Abstract
Cheetahs (Acinonyx jubatus) have a combination of ranging patterns and social system that is unique in mammals, whereby male coalitions occupy small territories less than 10% of the home range of solitary females. This study uses non-invasive genetic sampling of a long-term study population of cheetah in the Serengeti National Park in Tanzania to infer the mating system. Individual cheetah genotypes at up to 13 microsatellite loci were obtained from 171 faecal samples. A statistical method was adapted to partition the cubs within each litter (n=47) into full-sibling clusters and to infer the father of each cluster using these loci. Our data showed a high rate of multiple paternity in the population; 43% of litters with more than one cub were fathered by more than one male. The results also demonstrated that female fidelity was low, and provided some evidence that females chose to mate with unrelated males within an oestrus cycle. The low rate of paternity assignments indicated that males living outside the study area contributed substantially to the reproduction of the cheetah population.
Keywords: microsatellites, multiple paternity, mating system, cheetahs, paternity analysis
1. Introduction
The study of mating systems is fundamental to our understanding of the evolutionary and ecological forces underpinning sociality, yet disproportionate attention has focused on a relatively small number of systems (Clutton-Brock 1988), while others, often more unusual, have been neglected despite the fact that these systems are a potential key to a complete understanding. In wild populations, it is widely assumed that males are promiscuous while females are coy (Shuster & Wade 2003). This is because male reproductive success increases with multiple mating, while the potential benefits that females gain from mating with more than one male have long been debated (Jennions & Petrie 2000).
In carnivores, mating tactics are usually linked to their spacing patterns, and felids, in particular, typically exhibit a social system of intrasexual territoriality with males occupying larger ranges that encompass those of several females (Sandell 1989). This pattern of territoriality facilitates male access to several sexually receptive females. In contrast, adult female cheetahs (Acinonyx jubatus) in the Serengeti National Park in Tanzania occupy extensive home ranges (average 833 km2), while males hold considerably smaller territories (37 km2). Solitary males that are unable to retain a territory move over to an area of similar scale to the females (777 km2). Male competition for territories is intense, and coalitions of 2–3 males, usually brothers, have greater success in taking over occupied territories, while single males are more frequently non-territorial or ‘floaters’ (Caro 1994). The formation of long-term male coalitions is unusual among mammals, with only a handful of species forming male coalitions that are not associated with sociality in females (Waterman 1997); however, in all these species, male home ranges are of the same size or larger than female home ranges. The combination of permanent male coalitions together with male defence of a small portion of a female's home range has only been recorded in cheetahs.
Male cheetahs only associate with females at mating and provide no parental care. Sexual selection theory predicts that when there is no parental investment, males should mate with as many females as possible (Trivers 1972). To achieve this, males should exert control over females by monopolizing them during oestrus to prevent multiple matings with other males and reduce competition for paternity. Therefore, it has been suggested that intense intraspecific competition in males promotes formation of male coalitions that have greater success in territory acquisition and maintenance, and consequently are able to monopolize visiting females to gain benefits in reproductive success (Caro 1994). Coalition partners might be expected to cooperate to monopolize a female, losing some reproductive benefits through unequal access to the female but gaining benefits through prolonged access to the female and through kin selection when coalition partners are brothers.
Male manipulation of female mate choice can be through direct harassment of potentially receptive females or by indirect male–male competition over territories or access to females. Either way, if male manipulation compromises female inclusive fitness, then females should evolve ways to avoid this tactic, as predicted by sexual-conflict theory (Parker 1979), and polyandry is likely to arise. Females can mate with multiple males to confuse paternity, which in turn deters infanticide, or to reduce male harassment that can cause injury or death (Wolff & Macdonald 2004). Alternatively, females can obtain genetic benefits from polyandry if they are prevented from mating with the best partner, or if multiple mating results in multiply sired litters increasing the genetic diversity of the offspring (Jennions & Petrie 2000). Mating with multiple males and preventing monopolization of paternity could also help to reduce male–male conflict and favour post-copulatory sperm competition to enhance female reproductive success. Large female home ranges, which encompass several male territories, provide females with access to many males at relatively low cost and thus a high incidence of multiple mating is possible within this system.
An increasing number of studies demonstrate that polyandry is more common than previously thought, suggesting that the pay-offs from this mating strategy are likely to be significant. Polyandry in species with male coalitions might be expected to occur if females mate with each member of the coalition. Males therefore gain potential reproductive advantages from remaining in the coalition, while coalition formation should be stable provided the coalition can reap benefits through increased access to females or increased survival, both of which translate into overall reproductive success.
As cheetah matings are rarely observed in the wild, it has not been possible to develop an explicit understanding of the mating system and the potential reproductive pay-offs to either males or females through observation alone. In this study, we used microsatellite analysis of DNA from faecal samples of known individual cheetahs collected over a 9-year period, together with accurate long-term field records, to test whether polyandry among Serengeti female cheetahs is pervasive, as is predicted if females are able to counterbalance male manipulation and maximize female inclusive fitness. Our aim hereby is to use this dataset to shed further light on their unusual mating system.
2. Material and methods
(a) Study area and sampling
Cheetahs were studied on the plains of the Serengeti National Park in Tanzania (see Sinclair 1979 for a full description of the study area). Variation in seasonal rainfall drives the migratory patterns of the many herbivores in the ecosystem. Thomson's gazelle (Gazella thomsoni), the main prey species for cheetahs (Caro 1994), move up to the woodland border to the north and west of the plains during the dry season, and out onto the short grass plains at the start of the rains (Durant et al. 1988). Female and non-territorial male cheetahs follow this migration (Durant et al. 1988).
The collection of samples used in this study started in 1996. Samples were collected from cheetahs individually identified by photographic records and all samples were stored in 96% ethanol. When collecting faecal samples, cheetahs were individually identified and followed continuously until they defecated. After defecation, the observer waited until the cheetah moved away, then drove up slowly to the sample and placed a 1.5 cm2 subsample into a tube with ethanol. Cheetahs normally moved away within minutes of defecation and hence samples were collected very quickly. In a small number of situations, cheetahs did not move for a while, and these samples were collected up to an hour after defecation. The time delay from defecation to sample collection was recorded and did not affect DNA extraction. In no situation did any cheetah in this study defecate on top of existing faeces, cheetah or otherwise.
A total of 176 samples were collected; the majority were faecal samples (n=171), but a few tissue samples (n=5) were taken from carcasses. Most samples (n=173) came from within the study area. An additional three samples came from a woodland area in the north of the park (Towe, Roho and Pepo). We attempted to extract DNA from all samples; however, nine faecal samples failed to produce reliable genotypes at five or more loci and were removed from the analysis. The percentage of known adult males (candidate fathers) sampled increased steadily during the study from 18.2% in 1996 to almost 79% in 2004 (table 1). Three litters in the 47 mother–cub units included adopted cubs, i.e. cubs born to another female but accepted into a mother's existing litter. Adoptions are an occasional feature of cheetah reproduction (Caro 1994; Durant et al. 2004). DNA from faecal and tissue samples was independently extracted two or three times using the QIAGEN stool or QIAGEN tissue extraction kits according to the manufacturer's protocols. Thirteen microsatellite loci were selected from the domestic cat library on the basis of high polymorphism, reliable amplification and easy scorability (Menotti-Raymond et al. 1999).
Table 1.
Percentage of adult males, number of litters sampled and successful number of assigned paternities per year.
| year | study population sizea | adult males sampled (%)b | litters sampled | paternity assigned | |
|---|---|---|---|---|---|
| adult females | adult males | ||||
| 1996 | 38 | 18 | 18.18 | 2 | 0 |
| 1997 | 46 | 24 | 10.71 | 0 | 0 |
| 1998 | 46 | 29 | 14.29 | 5 | 0 |
| 1999 | 49 | 24 | 15.79 | 0 | 0 |
| 2000 | 62 | 26 | 45.45 | 1 | 0 |
| 2001 | 64 | 22 | 45.71 | 8 | 1 (12.5%) |
| 2002 | 65 | 26 | 61.29 | 13 | 4 (30%) |
| 2003 | 59 | 31 | 68.42 | 10 | 4 (40%) |
| 2004 | 59 | 35 | 78.95 | 5 | 1 (20%) |
Calculated as number of adults estimated to be in the population at the end of the year.
Percentage was a conservative estimate and was calculated as the percentage of males sampled out of the total number of males estimated to be in the population throughout the year, and hence includes individuals who died during the year.
Genotyping was carried out using fluoro-labelled primers and by performing 40 cycles of PCR amplification in a 6 μl reaction volume containing 1 μl (≤50 ng) DNA, 0.5 μl (0.2 μM) of each primer and 4 μl of PCR Master Mix, QIAGEN (provides a final concentration of 3 mM MgCl2). This hot start Taq DNA polymerase enables the PCR amplification of two or more products in a single reaction. Three primer pairs labelled with different dyes and with different allele sizes were amplified and run together with a size standard (Tamra 350) on a ABI 377 Automatic Sequencer (Applied Biosystems) running Genescan v. 2.1. Reference samples were included on each electrophoresis gel to allow the standardization of allele size measurements across gels. Microsatellite data were analysed using Genotyper v. 2.1 software (Applied Biosystems). Faecal samples were individually amplified at least three times for heterozygous and five for homozygous per locus to avoid genotyping errors due to allelic drop out and false alleles (Navidi et al. 1992; Taberlet et al. 1996).
(b) Statistical analysis
For each of the 13 loci of each cub, one allele is from the mother and the other from the father. However, owing to the lack of genetic variability in our cheetah population (table 2), it may be common for a father and mother to share the same alleles at several loci. In these circumstances, determination of paternity is not straightforward. The genetic data at the 13 microsatellite loci were used to partition the cubs within each litter into full-sib clusters and to infer the father of each cluster, based on a statistical method adapted from Wang (2004). The modifications of the method are described briefly below.
Table 2.
Number of alleles and observed and expected heterozygosity (%) of 13 domestic cat loci in our cheetah population.
| locus | no. of alleles | Ho | He |
|---|---|---|---|
| Fca 8 | 8 | 69 | 66 |
| Fca 45 | 6 | 67 | 57 |
| Fca 52 | 6 | 64 | 69 |
| Fca 84 | 4 | 63 | 57 |
| Fca 126 | 8 | 79 | 72 |
| Fca 133 | 9 | 68 | 64 |
| Fca 152 | 5 | 51 | 48 |
| Fca 193 | 9 | 82 | 82 |
| Fca 205 | 6 | 59 | 62 |
| Fca 212 | 6 | 72 | 72 |
| Fca 247 | 8 | 84 | 78 |
| Fca 254 | 5 | 75 | 69 |
| Fca 339 | 6 | 55 | 53 |
Consider a mother–cub unit consisting of a mother and its n cubs. The mother and cubs are genotyped at a locus with k codominant alleles. The father of each cub is unknown, but a number of N candidate fathers with known genotypes are identified. The prior probability that the true father of a cub is included in the candidates is u. The method aims to partition the n cubs into full-sib clusters and to infer the father of each cluster, using the genotype data of the mother, cubs and the candidates, and the population allele frequencies.
There are many possible configurations into which the total n+N+1 individuals can be arranged, and we are interested in finding the most probable one with the highest likelihood as our best inference. Suppose for a particular configuration, the n cubs are partitioned into f (1≤f≤n) full-sib families with family j (j=1, 2, …, f) containing dj cubs, and a number of m (0≤m≤f) candidates are inferred to be the fathers. Under random mating, the likelihood of the configuration is proportional to
The first part of the function is proportional to the prior probability of the configuration assuming that each candidate male is equally likely to be the father of a cub. In the second part of the function, Sy,z,i≡1/k2 if candidate i is inferred to be a father and if otherwise, where py and pz are the frequencies of allele y and z (=1, 2, …, k) and is the probability of observing the genotype of candidate i (Ri), given its true genotype is Gy,z. is calculated by assuming a prior probability of genotyping errors (Wang 2004). In the third part, , where T0 is the observed genotype of the mother, if the observed genotype of father j (Tj) is known and Qy,z,j=pypz if otherwise. The probability of observing the genotype of the ith cub in full-sib family j given parental genotypes, , was calculated following Mendelian segregation law and allowing genotyping errors (Wang 2004). For independent loci, the likelihood is simply the product of single-locus likelihoods. A simulated annealing algorithm similar to that of Wang (2004) is adapted to search for the best configuration with the maximum likelihood.
When both the father(s) and mother were unknown for a litter of cubs and candidate fathers and candidate mothers were available, we derived a similar likelihood function to partition the cubs into full-sib families and infer the fathers and the mother simultaneously.
A Bayesian method proposed in Emery et al. (2001) can be applied to the case of inferring paternity for cubs with a known mother, but not to the case of inferring both paternity and maternity for a litter of cubs. We used this method to double-check our results. For our simulated and cheetah datasets, this Bayesian method (where applicable) yields results very similar to those of our likelihood method. For simplicity and generality, therefore, we present only the analysis results obtained using the likelihood method. The data were also analysed using the more traditional program Cervus (Kalinowski et al. 2006) as another check of our novel method.
(c) Analyses of simulated data
To check the adequacy of the marker information and the performance of the proposed method in realistic situations, we analysed simulated data mimicking the cheetah dataset. We considered 17 types of mother–cub units varying in the number of cubs and their distribution among full-sib families with true fathers included in, or excluded from, a total of 50 candidates. For each mother–cub structure, the mother was assumed to be (i) known and genotyped, (ii) unknown but genotyped and included in the candidates, or (iii) unknown and absent from the candidates. In the latter two cases, the mother of the cubs was inferred together with the fathers of, and full sibships among, the cubs. A relatively uninformative prior, u=0.5, is used in the analyses to indicate that the true parent is equally likely to be included in and excluded from the candidates.
The genotypes of unrelated parents and candidates were generated, assuming the same allele frequency distributions of the 13 markers as in the cheetah dataset, and cub genotypes were generated from parental genotypes following Mendelian segregation. All these genotypes were then changed, at a given rate of 0.01, following the model of class II typing errors to give their corresponding observed genotypes (Wang 2004), which were analysed by the proposed method. A total of 1000 independent datasets were generated and analysed for each possible mother–cub structure: with a known mother; unknown but included mother; or unknown and excluded mother. The estimation accuracy was indicated by the proportion of replicates in which the simulated relationship structure (fathers, mother and full sibship among cubs) was fully recovered.
(d) Analysis of cheetah dataset
Among the 47 mother–cub units, 43 units had mothers with known genotypes at the 13 marker loci, one unit had a mother known to be dead whose genotype was unavailable and the remaining three units contained cubs with both known and unknown mothers (i.e. adopted cubs). For the 44 units with known mothers and the unadopted cubs of the remaining three units, we inferred paternity among a number of candidate fathers. For the three adopted cubs, we inferred both paternity and maternity simultaneously. Males (and females) who were estimated to be less than 18 months old at the time of the cubs' birth were excluded from the analysis. The number of candidate fathers (or mothers) varied among the 47 litters, from 19 to 65, with an increase in the number of candidates towards the end of the study. The genotyping error rates of the 13 loci are unknown, but are expected to be very low because each individual locus was genotyped three to five times. However, to avoid false exclusion of paternity, the data were analysed assuming a genotyping error rate of 1% per locus as described above. For each of the 47 mother–cub units, multiple configurations of the relationship structure with corresponding posterior probabilities can be obtained from the method. For brevity, however, we show only the best configuration with the maximum likelihood, whose reliability is indicated by its posterior probability. The second best configuration is shown only when its likelihood is greater than 90% of the maximum likelihood.
3. Results
Multilocus genotypes were 98.9% complete with full multilocus genotypes at 13 loci in 87.4% (146 of 167) of our genetic profiles. Ten genotypes had missing data at one locus, six at two loci, four at three loci and two at four loci. The observed heterozygosity for the 13 loci varied from 51% (Fca 152) to 84% (Fca 247) and the mean number of alleles per locus was 6.62 (table 2).
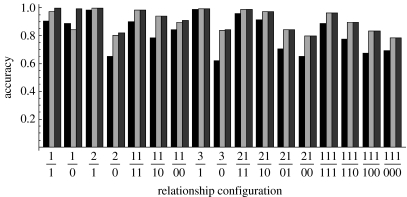
The 13 microsatellite markers showed relatively low polymorphism in our cheetah population. To check whether the information provided by the markers is sufficient for inferring paternity, simulated data were generated mimicking the cheetah data structures and were analysed by our statistical method (figure 1). It can be seen that using the 13 microsatellites, the method reconstructed completely the true relationship structures (full sibship within a litter, and parentage) in more than 80% of the replicates when the mother was either known or unknown but included in the candidate pool. When the mother was unknown and excluded from the candidates, then accuracy could fall to approximately 60%. The accuracy decreased with an increasing number of cubs whose fathers were absent from the candidate pool. Except for the case of a single offspring per litter, the inference accuracy when mothers were unknown but included in the candidates was indistinguishable from that when mothers were known. This was understandable because mothers included in the candidates were accurately inferred when the litter size was greater than 1. In contrast, the situation of the mothers unknown and absent from the candidate pool resulted in a substantial decrease in accuracy. For a given litter size, the accuracy increased with a decreasing number of full sibships nested in a maternal half sibship.
Figure 1.
Accuracy of relationship inference for 17 simulated datasets. For each dataset, the label on the x-axis indicates the full sibship and parentage structure. Taking 21/10 as an example, the numerator indicates that the maternal halfsib family of cubs consists of two full-sib families that contain 2 and 1 offspring, respectively. The denominator indicates that the father of the first full sibship with 2 members is included in the pool of candidates, and the father of the second full sibship with 1 member is excluded from the pool of candidates. For each dataset, the three bars from the left to the right show the inference accuracy when the mother is unknown and excluded from the candidate pool, unknown and included in the candidate pool, and known. Inference accuracy is the percentage of replicates in which the relationship structure (full sibship among cubs and parentage) is completely reconstructed from genetic data.
(a) Allocation of paternity and multiple paternity litters
When we used our method to identify the father(s) for each litter of cubs among the sampled candidate males, paternity was successfully assigned to a candidate male in only 10 litters (21.3%; table 3). For each litter, table 3 gives the best (maximum likelihood) inferred relationship configuration consisting of both sibship and parentage, and corresponding probability. In the case where the best inference is not much better (measured by likelihood) than the others, the second best configuration and probability are also listed. It should be noted that when one is concerned with parentage only (which is just a component of the entire relationship configuration), the two best configurations may be the same, so the relevant probability should be the sum of the two probabilities. As an example, consider mother–cubs unit 21. In the two best configurations, male Ben is the father of cubs 154C2, 154C3 and AmC4. The probability that male Ben is the father of the three cubs is therefore 0.55+0.44=0.99. In contrast, male Jerry is inferred as the father of cub 154C1 in the first configuration with probability 0.55, but not in the second configuration. Among the 10 litters (units 7, 9, 19, 21, 22, 29, 31, 37, 38 and 45) with assigned paternity, only two assignments (units 7 and 45) seemed to be dubious because the alternative (unassigned paternity) had a probability not much smaller than the assignment probability. The paternities for the remaining 8 litters were reliable. In addition to inferring relationships, our method also identifies genotyping errors (or mutations) at each locus in each mother–cub unit. Among the 47 separately analysed mother–cub units, 13 units have mismatches in genotypes given the known and inferred relationships. Among the 13 units, one unit has four, one has three, one has two, and each of the remaining 10 units has one mismatch.
Table 3.
Paternity (and maternity) assignments for 47 cheetah cub litters. (Note: column one lists mother–cub unit number, mother ID (or the numbers of candidate mothers for the last 3 litters) and litter's date of birth. Columns 4 and 6 give the best and second best inferences about parentage of, and sibship between, cubs in a litter, inferred full-sib cubs are placed in brackets, with their inferred parent placed before the bracket (N before a bracket indicates parentage assigned to an individual not included among candidates). Columns 5 and 7 give corresponding posterior probabilities of the two inferences. For the first 44 litters of cubs, the mothers are known and inferences are made about paternity and full sibship of cubs. For litters numbered 45, 46 and 47, both maternity and paternity are inferred. Paternity inferred with relatively high confidence is shown in bold. Rows shown in italics show the analysis results for combined litters from the same mother for different reproductive cycles.)
| group—mother ID—dob | no. of cubs | no. of male candidates | inference 1 | prob. 1 | inference 2 | prob. 2 |
|---|---|---|---|---|---|---|
| 1—Rays—9/02 | 3 | 47 | N(190C1, 190C2, 190C3) | 0.97 | ||
| 2—Prune—6/02 | 3 | 38 | N(PruFC2, PruMC); N(PruFC1) | 0.59 | ||
| 3—Meryl—6/02 | 3 | 39 | N(MerMC1, MerMC2, MerC) | 0.99 | ||
| 4—Chaumes—8/01 | 3 | 33 | N(Gus, ChaFC1, ChaFC2) | 0.70 | ||
| 5—Char—9/96 | 2 | 23 | N(CharC2, CharC1) | 1.00 | ||
| 6—Pista—9/01 | 2 | 37 | N(Pean, Braz) | 0.63 | N(Pean); N(Braz) | 0.28 |
| 7—Pean—10/03 | 1 | 55 | 57M(Almo) | 0.55 | N(Almo) | 0.45 |
| 8—Mou—6/02 | 2 | 40 | N(3417C1, 3417C2) | 1.00 | ||
| 9—Mou—3/01 | 1 | 33 | Mr Big(3327MC) | 0.66 | N(3327MC) | 0.34 |
| 10—Mou—11/03 | 1 | 65 | N(3601UC1) | 0.99 | ||
| 8-9-10—Mou | 4 | 65 | N(3417C1, 3417C2); N(3327MC); N(3601UC1) | 0.48 | N(3417C1, 3417C2); Mr Big(3327MC); N(3601UC1) | 0.48 |
| 11—Coco—11/98 | 4 | 26 | N(Ben, Haag, Jerry, Tira) | 0.88 | ||
| 12—Snuf—2/02 | 1 | 40 | N(SnufC1) | 0.99 | ||
| 13—Hazel—5/98 | 3 | 24 | N(HazFC1, Pecan, Pista) | 0.60 | N(HazFC1); N(Pecan, Pista) | 0.18 |
| 14—Pecan—8/01 | 1 | 37 | N(Pine) | 1.00 | ||
| 15—Pecan—5/03 | 2 | 50 | N(Tita); N(Ober) | 0.40 | YPM(Tita); N(Ober) | 0.16 |
| 14-15—Pecan | 3 | 49 | N(Pine); N(Tita); N(Ober) | 0.35 | YPM (Pine); N(Tita); N(Ober) | 0.14 |
| 16—Angie—12/01 | 1 | 40 | N(Sika) | 0.87 | ||
| 17—Siku—5/95 | 1 | 22 | N(Angie) | 0.99 | ||
| 18—Siku—4/92 | 1 | 19 | N(Talis) | 0.71 | 57M(Talis) | 0.29 |
| 17-18—Siku | 2 | 22 | N(Angie); N(Talis) | 0.75 | N(Angie); 57M(Talis) | 0.24 |
| 19—Angie—5/03 | 1 | 50 | 184M2(AnFC1) | 1.00 | ||
| 16-19—Angie | 2 | 50 | N(Sika); 184M2(AnFC1) | 0.91 | 184M2(AnFC1); 3037(Sika) | 0.04 |
| 20—Talis—4/98 | 1 | 24 | N(Amar) | 0.99 | ||
| 21—Amar—5/02 | 4 | 40 | Ben(154C2, 154C3, AmC4); Jerry(154C1) | 0.55 | Ben(154C2, 154C3, AmC4); N(154C1) | 0.44 |
| 22—Amar—4/04 | 1 | 62 | Tisa(Musc) | 0.74 | ||
| 21-22—Amar | 5 | 62 | Ben(154C2, 154C3, AmC4); N(154C1); Tisa(Musc) | 0.43 | Tisa(Musc); Ben(154C2, 154C3, AmC4); Jerry(154C1) | 0.30 |
| 23—Lemu—5/02 | 1 | 40 | N(LemuC) | 0.99 | ||
| 24—Towe—4/96 | 2 | 23 | N(Pepo); N(Roho) | 0.46 | N(Pepo, Roho) | 0.22 |
| 25—Came—1/98 | 1 | 24 | N(Camba) | 1.00 | ||
| 26—Twirl—4/98 | 1 | 24 | N(Val) | 0.99 | ||
| 27—Twirl—6/95 | 2 | 22 | N(Prune); N(Rays) | 0.35 | N(Prune, Rays) | 0.35 |
| 26-27—Twirl | 3 | 24 | N(Val); N(Prune); N(Rays) | 0.34 | N(Val); N(Prune, Rays) | 0.33 |
| 28—SB178—7/02 | 4 | 40 | N(178C1, 178C2, 178C3, 178C4) | 0.98 | ||
| 29—Fusi—9/02 | 3 | 46 | N(FusC1); Brad(FusC2, FusiC3) | 0.99 | ||
| 30—Tira—1/03 | 1 | 46 | N(TiraFC) | 0.84 | ||
| 31—Pina—4/03 | 1 | 55 | Owen (PinaC) | 0.91 | ||
| 32—Devo—12/02 | 5 | 46 | N(DevC2, DevC4, DevC5); N(DevC3); N(DevC6) | 0.41 | N(DevC2, DevC4, DevC5); N(DevC3, DevC6) | 0.24 |
| 33—Aman—10/03 | 3 | 55 | N(AmaC1, AmaC2, AmaC3) | 0.99 | ||
| 34—Elai—1/04 | 1 | 58 | N(3400C) | 0.97 | ||
| 35—Stre—4/01 | 1 | 33 | N(StreC1) | 0.60 | ||
| 36—Stre—2/04 | 3 | 62 | N(StreC2, StreC3); N(StreC4) | 0.32 | N(StreC2); Joey(StreC3, StreC4) | 0.19 |
| 35-36—Stre | 4 | 62 | N(StreC1); N(StreC2, StreC3); N(StreC4) | 0.20 | N(StreC1); Joey(StreC4, StreC3); N(StreC2) | 0.13 |
| 37—Bell—3/02 | 2 | 40 | 3575M2(3148FC, 3148MC) | 0.88 | ||
| 38—Frasca—3/02 | 1 | 40 | Pepo(Tequ) | 1.00 | ||
| 39—Aadje—12/01 | 3 | 40 | N(AdMC, Jojo, AdFC) | 0.98 | ||
| 40—Aadje—4/00 | 1 | 27 | N(Tisa) | 0.77 | ||
| 41—Aadje—4/03 | 3 | 50 | N(AdC1, AdC2, AdC3) | 0.92 | N(AdC3, AdC1); 57M(AdC2) | 0.02 |
| 39-40-41—Aadje | 7 | 49 | N(AdMC, Jojo, AdFC); N(Tisa); N(AdC3, AdC1, AdC2) | 0.77 | N(AdMC, Jojo, dFC); 3575M2(Tisa); N(AdC3, AdC1, AdC2) | 0.12 |
| 42—Carrie | 3 | 46 | N(CaMC, CaFC1); N(CaFC2) | 0.37 | N(CarMC, CarFC1); Mick(CarFC2) | 0.28 |
| 43—Ginger | 1 | 55 | N(GiMC) | 0.48 | Mick(GiMC) | 0.46 |
| 44—3315—2/01 | 3 | 33 | N(3315C2, 3315C3); N(3315C1) | 0.30 | N(3315C2, 3315C1); N(3315C3) | 0.21 |
| 45-(30)—5/03 | 1 | 50 | 3575M2(AngCa) | 0.47 | N(AngCa) | 0.46 |
| 45—maternity (30) | 1 | 50 | Stre(AngCa) | 0.97 | ||
| 46-(32)—2/02 | 1 | 40 | N(SnufCa) | 0.68 | 57M(SnufCa) | 0.19 |
| 46—maternity (32) | 1 | 40 | N(SnufCa) | 0.87 | Twirl(SnufCa) | 0.08 |
| 47-(32)—12/02 | 1 | 46 | N(DevCa) | 0.95 | ||
| 47–maternity (32) | 1 | 46 | N(DevCa) | 0.95 |
Adopted cubs.
The fact that paternity allocation was low, despite the supposedly high proportion of potential fathers sampled in the study site, particularly in the later years of the study (table 1), suggests that oestrous females might have wandered across larger areas than previously thought, frequently venturing outside the study area into areas where male cheetahs were not sampled. The low paternity assignments could also be due to males, that usually reside outside the study area, venturing inside looking for oestrous females. Potential areas with cheetah outside our study site include areas within the park; from the woodland areas to the north and west of the park; or from outside the park to the south and east. Sightings of unknown adult males are not unusual but, due to the short period they spend within the study site, they are difficult to sample. Evidence for matings outside the study area comes from one of our assigned paternities which placed the father of litter 38, born within the study area, to a cheetah (Pepo) whose natal range was known to have been more than 50 km north of the study area. The 47 mother–cub units sampled included 23 litters with two or more genotyped cubs. Of these, 10 litters (43%) were sired by two or three males, showing clear evidence of multiple paternity and hence polyandry. The average litter size of this subsample was 2.96 cubs (range 2–5).
(b) Reproductive skew within male coalitions
Among the 10 mother–cub units with evidence of genetic polyandry, paternity was assigned to candidate fathers on two occasions. In mother–cub unit 21, two males fathered four cubs, and the analysis showed that Ben, a territorial male, sired three of these cubs. Paternity of the one remaining cub, 154C1, was allocated to either male Jerry (with probability 0.55) or a male not listed in the candidates (with probability 0.44). Despite the inclusion of complete 13 loci genotypes of the two other males in Ben's coalition (brothers Haagandez and Jerry) among the candidate fathers, the paternity of 154C1 was not conclusively allocated to either of them. A similar pattern was observed in mother–cub unit 29, where paternity of the three cubs in this litter was shared by the territorial male Brad and another unknown male, and none of the cubs were sired by Brad's brother and sole coalition partner, Pitt, who was present among the candidate males.
(c) Female fidelity
We analysed the paternity of cubs pooled over different litters from the same mother but from different reproductive cycles to search for evidence of female fidelity. The analysis of pooled data also acts to check the reliability of the method and the sufficiency of the marker information. If the marker information is sufficient and the statistical method reliable, then the results should be similar whether different litters from the same mother are analysed separately or jointly. Furthermore, the pooled data allow us to assess whether separate litters from the same female are sired by the same males. Our dataset contains eight females who bred successfully more than once during the study period, five of these produced two separate litters and three produced three separate litters. Separate and joint analyses gave almost identical results in terms of both full-sib sibship and paternity inferences (table 3). None of the litters from the same female were fathered by the same male, and hence none of the eight females returned to the same male(s) that sired previous cubs for subsequent matings (table 3).
(d) Adoptions
We tried to assign the genetic mother to three adopted cubs, units 45, 46 and 47 (table 3). In only one case, mother–cub unit 45, maternity was allocated to a female present among the candidate sampled females. The mother of this adopted cub (Streep) was not a close maternal relative of the adoptive mother (Angie). Streep can be traced back two generations to her grandmother Lucy, and shows no evidence of being linked to Angie's pedigree that can be traced back three generations to her great grandmother. For each of the three mother–cub units, the adoptive mother was included in the candidate mothers but was not assigned the mother by the genetic analysis, suggesting that adoptions are being correctly identified by field observations and the method does not assign false parentage.
4. Discussion
Our results demonstrate that DNA extracted from faecal samples can be used to uncover detailed information about the mating systems in our cheetah population. Sampling of cheetahs in the Serengeti plains is very difficult due to the vast areas that need to be covered, and considerable time and effort were put into obtaining the samples used in this study. Despite this, the number of assigned paternities was low (21.3%). These low paternity assignments could be in part attributed to the difficulty involved in sampling cheetahs, which are extremely mobile and can cover large distances in a matter of days. However, there was strong evidence of an improvement in paternity assignment rate during the study from 0% in the first 3 years to 32% in the last 3 years (table 1) as a result of increased sampling of males in the population. However, a number of potential fathers, known to be residents in the study area, were missing from the dataset. During the last 3 years of the study, 21–38% of males within the study area were not sampled. Nonetheless, it is unlikely that these missing males are responsible for the 68% of non-assigned paternities over this period. Instead, at least some of the candidate fathers are likely to reside outside the study site.
To cross check the accuracy of our method, the data were also analysed using Cervus v. 3.0 (Kalinowski et al. 2006), a likelihood-based method that infers paternity of each offspring separately. All paternity assignments found by our method were confirmed by Cervus at 95% confidence level. In addition, Cervus allocated five more paternities at the lower 80% confidence limit which were not inferred by our method. However, all these paternity assignments had one (n=2) or two (n=3) mismatches among the genotypes of the mother–cub-assigned father trio.
We tested the performance of Cervus by inferring the maternity of one of the largest litters in our dataset for which we had assigned paternity: the four cubs in group 21 (table 3). The known mother (Amar) of the cubs was either excluded from (case 1) or included in (case 2) the candidate females. In the first case where Amar was excluded from the candidates, Cervus assigned maternity of the four cubs to three females; all assignments were at the 80% confidence level with a single-locus mismatch. In the second case where Amar was included in the candidates, the real mother was assigned for two cubs (0 mismatch), while two other females were assigned maternity for the other two cubs (0 and 1 mismatch), all at an 80% confidence level. Interestingly, two of the falsely assigned females in both analyses were Talis and Tira, the maternal grandmother and paternal aunt of the litter. In contrast, our method yielded the same results for sibship among, and paternity of, the cubs as listed in table 3, no matter whether the true mother, Amar, was included in or excluded from the candidate mother list. When Amar was included in the candidates, she was assigned as the mother of all four cubs with a probability of 1. When she was excluded from the candidates, maternity was allocated to one unsampled female with a probability of 0.61. The second best maternity inference suggested that Talis (the maternal grandmother) was the mother of all four cubs with a probability of 0.38. The striking difference in power between the two methods is understandable because our method uses information of all the cubs to infer parentage, while Cervus uses the information of a single cub.
The data show strong support for our prediction that multiple paternity was common within litters. We found evidence of multiple paternity in 43% of litters comprising two or more cubs, confirming that in the Serengeti, female cheetahs are promiscuous and polyandry is common. High levels of mixed paternity within litters have been found among Lake Malawi cichlids (Kellog et al. 1998), tree swallows (Lifjeld et al. 1993) and insects (Zeh et al. 1997). Among mammals, multiple paternity has been described in species where females are likely to mate with close relatives like the European shrew (Sorex araneus Stockley et al. 1993) and Ethiopian wolf (Canis simiensis Sillero-Zubiri et al. 1996) and could have been selected as an inbreeding avoidance mechanism. High levels of polyandry have also been found among the Serengeti spotted hyena population (Crocuta crocuta East et al. 2003). However, apart from the domestic cat (Felis catus Say et al. 1999), there is no published evidence of multiple paternity in felids, which are usually territorial, with strong mate-guarding behaviours. Furthermore, polyandrous matings may have been underestimated by this study. First, the sampling of large/entire litters is difficult due to high early cub mortality suffered by cheetahs in our study area (Laurenson 1994). Second, some apparently single paternity litters might be the product of sperm competition or cryptic female choice.
Cheetah female receptivity to males varies from 1 to 3 days (Bertschinger et al. 1984); however, there is also evidence that receptivity can last up to 14 days (Seager & Demorest 1978). Selection to prolong the period of female receptivity would help promote multiple matings (Shuster & Wade 2003), as female cheetahs with even a slightly prolonged oestrous could cover areas including several male territories and also encounter other, non-territorial, males.
Seeking out additional males is energetically costly and increases the risk of predation. Multiple mating also increases the risk of exposure to parasites and sexually transmitted diseases (Koga et al. 1998). Therefore, there should be fitness advantages for female cheetahs that offset these costs. Multiple mating that results in multiply sired litters will increase the genetic diversity of the progeny (bet-hedging), which could be advantageous in an unpredictable environment, like the Serengeti, where female cheetahs that produce a genetically diverse set of offspring could increase the likelihood that some of them will survive and breed in the next generation. However, it has been argued that increased genetic diversity could be the result of multiple matings rather than its selective force (Wolff & Macdonald 2004). Multiple mating could have primarily evolved as a deterrent to infanticide or to avoid sexual harassment, the latter of which has been documented in cheetahs (Caro 1994). While female cheetahs are receptive, males try to monopolize them, a behavioural strategy common in many mating systems that use variations of mate-guarding adapted to the spatial and temporal distribution of oestrous females (Clutton-Brock 1989). However, our results demonstrate that males are often not successful in monopolizing a female and polyandry may have been selected to counterbalance this male manipulation by increasing female mate choice (Birkhead 2000). Our results also indicate low levels of female fidelity between different reproductive cycles, since all eight females who produced more than 1 litters during this study mated with different males in each reproductive season. This is likely to diminish the variance in male reproductive success within the population and increase the overall genetic variability of female progeny over their lifetime.
Our data were too limited to test reproductive skew among coalition partners. However, the analysis of mother–cubs units 21 and 29 did not provide support to the hypothesis that males from the same coalition have an equal share in paternity within litters. Further paternity analysis is needed for a more accurate test of this hypothesis.
Finally, our results shed some light on a poorly understood aspect of cheetah behaviour. Female cheetahs are known to adopt cubs, provided they have a litter of similar age. This behaviour has been shown to result in reproductive success benefits in terms of increased survival of male adolescents through the presence of a sister (Durant et al. 2004). It has also been suggested that single male cubs reap future reproductive success benefits in terms of increased mating success by having an adoptive brother through future coalition formation (Caro 1994). However, if adopted cubs are from related litters, female cheetahs may also reap benefits in reproductive success by helping to rear offspring of close relatives. Out of three known females with an adoptive cub in this study, maternity of the offspring was allocated in only one case, where both females were clearly not close relatives through the maternal line. This suggests that adoptions between unrelated animals do occur, and lends further support to the suggestion that adoptions are likely to result in real survival or reproductive benefits to cubs, rather than genetic benefits through kin selection.
The mating system of cheetahs and the transmission of paternal genes from generation to generation are not just of academic interest. Cheetahs are a threatened species and are declining throughout their range (Marker 1998). Large carnivores, in general, live at low density, but some of the less competitive species, such as cheetah, are threatened by larger carnivores in the ecosystem. Cheetah densities are limited by lions (Panthera leo Laurenson 1995) and probably hyaenas, and the species never reaches high numbers within even the largest protected areas. As populations become increasingly fragmented with the global acceleration in land use change and habitat degradation, they become vulnerable to losses of genetic diversity (Lande 1994, 1995; Lacy 1997). Such losses can be prevented through a sound genetic management strategy, but this needs to be based on accurate knowledge of how genetic diversity is maintained in wild populations. It is known that reproduction in cheetahs is dominated by a few individual females (Kelly 2001); however, the contribution of males to the gene pool is not known. Generally, it is assumed that a smaller proportion of males than females contribute to the gene pool owing to high levels of competition among males for mates (Clutton-Brock 1989). Our results demonstrate that in cheetah, a much higher number of males than expected contribute to the gene pool, which suggests that rates of genetic diversity loss should be lower than anticipated by Kelly (2001).
In conclusion, our results indicate that female cheetahs are promiscuous, with high levels of multiple paternity and no evidence of mate fidelity between reproductive seasons. Females also appear to mate with unrelated males within the same reproductive cycle. The low success rate in paternity assignments indicates that males living outside the study area, and potentially outside the park, contribute substantially to cheetah reproduction. This finding reinforces the crucial role that high mobility plays in cheetah ecology and their conservation. The understanding of the mating system of this cheetah population will aid in the development of future management plans aimed at maintaining genetic diversity of cheetahs within and outside protected areas.
Acknowledgments
We would like to thank TANAPA, TAWIRI and the Tanzania Commission for Science and Technology for providing permission for the long-term study. We would also like to thank Anne Hilborn and John Shemkunde for their help in collecting samples and relevant demographic data for this study. We thank Nathalie Pettorelli, Nicola Jenner, Marcela Kelly and an anonymous referee for comments on the manuscript. We are also grateful to many organizations for funding this study, principally the Royal Society, UK, Howard G. Buffett Foundation, Wildlife Conservation Society and Frankfurt Zoological Society (FZS). Finally, we thank numerous people and organizations that have provided much needed logistical support (and the occasional faecal sample), including O. Newman, A. Barrett, J. Driessen, J. Jackson, A. Geertsema, P. and L. White and the staff and management of Ndutu Safari Lodge, fellow scientists at SWRC, and M. Borner and staff at FZS.
References
- Bertschinger H.J, Meltzer D.G.A, van Dijk A, Courbroughl R.I, Soley J.T, Collett F.A. Cheetah lifeline. Nucl. Act. 1984;30:2–7. [Google Scholar]
- Birkhead T. Harvard University Press; Cambridge, MA: 2000. Promiscuity. [Google Scholar]
- Caro T.M. Chicago University Press; Chicago, IL; London, UK: 1994. Cheetahs of the Serengeti plains. [Google Scholar]
- Clutton-Brock T.H. Reproductive success. In: Clutton-Brock T.H, editor. Reproductive success: studies of individual variation in contrasting breeding systems. University of Chicago Press; Chicago, IL: 1988. pp. 472–485. [Google Scholar]
- Clutton-Brock T.H. Mammalian mating systems. Proc. R. Soc. B. 1989;236:339–372. doi: 10.1098/rspb.1989.0027. doi:10.1098/rspb.1989.0027 [DOI] [PubMed] [Google Scholar]
- Durant S.M, Caro T.M, Collins D.A, Alawi R.M, FitzGibbon C.D. Migration patterns of Thompson's gazelles and cheetahs on the Serengeti plains. Afr. J. Ecol. 1988;26:257–268. [Google Scholar]
- Durant S.M, Kelly M, Caro T.M. Factors affecting life and death in Serengeti cheetahs: environment, age and sociality. Behav. Ecol. 2004;15:11–22. doi:10.1093/beheco/arg098 [Google Scholar]
- East M.L, Burke T, Wihelm K, Greig C, Hofer H. Sexual conflicts in spotted hyenas: male and female mating tactics and their reproductive outcome with respect to age, social status and tenure. Proc. R. Soc. B. 2003;270:1247–1254. doi: 10.1098/rspb.2003.2363. doi:10.1098/rspb.2003.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery A.M, Wilson I.J, Craig S, Boyle P.R, Noble L.R. Assignment of paternity groups without access to parental genotypes: multiple mating and developmental plasticity in squid. Mol. Ecol. 2001;10:1265–1278. doi: 10.1046/j.1365-294x.2001.01258.x. doi:10.1046/j.1365-294X.2001.01258.x [DOI] [PubMed] [Google Scholar]
- Jennions M.D, Petrie M. Why do females mate multiply? A review of genetic benefits. Biol. Rev. 2000;75:21–64. doi: 10.1017/s0006323199005423. doi:10.1017/S0006323199005423 [DOI] [PubMed] [Google Scholar]
- Kalinowski S.T, Taper M.L, Marshall T.C. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2006;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. doi:10.1111/j.1365-294X.2007.03089.x [DOI] [PubMed] [Google Scholar]
- Kellog K.A, Markert J.A, Stuaffer J.R, Kocher T.D. Intraspecific brood mixing and reduced polyandry in a maternal mouth-brooding cichlid. Behav. Ecol. 1998;9:309–312. doi:10.1093/beheco/9.3.309 [Google Scholar]
- Kelly M. Lineage loss in Serengeti cheetahs: consequences of high reproductive variance and heritability of fitness on effective population size. Conserv. Biol. 2001;15:137–147. doi:10.1046/j.1523-1739.2001.99033.x [Google Scholar]
- Koga T, Backwell P.R.Y, Jennions M.D, Christy J.H. The effect of predation risk on mating behaviour in the fiddler crab. Proc. R. Soc. B. 1998;265:1385–1390. doi:10.1098/rspb.1998.0446 [Google Scholar]
- Lacy R.C. Importance of genetic variation to the viability of mammalian populations. J. Mammal. 1997;78:320–335. doi:10.2307/1382885 [Google Scholar]
- Lande R. Risk of population extinction from fixation of new deleterious mutations. Evolution. 1994;48:1460–1469. doi: 10.1111/j.1558-5646.1994.tb02188.x. doi:10.2307/2410240 [DOI] [PubMed] [Google Scholar]
- Lande R. Mutation and conservation. Conserv. Biol. 1995;9:782–791. doi:10.1046/j.1523-1739.1995.09040782.x [Google Scholar]
- Laurenson M.K. High juvenile mortality in cheetahs (Acinonyx jubatus) and its consequences for maternal care. J. Zool. Lond. 1994;234:387–408. [Google Scholar]
- Laurenson M.K. Implications of high offspring mortality for cheetah population dynamics. In: Sinclair A.R.E, Arcese P, editors. Serengeti II: dynamics, management and conservation of an ecosystem. University of Chicago Press; Chicago, IL: 1995. pp. 385–399. [Google Scholar]
- Lifjeld J, Dunn P.O, Robertson R.J, Boag P.T. Extra-pair paternity in monogamous tree swallow. Anim. Behav. 1993;45:213–229. doi:10.1006/anbe.1993.1028 [Google Scholar]
- Marker, L. 1998 Current status of the cheetah (Acinonyx jubatus). In A Symposium on cheetahs as game ranch animals (ed. Penzhorn, B. L.), pp. 1–17, Onderstepoort, South Africa.
- Menotti-Raymond M.A, David V.A, Lyons L.A, Schoffer A.A, Tomlin J.F, Hutton M.K, O'Brien S.J. A linkage map of microsatellites in the domestic cat. Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. doi:10.1006/geno.1999.5743 [DOI] [PubMed] [Google Scholar]
- Navidi W, Arnheim N, Waterman M.S. A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am. J. Hum. Genet. 1992;50:347–359. [PMC free article] [PubMed] [Google Scholar]
- Parker G.A. Sexual selection and sexual conflict. In: Blum M.S, Blum N.A, editors. Sexual selection and reproductive competition in insects. Academic; New York, NY: 1979. pp. 123–166. [Google Scholar]
- Sandell M. The mating tactics and spacing patterns of solitary carnivores. In: Gittleman J.L, editor. Carnivore behavior, ecology and evolution. vol. 1. Cornell University Press; New York, NY: 1989. pp. 164–182. [Google Scholar]
- Say L, Pontier D, Natoli E. High variation in multiple paternity of domestic cats (Felis catus L.) in relation to environmental conditions. Proc. R. Soc. B. 1999;266:2071–2074. doi: 10.1098/rspb.1999.0889. doi:10.1098/rspb.1999.0889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seager S.W, Demorest C.N. Reproduction of captive wild carnivores. In: Fowler M.E, editor. Zoo and wild animal medicine. W. B. Sounders and Co.; Philadelphia, PA: 1978. pp. 667–706. [Google Scholar]
- Shuster S.M, Wade M.J. Mating systems and strategies. Princeton University Press; Princeton, NJ; Oxford, UK: 2003. Multiple matings and postcopulatory, prezygotic sexual selection; pp. 109–127. [Google Scholar]
- Sillero-Zubiri C, Gottelli D, Macdonald D.W. Male philopatry, extra pack copulations and inbreeding avoidance in Ethiopian wolves. Behav. Ecol. Sociobiol. 1996;38:331–340. doi:10.1007/s002650050249 [Google Scholar]
- Sinclair A.R. The Serengeti environment. In: Sinclear A.R, Norton-Griffiths M, editors. Serengeti: dynamics of an ecosystem. University of Chicago Press; Chicago, IL: 1979. pp. 31–43. [Google Scholar]
- Stockley P, Searle J.B, Macdonald D.W, Jones C.S. Female multiple mating behaviour in the common shrew as a strategy to reduce inbreeding. Proc. R. Soc. B. 1993;254:173–179. doi: 10.1098/rspb.1993.0143. doi:10.1098/rspb.1993.0143 [DOI] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits L.P, Bouvet J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. doi:10.1093/nar/24.16.3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivers R.L. Parental investment and sexual selection. In: Campbell B, editor. Sexual selection and the descendent of man 1871–1971. Aldine; Chicago, IL: 1972. pp. 136–179. [Google Scholar]
- Wang J. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. doi:10.1534/genetics.166.4.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman J.M. Why do male Cape ground squirrels live in groups? Anim. Behav. 1997;53:809–817. doi:10.1006/anbe.1996.0346 [Google Scholar]
- Wolff J.O, Macdonald D.W. Promiscuous females protect their offspring. Trends Ecol. Evol. 2004;19:127–134. doi: 10.1016/j.tree.2003.12.009. doi:10.1016/j.tree.2003.12.009 [DOI] [PubMed] [Google Scholar]
- Zeh D.W, Zeh J.A, Bermingam E. Polyandrous sperm-storing females: carriers of male genotypes through episodes of adverse selection. Proc. R. Soc. B. 1997;264:119–125. doi:10.1098/rspb.1997.0018 [Google Scholar]