPase α-subunit have been cloned in vertebrates (reviewed in [31-34]). Evidence to date indicates that α-subunit isoforms with low affinity (certain α1 isoforms) and high affinity (α1, α2, α3 and α4 isoforms) for the steroid, ouabain, are present in a variety of animal species (reviewed in [32-34]). Isoform-1 of the α-subunit is ubiquitous and has been considered the “housekeeping” form of the enzyme and it has been proposed that the high affinity ouabain binding α1-subunit of Na/K-ATPase may serve to maintain low “bulk” cytosolic Na+ concentrations [9]. The α4-subunit has been reported only in testis [34]. The neonate and the adult rat hearts exhibit both low and high affinity ouabain binding sites (e.g. [35]), although the low affinity isoform constitutes about 80-90% of the total; thus the rat heart is relatively insensitive to ouabain. The rat kidney and several other rat tissues contain only the low affinity isoform [32]. Thus, the adult rat heart and kidney are relatively insensitive to ouabain and may be used as a sources for the low affinity ouabain binding α1-isoform.
Pralong-Zamofing et al. [33] have identified the α1 isoform in a plasma membrane-rich sucrose gradient fraction prepared from Stage VI (fully grown) X. laevis oocytes using anti-α1-serum. Ouabain sensitivity of endogenous X. laevis ovarian α1 isoform was not measured., although ouabain binding to intact oocytes following expression of cRNA's encoding kidney α1-isoform has been reported [36]. No published information was found relating to the α2, α3 and/or α4 isoforms in vertebrate oocytes or ovaries.
5.2 The Structure of the α1-Subunit of the Na/K-ATPase
Figure 2 illustrates the primary amino acid sequence of the α1 subunit of X. laevis Na/K-ATPase (primary accession # Q92123) in Scalable Vector graphics (SVG) format [17]. Projection A (left) shows the complete α1-subunit, and projection B (right) represents a truncated version with emphasis on the external loops and transmembrane helices. The N and C termini of the α-subunit of the Na/K-ATPase are intracellular. The peptide has the 10 transmembrane domains and the 2 large intracellular loops found in all α-isoforms (reviewed in [30-32]). Transmembrane domains are α-helices composed primarily of non-polar amino acids. The transmembrane domain is stable across the fluid lipid bilayer because the hydrophobic effect operating on the side chains provides ∼40 lcal mol-1 of favorable free energy [36].
Figure 2.

The primary amino acid sequence of the complete (A) and truncated (B) graphics of the α-1 subunit of Xenopus laevis Na/K-ATPase in Scalable Vector graphics (SVG) format [17]. The sequence (primary accession # Q92127) was obtained from the nucleotide sequence of Xenopus laevis kidney epithelium [43] and neural plate cells [44]. The N and C termini of the α-subunit of the Na/K-ATPase are located intracellularly and the protein has 10 transmembrane domains and 2 large intracellular loops (see text). The intracellular N-terminal and C-terminal ends contain a white spacer to shorten the terminal sequence due to space limitations.
Conventional α-helices also occur within the loops between the transmembrane domains and will be considered separately as progesterone binding sites in section 6.1, below.
The primary amino acid sequence (1023 residues) of the α1-subunit of X. laevis has a 93% sequence homology with that from both sheep (primary accession # P04074) and human (primary accession #P05023). For comparison, Bufo marinus (Giant toad) (primary accession #P30714) has a 94% sequence homology with the Na/K-ATPase α1-subunit of X. laevis. An amino acid sequence for the Na/K-ATPase α1-subunit from R. pipiens has not been published. Since R. pipiens (Leopard frog) is closely related to X. laevis (African clawed frog), the amino acid sequence for the α1-subunit of X. laevis is taken as representative of the frog.
5.3 The Ouabain Binding Site on the α1-Subunit
Three general approaches have been used to describe the high affinity binding site on the ouabain-sensitive α1-subunit of the Na/K-ATPase. Mutagenesis of an α1-subunit Na/K-ATPase gene of the H1C1 cell line [37] and random mutagenesis of the high affinity ouabain binding sheep α1 isoform [13], coupled with expression, has shown that the amino acids which alter ouabain binding and/or sensitivity cluster in two major regions. The first ouabain binding region comprises the extracellular loop joining the first two membrane spanning domains in the N-terminal region. A second ouabain-binding region is formed by the extracellular portions of transmembrane domains four, five, six and seven. In a second approach, the 2.6 Å resolution X-ray crystalographic structure of the Ca2+-ATPase of skeletal muscle sarcoplasmic reticulum (10 transmembrane domains) was used to infer the ouabain binding site in the Na/K-ATPase [38]. The luminal and cytoplasmic loop topology of the two peptides is remarkably similar (compare primary accession numbers O14983 (Ca-pump) and P05023 (Na-pump). However, the Ca2+-ATPase does not bind ouabain, and except for a seven residue phosphorylation site on a cytoplasmic loop, there is a relatively low (20-25%) sequence homology between the Ca2+- and the Na/K-ATPase with less than 10% sequence homology in the first and second luminal loops identified as putative ouabain binding sites. The crystallographic data may, however, predict an arrangement of the ten α1-subunit helices within the plane of the lipid bilayer that is common to both enzymes. In a third approach, chimeras of individual Na/K-ATPase loop regions were inserted into either the skeletal muscle sarcoplasmic reticulum/endoplasmic reticulum Ca2+-ATPase (SERCA1) [39,40] or the gastric H/K-ATPase [41,42]. The results suggest that M1-M2, M3-M4 and M5-M6 extracellular loops (see Figure 2) are important for ouabain sensitivity.
Few studies have been designed to elucidate the ouabain binding site on the low affinity ouabain binding isoform of the α1-subunit. Lingrel et al. [13] suggested that the M1-M2 region of the α1-subunit of sheep Na/K-ATPase, when dephosphorylated, is the low affinity ouabain-binding site, whereas in the high affinity (phosphorylated) conformation a single ouabain molecule spans both the M1-M2 and M4-M5-M6 regions. However, as outlined above, the concept of two unique α1 proteins with different sensitivities to ouabain best fits the data from a number of laboratories (reviewed in [9]). This implies that the low affinity ouabain binding region is restricted to the extracellular loop joining the first two membrane spanning domains in the N-terminal region.
6. Characterization of Progesterone Binding to the α1-Subunit of Na/K-ATPase
6.1 Progesterone Binding Sites(s) on the In Situ α1-Isoform
If progesterone selectively binds to the low affinity ouabain site on the α-subunit, a comparison of the amino acid sequence of the α1 isoform with low ouabain affinity (rat; primary accession #P06685) with that for an α1 isoform with high ouabain affinity (sheep; primary accession #P04074) could indicate the loci at which progesterone binding occurs. The amino acid sequence for the successive external loops of the α1 isoforms from rat and sheep is illustrated in Table 3 (the sequence for the α1 isoform from X. laevis kidney epithelial cells [43] and neural plate cells [44] (primary accession #Q92123) is shown for comparison). The amino acid sequence of the α1-subunit in R. pipiens and X. laevis should be highly conserved. The amino acids highlighted in red indicate those residues identified as determinants of ouabain binding in the sheep enzyme [13, 45], whereas those highlighted in green indicate residues that differ in rat (low affinity) compared to sheep (high affinity). Underlined residues in the C-terminal region of the M5-M6 loop indicate residues that are part of the transmembrane helix.
Table 3.
Variations1 in the External Loops of the α1-Subunit of the Na/K-ATPase of Sheep, Xenopus and Rat
| M1-M2 External Loop | |
| Sheep | |
| Xenopus | |
| Rat | |
| M3-M4 External Loop | |
| Sheep | |
| Xenopus | |
| Rat | |
| M5-M6 External Loop | |
| Sheep | |
| Xenopus | |
| Rat | |
| M7-M8 External Loop | |
| Sheep | |
| Xenopus | |
| Rat | |
| M9-M10 External Loop | |
| Sheep | CPGMGAALRMYPLKP |
| Xenopus | CPGMDVALRMYPLKP |
| Rat | CPGMGAALRMYPLKP |
Amino acids highlighted in red indicate residues identified as determinants of ouabain binding in the sheep enzyme (e.g. [13]), whereas those highlighted in green indicate residues that differ in Rat compared to sheep. Underlined sequences in C-terminal region of the M5-M6 loop indicate residues that are part of the transmembrane helix.
Based on RbDe and UniprotKB/Swiss-Prot/EMBL projections, the five external loops illustrated in Figure 2 contain 23, 12, 10, 51 and 15 residues going respectively from N to C terminal ends. The amino acid sequences of the 5 external loops of the α1 isoforms of rat, sheep and X. laevis differ primarily in the 23 amino acid external loop between M1 and M2. The residues identified as determinants of cardiac glycoside binding in the sheep and human enzymes [45, 46]; are C109, Y113, Q116, A114, P123, D126, N127, Y313, W315, T802, and R885 (sheep numbering system; highlighted in red, see Table 3). In the first external loop, 5 of the 23 amino acids in rat differ from sheep, 3 of which have been identified as determinants of ouabain binding. In loops 2 and 3, all 3 amino acids identified by site-directed mutagenesis are the same in rat and sheep. In the largest external loop (M7-M8), only one residue has been identified by site-directed mutagenesis. It is the same in rat, sheep and X. laevis. Finally, in loop 5, no amino acids have been identified by site- directed mutagenesis. Loop 3 is unique in that the one residue identified by mutagenesis (T) is within the outer part of the M6 transmembrane helix. A comparison of the amino acids required for ouabain binding in Table 2 suggest that: 1) if ouabain binds to a site on the external loops of the α1 isoform, and 2) if rat and sheep α1 isoforms represent low and high affinity ouabain binding forms, respectively, then the primary low affinity ouabain binding site may lie in the external loop between M1 and M2.
6.2 Topology of the First External Loop of the α1-Subunit
Figure 3 compares the 3D projection of the first external loop of the human α1 isoform (upper) with that of the rat α1 isoform (lower) of the Na/K-ATPase using Chem3D Ultra v. 10.0. As can be seen, the first external loop of the isoform that binds ouabain with low affinity (rat, Kd = 10−6 M) is more tightly organized, with a large α-helical content. In addition, there is a hook in the c-terminal region due to the Pro127-Pro128 sequence in the low affinity ouabain binding form. In contrast, the first external loop of the isoform that binds ouabain with high affinity (sheep, human, Kd = 10−9 M) contains only two small helical elements.
As noted above, an analysis of differences in the amino acid sequences between low and high affinity ouabain binding isoforms should indicate the peptide sequence important in progesterone binding. To this end, an expanded amino acid SVG plot of the M1 and M2 helices from the high ouabain affinity α1 subunit, together with the extracellular loop that joins the two transmembrane domains is illustrated in Figure 4. The diamonds indicate the positions of the random amino acid substitutions that confer the greatest ouabain resistance: which include Cys113, Tyr117, Gln120, Pro127, Asp130, and Asn131 [45] (positions indicated in Figure 4 are based on X. laevis α1-subunit numbering). In the low affinity ouabain form (rat) Gln120 becomes Arg, Ala121 becomes Ser, and Asn131 becomes Asp. Based on the M1-M2 rat sequence shown in Table 3, a consensus sequence for the progesterone binding site in the α1-subunit may therefore include the following CFLAYGIRSATEEEPPNDD (residues 113 to 131).
Figure 4.

An expanded amino acid Scalable Vector Graphics (SVG) plot of the M1-M2 domain with the extracellular loop that joins the two transmembrane domains for the α1-isoform from X. laevis (Primary accession #Q92127). Arrows indicate residues identified as determinants of ouabain sensitivity for the sheep enzyme [13]; Cys113, Tyr117, Gln121, Pro127, Asp130, and Asn131. Residue numbers are based on the sequence for X. laevis [43,44]. The N-terminal intracellular domain (80 residues, primary accession # P04074)of the sheep α1 subunit is about 9 residues shorter than that of the Xenopus α1 subunit (89 residues).
6.3 Hydrophobicity and Polarity Gradients within the First External Loop
Structure-function studies indicate that the spatial arrangement of the polar substituents on the upper or β face of the steroid molecule is of critical importance for progesterone binding [47]. The 3, 20-dione, 21-ol configuration is the most active. An unsubstituted α or lower surface is also required for activity; introduction of a polar group diminishes or abolishes activity. The upper or β surface may interact with a polar region, whereas the lower or α surface may interact with a hydrophobic region. Figure 5 compares the hydrophobicity (upper panel) polarity (lower panel) gradients as a function of position from residue 110 to residue 134 (abscissa). The hydrophobicity and polarity scores were calculated using the ProtScale Tool (ExPASy) based on Kyte and Doolittle [18] and Grantham [19], respectively. The plots indicate that a steep gradient exists within the loop. The N-terminal region of the loop is hydrophobic whereas the C-terminal region is highly polar.
Figure 5.

The hydrophobicity (upper panel) and polarity (lower panel) gradients within the first external loop of the gradient for the α1 isomer illustrated in Figure 5. The sequence (residues 110-132, abscissa) was analyzed using ProtScale (expasy.org) and is shown as a function of the hydrophobicity or polariety score on the ordinate. The window was set at 5 and the algorithm of Kyte and Doolittle [18] was used to estimate hydrophobicity whereas the algorithm of Grantham [19], was used to measure polarity.
The 23 amino acid sequence in the first external loop in rat can also be written as a classical α-helix (3.6 amino acids per turn) on the surface of a cylinder. After 5 turns, residues I109 and P126 have similar positions parallel to the axis of the cylinder. Gaboriaud et al. [20] have shown that if the cylinder is cut parallel to its axis and unrolled, sets of adjacent hydrophobic residues can be encircled and termed hydrophobic clusters (see Figure 6). As some adjacent amino acids (e.g. G116 and I117) are widely separated by the unfolding of the cylinder, the representation is duplicated in the lower projection, making the sequence easier to follow and giving a better impression of the microenvironment of individual amino acids.
Figure 6.

Protein sequence analysis of the first external loop (M1-M2) using hydrophobic cluster analysis [20]. The upper projection illustrates the sequence I109 …Y131 (rat Primary accession # P06685) written on a classical α-helix (3.6 amino acids per turn). The lower projection indicates the hydrophobic cluster analysis after unrolling the helix (see text) with the hydrophobic clusters circled.
Sets of adjacent hydrophobic residues in the pattern are encircled and termed hydrophobic clusters (Figure 6). I, L, F, and Y were considered to be hydrophobic amino acids, whereas P (star) is primarily considered as a breaker of these clusters, and A and C as mimetic, i.e. hydrophobic only in a hydrophobic environment. Consistent with the method of Gaboriaud et al. [20], different symbols are used for proline (star), glycines (square), which are often present in loops, and cysteines (C) which may be involved in disulfide bond formation. As shown in the lower portion of Figure 6, a large hydrophobic cluster exists in the N-terminal region of the first external loop. This predicts that the lower or α surface of the steroid molecule interacts with the hydrophobic region (residues 110-120). The upper or β face of progesterone would be expected to interact with the more polar portion of the loop (residues 121-134).
6.4 Changes in Flexibility within the First External Loop
Figure 7 illustrates the average flexibility score [48] as a function of position along the first external loop in the Xenopus laevis α1-subunit of Na/K-ATPase. The average flexibility index for the sequence (residues 110-132, abscissa) was calculated using the algorithm of Bhaskaran and Ponnuswamy [48] with ProtScale (expasy.org). The positions of the amino acids (Arg118, Asp129, Cys111) at which site-directed mutagenesis studies indicate that substitutions confer the greatest ouabain resistance, are indicated by arrows. Regions of low flexibility occur in the hydrophobic region whereas the highest flexibility occurs near the mid-point of the loop, suggesting that folding occurs within the loop, further stabilizing the progesterone binding site.
Figure 7.

The average flexibility index along the first external loop is illustrated as a function of amino acid position in Xenopus laevis α1-subunit of Na/K-ATPase. The average flexibility index for the sequence (residues 110-132, abscissa) was calculated by the algorithm of Bhaskaran and Ponnuswamy [48] using ProtScale (expasy.org).
7.0 Progesterone Binding to the α1-Subunit of the Na/K-ATPase
7.1 Molecular Modeling of the Progesterone Binding Site
Figure 8 illustrates the proposed progesterone binding site within the first external loop of the low affinity ouabain binding isoform of the α1-subunit. As mentioned above, X-ray crystallographic data for the α1-subunit of the Na/K-ATPase are not available. It is not possible, therefore, to develop a molecular model similar to that published for the cytosolic progesterone receptor [49]. The graphic in Figure 8 represents a view looking down on the cell surface, with an end view of the first (M1) and second (M2) transmembrane helices and the interconnecting loop of 23 amino acids. The peptide bonds between the helices and the external loop amino acids are discontinuous to allow for 90 degree rotation of the loop sequence. The amino acids thought to be critical for ouabain binding are indicated in single letter codes (C111, Y115, R118, S122, P128, D129, D131). The planar progesterone is shown on its side with the α hydrophobic face upward facing left and the polar β surface facing down and to the right. The arrows indicate the direction of insertion of the planar progesterone molecule.
The ouabain binding domain of Na/K-ATPase and the classical cytosolic steroid receptors appear to share amino acid sequence homologies, as reported by LaBella and Templeton [50] and Seccombe et al. [51]. In the ouabain radioligand assay used by Seccombe et al. [51], the potencies of a series of steroid moieties suggest that the steroid-binding domain of the Na/K-ATPase can accommodate compounds with planar configurations. Progesterone, dehydroepiandosterone, 11-hydroxycortisol and 18-hydroxy-11-deoxycorticosterone (18-OH-DOC) were found to have digoxin-like immunoreactivity at low concentrations in mammals. (Digoxin differs from ouabain in that it has 2 additional sugar moieties in the 3-position and lacks three hydroxyl groups). Furthermore, progesterone and 18-OH-DOC were most efficient in displacing radiolabeled-ouabain from canine kidney Na/K-ATPase. These observations are consistent with our previous ouabain competition studies [8], as well as with our earlier findings of the order of efficiency of different steroids in inducing amphibian meiosis [47].
Previous studies in this laboratory have shown that three structural features of the steroid molecule are essential for induction of meiosis [47]: 1) a planar ring system, 2) a hydrophobic lower or α surface of the steroid, and 3) an angular methyl group at C-10. Thus, testosterone or androstanediol are effective inducers, 19-nor-testosterone or 19-nor-androstanediol are inactive (Table 2). Although the 3,20-dione, 21-ol pregnane configuration is the most active in meiotic induction [47], steroids with a lactone in the 17-position will also induce nuclear membrane breakdown (digitoxigenin, Table 2).
Steroids with a a nitroxide ring (DOXYL) in the 3-position will also induce meiosis (3-DOXYL-17β-OH-5α-androstane, Table 2). 3-DOXYL-17β-OH-5α-androstane (Cat. #25,354-5, Aldrich Chemical Co., Milwaukee, WI) is readily taken up by isolated R. pipiens plasma-vitelline membranes. The order parameter, S, can be measured using electron spin resonance techniques [52] [the order parameter is a general concept, and ranges from a value of 1.0 (no motion) to 0 (complete averaging)]. In contrast to an S value of 0.681±0.010 (N = 5) for the 5-DOXYLstearic acid probe taken up by the isolated plasma membrane, the EPR signal for 3-DOXYL-17β-OH-5α-androstane indicated no motion. In other words, the 3-DOXYL moiety of the steroid was inserted into a totally restricted environment in the oocyte plasma membrane. The 3-DOXYL-17β-OH-5α-androstane spin probe could be recovered from the isolated plasma membranes by extraction with CHCl3:CH3OH (2:1)
If a strict geometric fit is assumed between the shape of the steroid molecule and the receptor, then the structure-function restrictions outlined here and elsewhere [47] suggest that the steroid is inserted into the first external loop with the upper or β face conforming to a polar region (residues 123-132, Figure 5) and the lower or α face closely associated with a hydrophobic region (residues 113-122, Figure 5). Replacement of Gln126 with Pro126 in the low affinity ouabain binding isoform introduces a sharp bend in the peptide chain at the N-terminal end of the first external loop (lower cartoon, Figure 3). We propose (Figure 8) that the hydrophobic α surface interacts with the hydrophobic clusters in the N-terminal region (Figure 6), and that the angular methyl group at C-10 interacts with the Leu132-Gly133-Val134 sequence at the beginning of the M2 helix.
The sharp bend produced by the Pro125-Pro126 sequence would also place the 3-keto group in juxtaposition to a Glu122-Glu123-Glu124 sequence on one side of the bend, and an Asp128- Asp129 sequence on the other. The optimal inducing activity of the 3,20-keto steroids such as progesterone could be due to hydrogen bonding between the carbonyl dipole and adjacent polar amino acids (Glu122-Glu123-Glu124 and Asn127-Asp128) within the bend. Such bonds are highly directional and would project above the β-plane of the steroid. Cys111 at the interface with the external loop, M1, is known to be essential for ouabain binding/inhibition [45] and could participate in hydrogen bonding to the 20-keto group of progesterone, as has been proposed for progesterone binding to the cytosol receptor [49]. As noted above, maximum flexibility occurs near the midpoint of the external loop (Figure 8), suggesting that if M1 and M2 moved towards each other, folding would occur within the loop and further stabilize progesterone binding.
7.2 Role of Plasma Membrane Lipids in Progesterone Binding
The molecular model illustrated in Figure 8 represents an integral membrane protein with multiple transmembrane domains within the lipid bilayer. In non-dividing cells, phosphatidylcholine (PC), sphingomyelin (SM) and glycosphingolipids are primarily localized in the outer (exoplasmic) lipid leaflet, whereas phosphatidyethanolamine (PE), phosphatidylserine and phosphatidylinositol are generally enriched in the cytoplasmic leaflet [53]. The inner and outer regions of the α1-subunit are therefore exposed to different lipid environments.
The model in Figure 8 does not consider possible contributions by other hydrophobic membrane constituents (e.g. PE, SM, cholesterol) on progesterone binding to an exoplasmic peptide loop associated with the outer bilayer. A number of reports indicate that the topological organization, and thus the function of many membrane proteins, are not only determined by the amino acid sequence of the protein but also by the lipid composition of the membrane (reviewed in [54]). Major changes in the functional properties and topological organization of transport proteins such as GABA permease and the phenylalanine transporter of E. Coli have been reported to occur when membrane PE is altered [54].
We have found that progesterone binding to the plasma membrane increases several fold during the first 4-5 h, during which time PE is converted to PC and there is rise in SM (reviewed in [3]). These changes could alter the microenvironment of the external peptide loops of the α1-subunit of Na/K-ATPase and contribute to increased progesterone binding. Changes in the lipid microenvironment could also modulate α1-subunit interaction with other integral membrane proteins. Phospholipid changes could contribute to the observed progesterone-induced increases in integral membrane enzymes such as PE-N-methyl transferase (4 transmembrane domains) and sphingomyelin synthase (1 transmembrane domain) (reviewed in [3]).
8.0 Comparison of the α1-Subunit Progesterone Binding Site with Other Putative Progesterone Receptors
As noted in the introduction, at least two other plasma membrane receptors have been proposed to explain steroid induction of the meiotic divisions in fish and Xenopus. A 40 kDa G-protein linked membrane steroid receptor has been described in fish ovary [55]. More recently, it has been proposed that the classical Xenopus progesterone receptor (named xPR-1) can interact with the plasma membrane through its ligand-binding domain [11]. Overexpression of xPR-1 accelerates progesterone-induced oocyte maturation (as does ouabain, Table 2). Injection of antisense oligonucleotides almost completely blocks progesterone-induced nuclear membrane breakdown.
Steroid induction of meiosis and/or binding in fish and Xenopus has been generally described as if the rapid response to progesterone (or a progesterone metabolite) was sufficient to complete the first meiotic division. However, it was first shown by Detlaff [56] that gonadotropin (responsible for progesterone synthesis and release from the follicle cells) must be present throughout the period leading to nuclear membrane breakdown in Rana temporia follicles. Dettlaff termed this the “hormone-dependent period”. We confirmed this finding with Rana pipiens follicles [57] and found that at physiological (nmolar) concentrations, extracellular progesterone must be present throughout the first 4-5 h for nuclear membrane breakdown to occur 7 - 8 h after the initial exposure [8,57]. Similar experiments with Stage VI X. laevis follicles (unpublished) indicate that a continuous exposure to low levels of progesterone for at least 90 min is necessary for nuclear membrane breakdown to occur at about 3 h.
We have found that, in the gonadotropin-stimulated Rana pipiens ovarian follicle, progesterone levels increase several-fold during the hormone-dependent period both in vivo and in vitro [8]. The progesterone continuously synthesized by the follicle cells becomes largely bound to the oocyte plasma membrane over the first 4-5 h.. At 5-6 h, about 95% the membrane-bound progesterone is internalized along with about 60% of the plasma membrane. The internalized progesterone is rapidly metabolized to highly polar (polyhydroxylated) derivatives [58]. Since plasma membrane bound-progesterone is internalized prior to nuclear membrane breakdown in Rana oocytes, the appearance of xPR-1-bound steroid could reflect a transfer between ligands following steroid internalization.
A broad range of steroids, including progesterone, induce nuclear membrane breakdown in human chorionic gonadotropin (hCG)-primed fish oocytes [55]. The finding that radiolabeled progesterone is converted to 17α,20β,21-trihydroxy-4-pregnen-3-one (20β-S) (as well as to at least one other unidentified polar steroid) by ovarian segments (taken from the Atlantic Croaker) has been cited as evidence that 20β-S is the maturation-inducing steroid in fish. However, 20β-S will not induce nuclear membrane breakdown unless the fish ovarian segments have been pretreated with hCG for 9-12 h. As noted by Patino and Thomas [55], “maximal response to 20β-S was seen in ovaries with oocytes showing the first signs of morphological (meiotic) maturation”. Furthermore, radiolabeled 20β-S was extracted and identified 1-2 h after completion of nuclear membrane breakdown in the fish ovarian segments. Thus, the steroid-dependent events monitored in the Atlantic croaker may occur after the hormone-dependent period and the appearance of 20β-S may coincide with conversion of internalized progesterone to polar steroids in Rana pipiens oocytes [58]. The finding that a polyhydroxylated steroid (ouabain) will potentiate progesterone induction of meiosis (Table 2) suggests that the polar (ouabain-like) steroids produced following internalization of membrane-bound progesterone may be necessary but are not sufficient by themselves to induce meiosis.
9.0 Summary and Conclusions
Progesterone induces the meiotic divisions in the amphibian oocyte by activating a signaling system in the plasma membrane. We present evidence that the progesterone receptor on the oocyte surface is located in an isoform of the α1-subunit of Na/K-ATPase, which is characterized by a low affinity for ouabain (Kd = 2 × 10−6 M, Table 1). Progesterone competes in vitro with ouabain for this low affinity site on the α1-isoform; however under physiological conditions the nmolar concentrations of circulating ouabain would not affect binding by μmolar levels of progesterone (Kd = 5 × 10−7 M for progesterone, Table 1). In comparing the low and high ouabain binding α-isoforms, we find a sequence difference of six amino acids localized in the M1-M2 N-terminal external loop. Seven amino acids in this loop of the high ouabain affinity isoform have been identified as determinants of ouabain binding in the high affinity isoform (Table 3 and Figure 4).
Based on these differences, we conclude that at least seven of the 23 amino acids in the first external M1-M2 loop of the low ouabain affinity form may be involved in progesterone binding. The findings predict that the polar β and the hydrophobic α surfaces of the planar progesterone molecule interact with opposite sides of the folded M1-M2 loop (Figure 8). This would be consistent with the countercurrent hydrophobicity and polarity gradients within the loop (Figure 5) as well as with the hydrophobic clusters within the N-terminal region of the loop (Figure 6). The molecular model (Figure 8) further suggests that the angular methyl group between the A and B rings forms hydrophobic bonds with the Leu132-Gly133-Val134 sequence at the N-terminal region of the M2 helix. The model also predicts that the 3-keto group of progesterone hydrogen-bonds to residues Arg118, Asp129 and possibly Glu122-124. Similarly, Cys111 could hydrogen bond to the 20-keto group.
We propose that the low affinity ouabain α1-isoform is a progesterone receptor, whereas the high affinity ouabain α1-isoform regulates Na+-pump activity. As noted above, even at maximal physiological ouabain concentrations in the blood (nmolar [21]), there would be no significant binding of the cardiotonic steroids to the low affinity ouabain α1-isoform. Our structure-function studies with the oocyte indicate that classical steroids other than progesterone may also bind to the first external loop of the α1-subunit of the Na/K-ATPase [47]. Since C-21 (pregnanes) and C-19 (androstanes) steroids often have overlapping biological activities, the low affinity ouabain α1-subunit of the Na/K-ATPase in the plasma membrane may serve a broader function in nongenomic (plasma membrane) responses to steroids by interacting with tissue-specific integral membrane proteins.
Acknowledgments
This research was supported in part by National Institutes of Health research grants HD-10463 and HL-36573 (NHLBI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li X, Lonard DM, O'Malley BW. A contemporary understanding of progesterone receptor function. Mech. Ageing Dev. 2004;125:669–78. doi: 10.1016/j.mad.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Watson CS, Gametchu B. Minireview: Proteins of multiple classes may participate in nongenomic steroid actions. Expl Biol & Med. 2003;228:1272–81. doi: 10.1177/153537020322801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrill GA, Kostellow AB. Progesterone induces meiotic division in the amphibian oocyte by releasing lipid second messengers from the plasma membrane. Steroids. 1999;64:157–67. doi: 10.1016/s0039-128x(98)00093-2. [DOI] [PubMed] [Google Scholar]
- 4.Cato ACB, Nestl A, Mink S. Rapid actions of steroid receptors in cellular signaling pathways. Sci. STKE. 2002:re9. doi: 10.1126/stke.2002.138.re9. 2002. [DOI] [PubMed] [Google Scholar]
- 5.Smith LD, Ecker RE. Role of the oocyte nucleus in physiological maturation in Rana pipiens. Dev Biol. 1969;19:281–309. doi: 10.1016/0012-1606(69)90065-7. [DOI] [PubMed] [Google Scholar]
- 6.Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation in frog oocytes. J Expl Zool. 1971;177:129–45. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- 7.Ziegler D, Morrill GA. Regulation of the amphibian oocyte plasma membrane ion permeability by cytoplasmic factors during the first meiotic division. Dev Biol. 1977;60:318–25. doi: 10.1016/0012-1606(77)90129-4. [DOI] [PubMed] [Google Scholar]
- 8.Morrill GA, Erlichman J, Gutierrez-Juarez R, Kostellow AB. The Steroid-Binding Subunit of the Na/K-ATPase as a Progesterone Receptor on the Amphibian Oocyte Plasma Membrane. Steroids. 2005;70:933–45. doi: 10.1016/j.steroids.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Juhaszova M, Blaustein MP. Na+ pump low and high affinity alpha subunit isoforms are differently distributed in cells. Proc Natl Acad Sci (USA) 1997;94:1800–05. doi: 10.1073/pnas.94.5.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pace MC, Thomas P. Steroid-induced oocyte maturation in Atlantic Croaker (Micropogonias undulatus) is dependent on activation of the phosphatidylinositol 3-kinase/Akt signal transduction pathway. Biol. Reprod. 2005;73:988–996. doi: 10.1095/biolreprod.105.041400. [DOI] [PubMed] [Google Scholar]
- 11.Martinez S, Pasten P, Suarez K, Garcia A, Nualart F, Montecino M, Hinrichs MV, Olate J. Classical Xenopus laevis progesterone receptor associates to the plasma membrane through its ligand binding. J. Cell. Physiol. 2007;211:560–7. doi: 10.1002/jcp.20964. [DOI] [PubMed] [Google Scholar]
- 12.Weinstein SP, Kostellow AB, Ziegler DH, Morrill GA. Progesterone-induced down-regulation of an electrogenic Na+, K+-ATPase during the first meiotic division in amphibian oocytes. J Memb Biol. 1982;69:41–48. doi: 10.1007/BF01871240. [DOI] [PubMed] [Google Scholar]
- 13.Lingrel JB, Cryole ML, Woo AL, Arguello JM. Ligand binding sites of Na,K-ATPase. Acta Physiol Scand. 1998;163(Suppl 643):69–77. [PubMed] [Google Scholar]
- 14.Morrill GA, Kostellow AB. Role of ions in oocyte function and the meiotic divisions. In: Kinne RKH, Kinne-Saffran E, Beyenbach KW, editors. Oogenesis, Spermatogenesis and Reproduction, Comp Physiol. Vol. 10. S. Karger; Basel: 1991. pp. 37–85. [Google Scholar]
- 15.Kostellow AB, Weinstein SP, Morrill GA. Specific binding of progesterone to the cell surface and its role in the meiotic divisions in Rana oocytes. Biochim Biophys Acta. 1982;720:356–63. doi: 10.1016/0167-4889(82)90112-4. [DOI] [PubMed] [Google Scholar]
- 16.Morrill GA, Ma G-Y, Kostellow AB. Progesterone binding to plasma membrane and cytosol receptors in the amphibian oocyte. Biochem Biophys Res Comm. 1997;232:213–7. doi: 10.1006/bbrc.1997.6190. [DOI] [PubMed] [Google Scholar]
- 17.Konvicka K, Campagne F, Weinstein H. Interactive construction of residue-based diagrams of proteins: the RbDe web service. Protein Engineering. 2000;13:395–96. doi: 10.1093/protein/13.6.395. [DOI] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle RF. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–64. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 20.Gaboriaud C, Bissery V, Benchetrit T, Mornon JP. Hydrophobic cluster analysis: an efficient new way to compare and analyse amino acid sequences. FEB. 1987;224:149–55. doi: 10.1016/0014-5793(87)80439-8. [DOI] [PubMed] [Google Scholar]
- 21.Schoner W. Endogenous cardiac glycosides, a new class of steroid hormones. Eur J Biochem. 2002;269:2440–48. doi: 10.1046/j.1432-1033.2002.02911.x. [DOI] [PubMed] [Google Scholar]
- 22.Hamlyn JM, Lu Z, Manunta P, Ludens JH, Kimura K, Shah JR, Laredo J, Hamilton JP, Hamilton MJ, Hamilton BP. Observations on the nature, biosynthesis, secretion and significace of endogenous ouabain. Clin. Expl. Hypertens. 1998;20:523–33. doi: 10.3109/10641969809053230. [DOI] [PubMed] [Google Scholar]
- 23.Komiyama Y, Nishimura N, Munakata M, Mori T, Okuda K, Nishino N, Hirose S, Kosaka C, Matsuyda M, Takahashi H. Identification of endogenous ouabain in culture supernatant of PC12 cells. J Hypertens. 2001;19:229–36. doi: 10.1097/00004872-200102000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Perrin A, Brasmes B, Chambaz EM, Defaye G. Bovine adrenocortical cells in culture synthesize an ouabain-like compound. Mol Cell Endocrinol. 1997;26:7–15. doi: 10.1016/s0303-7207(96)03964-0. [DOI] [PubMed] [Google Scholar]
- 25.Vitto A, Wallace RA. Maturation of Xenopus Oocytes I. Facilitation by ouabain. Expl. Cell Res. 1976;97:56–62. doi: 10.1016/0014-4827(76)90654-6. [DOI] [PubMed] [Google Scholar]
- 26.Cartaud A, Marcher K, Ozon R. Digitoxigenin, a digitalis steroid, induces meiotic maturation of Xenopus laevis oocytes. J Steroid Biochem. 1984;21:101–6. doi: 10.1016/0022-4731(84)90066-9. [DOI] [PubMed] [Google Scholar]
- 27.Weinstein SP, Morrill GA, Kostellow AB. Increased potassium conductance in Rana follicles after stimulation by pituitary extract. Develop Growth and Differ. 1983;25:11–21. doi: 10.1111/j.1440-169X.1983.00011.x. [DOI] [PubMed] [Google Scholar]
- 28.Kalimi M, Ziegler D, Morrill GA. Characterization of a progestin-binding macromolecule in the amphibian oocyte cytosol. Biochem Biophys Res Commun. 1979;86:560–567. doi: 10.1016/0006-291x(79)91750-9. [DOI] [PubMed] [Google Scholar]
- 29.Tian J, Kim S, Heilig E, Ruderman JV. Identification of XPR-1, a progesterone receptor required for Xenopus oocyte maturation. Proc Natl Acad Sci (USA) 2001;98:8–10. doi: 10.1073/pnas.250492197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blostein R. Structure-function studies of the sodium pump. Biochem Cell Biol. 1999;77:1–10. doi: 10.1139/o99-018. [DOI] [PubMed] [Google Scholar]
- 31.Blanco G, Mercer RW. Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am J Physiol. 1998;275:F633–F650. doi: 10.1152/ajprenal.1998.275.5.F633. [DOI] [PubMed] [Google Scholar]
- 32.Feraille E, Doucet A. Sodium-Potassium-Adenosinetriphosphase-dependent Sodium Transport in the kidney: Hormonal Control. Physiol Revs. 2001;81:345–418. doi: 10.1152/physrev.2001.81.1.345. [DOI] [PubMed] [Google Scholar]
- 33.Pralong-Zamofing D, Qi-Han Yi, Schmalzing G, Good P, Geering K. Regulation of α1-β3-Na+-K+-ATPase isozyme during meiotic maturation of Xenopus laevis oocytes. Am J Physiol. 1992;262:C1520–C1530. doi: 10.1152/ajpcell.1992.262.6.C1520. [DOI] [PubMed] [Google Scholar]
- 34.Shamraj OI, Lingrel JB. A putative fourth Na+, K+-ATPase α subunit gene is expressed in testis. Proc Soc Natl Acad Sci (USA) 1984;91:12952–56. doi: 10.1073/pnas.91.26.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucchesi PA, Sweadner KJ. Postnatal changes in Na,K-ATPase IsoformExpression in rat cardiac ventricle. J. Biol. Chem. 1991;266:9327–9331. [PubMed] [Google Scholar]
- 36.White SH, von Heijne G. Transmembrane helices before, during, and after insertion. Cur Opin Struct Biol. 2005;15:378–386. doi: 10.1016/j.sbi.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Askew GR, Lingrel JB. Identification of an amino acid substitution in Human α1 Na,KATPase which cobfers differentially reduced affinity for two related cardiac glycosides. J Biol Chem. 1994;269:24120–26. [PubMed] [Google Scholar]
- 38.Toyoshima T, Nakasako M, Nomura N, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–55. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 39.Ishi T, Takeyasu K. The amino-terminal 200 amino acids of the plasma membrane Na+, K+-ATPase alpha subunit confer ouabain sensitivity on the sarcoplasmic reticulum Ca2+-ATPase. Proc Natl Acad Sci (USA) 1993;90:8881–85. doi: 10.1073/pnas.90.19.8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishi T, Lemas MV, Takeyasu K. Na+, ouabain-, Ca2+-, and thapsigargin-sensitive ATPase activity expressed in chimeras between the calcium and the sodium pump alpha subunits. Proc Natl Acad Sci (USA) 1994;91:6103–7. doi: 10.1073/pnas.91.13.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koenderink JB, Hermsen HPH, Swarts HGP, Willems PHGM, De Pont JJHHM. High affinity ouabain binding by a chimeric gastric H+, K+-ATPase containing transmembrane hairpins M3-M4 and M5-M6 of the alpha 1-subunit of rat Na+, K+-ATPase. Proc Natl Acad Sci (USA) 2000;97:11209–14. doi: 10.1073/pnas.200109597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asano S, Kimura T, Ueno S, Kawamura M, Takeguchi N. A chimeric gastric H+, K+-ATPase inhibitable with both ouabain and SCH 28080. J Biol Chem. 1999;274:6849–54. doi: 10.1074/jbc.274.11.6848. [DOI] [PubMed] [Google Scholar]
- 43.Verrey F, Kairouz P, Schaerer E, Fuentes P, Geering K, Rossier BC, Kraehenbuhl J-P. Primary sequence of Xenopus laevis Na+-K+-ATPase and its localization in A6 kidney cells. Am J Physiol. 1989;256:F1034–F1043. doi: 10.1152/ajprenal.1989.256.6.F1034. [DOI] [PubMed] [Google Scholar]
- 44.Davies CS, Messenger NJ, Craig R, Warner AE. Primary sequence and developmental expression pattern of mRNAs and protein for an alpha1 subunit of the sodium pump cloned from the neural plate of Xenopus laevis. Dev Biol. 1996;174:431–47. doi: 10.1006/dbio.1996.0086. [DOI] [PubMed] [Google Scholar]
- 45.Croyle ML, Woo AL, Lingrel JB. Extensive random mutagenesis analysis of the Na+/K+-ATPase α subunit identifies known and previously unidentified amino acid residues that alter ouabain sensitivity. Implications for ouabain binding. Eur J Biochem. 1997;248:488–95. doi: 10.1111/j.1432-1033.1997.00488.x. [DOI] [PubMed] [Google Scholar]
- 46.Paula S, Tabet MR, Ball WJ., jr Interactions between cardiac glycosides and sodium/potassium-ATPase: three-dimensional structure-activity relationship models for ligand binding to the E2-Pi form of the enzyme versus activity inhibition. Biochemistry. 2005;44:498–510. doi: 10.1021/bi048680w. [DOI] [PubMed] [Google Scholar]
- 47.Morrill GA, Bloch E. Structure-function relationships of various steroids relative to induction of nuclear membrane breakdown and ovulation in isolated amphibian oocytes. J Steroid Biochem. 1977;8:133–39. doi: 10.1016/0022-4731(77)90036-x. [DOI] [PubMed] [Google Scholar]
- 48.Bhaskaran R, Ponnuswamy PK. Possible flexibilities of amino acid residues in globular proteins. Int J Peptide Prot Res. 1988;32:242–55. doi: 10.1111/j.1399-3011.1984.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 49.Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–96. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 50.LaBella FS, Templeton JF. Pregnanes that bind to the digitalis receptor. Clin Expl Hypertens. 1998;20:601–9. doi: 10.3109/10641969809053238. [DOI] [PubMed] [Google Scholar]
- 51.Seccombe DW, Pudek MR, Nowaczynski W, Humphries KH. Digoxin-like immunoreactivity, displacement of ouabain and inhibition Na+/K+-ATPase by four steroids known to be increased in essential hypertension. Clin Biochem. 1989;22:17–21. doi: 10.1016/s0009-9120(89)80064-5. [DOI] [PubMed] [Google Scholar]
- 52.Morrill GA, Doi K, Kostellow AB. Progesterone induces transient changes in plasma membrane fluidity of amphibian oocytes during the first meiotic division. Arch Biochem Biophys. 1989;269:690–4. doi: 10.1016/0003-9861(89)90153-7. [DOI] [PubMed] [Google Scholar]
- 53.Yamaji-Hasegawa A, Tsujimoto M. Assymetric distribution of phospholipids in biomembranes. Biol. Pharm. Bull. 2006;29:1547–1553. doi: 10.1248/bpb.29.1547. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane topology. Phosphatidylethanolamine is required for the proper topological organization of the γ-aminobutyric acid permease (GabP) of Escherivhia Coli. J. Biol. Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 55.Patino R, Thomas P. Characterization of membrane receptor activity for 17α,20β,21-trihydroxy-4-pregnen-3-one in ovaries of Spotted Seatrout (Cynoscion nebulosus) Gen Comp Endocrin. 1990;78:204–217. doi: 10.1016/0016-6480(90)90007-9. [DOI] [PubMed] [Google Scholar]
- 56.Dettlaff TA. Action of actinomycin and puromycin upon frog oocyte maturation. J. Embryol. Expl. Morphol. 1966;16:183–95. [PubMed] [Google Scholar]
- 57.Schatz F, Morrill GA. Studies on the relative roles of pituitary and progesterone in the induction of meiotic maturation in the amphibian oocyte. Differentiation. 1979;13:89–99. doi: 10.1111/j.1432-0436.1979.tb01571.x. [DOI] [PubMed] [Google Scholar]
- 58.Morrill GA, Schatz F, Kostellow AB, Bloch E. Gonadotropin stimulation of steroid synthesis and metabolism in the Rana pipiens ovarian follicle: Sequential changes in endogenous steroids during ovulation, fertilization and cleavage stages. J Steroid Biochem Molecular Biol. 2006;99:129–38. doi: 10.1016/j.jsbmb.2006.01.008. [DOI] [PubMed] [Google Scholar]