Abstract
Host–parasite associations are shaped by coevolutionary dynamics. One example is the complex fungus-growing ant–microbe symbiosis, which includes ancient host–parasite coevolution. Fungus-growing ants and the fungi they cultivate for food have an antagonistic symbiosis with Escovopsis, a specialized microfungus that infects the ants' fungus gardens. The evolutionary histories of the ant, cultivar and Escovopsis are highly congruent at the deepest phylogenetic levels, with specific parasite lineages exclusively associating with corresponding groups of ants and cultivar. Here, we examine host–parasite specificity at finer phylogenetic levels, within the most derived clade of fungus-growing ants, the leaf-cutters (Atta spp. and Acromyrmex spp.). Our molecular phylogeny of Escovopsis isolates from the leaf-cutter ant–microbe symbiosis confirms specificity at the broad phylogenetic level, but reveals frequent host-switching events between species and genera of leaf-cutter ants. Escovopsis strains isolated from Acromyrmex and Atta gardens occur together in the same clades, and very closely related strains can even infect the gardens of both ant genera. Experimental evidence supports low host–parasite specificity, with phylogenetically diverse strains of Escovopsis being capable of overgrowing all leaf-cutter cultivars examined. Thus, our findings indicate that this host–pathogen association is shaped by the farming ants having to protect their cultivated fungus from phylogenetically diverse Escovopsis garden pathogens.
Keywords: coevolution, leaf-cutting ants, host–parasite interactions, host switching
1. Introduction
Coevolutionary interactions shape the structure and diversity of biological communities (Thompson 1994, 2005), and are theoretically predicted to lead to cospeciation (Johnson et al. 2003). This expectation is especially pertinent for host–parasite interactions, where parasites depend on their hosts for survival and reproduction, at the expense of host fitness (Price 1980). Hosts are selected to defend against parasites, while parasites are selected to overcome host defences (Dybdahl & Storfer 2003; Wegner et al. 2003). This coevolutionary arms race should result in congruent host and parasite phylogenies (Hafner & Nadler 1990; Page & Charleston 1998), where a parasite is highly specialized for its matching host (Carius et al. 2001). These expectations have been thoroughly examined in numerous host–parasite interactions (see reviews by Klassen 1992; Page 2003).
Although there are many examples where there is some degree of phylogenetic congruence between hosts and parasites (Hafner & Nadler 1990; Schardl et al. 1997; Currie et al. 2003; Page 2003; Ricklefs et al. 2004; Weckstein 2004), perfect or near-perfect congruence is much less common (Hafner et al. 1994; Clayton & Johnson 2003; Gerardo et al. 2004). In addition, several parasites have wider host ranges than expected under coevolutionary theory (Harwood et al. 1998; Ricklefs et al. 2004). A further expectation of host–parasite coevolution is that geographical differentiation among parasites will reflect patterns of host differentiation across the geographical distribution, as parasites are predicted to specialize on locally common host genotypes (Thompson 1994, 2005; Lively 1999; Dybahl & Storfer 2003; Lively et al. 2004).
Host–parasite coevolutionary dynamics have been examined in the quadripartite attine ant–microbe symbiosis (Currie et al. 2003; Gerardo et al. 2004). Attine ants (i.e. fungus-growing ants; Formicidae, Attini) are a monophyletic group of 12 genera and over 210 described species (Hölldobler & Wilson 1990; Schultz & Meier 1995), which have a geographical range from southern Argentina to Long Island, USA (Weber 1972). Fungus-growing ants obligately cultivate fungi for food (Weber 1972; Hölldobler & Wilson 1990) by nourishing and fertilizing their fungal cultivars. The ant's fungal cultivars are frequently parasitized by Escovopsis (Hypocreales: Ascomycota), a monophyletic genus of microfungi that directly consumes the cultivar (Currie et al. 1999a, 2003; Reynolds & Currie 2004). The parasite is horizontally transmitted between nests (Currie et al. 1999a), although the mechanisms by which this occurs are unknown. Infections by Escovopsis result in a reduction in the growth rates of the fungus gardens, and occasionally force the ants to abandon their nests (Currie et al. 1999a; Currie 2001). To defend against Escovopsis, the ants employ specific behaviours and have a mutualistic association with actinomycetous bacteria that produce antibiotics that specifically inhibit the specialized parasite (Currie et al. 1999b; Currie & Stuart 2001).
The evolutionary histories of Escovopsis, cultivars and attine ants are highly congruent at the deepest phylogenetic levels (Currie et al. 2003). In contrast, Gerardo et al. (2004) demonstrated that Escovopsis strains from the nests of three sympatric species of Cyphomyrmex (a ‘lower’ genus of attines) are specifically associated with particular cultivar strains, despite the incongruence in the evolutionary histories of the ants and their fungal mutualists at lower (i.e. more recent) phylogenetic scales for some ‘lower’ attines (Green et al. 2002). However, since attine ants are very diverse, have a broad geographical distribution and vary in foraging and nest construction behaviours (Hölldobler & Wilson 1990), it is possible that different patterns of host-specificity occur across the phylogenetic diversity of attine ants.
Here, we examine whether Escovopsis strains specialize on particular species of leaf-cutting ants, a monophyletic group within the ‘higher’ attines that only uses fresh plant material, mostly leaves, as substrate for cultivating their fungus garden (Schultz & Meier 1995; Wetterer et al. 1998). The fungal mutualists cultivated by members of the leaf-cutting ant genera Atta and Acromyrmex are genetically and structurally distinct from the cultivars farmed by other groups of attine ants (Chapela et al. 1994). In contrast to lower attines, which farm a wider diversity of cultivar strains (Mueller et al. 1998), leaf-cutting ants cultivate closely related fungal clones that are apparently derived from a single unique ancestor (Chapela et al. 1994), although recent research suggests that horizontal transmission and recombination occur between cultivars in different nests (Bot et al. 2001; Mikheyev et al. 2006). We reconstructed a phylogeny of Escovopsis isolated from the gardens or refuse of Atta and Acromyrmex nests collected across their geographical range and examined this phylogeny for evidence of congruence with the ant phylogeny and for geographical patterns of host–parasite association. We evaluated whether Escovopsis strains from different leaf-cutting ant nests form well-supported clades, and tested whether members of the clades differ in their ability to overgrow Atta or Acromyrmex cultivars by challenging members of the different clades of Escovopsis with diverse garden material from leaf-cutter ant nests.
2. Material and methods
(a) Collections, isolation and DNA extraction
Strains of Escovopsis used in this study were isolated from a diverse collection of leaf-cutting ant species, including Acromyrmex coronatus, Acromyrmex echinatior, Acromyrmex heyeri, Acromyrmex hispidus, Acromyrmex hystrix, Acromyrmex laticeps, Acromyrmex octospinosus, Atta cephalotes, Atta colombica, Atta mexicana, Atta sexdens and Atta vollenweideri. For outgroup comparison, isolates of Escovopsis obtained from the gardens of the closely related genus Trachymyrmex were also included in this study. Collections were made in Mexico (Veracruz in Veracruz State), Panama (Pipeline Road and the Canal Zone in Panamá Province; Boquete, Fortuna and Boca Brava in Chiriquí Province; Isla Colón in Bocas del Toro Province; Santa Clara in Coclé Province; Fort Sherman in Colón Province; and Rancho Frío in Darién Province), Argentina (Parque Nacional Chaco and the road between Empedrado and Resistencia in Chaco Province; Parque Cruce Caballero, Iguazú and San Pedro in Misiones Province), Ecuador (La Selva Lodge and Biological Station and Tiputini Biodiversity Station in Orellana Province) and Guadeloupe (Basse-Terre island) from 1997 to 2004. In most cases, ant nests were excavated to sample Escovopsis directly from the fungus garden. In some cases, we isolated Escovopsis from colonies of A. colombica by collecting refuse material, following protocols outlined by Currie (2001). In each case, up to 100 pieces of refuse or fungus garden from each nest were placed on potato dextrose agar containing antibiotics (50 mg l−1 each of penicillin and streptomycin). When mycelium of Escovopsis appeared, it was sub-cultured onto fresh media and allowed to grow (following the protocol of Currie et al. 1999a). In addition, we obtained live cultures of Escovopsis weberi (an Escovopsis strain from an Atta sp. nest collected in Brazil) and Escovopsis aspergilloides (an Escovopsis strain from a Trachymyrmex ruthae nest collected in Trinidad) from the Centraalbureau voor Schimmelcultures (CBS 810.71 and 423.93). Samples of pure culture Escovopsis were stored at −20°C. DNA was extracted following a CTAB protocol modified from Bender et al. (1983).
(b) Amplifications, sequencing and alignment
DNA sequencing targeted four exons and two introns of the nuclear elongation factor-1α (EF-1α) gene, spanning a total of 1717 nucleotides. Three exons and both introns were amplified with primers EF1-3F (5′-CACGTCGACTCCGGCAAGTC-3′) and EF1-5R1 (5′-GTGATACCACGCTCACGCTC-3′; Gerardo et al. 2004), and the fourth exon was amplified with the Escovopsis-specific primers EF6-20F (5′-AAGAACATGATCACTGGTACCT-3′) and EF6-1000R (5′-CGCATGTCRCGGACGGC-3′). Escovopsis DNA was diluted from its original concentrations to lower concentrations (between 1 : 100 and 1 : 1 000 000) before polymerase chain reaction (PCR) amplification. PCR amplifications for both DNA fragments were performed following Gerardo et al. (2004). DNA purifications were performed using QIAquick PCR purification kits (Qiagen, Inc., Valencia, CA).
We sequenced a single Escovopsis isolate from each of one A. coronatus nest (from Orellana, Ecuador), three A. echinatior nests (two from Panamá Province, Panama and one from Chiriquí, Panama), one A. heyeri nest (from Misiones, Argentina), one A. hispidus nest (from Chaco, Argentina), four A. hystrix nests (all from Orellana, Ecuador), one A. laticeps nest (from Misiones, Argentina), five A. octospinosus nests (one from Basse-Terre, Guadeloupe, two from Panamá Province, Panama, one from Darién, Panama, one from Chiriquí, Panama) and one Acromyrmex sp. A nest (from Misiones, Argentina), five A. cephalotes nests (one from Orellana, Ecuador, three from Panamá Province, Panama and one from Bocas del Toro, Panama), four A. colombica nests (one from Orellana, Ecuador, one from Panamá Province, Panama, one from Chiriquí, Panama and one from Colón, Panama), one A. mexicana nest (from Veracruz, Mexico), five A. sexdens nests (two from Misiones, Argentina, one from Coclé, Panama and two from Chiriquí, Panama), one A. vollenweideri nest (from Chaco, Argentina) and one Atta sp. nest (from Brazil). In addition, isolates from four Trachymyrmex nests (one Trachymyrmex cornetzi from Ecuador, one Trachymyrmex diversus from Ecuador, one Trachymyrmex zeteki from Panama and one T. ruthae from Trinidad) were sequenced. Samples were sequenced at the Idaho State University Molecular Research Core Facility or at the University of Wisconsin-Madison Biotechnology Center. All sequences are deposited in GenBank (accession numbers EF589910–EF589949).
We aligned sequences in Sequencher v. 4.2 (Gene Codes Corp., Ann Arbor, Michigan, 2000), using a dirty data assembly algorithm with a 60% minimum match and a 10% minimum overlap. We manually edited the sequences in Maclade v. 4.06 (Maddison & Maddison 2003). We ran maximum parsimony analyses in PAUP* v. 4.0b10 (Swofford 2002) using the TBR branch swapping option and 1000 random taxon-addition replicates. We obtained bootstrap support using 1000 maximum parsimony pseudoreplicates.
We performed Bayesian analyses in MrBayes v. 3.1 (Ronquist & Huelsenbeck 2003), which calculates posterior probabilities (PP) using a metropolis-coupled Markov chain Monte Carlo analysis. We employed the GTR+Γ+I model (general time reversible with a proportion of sites invariant and gamma-distributed rates). All of the analyses employed one cold chain and three incrementally heated chains, where the heat of the ith chain is B=1/[1+ (i−1)T] and T=0.2; when i=1, B=1, corresponding to the cold chain. Four independent analyses, with two million generations each, were conducted. The program Tracer v. 1.3 (Rambaut & Drummond 2005) was used to analyse the log-likelihood scores, which rapidly and consistently stabilized after 9800 generations in all analyses. Therefore, the initial 10 000 generations from each run were discarded, and 1 of every 100 generations was sampled to calculate PP for each branch in the ML tree. Priors for the substitution rates were set to a flat distribution.
(c) Garden Escovopsis bioassay challenges
To experimentally examine the host specificity of Escovopsis within the leaf-cutting ant–microbe symbiosis, we selected 15 leaf-cutting ant colonies maintained in the laboratory at the University of Kansas, including nine Atta and six Acromyrmex species (table 1). Although no genetic information is available for the fungal mutualists cultivated by these colonies of leaf-cutting ants, we assume that they were genetically diverse because they were collected from the nests of different ant genera and species and from different locations (Panama and Argentina). The colonies were maintained in the laboratory for at least 5 days following collection to allow the ants to stabilize their fungus gardens. Pieces of fungus garden material (1 cm3) from each nest were placed in the centre of sterile Petri plates. All worker ants were removed from the garden pieces. The outer edge of each Petri chamber was lined with cotton moistened with distilled water.
Table 1.
Nests from which garden pieces were obtained for bioassay challenges. (Columns indicate the species of ant that constructed the nest, and the countries, provinces or states and localities that each nest was collected from.)
| ant species | country | locality, state/province | colony code |
|---|---|---|---|
| Atta sexdens | Argentina | Iguazú, Misiones | CC030329-01 |
| Atta sexdens | Argentina | Iguazú, Misiones | UGM030330-07 |
| Atta cephalotes | Panama | El llano, Panamá | CC020531-01 |
| Atta cephalotes | Panama | El llano, Panamá | A. ceph. no. 2 El llano |
| Atta cephalotes | Panama | Canal Zone, Panamá | CC031212-04 |
| Atta cephalotes | Panama | Canal Zone, Panamá | ST041101-02 |
| Atta colombica | Panama | Canal Zone, Panamá | ST040903-01 |
| Atta colombica | Panama | Canal Zone, Panamá | ST040903-02 |
| Atta colombica | Panama | Pipeline Road, Panamá | ST041004-02 |
| A. laticeps | Argentina | Iguazú, Misiones | UGM030330-05 |
| A. laticeps | Argentina | Iguazú, Misiones | UGM030330-04 |
| A. niger | Argentina | Salto Encantando, Misiones | CC030327-02 |
| A. niger | Argentina | Salto Encantando, Misiones | UGM030327-03 |
| A. octospinosus | Panama | Pipeline Road, Panamá | ST041004-01 |
| A. echinatior | Panama | Canal Zone, Panamá | ST041101-01 |
The workerless garden pieces were inoculated with approximately 10 mm3 pieces of agar with mature spores of different strains of Escovopsis following Gerardo et al. (2004). The strains of Escovopsis used in the bioassay challenges represent the phylogenetic diversity of Escovopsis as determined in this study. Specifically, we selected four strains from each distinct clade of Escovopsis associated with leaf-cutting ant fungus gardens. In addition, we selected one Escovopsis strain each from a T. zeteki nest in Gamboa, Panama, and a Trachymyrmex sp. nest in Rancho Frío, Panama. To look at host specificity, each garden piece from all 15 leaf-cutting ant colonies was infected with one of the 10 selected strains of Escovopsis. Assays involving every combination of garden pieces from the 15 leaf-cutting ant colonies and the 10 strains of Escovopsis were conducted. In each assay, the specific garden pieces infected with the selected strain of Escovopsis were randomly selected from each parent garden piece. Each pairing of garden and Escovopsis was replicated five times. In addition, as controls, five garden pieces from each colony were not inoculated. After inoculation, each plate was sealed with Parafilm, to prevent drying, and kept at 20°C.
Infected and control garden pieces were observed daily for 10 days post-inoculation. If Escovopsis hyphae were observed growing over the garden piece, they were scored as overgrown (figure 1). Pieces where no fungal growth was observed, or where Escovopsis grew only on the agar from the inoculum, were scored as suppressed. If a garden piece in the control treatment was overgrown by Escovopsis, it was assumed that the nest had a previous infection, and all treatments involving that nest were removed from the analysis. If a fungus other than Escovopsis was observed overgrowing the garden, that individual piece was removed from the analysis.
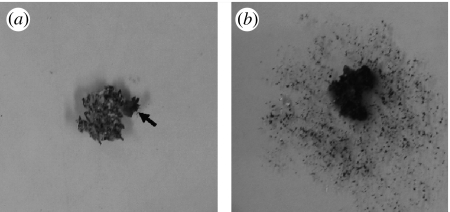
Figure 1.
Experimental setup of bioassay challenges between the fungus garden parasite Escovopsis and fungus garden pieces. (a) A garden piece freshly inoculated with Escovopsis (the parasite is indicated by an arrow). (b) An overgrown garden piece 5 days after infection; the garden piece is shrivelled and discoloured, and the parasite, which is growing on the garden piece, has sporulated (210×99 mm (600×600 DPI)).
3. Results
(a) Phylogenetic relationships of leaf-cutting Escovopsis
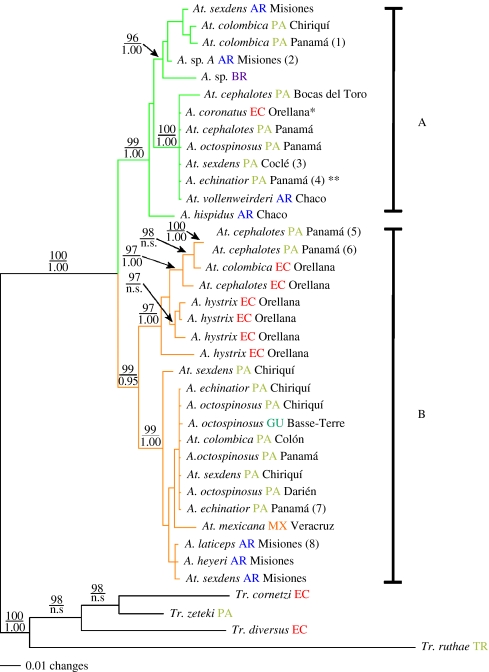
EF-1α introns were not alignable, and were excised from the sequences prior to phylogenetic analysis. Of the 1154 aligned base pairs, 351 bp were variable and 189 of these were parsimony informative. Escovopsis from leaf-cutting ant nests form a monophyletic group (figure 2), indicating a single origin of the Escovopsis parasite associated with the leaf-cutting ants. Escovopsis strains associated with leaf-cutting ants form two main clades (labelled A and B in figure 2), supported by parsimony bootstrap values of 90 or higher and Bayesian PP of 0.95 or higher. Isolates of Escovopsis obtained from the nests of both leaf-cutting ant genera (Atta and Acromyrmex) occur in both clade A and clade B. In addition, based on the complete EF-1α sequences generated (i.e. including the introns excised from the full analyses), several isolates of Escovopsis obtained from Acromyrmex and Atta nests are nearly identical (i.e. 1 or 2 bp differences, e.g. an A. sexdens nest collected in Fortuna, Panama and an A. octospinosus nest collected in Rancho Frío, Panama). In addition to the lack of correlation with their ant hosts, the two main Escovopsis clades are also uncorrelated with geography, as both clades contain isolates from Argentina, Panama and Ecuador. In fact, the entire EF-1α gene fragment, including the introns, is identical for some strains isolated from very different geographical locations. For example, an isolate from an A. coronatus nest collected in Ecuador (colony code AGH030518-14, GenBank accession number EF589948, indicated in figure 2 by ‘*’) and that from an A. echinatior nest collected in Panama (colony code CC020610-02, GenBank accession number EF589949, indicated in figure 2 by ‘**’) have identical sequences.
Figure 2.
Phylogenetic tree of 34 Escovopsis strains from leaf-cutting ant gardens and four outgroup strains of Escovopsis from Trachymyrmex gardens. Each parasite strain is indicated by the species name of the ant host garden from which Escovopsis was isolated. Upper node values correspond to maximum parsimony bootstrap proportions (only values greater than 90 are shown), while lower node values correspond to Bayesian PP (only values greater than 0.95 are shown). Branch colours correspond to two distinct Escovopsis clades (green, clade A; orange, clade B). The countries from which each strain was isolated are indicated by coloured abbreviations (PA, Panama; AR, Argentina; EC, Ecuador; MX, Mexico; GU, Guadeloupe; BR, Brazil; TR, Trinidad), followed by province or state names when available. Numbers in parentheses indicate strains that were used for bioassay challenges (table 2). * indicates a strain from an A. coronatus nest collected in Ecuador, colony code AGH030518-14; ** indicates a strain from an A. echinatior nest collected in Panama, colony code CC020610-02 (§3).
Within each of the two main clades, there is support for additional monophyletic groups. Clade A is composed of two subclades, although one subclade has low bootstrap support (less than 90). Clade B is divided into two subclades with bootstrap support values of 97 and 99. These subclades vary in their diversity, with two subclades (the upper subclade in clade A, and the upper subclade in clade B, figure 2) being divided into several well-supported clades (i.e. with bootstrap values greater than 90), and the other subclades containing lower genetic diversity. The upper subclade in clade B contains six (out of seven) Escovopsis strains from Ecuador, suggesting that there may be a slight geographical pattern within the phylogeny. However, this subclade also contains two isolates from A. cephalotes nests collected in Panama, and one Escovopsis isolate from an A. coronatus nest collected in Ecuador occurs outside of this subclade.
(b) Garden bioassays
Virtually all 675 garden pieces inoculated with Escovopsis were overgrown by the parasite (table 2). Only three garden pieces were not, but they were overgrown by another fungus (indicated on table 2 by ‘*’). The control garden pieces of three Atta nests (one A. sexdens from Argentina and two A. colombica from Panama) were overgrown by Escovopsis, and thus were removed from the analysis. Four leaf-cutting ant gardens (indicated on table 2 by ‘—’) were not tested with the Escovopsis isolates from the Trachymyrmex nests due to the unavailability of the parasite strain. Excluding the pieces overgrown by a fungus other than Escovopsis, each inoculated garden piece was visibly overgrown by Escovopsis no later than 6 days post-inoculation.
Table 2.
Infection experiments of leaf-cutting ant fungus garden fragments with the garden parasite Escovopsis. (The first and second columns indicate the species of ant and country of origin for the colonies from which the garden pieces were isolated from, respectively. The remaining columns represent different combinations of Escovopsis strains and garden pieces. Each cell represents the number of pieces that the parasite overgrew (out of five). Escovopsis strains from clade A and clade B are coded by number (the numbers in table 2 correspond with the numbers indicated on figure 2). Escovopsis strains from Trachymyrmex nests are labelled as follows: (I) an isolate from a T. zeteki colony from Panama and (II) an isolate from a Trachymyrmex sp. colony from Panama. Colonies where any control pieces were overgrown by Escovopsis were assumed to have been previously infected by the parasite, and are not shown. *Escovopsis-garden combinations where one piece was overgrown by a fungus other than Escovopsis.)
| Escovopsis isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| clade A | clade B | Trachymyrmex | |||||||||
| ant species | country | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | (I) | (II) |
| Atta sexdens | Argentina | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Atta cephalotes | Panama | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Atta cephalotes | Panama | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Atta cephalotes | Panama | 5 | 5 | 5 | 5 | 5 | 5 | 4* | 5 | 5 | 4* |
| Atta cephalotes | Panama | 4* | 5 | 5 | 5 | 5 | 5 | 5 | 5 | — | — |
| Atta colombica | Panama | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | — | — |
| A. laticeps | Argentina | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| A. laticeps | Argentina | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| A. niger | Argentina | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| A. niger | Argentina | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| A. octospinosus | Panama | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | — | — |
| A. echinatior | Panama | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | — | — |
4. Discussion
Our molecular phylogenetic analyses reveal that Escovopsis strains isolated from the gardens of leaf-cutting ants form a monophyletic group that is subdivided into two well-supported clades. The strong support for the monophyly of Escovopsis isolated from these ant genera supports the conclusion by Currie et al. (2003) that the group of Escovopsis parasites found in leaf-cutting ant nests is specific to leaf-cutting ant gardens. The growth patterns and morphological characteristics of the different Escovopsis strains are consistent with the more than 600 Escovopsis isolates from leaf-cutting ant nests that we have examined (S. J. Taerum & C. R. Currie 2004, personal observations). This study also demonstrates that the gardens of leaf-cutting ants are host to a phylogenetically diverse assemblage of Escovopsis parasites.
The phylogeny generated in this study indicates that the evolutionary history of leaf-cutting Escovopsis pathogens has not tracked the evolutionary history of the leaf-cutting ants. We found the same strains of Escovopsis, representing both major leaf-cutting Escovopsis clades, parasitizing the gardens of both Atta and Acromyrmex. If Escovopsis species had cospeciated with their ant hosts, then Escovopsis strains from the nests of Acromyrmex and Atta species would group into distinct clades. Identical strains of Escovopsis appear to frequently infect the gardens of both Atta and Acromyrmex. In this study, certain Escovopsis strains isolated from Atta and Acromyrmex nests had identical or nearly identical sequences, including the highly variable regions such as the introns. These findings further confirm the lack of ant–Escovopsis specificity in the leaf-cutting ant–microbe symbiosis, since, if there was cospeciation between the ants and the parasites, the gardens cultivated by the two different leaf-cutting ant genera would not be attacked by such genetically similar Escovopsis strains. Our results correspond to those of Gerardo et al. (2004), who found that there was no phylogenetic congruence between ants of the ‘lower’ attine genus Cyphomyrmex and their Escovopsis parasites.
Furthermore, our results indicate that there is no host–parasite specificity between fungal cultivars and Escovopsis pathogens in leaf-cutting ant gardens. Bioassay challenges between the garden material of diverse leaf-cutting ants and a wide diversity of Escovopsis strains, including those that were isolated from Trachymyrmex nests, established no difference in the ability of the Escovopsis strains to overgrow Atta or Acromyrmex garden pieces. Inversely, there were no differences in the abilities of the diverse cultivar pieces to defend themselves against the diverse Escovopsis strains. These results indicate that Escovopsis strains are not specialized on particular leaf-cutting garden strains. We conclude, therefore, that Escovopsis specificity is not mediated by garden cultivar defences in the absence of the ants. However, future studies are required to reconstruct the phylogeny of leaf-cutting ant cultivars in order to completely rule out whether the evolutionary history of the cultivar is congruent with that of leaf-cutting Escovopsis pathogens.
The absence of specificity between cultivars and Escovopsis in the leaf-cutting ant–microbe symbiosis could indicate that either all strains of leaf-cutting ant Escovopsis have virulence alleles that match the cultivar's inhibitory alleles (Agrawal & Lively 2002) or that the cultivar lacks inhibitory genes entirely. Under either scenario, Escovopsis is apparently able to detect and parasitize diverse leaf-cutting cultivar types. However, it remains possible that, although overgrowth of all of the garden pieces occurred in roughly the same amount of time, there are still subtle and unobservable differences in the rate at which the various Escovopsis clades recognize and/or consume the different cultivar strains. In situations where Escovopsis faces inhibition from a combination of leaf-cutting ants, cultivar and actinomycetous bacterium, any slight differences in the speed or ability of Escovopsis to use cultivar may affect the parasites' fitness.
Although Gerardo et al. (2004) found evidence of specificity between Escovopsis and fungal cultivars in the lower attine ant genus Cyphomyrmex; their findings are not necessarily incongruent with the results we obtained in this study. Cyphomyrmex is a phylogenetically intermediate genus of attine ants that belongs to the group commonly referred to as ‘lower’ attines. Fungiculture in this group of ants has some important similarities to the leaf-cutters; both cultivate monocultures within individual colonies and the primary mode of transmission of the fungal mutualist is vertical. However, unlike in the leaf-cutters where the genetic diversity of cultivar is relatively low between nests (Chapela et al. 1994), different species of Cyphomyrmex are specialized on cultivating genetically distinct clades of cultivar (Mueller et al. 1998). More specifically, Cyphomyrmex muelleri and Cyphomyrmex costatus are both specialized on fungal cultivars from the same monophyletic clade, while Cyphomyrmex longiscapus colonies associate with fungal symbionts from a separate and relatively genetically distantly related clade of cultivars. Gerardo et al. (2004) found evidence for two separate clades of Escovopsis associated with these three ant species, with each clade specializing on one of the two specific clades of cultivar in these ants. Since leaf-cutter ants cultivate genetically less diverse fungi, all of which share a single common ancestor, this finding of specificity at the clade level corresponds to our finding that Escovopsis from leaf-cutters is broadly specialized to leaf-cutter cultivar. Therefore, although Gerardo et al. (2004) identified specific clades of Escovopsis specialized on specific clades of fungal cultivar in Cyphomyrmex, it is unclear if within each of these clades specific strains of Escovopsis can parasitize diverse strains of cultivar, as we found within the leaf-cutter Escovopsis. Future work should explore whether Escovopsis parasitizing the gardens of other groups of attine ants shows specificity at finer phylogenetic levels.
In our study, there were no strong phylogeographical patterns evident in leaf-cutter Escovopsis. The one exception was within clade B, where Escovopsis isolates from Panama and Ecuador formed two distinct groups. However, this pattern is insufficient to conclude that Escovopsis generally evolved in response to geographical variation. The lack of geographical patterning across the phylogeny of leaf-cutting ant Escovopsis suggests that localized adaptation has not occurred. In addition, this symbiosis does not support the geographical mosaic theory of host–parasite adaptation (Thompson 1994, 2005). Possible explanations for these findings are that there is too little variation in cultivar genotypes between the different locations to select for geographical patterns in the Escovopsis phylogeny, or that the parasite migrates too far and too quickly to specialize on local populations (Lively 1999). To test the latter hypothesis, life-history studies are required to determine the mechanisms by which Escovopsis is transmitted between nests.
The inability of leaf-cutting cultivars to defend themselves against Escovopsis demonstrates that the gardens rely heavily on the ants and actinomycetous bacteria to suppress parasite infections. However, the lack of phylogenetic congruence between the ant and Escovopsis phylogenies demonstrates that specialized ant behaviours and nest structures alone are not responsible for the phylogenetic pattern observed in leaf-cutting ant Escovopsis. This could indicate that the parasite is sufficiently able to evade detection by the ants to allow transmission between nests, or that the ants' weeding behaviours and actinomycetous bacteria are insufficient to prevent some critical level of growth and transmission of the parasite. This may explain why Escovopsis is able to persist even in relatively healthy, mature leaf-cutting ant colonies (S. J. Taerum & C. R. Currie, personal observations).
Escovopsis evolution is probably driven by a combination of inhibition by the ants, cultivar and actinomycetous bacteria. This could be a modified form of gene-for-gene evolution (Agrawal & Lively 2002), in which certain clades of Escovopsis may be better suited to overcome particular combinations of the three mutualists' genotypes than others. Testing this hypothesis would require experiments that simultaneously involve all four symbionts. Future work should explore the relative contribution of the ants, cultivar and actinomycetous bacteria in host defence against Escovopsis, as well as exploring the possible phylogenetic congruence and specificity between the garden parasite Escovopsis and ant-associated actinomycetes.
Acknowledgments
This work was supported by NSF award DEB-01 10073 to C.R.C. and T.R.S. and a Smithsonian Tropical Research Institute short-term fellowship to S.J.T. and a University of Kansas Plant Biology research fellowship to S.J.T. We thank the Smithsonian Tropical Research Institute and the Autoridad Nacional del Ambiente of the Republic of Panamá for research permits. We also thank H. Alexander, O. Arosemena, A. Bilgray, K. Bolton, P. Cartwright, H. Fernandez, N. Gerardo, A. Herre, H. Herz, A. Himler, S. Ingram, G. Jarome, M. Leone, R. Lichtwardt, C. Martin, E. McClain, E. McGee, U. Mueller, M. Poulsen, S. Price, S. Rehner, H. Reynolds, J. Scott, T. Scott, D. Smith, S. Solomon, W. Wcislo and M. White for their research assistance and/or invaluable comments.
References
- Agrawal A.F, Lively C.M. Infection genetics: gene-for-gene versus matching-alleles models and all points in between. Evol. Ecol. Res. 2002;4:79–90. [Google Scholar]
- Bender W, Spierer P, Hogness D.S. Chromosomal walking and jumping to isolate DNA from the Ace and Rosy loci and the bithorax complex in Drosophila melanogaster. J. Mol. Biol. 1983;168:17–33. doi: 10.1016/s0022-2836(83)80320-9. doi:10.1016/S0022-2836(83)80320-9 [DOI] [PubMed] [Google Scholar]
- Bot A.N.M, Rehner S.A, Boomsma J.J. Partial incompatibility between ants and symbiotic fungi in two sympatric species of Acromyrmex leaf-cutter ants. Evolution. 2001;55:1980–1991. doi: 10.1111/j.0014-3820.2001.tb01315.x. [DOI] [PubMed] [Google Scholar]
- Carius H.J, Little T.J, Ebert D. Genetic variation in a host–parasite association: potential for coevolution and frequency-dependent selection. Evolution. 2001;55:1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. [DOI] [PubMed] [Google Scholar]
- Chapela I.H, Rehner S.A, Schultz T.R, Mueller U.G. Evolutionary history of the symbiosis between fungus-growing ants and their fungi. Science. 1994;266:1691–1694. doi: 10.1126/science.266.5191.1691. doi:10.1126/science.266.5191.1691 [DOI] [PubMed] [Google Scholar]
- Clayton D.H, Johnson K.P. Linking coevolutionary history to ecological process: doves and lice. Evolution. 2003;57:2335–2341. doi: 10.1111/j.0014-3820.2003.tb00245.x. [DOI] [PubMed] [Google Scholar]
- Currie C.R. Prevalence and impact of a virulent parasite on a tripartite mutualism. Oecologia. 2001;128:99–106. doi: 10.1007/s004420100630. doi:10.1007/s004420100630 [DOI] [PubMed] [Google Scholar]
- Currie C.R, Stuart A.E. Weeding and grooming of pathogens in agriculture by ants. Proc. R. Soc. B. 2001;268:1033–1039. doi: 10.1098/rspb.2001.1605. doi:10.1098/rspb.2001.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C.R, Mueller U.G, Malloch D. The agricultural pathology of ant fungus gardens. Proc. Natl Acad. Sci. USA. 1999a;96:7998–8002. doi: 10.1073/pnas.96.14.7998. doi:10.1073/pnas.96.14.7998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C.R, Scott J.A, Summerbell R.C, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999b;398:701–704. doi:10.1038/19519 [Google Scholar]
- Currie C.R, Wong B, Stuart A.E, Schultz T.R, Rehner S.A, Mueller U.G, Sung G, Spatafora J.W, Straus N.A. Ancient tripartite coevolution in the attine ant–microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. doi:10.1126/science.1078155 [DOI] [PubMed] [Google Scholar]
- Dybdahl M.F, Storfer A. Parasite local adaptation: Red Queen vs. Suicide King. Trends Ecol. Evol. 2003;18:523–530. doi:10.1016/S0169-5347(03)00223-4 [Google Scholar]
- Gerardo N.M, Mueller U.G, Price S.L, Currie C.R. Exploiting a mutualism: parasite specialization on cultivars within the fungus-growing ant symbiosis. Proc. R. Soc. B. 2004;271:1791–1798. doi: 10.1098/rspb.2004.2792. doi:10.1098/rspb.2004.2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A.M, Mueller U.G, Adams R.M.M. Extensive exchange of fungal cultivars between sympatric species of fungus-growing ants. Mol. Ecol. 2002;11:191–195. doi: 10.1046/j.1365-294x.2002.01433.x. doi:10.1046/j.1365-294X.2002.01433.x [DOI] [PubMed] [Google Scholar]
- Hafner M.S, Nadler S.A. Cospeciation in host–parasite assemblages: comparative analysis of rates of evolution and timing of cospeciation events. Syst. Zool. 1990;39:192–204. doi:10.2307/2992181 [Google Scholar]
- Hafner M.S, Sudman P.D, Villablanca F.X, Spradling T.A, Demastes J.W, Nadler S.A. Disparate rates of molecular evolution in cospeciating hosts and parasites. Science. 1994;265:1087–1090. doi: 10.1126/science.8066445. doi:10.1126/science.8066445 [DOI] [PubMed] [Google Scholar]
- Harwood S.H, McElfresh J.S, Nguyen A, Conlan C.A, Beckage N.E. Production of early expressed parasitism-specific proteins in alternate sphingid hosts of the braconid wasp Cotesia congregata. J. Invertebr. Pathol. 1998;71:271–279. doi: 10.1006/jipa.1997.4745. doi:10.1006/jipa.1997.4745 [DOI] [PubMed] [Google Scholar]
- Hölldobler B, Wilson E.O. Springer; Berlin, Germany: 1990. The ants. [Google Scholar]
- Johnson K.P, Adams R.J, Page R.D.M, Clayton D.H. When do parasites fail to speciate in response to host speciation? Syst. Biol. 2003;52:37–47. doi: 10.1080/10635150390132704. doi:10.1080/10635150390132704 [DOI] [PubMed] [Google Scholar]
- Klassen G.J. Coevolution: a history of the macroevolutionary approach to studying host–parasite associations. J. Parasitol. 1992;78:573–587. doi:10.2307/3283532 [PubMed] [Google Scholar]
- Lively C.M. Migration, virulence, and the geographic mosaic of adaptations by parasites. Am. Nat. 1999;153:S34–S47. doi: 10.1086/303210. doi:10.1086/303210 [DOI] [PubMed] [Google Scholar]
- Lively C.M, Dybahl M.F, Jokela J, Delph L.F. Host sex and local adaptation by parasites in a snail–trematode interaction. Am. Nat. 2004;164:S6–S18. doi: 10.1086/424605. doi:10.1086/424605 [DOI] [PubMed] [Google Scholar]
- Maddison, D. R. & Maddison, W. P. 2003 Maclade 4: analysis of phylogeny and character evolution v. 4.06. Sunderland, MA: Sinauer.
- Mikheyev A.S, Mueller U.G, Abbot P. Cryptic sex and many-to-one coevolution in the fungus-growing ant symbiosis. Proc. Natl Acad. Sci. USA. 2006;103:10 702–10 706. doi: 10.1073/pnas.0601441103. doi:10.1073/pnas.0601441103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U.G, Rehner S, Schultz T.R. The evolution of agriculture in ants. Science. 1998;281:2034–2038. doi: 10.1126/science.281.5385.2034. doi:10.1126/science.281.5385.2034 [DOI] [PubMed] [Google Scholar]
- Page, R. D. M. (ed.) 2003 Tangled trees: phylogeny, cospeciation, and coevolution Chicago, IL: University of Chicago Press.
- Page R.D.M, Charleston M.A. Trees within trees: phylogeny and historical associations. Trends Ecol. Evol. 1998;13:356–359. doi: 10.1016/s0169-5347(98)01438-4. doi:10.1016/S0169-5347(98)01438-4 [DOI] [PubMed] [Google Scholar]
- Price P.W. Princeton University Press; Princeton, NJ: 1980. Evolutionary biology of parasites. [Google Scholar]
- Rambaut, A. & Drummond, A. 2005. Tracer v1.3 2003–2005. MCMC trace file analyser Oxford, UK: University of Oxford. See (http://evolve.zoo.ox.ac.uk/beast/).
- Reynolds H.T, Currie C.R. Pathogenicity of Escovopsis: the parasite of the attine ant–microbe symbiosis directly consumes the ant cultivated fungus. Mycologia. 2004;96:955–959. [PubMed] [Google Scholar]
- Ricklefs R.E, Fallon S.M, Bermingham E. Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Syst. Biol. 2004;53:111–119. doi: 10.1080/10635150490264987. doi:10.1080/10635150490264987 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. doi:10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Schardl C.L, Leuchtmann A, Chung K.R, Penny D, Siegel M.R. Coevolution by common descent of fungal symbionts (Epichloë spp.) and grass hosts. Mol. Biol. Evol. 1997;14:133–143. [Google Scholar]
- Schultz T.R, Meier R. A phylogenetic analysis of the fungus-growing ants (Hymenoptera: Formicidae: Attini) based on morphological characters of the larvae. Syst. Entomol. 1995;20:337–370. [Google Scholar]
- Swofford, D. L. 2002 PAUP*phylogenetic analysis using parsimony (*and other methods) v. 4.b10. Sunderland, MA: Sinauer.
- Thompson J.N. The University of Chicago Press; Chicago, IL: 1994. The coevolutionary process. [Google Scholar]
- Thompson J.N. The University of Chicago Press; Chicago, IL: 2005. The geographic mosaic of coevolution. [Google Scholar]
- Weber N.A. Gardening ants: the attines. vol. 92. Memoirs of the American Philosophical Society; Philadelphia, PA: 1972. [Google Scholar]
- Weckstein J.D. Biogeography explains cophylogenetic patterns in toucan chewing lice. Syst. Biol. 2004;53:154–164. doi: 10.1080/10635150490265085. doi:10.1080/10635150490265085 [DOI] [PubMed] [Google Scholar]
- Wegner K.M, Reusch T.B.H, Kalbe M. Multiple parasites are driving major histocompatibility complex polymorphism in the wild. J. Evol. Biol. 2003;16:224–232. doi: 10.1046/j.1420-9101.2003.00519.x. doi:10.1046/j.1420-9101.2003.00519.x [DOI] [PubMed] [Google Scholar]
- Wetterer J, Schultz T.R, Meier R. Phylogeny of fungus-growing ants (Tribe Attini) based on mtDNA sequence and morphology. Mol. Phyl. Evol. 1998;9:42–47. doi: 10.1006/mpev.1997.0466. doi:10.1006/mpev.1997.0466 [DOI] [PubMed] [Google Scholar]