Abstract
Batesian mimics—benign species that predators avoid because they resemble a dangerous species—often vary geographically in resemblance to their model. Such geographical variation in mimic–model resemblance may reflect geographical variation in model abundance. Natural selection should favour even poor mimics where their model is common, but only good mimics where their model is rare. We tested these predictions in a snake-mimicry complex where the geographical range of the mimic extends beyond that of its model. Mimics on the edge of their model's range (where the model was rare) resembled the model more closely than did mimics in the centre of their model's range (where the model was common). When free-ranging natural predators on the edge of the model's range were given a choice of attacking replicas of good or poor mimics, they avoided only good mimics. By contrast, those in the centre of the model's range attacked good and poor mimics equally frequently. Generally, although poor mimics may persist in areas where their model is common, only the best mimics should occur in areas where their model is rare. Thus, counter-intuitively, the best mimics may occur on the edge of their model's range.
Keywords: Batesian mimicry, geographical variation, predation, Micrurus fulvius, Lampropeltis triangulum elapsoides
1. Introduction
Batesian mimicry evolves when a palatable species (the ‘mimic’) co-opts a warning signal from a dangerous species (the ‘model’) and thus deceives its potential predators (Bates 1862). Such resemblances can be favoured by natural selection if predators confuse models with lookalikes (the evidence for and the principles of mimicry are reviewed in Wickler (1968), Edmunds (1974), Pough (1988), Endler (1991), Mallet & Joron (1999), Brodie & Brodie (2004) and Ruxton et al. (2004)).
It is generally assumed that selection favours mimics that most closely resemble their model. Yet, mimics often vary geographically in resemblance to their model. For example, in the New World, many species of non-venomous snakes mimic several species of highly venomous coral snakes (Greene & McDiarmid 1981; Brodie & Brodie 2004). Although these mimics may bear a striking resemblance to their model in some areas, the same mimic species may only vaguely resemble their model in other areas (e.g. Pfennig et al. 2007).
Geographical variation in mimic–model resemblance may reflect variation in model abundance. If a model is rare (relative to its mimics), selection to avoid the model (and any lookalikes) should be weak (Huheey 1964; Oaten et al. 1975; Getty 1985; Pfennig et al. 2001). In such situations, only those mimics that most closely resemble the model should receive any protection. Thus, in geographical areas where the model is rare, natural selection should favour only the best mimics. By contrast, if a model is relatively common, predators' risk of encountering the model is high, and they should therefore be under strong selection to avoid any species that remotely resembles the model. Thus, in geographical areas where the model is common, even crude lookalikes (‘poor’ mimics) may be protected from predation, especially if the model is highly noxious (Getty 1985; Pough 1988; Brodie & Janzen 1995; Caley & Schluter 2003).
Additionally, poor mimicry may actually be selectively favoured when the model is common. Poor mimics might benefit by resembling several models simultaneously, they may be more cryptic than their model, they may have thermoregulatory advantages (e.g. more black coloration could allow individuals to heat up quicker, whereas more white may keep individuals cooler in exposed environments) and/or they could avoid investing in costly conspicuous signals (e.g. colour pigments). Thus, in geographical areas where the model is common, poor mimicry may evolve owing to a relaxation of selection to closely resemble the model and because selection might actually favour poor mimics in such areas.
Here, we focus on a coral-snake-mimicry complex to ask whether mimics resemble their model more closely in regions where models are relatively rare than in regions where they are common. We conducted (i) morphometric analyses to determine whether mimics vary in their resemblance to the local model in different geographical areas, (ii) population censuses to ascertain whether model-to-mimic abundance differs in these same geographical areas, and (iii) field experiments to establish whether the strength of selection on mimic–model resemblance varies in different geographical areas. Our results suggest that selection favours only the best mimics where their model is rare.
2. Material and methods
(a) Study system
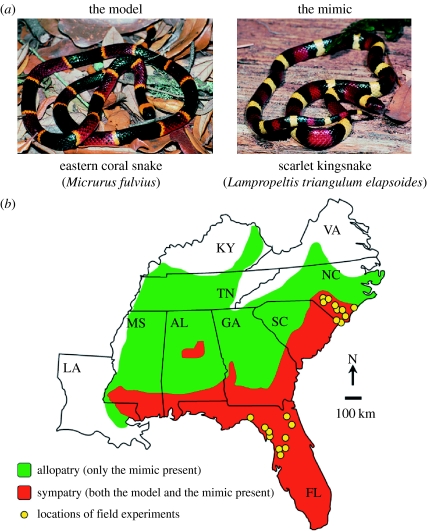
In the southeastern USA, eastern coral snakes (Micrurus fulvius)—highly venomous, aposematically coloured elapids—serve as models for non-venomous scarlet kingsnakes (Lampropeltis triangulum elapsoides). Both species co-occur from Florida (‘deep sympatry’) to southern North Carolina (‘edge sympatry’), but the geographical range of L. t. elapsoides (the mimic) extends beyond that of M. fulvius (its model; figure 1). Previous studies revealed that free-ranging natural predators avoid replicas of L. t. elapsoides in sympatry with M. fulvius but not in allopatry (Pfennig et al. 2001, 2007).
Figure 1.
(a) The non-venomous scarlet kingsnake (L. t. elapsoides) mimics the highly venomous eastern coral snake (M. fulvius). (b) The geographical range of L. t. elapsoides (the mimic) greatly exceeds that of its model.
We conducted three separate studies. First, to determine whether mimics vary in resemblance to their local model in different regions, we compared colour patterns of models and mimics in edge sympatry and deep sympatry. Second, to establish that the relative abundance of models differs between edge sympatry and deep sympatry, we tallied published accounts and museum specimens of each species (such data were used as a proxy for population estimates). Finally, to determine whether the strength of selection on mimic–model resemblance probably varies in different geographical areas, we conducted a field experiment to assess predation on replicas of ‘good’ mimics (i.e. mimics that closely matched their model) and ‘poor’ mimics (i.e. mimics that matched their model less closely but that were still within the range of variation of actual mimics).
(b) Mimic–model resemblance in different areas
First, we conducted morphometric analyses of both M. fulvius (the model) and L. t. elapsoides (the mimic) to determine whether mimics vary in their resemblance to the local model in different geographical areas. We began by photographing preserved specimens of each species by using a digital camera (see electronic supplementary material for the locations of the specimens). All snakes were photographed on the same background material.
We then used these photographs to measure two pattern characteristics that, we had determined a priori, differentiated good mimics from poor mimics (Harper 2006): (i) the proportion of the snake's mid-dorsum that is black and (ii) the proportion of the snake's mid-dorsum that is red. Specifically, we calculated for each snake the proportion of its mid-dorsum (i.e. the entire back of the snake, from its head to its cloaca) that was black or red, respectively. We chose these two characteristics, because previous morphometric analyses had revealed that these characteristics change the most as the mimetic pattern breaks down in allopatry. In particular, compared with M. fulvius and L. t. elapsoides in sympatry, L. t. elapsoides in allopatry tend to have less black and more red on their dorsum (Harper 2006; Pfennig et al. 2007). We limited our analysis to black and red, because these are the predominant colours on both models and mimics, and including all three colours (black, red and yellow) would remove the independence of the characteristics.
We compared these characteristics between the four categories of snakes: (i) edge sympatry models (M. fulvius, N=22), (ii) deep sympatry models (M. fulvius, N=25), (iii) edge sympatry mimics (L. t. elapsoides, N=41), and (iv) deep sympatry mimics (L. t. elapsoides, N=46). We asked whether mimics were more similar phenotypically to their local models in edge sympatry than in deep sympatry. Additionally, we performed discriminant analyses on the colour patterns of models and mimics from each region to determine how often mimics were mistaken for models and vice versa.
(c) Relative abundances of models and mimics in different areas
We next asked whether the relative abundance of the model varies geographically. We assembled data on numbers of L. t. elapsoides and M. fulvius collected from Florida (deep sympatry) by noting the numbers of each species present in museum collections (Harper 2006). For North Carolina (edge sympatry), we also used data published by Palmer & Braswell (1995). Although such information provides a rough estimate of the actual abundances of the two species, we used these data for qualitative purposes only. In particular, we sought to confirm the often-stated view that both the mimic and the model are locally abundant in Florida (Kenny Krysko, personal communication, 2006), but that only the mimic is relatively abundant in North Carolina (e.g. Palmer & Braswell (1995) assert that in North Carolina, L. t. elapsoides ‘are locally common’ but M. fulvius are ‘extremely rare’). Museum collections and published accounts also have the advantage of providing estimates of species abundances over many decades.
We tallied the number of individuals of each species by county and used only counties that had at least one mimic sample. We calculated the ratio of models to mimics for each county and used these ratios to calculate model-to-mimic ratios for the two regions (edge and deep sympatry). We then used a non-parametric Wilcoxon rank-sum test to compare the median ratio of models to mimics in each region.
(d) Predation pressure on good and poor mimics in different areas
Finally, we sought to determine whether selection on good and poor mimics varies in geographical regions that differ in the relative abundance of the model. We specifically asked whether poor mimics were more likely to be attacked than good mimics in regions where models were relatively rare (i.e. in edge sympatry), and whether poor mimics were as likely to be attacked as good mimics in regions where models were relatively abundant (i.e. in deep sympatry). To address this issue, we used Plasticine replicas of snakes to estimate selection on different colour patterns by exposing different types of replica to free-ranging predators.
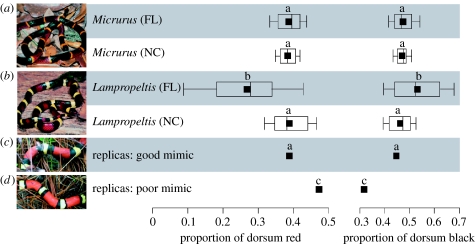
We constructed artificial models of snakes (replicas) similar to those used in two recent studies (Pfennig et al. 2001, 2007). We created snake replicas (1.5×18 cm) of precoloured, non-toxic Plasticine with a tricolour ringed pattern that had proportions of red, black and yellow similar to those of M. fulvius (the good mimic), a tricolour ringed pattern with more red and yellow and less black than the average M. fulvius (the poor mimic), and a plain brown pattern (the latter served as controls). Good and poor mimic replicas resembled naturally occurring L. t. elapsoides in size, colour hue, colour order and ring width (colours were matched by human eye). The poor mimic contained 8% more red, 4% more yellow and 12% less black than the good mimic (figure 2). The good mimic was modelled after a typical L. t. elapsoides from southern North Carolina (edge sympatry). The poor mimic was modelled after a typical L. t. elapsoides from northern North Carolina, an allopatric region where selection does not favour mimicry and where the mimetic phenotype has begun to break down (Pfennig et al. 2007). Although replicas of poor mimics differed significantly from the typical L. t. elapsoides found in deep sympatry (figure 2), we nevertheless used these replicas in both edge and deep sympatry because (as previously mentioned) previous research had shown that this phenotype could be regarded as a poor mimic.
Figure 2.
Comparison of two diagnostic traits for (a) the model (Micrurus) and (b) the mimic (Lampropeltis) from Florida (FL, deep sympatry) and North Carolina (NC, edge sympatry) and for (c) the good and (d) poor mimic replicas. Black squares, means. Box plots show 10th, 25th, 50th (median), 75th and 90th percentiles. Means with different superscripts are significantly different (p<0.05; Tukey–Kramer HSD).
We conducted experiments during April and May 2006 at 10 sites in North Carolina and 10 sites in Florida (figure 1). At each site, we arranged three different replicas (good mimic, poor mimic and control; the three replicas formed a ‘triplet’) 2 m apart in natural habitat. We then walked in a straight line for approximately 75 m and positioned another triplet, repeating the procedure until we had formed a 750 m long transect containing 10 triplets (30 replicas). This design gave predators a choice of attacking different phenotypes. Within each region (edge and deep sympatry), there were 10 transects and 300 replicas in total, 100 of each type. Each replica was used only once.
We collected replicas four weeks after their placement. Following collection, a person without knowledge of the replicas' location scored attacks by noting any impressions corresponding to a predator. We scored a replica as having been ‘attacked’ only if it contained teeth marks of a carnivore (e.g. black bear, bobcat, coyote, fox, raccoon; multiple attacks on the same replica were scored as one attack). There were no bird attacks. Impressions made by rodents or insects were excluded from the analysis, because these animals would not have represented a threat to a live snake. In addition, in one transect in Florida, replicas attacked by a feral pig were excluded.
For the analyses, the response measure was the proportion of good or poor mimic replicas attacked within each transect (equal to the number of attacks on good or poor mimics, respectively, divided by the total number of attacks on all replicas). Within each region, different transects were considered replicates (N=10 replicates per region). For the statistical analyses, we compared, separately for each region, the proportion of good and poor mimics attacked with the proportion expected if attacks were random with respect to phenotype (=0.33). We used a non-parametric, one-sample, Wilcoxon signed-rank test to compare the mean proportion of each type of replica attacked in each region.
3. Results
(a) Mimic–model resemblance in different areas
Mimics resembled their local model in edge sympatry but not in deep sympatry. In particular, an ANOVA of diagnostic phenotypes revealed that M. fulvius (the model) from deep sympatry did not differ significantly from M. fulvius in edge sympatry, nor did M. fulvius differ from L. t. elapsoides (the mimic) in edge sympatry (figure 2). However, L. t. elapsoides from deep sympatry differed significantly from the other three groups in proportion of dorsum red (ANOVA: F3,118=18.16, p<0.0001; Tukey–Kramer HSD: p<0.05) and black (ANOVA: F3,118=6.87, p=0.0003; Tukey–Kramer HSD: p<0.05; figure 2).
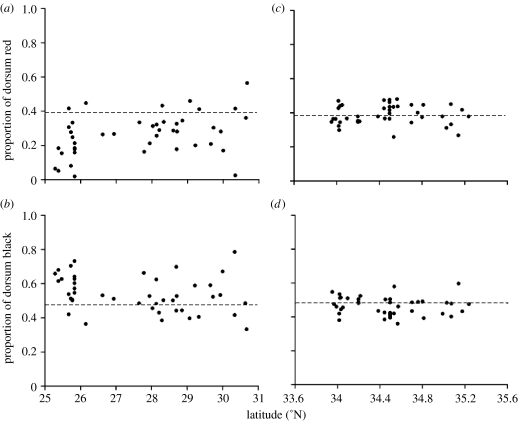
Mimics in deep sympatry were more phenotypically variable than were mimics in edge sympatry (figure 3). Compared with individuals from edge sympatry, L. t. elapsoides from deep sympatry were more variable in the proportion of both dorsum black (Levene's test, F=16.129, p=0.0001) and dorsum red (F=16.554, p=0.0001).
Figure 3.
Geographical variation in two diagnostic traits for L. t. elapsoides (mimics) from (a,b) Florida (deep sympatry) and (c,d) North Carolina (edge sympatry). Each dot represents the trait value for an individual L. t. elapsoides. The dashed horizontal line indicates the mean trait value for the local model.
Finally, a discriminant analysis based on the two pattern characteristics misclassified significantly more models and mimics in edge sympatry than in deep sympatry (34.93 versus 18.31%; two-tailed Fisher's exact test, p=0.0322). Thus, these data again suggest that mimics in edge sympatry were more phenotypically similar to their model than were mimics in deep sympatry.
(b) Relative abundances of models and mimics in different areas
Our estimates of each species' abundance in different parts of their range confirmed our a priori expectation that the model was relatively common in the centre of its range (deep sympatry), but relatively rare on the edge of its range (edge sympatry). In particular, 48 counties in Florida (72% of counties in the state) and 25 counties in North Carolina (25%) had at least one record for L. t. elapsoides. The ratios of M. fulvius (the model) to L. t. elapsoides (the mimic) for Florida (deep sympatry) and North Carolina (edge sympatry) were significantly different (Wilcoxon rank-sum test, S=548, p<0.0001). Models outnumbered mimics in Florida (mean±s.d.=2.61±4.69; median=1 model per mimic). By contrast, mimics outnumbered models in North Carolina (mean±s.d.=0.43±1.40; median=0 model per mimic).
(c) Predation pressure on good and poor mimics in different areas
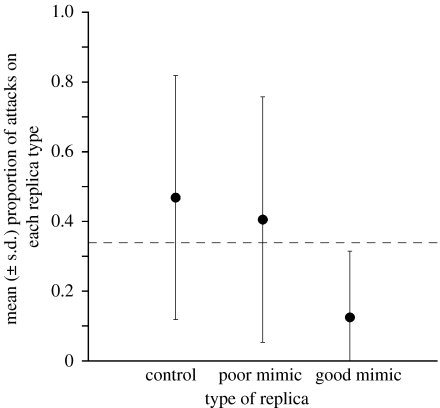
When free-ranging natural predators in edge sympatry were given a choice of attacking replicas of good or poor mimics, they avoided only good mimics. Of the 300 replicas placed in North Carolina, 21 were attacked (7%). Moreover, at least one replica was attacked in eight of the 10 transects. Predators attacked significantly fewer good mimics than the proportion expected (0.33) had they shown no colour pattern preference (figure 4; mean proportion of good mimic replicas attacked=0.125, N=8 transects, two-tailed Wilcoxon signed-rank test, p=0.039). Predators did not, however, avoid poor mimics (mean proportion attacked=0.406, N=8 transects, p=0.633) or controls (mean proportion attacked=0.469, N=8 transects, p=0.438).
Figure 4.
Mean (±s.d.) proportion of total attacks on each replica type in edge sympatry (for deep sympatry data, see text). The observed proportion of attacks on replicas of good mimics was significantly (p<0.05) less than 0.33 (dashed horizontal line), the value expected if predation attempts were random with respect to colour pattern.
By contrast, predators in deep sympatry attacked good and poor mimics equally frequently. Of the 300 replicas placed in Florida, only nine were attacked (3%). Indeed, significantly fewer replicas were attacked in Florida than in North Carolina (Fisher's exact test, two-tailed p=0.0377). Moreover, attacks were clustered in Florida: only four of the 10 transects had attacks. The combination of low attack rates and heterogeneity of attacks eliminated the power needed for statistical analysis. Nevertheless, of those replicas attacked, predators did not avoid either the poor mimic (mean proportion attacked=0.5, N=4 transects, p=0.789) or the good mimic (mean proportion attacked=0.5, N=4 transects, p=0.789). However, good and poor mimics were attacked equally frequently (mean proportion attacked=0.5 for both types of replicas).
4. Discussion
Batesian mimics often vary geographically in how faithfully they match their model (e.g. Pfennig et al. 2007). Here, we asked whether geographical variation in mimic–model resemblance reflects geographical variation in the relative abundance of the model. Batesian mimicry theory predicts that natural selection should favour mimics that bear even a crude likeness to their model as long as the model is relatively common (Ruxton et al. 2004), especially when the model is highly toxic (Getty 1985; Pough 1988; Brodie & Janzen 1995; Caley & Schluter 2003). In such situations, predators' risk of encountering the dangerous model is high. Consequently, predators should be under strong selection to avoid any species that remotely resembles the model. Thus, in areas where the model is common, even crude lookalikes may gain protection from predation. Poor mimics might be especially likely to evolve in such areas, if closely resembling the model is costly (e.g. producing and maintaining aposematic phenotypes may be physiologically taxing; Ruxton et al. 2004). By contrast, when the model is relatively rare, the likelihood of a predator encountering the model is low, and, consequently, selection on predators to avoid the model (and any lookalikes) will be relaxed (Huheey 1964; Oaten et al. 1975; Getty 1985). Thus, only mimics that closely resemble their model are likely to receive any protection from predation; poor mimics are unlikely to be protected.
In a snake-mimicry complex where the geographical range of the mimic extends beyond that of its model (figure 1), we found that coral-snake mimics, L. t. elapsoides, vary geographically (figure 2), such that mimic–model resemblance is higher on the edge of the model's range (edge sympatry), where models are relatively rare, and lower in the centre of the model's range (deep sympatry), where models are relatively common. Moreover, our field experiment revealed that predators on the edge of the model's range discriminated between good and poor mimics, tending to avoid only the former (figure 4). Thus, on the edge of their model's range, natural selection appears to favour only mimics that closely resemble their model.
Additional evidence that mimics on the edge of their model's range are under selection to closely resemble the models comes from our analysis of variation in colour patterns. Compared with mimics from edge sympatry, mimics from deep sympatry were significantly more variable in both phenotypic characteristics (figures 2 and 3). One possible explanation for this result is that in deep sympatry (but not in edge sympatry), between-population variation is greater than within-population variation. Yet, in deep sympatry, we observed no more variation between populations than within populations: the range of phenotypes among snakes from a given latitude (i.e. within a population) was often as great as that among snakes from different latitudes (i.e. between populations; figure 3a,b). Our findings therefore point to a relaxation of selection for close resemblance between the mimic and the model in deep sympatry.
In contrast to the situation in edge sympatry (figure 4), our attempts to measure predation pressure in deep sympatry met with limited success owing to low attack rates. These low attack rates may have reflected a general tendency for predators in deep sympatry to avoid all snakes (overall, there were significantly fewer snake replicas attacked in Florida than in North Carolina). Indeed, because venomous snakes are highly noxious models, and because many venomous snakes in Florida resemble both the control and mimic replicas, predators may have generalized the characteristics of these venomous snakes and avoided any object that resembled ‘a snake’ (e.g. Pough 1988). Despite the small sample size, however, we found that good and poor mimics were attacked equally frequently in deep sympatry, in accordance with our expectation.
It might be asserted that L. t. elapsoides differ in colour patterns in edge sympatry and deep sympatry, not owing to geographical variation in model abundance, but owing to geographical variation in some other (unmeasured) aspect of their environment. While we cannot completely rule out this possibility, it seems improbable for two reasons. First, mimics inhabit the same microhabitat as their models. Yet, models do not differ between regions (figure 2). Second, our field experiment provides a causal explanation for the observed geographical variation in mimic–model resemblance. In particular, our field experiment revealed that poor mimics would probably be selected against only in edge sympatry, thereby explaining the absence of poor mimics in this region.
More generally, our data allow us to reject five alternative hypotheses that can explain geographical variation in mimic–model resemblance. First, in systems where the range of the mimic exceeds that of the model, poor mimics may occur on the edge of the model's range if there is a ‘breakdown’ of mimicry in regions where mimics become more abundant (Brower & Brower 1962). This hypothesis cannot account for our results, however, because we found the opposite trend: the best mimics were on the edge of the model's range. Second, poor mimics may evolve in some geographical areas (but not in others) as a consequence of selection to resemble more than one model inhabiting separate areas (Edmunds 2000; Sherratt 2002; Darst & Cummings 2006). Such dual mimicry is improbable in our system: there are no dangerous species other than M. fulvius that L. t. elapsoides could conceivably mimic.
Third, seemingly poor mimics may evolve in some regions through antagonistic coevolution between the mimic and the model. Specifically, because models may suffer increased predation as mimics become more numerous (Oaten et al. 1975; Fisher 1999), selection may favour models that evolve away from their mimics. Selection for such ‘chase-away evolution’ (Gavrilets & Hastings 1988; Holmgren & Enquist 1999), which converts good mimics into poor mimics, may be stronger in some regions than in others. Contrary to the predictions of this hypothesis, however, we found that mimics in edge sympatry closely resemble models from both regions, whereas mimics in deep sympatry do not. Perhaps more critically, models in the two regions did not differ significantly in phenotype (figure 2), indicating that chase-away evolution has not occurred separately in the two regions.
Fourth, mimics may have been evolving with models longer in some regions than in others allowing more time for good mimicry to evolve in these regions. This hypothesis does not explain variation in mimic–model resemblance in our system, however. Genetic diversity is greater among L. t. elapsoides in Florida than in North Carolina (Harper 2006), indicating that L. t. elapsoides have probably been in sympatry at least as long in Florida as in North Carolina, and probably longer. Thus, there should have been sufficient time for good mimicry to evolve in Florida.
Finally, our analysis assumes that the cost for mistakenly attacking a model is identical in both regions. Yet, because spatial variation in venom toxicity is common in snakes (Chippaux et al. 1991; Alapegiron et al. 1994), it might be contended that mimics resemble models more closely in edge sympatry than in deep sympatry because models are more dangerous in deep sympatry. Variation in toxicity is unlikely to explain our results, however: coral snake venom is deadly, regardless of the population from which they derive (Roze 1996).
In sum, although poor mimics may persist in areas where their model is common, only the best mimics should occur in areas where their model is rare. Thus, counter-intuitively, the best mimics may occur on the edge of their model's range. Generally, variation in model abundance may critically determine the nature of selection on mimetic phenotypes and thereby explain why mimics often vary in resemblance to their model.
Acknowledgments
We thank K. Pfennig, J. Kingsolver, P. Marko, R. Martin, A. Rice, M. Servedio and three anonymous reviewers for their comments on the manuscript, J. Harper and M. Landstrom for their help in the laboratory and the field, and W. Van Devender for providing the photos used in figures 1 and 2. Thanks also to the Harvard Museum of Comparative Zoology, the National Museum of Natural History, the North Carolina Museum of Natural Sciences and the Yale Peabody Museum for access to their research collections. This research was funded by the University of North Carolina at Chapel Hill and the US National Science Foundation. All experimental procedures complied with applicable state and US federal regulations.
Supplementary Material
Information on snake specimens used in the analysis
References
- Alapegiron A, Lomonte B, Gustafsson B, Dasilva N.J, Thelestam M. Electrophoretic and immunochemical studies of Micrurus snake venoms. Toxicon. 1994;32:713–723. doi: 10.1016/0041-0101(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Bates H.W. Contributions to an insect fauna of the Amazon valley. Lepidoptera: Heliconidae. Trans. Linn. Soc. Lond. 1862;23:495–566. [Google Scholar]
- Brodie III, E. D. & Brodie Jr, E. D. 2004 Venomous snake mimicry. In The venomous reptiles of the western hemisphere, vol. II (eds J. A. Campbell & W. W. Lamar), pp. 617–633. Ithaca, NY: Comstock Publishing Associates.
- Brodie E.D, III, Janzen F.J. Experimental studies of coral snake mimicry: generalized avoidance of ringed snake patterns by free-ranging avian predators. Funct. Ecol. 1995;9:186–190. doi:10.2307/2390563 [Google Scholar]
- Brower L.P, Brower J.V.Z. The relative abundance of model and mimic butterflies in natural populations of the Battus philenor mimicry complex. Ecology. 1962;43:154–158. [Google Scholar]
- Caley M.J, Schluter D. Predators favour mimicry in a tropical reef fish. Proc. R. Soc. B. 2003;270:667–672. doi: 10.1098/rspb.2002.2263. doi:10.1098/rspb.2002.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux J.P, Williams V, White J. Snake venom variability: methods of study, results and interpretation. Toxicon. 1991;29:1279–1303. doi: 10.1016/0041-0101(91)90116-9. doi:10.1016/0041-0101(91)90116-9 [DOI] [PubMed] [Google Scholar]
- Darst C.R, Cummings M.E. Predator learning favours mimicry of a less-toxic model in poison frogs. Nature. 2006;440:208–211. doi: 10.1038/nature04297. doi:10.1038/nature04297 [DOI] [PubMed] [Google Scholar]
- Edmunds M. Longmans; London, UK: 1974. Defense in animals. A survey of anti-predator defenses. [Google Scholar]
- Edmunds M. Why are there good and poor mimics? Biol. J. Linn. Soc. 2000;70:459–466. doi:10.1006/bijl.1999.0425 [Google Scholar]
- Endler J.A. Interactions between predators and prey. In: Krebs J.R, Davies N.B, editors. Behavioural ecology. An evolutionary approach. Blackwell; London, UK: 1991. pp. 169–196. [Google Scholar]
- Fisher R.A. Oxford University Press; New York, NY: 1999. The genetical theory of natural selection. A complete variorum edition. [Google Scholar]
- Gavrilets S, Hastings A. Coevolutionary chase in two species systems with applications to mimicry. J. Theor. Biol. 1988;191:415–427. doi: 10.1006/jtbi.1997.0615. doi:10.1006/jtbi.1997.0615 [DOI] [PubMed] [Google Scholar]
- Getty T. Discriminability and the sigmoid functional response: how optimal foragers could stabilize model–mimic complexes. Am. Nat. 1985;125:239–256. doi:10.1086/284339 [Google Scholar]
- Greene H.W, McDiarmid R.Y. Coral snake mimicry: does it occur? Science. 1981;213:1207–1212. doi: 10.1126/science.213.4513.1207. doi:10.1126/science.213.4513.1207 [DOI] [PubMed] [Google Scholar]
- Harper, G. R. J. 2006 Evolution of a snake mimicry complex. PhD thesis, University of North Carolina at Chapel Hill.
- Holmgren N.M.A, Enquist M. Dynamics of mimicry evolution. Biol. J. Linn. Soc. 1999;66:145–158. doi:10.1006/bijl.1998.0269 [Google Scholar]
- Huheey J.E. Studies of warning coloration and mimicry. IV. A mathematical model of model–mimic frequencies. Ecology. 1964;45:185–188. doi:10.2307/1937125 [Google Scholar]
- Mallet J, Joron M. Evolution of diversity in warning color and mimicry: polymorphisms, shifting balance, and speciation. Annu. Rev. Ecol. Syst. 1999;30:201–233. doi:10.1146/annurev.ecolsys.30.1.201 [Google Scholar]
- Oaten A, Pearce C.E.M, Smyth M.E.B. Batesian mimicry and signal detection theory. Bull. Math. Biol. 1975;37:367–387. doi: 10.1007/BF02459520. [DOI] [PubMed] [Google Scholar]
- Palmer W.M, Braswell A.L. University of North Carolina Press; Chapel Hill, NC: 1995. Reptiles of North Carolina. [Google Scholar]
- Pfennig D.W, Harcombe W.R, Pfennig K.S. Frequency-dependent Batesian mimicry. Nature. 2001;410:323. doi: 10.1038/35066628. doi:10.1038/35066628 [DOI] [PubMed] [Google Scholar]
- Pfennig D.W, Harper G.R.J, Brumo A.F, Harcombe W.R, Pfennig K.S. Population differences in predation on Batesian mimics in allopatry with their model: selection against mimics is strongest when they are common. Behav. Ecol. Sociobiol. 2007;61:505–511. doi:10.1007/s00265-006-0278-x [Google Scholar]
- Pough F.H. Mimicry in vertebrates: are the rules different? Am. Nat. 1988;131:S67–S102. doi:10.1086/284767 [Google Scholar]
- Roze J.A. Krieger Publishing Company; Malabar, FL: 1996. Coral snakes of the Americas: biology, identification, and venoms. [Google Scholar]
- Ruxton G.D, Sherratt T.N, Speed M.P. Oxford University Press; Oxford, UK: 2004. Avoiding attack: the evolutionary ecology of crypsis, warning signals and mimicry. [Google Scholar]
- Sherratt T.N. The evolution of imperfect mimicry. Behav. Ecol. 2002;13:821–826. doi:10.1093/beheco/13.6.821 [Google Scholar]
- Wickler W. McGraw-Hill; New York, NY: 1968. Mimicry in plants and animals. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information on snake specimens used in the analysis