Abstract
Understanding the nature of species” boundaries is a fundamental question in evolutionary biology. The availability of genomes from several species of the genus Aspergillus allows us for the first time to examine the demarcation of fungal species at the whole-genome level. Here, we examine four case studies, two of which involve intraspecific comparisons, whereas the other two deal with interspecific genomic comparisons between closely related species. These four comparisons reveal significant variation in the nature of species boundaries across Aspergillus. For example, comparisons between A. fumigatus and Neosartorya fischeri (the teleomorph of A. fischerianus) and between A. oryzae and A. flavus suggest that measures of sequence similarity and species-specific genes are significantly higher for the A. fumigatus - N. fischeri pair. Importantly, the values obtained from the comparison between A. oryzae and A. flavus are remarkably similar to those obtained from an intra-specific comparison of A. fumigatus strains, giving support to the proposal that A. oryzae represents a distinct ecotype of A. flavus and not a distinct species. We argue that genomic data can aid Aspergillus taxonomy by serving as a source of novel and unprecedented amounts of comparative data, as a resource for the development of additional diagnostic tools, and finally as a knowledge database about the biological differences between strains and species.
Keywords: comparative genomics, genetic diversity, genome sequences, identifcation of species boundaries
INTRODUCTION
The diversity of life on earth has two fundamental properties: the diversity is discontinuous and the discontinuity is hierarchically organised (Dobzhansky 1971). Understanding the nature and extend of this discontinuity, especially for microbial communities, is one of the great challenges in evolutionary biology. For example, how can we best identify the discontinuity breaks that demarcate microbial species boundaries? Are those species boundaries similar in kind across clades of the tree of life? Addressing these questions is important because the means by which microbial species are identified can have important consequences in understanding species” biomedically-important characteristics, such as their pathogenicity (Kasuga et al. 1999; Balajee et al. 2006) and their geographical distribution (Taylor et al. 2006).
A growing body of research is focused on investigations of microbial species boundaries and the design of appropriate methodologies for delimitating them (Taylor et al. 2000; Taylor et al. 2006; Ward et al. 2007). Briefly, three species concepts are prevalent in the fungal literature; the morphological species concept (MSC - species are recognised based on morphological characters); the biological species concept (BSC - recognition is based on the establishment of reproductive isolation), and the phylogenetic species concept (PSC - species are diagnosed on the basis of shared ancestry). The MSC has been the concept by which fungal species are classified, largely due to the ease of its application. In contrast, the lack of a recognisable sexual stage in many fungal species, such as Aspergillus, makes the application of the BSC across fungi impractical. The problems associated with the application of the MSC and BSC in fungi, coupled with the emergence of theoretical and experimental molecular systematics in the last two decades, have resulted in the increasing popularity of the PSC.
The nature of species boundaries has been experimentally investigated under the PSC for two Aspergillus species of relevance to human health, A. fumigatus and A. flavus. A. fumigatus is considered by many to be the world”s most harmful mould (Pringle et al. 2005), being both a primary and opportunistic pathogen as well as a major allergen (Denning 1998; Latgé 1999). Most A. fumigatus isolates belong to one phylogenetic subspecies with a global distribution (Pringle et al. 2005). A. fumigatus is characterised by surprisingly low levels of genetic diversity, a characteristic that sharply contrasts with the high levels observed in some of its closest sexual relatives, such as Neosartorya fischeri (Rydholm et al. 2006). A. flavus is one of the primary producers of aflatoxin, one of the most potent carcinogenic substances known (Geiser et al. 1998). Species boundaries in the A. flavus clade are more complex (Geiser et al. 1998). Examination of Australian A. flavus strains suggests the presence of at least two cryptic species, and provides convincing evidence that A. oryzae is a domesticated ecotype of A. flavus (Geiser et al. 1998).
The recent availability of several genomes from the genus Aspergillus (Galagan et al. 2005; Machida et al. 2005; Nierman et al. 2005; Baker 2006; Payne et al. 2006; Pel et al. 2007; Rokas & Galagan 2007) allows us for the first time to examine the demarcation of fungal species at the whole-genome level. Below, we discuss four case studies involving taxa from the genus Aspergillus. Two case studies focus on genome-scale intraspecific differences whereas the other two deal with genomic comparisons between closely related but taxonomically distinct species (interspecific).
MATERIALS AND METHODS
Genome sequences. The genome sequences discussed in this paper are available in public databases under the accession numbers: A. clavatus NRRL 1: AAKD00000000 (GenBank); N. fischeri NRRL 181: AAKE03000000 (GenBank); A. fumigatus A1163: ABDB01000000 (GenBank); A. fumigatus Af293: AAHF01000000 (GenBank); A. niger CBS 513.88: AM270980-AM27998 (GenBank); A. niger ATCC 1015: http://genome.jgi-psf.org/Aspni1/Aspni1.home.html (JGI); A. flavus: AAIH01000000; A. oryzae: AP007150-AP007177 (DDJB).
Sequence identity at the genome level. The assemblies larger than 5 Kb were aligned using the MUMmer package (http://mummer.sourceforge.net/) (Delcher et al. 1999). Alignments longer than 100 Kb were used to determine average sequence identity to avoid highly repetitive and duplicated regions.
Sequence identity at the protein level. First orthologous groups in the Aspergillus genomes were identified by bidirectional best BLASTp clustering using a cut-off of 1e-05 and were analyzed using the Sybil software package (http://sybil.sourceforge.net/) and the Aspergillus Comparative database (http://www.tigr.org/sybil/asp). Average sequence identity between reciprocal best matches, which were considered putative orthologs.
Sequence identity at the gene level. The PASA pipeline, initially developed to align EST data onto genomic sequences (Haas et al. 2003), was used to coding sequences (CDSs) from one strain to assemblies of another. The alignments were generated by the gmap program implemented in PASA. Average sequence identity was calculated only for CDSs that produced high quality alignments (more then 95 % identity and 90 % coverage).
Strain- and isolate-specific genes. The PASA pipeline was also applied to identify three classes of genes: (i) orthologous genes, which aligned well and passed validation criteria (more then 95 % identity and 90 % coverage); (ii) variable genes, which aligned poorly (less then 95 % identity and 90 % coverage); (iii) strain- or isolate-specific genes, which did not produce any alignments.
FOUR CASE STUDIES FROM THE GENUS ASPERGILLUS
To compare the extent of diversity within and between species we used three computational approaches based on comparisons between: (i) genomic alignments, (ii) coding sequences (CDS) and genomic alignments, and (iii) protein alignments (Tables 1, 2). Interestingly, whereas the highest similarity values (and presumably the most accurate ones) between pairs of A. fumigatus and A. niger strains, and between A. oryzae and A. flavus were obtained from the genomic alignment comparisons, the highest similarity value in the A. fumigatus - N. fischeri genomic comparison was produced by the protein alignments (Table 2). The most plausible explanation for the observed discrepancies is that many gene models are likely mis-annotated in Aspergillus genomes (annotation was done by six different sequencing centres, Rokas & Galagan 2007).
Table 1.
Major characteristics and genetic diversity of the Aspergillus genomes.
| Species | A. fumigatus | A. niger | A. flavus | A. oryzae | A. fumigatus | N. fischeri | ||
|---|---|---|---|---|---|---|---|---|
| Strain/Isolate | Af293 | A1163 | ATCC 1015 | CBS 513.88 | NRLL 3357 | ATCC 42149 | Af293 | NRRL 181 |
| Genome | ||||||||
| Size (Kb) | 28,81 | 29,133 | 36,83 | 33,976 | 36,51 | 37,111 | 28,81 | 33,289 |
| Difference in size (Kb) | 323 | 2,854 | 601 | 4,479 | ||||
| No assemblies > 5 Kb | 19 | 20 | 49 | 19 | 20 | 26 | 19 | 74 |
| No assemblies > 100 Kb | 18 | 11 | 23 | 18 | 16 | 19 | 18 | 14 |
| Shared genome % | 98.8 % | 97.7 % | 89 % | 96.5 % | 97.4 % | 95.8 % | n/a | n/a |
| Unique genome % | 1.20 % | 2.30 % | 11 % | 3.5 % | 2.6 % | 4.2 % | n/a | n/a |
| Number of genes | ||||||||
| Total | 9630 | 9930 | 11200 | 14097 | 13515 | 12074 | 9630 | 10415 |
| Shared (blastp) | 9366 | 9366 | 10158 | 10158 | 10620 | 10620 | 8676 | 8676 |
| Aligned (gmap) | 9422 | 9610 | 10279 | 13242 | 13086 | 11507 | 8606 | 8649 |
| Divergent (80-95 %) | 60 | 102 | 396 | 386 | 151 | 236 | 365 | 340* |
| Unique (gmap) | 148 | 218 | 525 | 469 | 278 | 331 | 954 | 1766 |
| Unique (gmap) % | 1.5 % | 2.4 % | 4.7 % | 3.3 % | 2.1 % | 2.7 % | 9.9 %* | 16.7 %** |
Alignments with identity from 80 % to 90 % were considered divergent
Values were calculated for reciprocal best BLAST matches
Table 2.
Percent identity of the Aspergillus genomes at the genome, gene and protein level.
| Species | Within A. fumigatus Af293 vs. A1163 | Within A. niger ATCC 1015 vs. CBS 513.88 | A. flavus vs. A. oryzae | A. fumigatus Af293 vs. N. fischeri |
|---|---|---|---|---|
| Genome vs. genome | 99.8 % | 99.3 % | 99.5 % | 92.4 % |
| CDS vs. genome | 99.6 % | 99.1 % | 99.1 % | 94.3 % |
| Protein vs. protein | 99.5 % | 96.7 % | 98 % | 93.4 % |
Intraspecific case study I: A. niger CBS 513.88 versus A. niger ATCC 1015
Aspergillus niger is a workhorse organism in the field of biotechnology and industrial mycology. This organism is a source of and product host for industrial enzymes as well as the basis for the most efficient filamentous fungal fermentation known, the production of citric acid. In addition to its role in the enzyme and chemical industries, A. niger is an important experimental model organism for the study of protein secretion, organic acid production and others areas of basic fungal biology. Genome sequence data have been generated for three different A. niger strains, ATCC 1015, ATCC 9029 and CBS 513.88 (Baker 2006). Strains ATCC 1015 and ATCC 9029 are wild type strains, while strain CBS 513.88 has been through limited mutagenesis. A detailed analysis of A. niger strain CBS 513.88 has recently been published (Pel et al. 2007).
The US Department of Energy Joint Genome Institute (JGI) made available a draft sequence of A. niger strain ATCC 1015 in April 2006. Following the release of the draft genome, the JGI-Stanford finishing team began a genome improvement project. Their work has resulted in a nearly complete sequence consisting of 24 scaffolds. At the time of this writing, there are not any internal gaps within these scaffolds that encompass 34.9Mb, 1Mb more than the published nucleotide content of the strain CBS 513.88 genome. While the size difference between the genome can be accounted for by the sequencing approaches and a larger number of intra-scaffold gaps in the strain CBS 513.88 sequence, there are significant differences between strains which are not due to uncaptured genomic sequence. The nucleotide sequence identity between the two strains is 99.3 % (Table 2).
The sequenced A. niger strains also differ in their morphology when grown on solid media. A. niger ATCC 1015 is characterised by dark black conidiophores with long strings of connected spores. In contrast, A. niger CBS 513.88 is brown in colour, has conidiophores bearing only a limited number of spores and has a sectored colony morphology. The phenotypic behaviour of the two strains is also different; A. niger ATCC 1015 was originally described in 1917 as a citric acid producing strain (Currie 1917) while CBS 513.88 is descended from NRRL 3122 which is described as an induced mutant with high glucoamylase production (Van Lanen & Smith).
The behaviour, morphology and sequence differences as well as others between strains are evidence that organisms classified as A. niger can differ considerably at both the levels of growth morphology and genome sequence. While for strains ATCC 1015 and CBS 513.88, it may not be readily obvious whether the differences between them are due to speciation, induced mutagenesis or both, the availability of genome sequences for these closely related strains raises the important issue of how differences in genomic and phenotypic characteristics can be used to define species within the black aspergilli.
An international team of researchers associated with the JGI strain ATCC 1015 project as well as the DSM researchers who led the strain CBS 513.88 genome project are currently analyzing and comparing the two genome sequences. One goal of this comparison will be the characterisation of phenotypic, genome organisation and sequence dissimilarities between the two strains. We anticipate cataloguing differences in morphology and extrolite profiles, single nucleotide polymorphisms, insertions, deletions and chromosomal rearrangements. While a definitive answer may not result from the study, the question of whether strains ATCC 1015 and CBS 513.88 are the same species or represent an early speciation event will help to guide the analysis.
Intraspecific case study II: A. fumigatus Af293 versus A. fumigatus A1163
Aspergillus fumigatus is the main causative agent of invasive aspergilliosis, the most common fungal infection worldwide, and can also cause mycotoxicosis and severe allergic reactions in humans (Denning 1998). The most frequent source of A. fumigatus, a thermotolerant mesophile, is spent compost prepared for growing mushrooms. Aspergillosis, also known as “Mushroom Worker”s Lung Disease”, is a highly lethal invasive disease that affects people with compromised immune function and mushroom pickers working with mouldy compost, but is pathogenic to all humans in concentrated quantities. The therapeutic management options for invasive aspergillosis are limited due to high toxicity, low efficacy rates, and growing drug resistance and even with antifungal therapy the mortality rate is approximately 50 %. Despite medical and agricultural importance of this species, the biology of this human pathogen has only recently been actively investigated by more than a few groups.
Sequencing of the first isolate, strain Af293 (Nierman et al. 2005), has brought many unexpected discoveries including the possibility of a hidden sexual cycle and the presence of an impressive array of secondary metabolism clusters. The A. fumigatus clinical isolates have been shown to vary significantly in their pathogenicity (Paisley et al. 2005) and resistance to antifungals (Denning, unpubl. data). Therefore the availability of the second sequenced strain, A1163, which is a derivative of another clinical isolate CBS144-89/CEA10 (made available through Merck), will provide a unique opportunity to gain new insight into the nature of fungal pathogenicity. A1163 was obtained from CEA17, which is a uridine/uracil auxotroph pyrG mutant of CEA10 (d”Enfert 1996), via the ectopic insertion of the A. niger pyrG gene.
Preliminary comparative genomic analysis shows that the “core” A. fumigatus genome is very conserved as evidenced by the high identity between Af293 and A1163 orthologous sequences and the low number of isolate-specific genes (Tables 1, 2). In addition to this core genome, both isolates contain several small variable loci and large unique regions ranging in size from 10 to 400 Kb. The variable loci are orthologous and highly syntenic, but share very little sequence identity (37-90 % at the protein level). They contain several genes that encode encode NACHT, NB-ARC and Pfs domains predicted to function and non-self recognition and programmed cell death during hyphal fusion between genetically incompatible individuals (Fedorova et al. 2005).
While variable loci in Af293 and A1163 are randomly distributed along the A. fumigatus chromosomes, unique regions are located on all chromosomes and display a clear subtelomeric bias. In Af293, the five out of 9 largest genomic islands are located within 300 Kb from chromosome ends. The unique regions contain mostly isolate-specific genes and numerous repeat elements. Af293-specific islands 1 and 2 house two recent segmental duplications, containing several rapidly evolving genes and apparent pseudogenes.
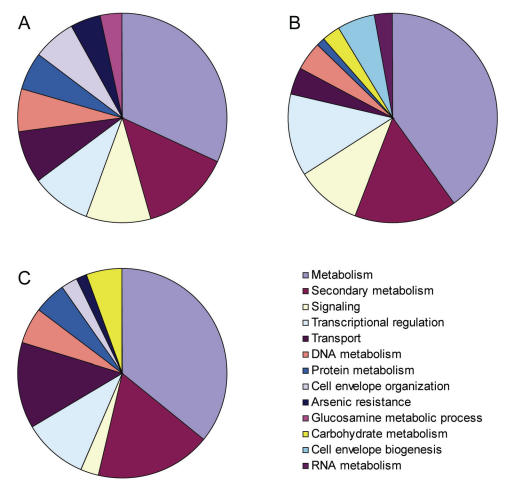
The Af293 and A1163 genomes have 208 and 320 unique genes, respectively, that aligned poorly or not at all to the other strain”s assemblies (Table 1). About 80 % of these genes are clustered together in isolate-specific genomic islands containing from 5 to 90 genes. Unexpectedly, more than 60 % of Af293-specific genes can be assigned a biological function such as cellular metabolism, secondary metabolism, signalling and transcriptional regulation (Fig. 1A). The high percentage can be explained in part by the fact that the list of specific genes includes one arsenic resistance gene cluster and two putative secondary metabolite biosynthesis gene clusters. A similar pattern is observed for the A1163-specific genes (Fig. 1B).
Fig. 1.
Functional characterisation of strain- and species-specific A. fumigatus genes. (A) Functional characterisation of 88 A. fumigatus strain Af293-specific genes absent in strain A1163; (B) Functional characterisation of 70 A. fumigatus strain A1163-specific genes absent in strain Af293; (C) Functional characterisation of 322 A. fumigatus strain Af293-specific genes absent in N. fischeri. Only the ten largest functional categories are shown and genes of unknown function are excluded.
Interspecific case study III: A. oryzae versus A. flavus
Aspergillus Section Flavi contains two economically important species. Accordingly, species within this section have received considerable attention, making it one of the best-studied groups of filamentous fungi. A. flavus is responsible for opening the modern field of mycotoxicology by its production of the potent carcinogen, aflatoxin. This toxin is responsible for millions of dollars in losses in the world and for significant health issues in developing countries, including human mortality. More recently, A. flavus has received notoriety as the second leading cause of aspergillosis in immunocompromised individuals. A. oryzae, on the other hand, has been widely used in Japanese fermentation industries to produce, sake (Japanese alcohol beverage), miso (soy bean paste) and shoyu (soy sauce) for more than a thousand years. A. oryzae secretes large quantities of diverse hydrolytic enzymes important for these fermentations. The extensive use of A. oryzae in food fermentation industries, and its long history as a safe food microbe, has lead to industrial applications of A. oryzae being listed as Generally Recognised as Safe (GRAS) by the Food and Drug Administration (FDA) in the United States of America (Tailor & Richardson 1979). A. oryzae is not considered to be either a plant on animal pathogen or to produce aflatoxin (Murakami et al. 1967). The safety of this organism is also supported by the World Health Organisation (FAO/WHO 1987).
While morphological differences can be used to distinguish between the two species (Klich & Pitt 1985), these differences can be subtle (Thom & Raper 1945; Hesseltine et al. 1970; Wicklow 1983) and controversy remains as whether these two fungi represent separate species or different ecotypes of the same species. It has been suggested that A. oryzae is a domesticated ecotype of A. flavus. While this is an appealing hypothesis, and genetic and molecular evidence support this hypothesis (Kurtzman et al. 1986; Klich & Mullaney 1987; Cruickshank & Pitt 1990; Geiser et al. 1998; Kumeda & Asao 2001; Montiel et al. 2003), there are little genomic data to support such a close relationship between the two species. The recently available whole genome sequences of A. flavus and A. oryzae present an opportunity to carefully examine two fungi with different ecologies but very similar morphology. We have initiated a comparative genomic analysis of these two species and our preliminary results are revealing some interesting similarities as well as differences. The sequence reads for A. flavus and A. oryzae have been deposited at National Center for Biotechnology Information (NCBI).
The results presented here represent an early peek into the similarities and differences between these two fungi based on a comparison of their genomes. As may have been predicted from earlier morphological and genetic studies, these two fungi appear similar at the genomic level. The estimated A. flavus genome size of 36.8 Mb is similar to that for A. oryzae (36.7 Mb). Interestingly, the genomes of these two fungi are larger than those of either A. nidulans or A. fumigatus and contain several lineage-specific genes. These lineage-specific genes are extra copies of particular genes specifically existing in the A. oryzae and A. flavus genomes as compared to A. nidulans and A. fumigatus (Machida et al. 2005). In general, these extra homologs are located in non-syntenic blocks, which are less conserved among species, rather than in syntenic blocks (Machida et al. 2005). The high conservation of genes from these two species in the non-syntenic blocks indicates that the two species are genetically close relatives. A comprehensive comparison between the two genomes revealed that these A. flavus/A. oryzae specific genes were acquired before domestication of A. oryzae but not during breeding strains for fermentation. The non-syntenic blocks appear to have been acquired by horizontal gene transfer.
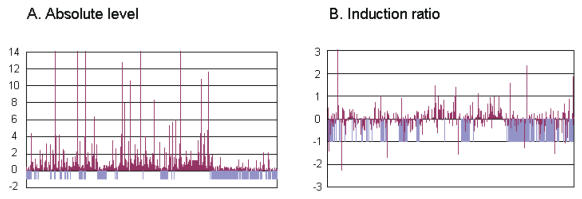
The genes on the non-syntenic blocks of A. oryzae generally have lower transcriptional levels than those on the syntenic blocks (Machida et al. 2005). This has been confirmed by the preliminary DNA microarray analysis (Fig. 2A). Further, the genes on the two blocks are regulated in a different manner as shown in Fig. 2B; most of the gene on syntenic and non-syntenic blocks were induced and repressed at heat shock, respectively. These results suggest that the genes on the two blocks may be regulated by the mechanisms specific to each block.
Fig. 2.
Gene expression analysis and of chromosome 6 of A. oryzae. Expression levels (A) and induction ratios (B) of all the genes in chromosome 6 are shown. The A. oryzae mycelia used in the oligonucleotide microarray analysis were incubated at 42 °C, using as a reference a mycelium incubated at 30 °C. Light blue bars indicate genomic regions (blocks) of A. oryzae that are absent in A. fumigatus and A. nidulans. The scale of the vertical axis is logarithmic.
Comparative analysis between the A. oryzae and A. flavus genomes revealed extremely high similarity in their nucleotide sequences and the amino acid sequences. Sequence analysis comparing genes predicted to code for proteins with over 100 amino acids revealed 306 genes unique to A. flavus and 332 genes unique to A. oryzae. Most of the genes unique to the species have no predicted function. Some, however, are involved in secondary metabolism. A. flavus has 34 polyketide synthases, two more than A. oryzae. While they have the same number of predicted non-ribosomal peptide synthases (24), each has two that are unique to the species. It remains unclear if these differences observed between the sequenced strains mirror that observed in population structure of these organisms. Natural variation is known to occur in populations of each fungus. For example, in each species there are strains that differ in the completeness of the aflatoxin biosynthetic cluster. As an example, the sequenced strain of A. oryzae contains the putative homolog of the entire gene cluster for aflatoxin biosynthesis, while some strains lack a part or the entire gene cluster. Overall, approximately half of the A. oryzae strains have nearly intact gene clusters. Strains of A. flavus are also known to differ in the completeness of their aflatoxin biosynthesis clusters (Chang et al. 2006).
There is also evidence of differences in gene expression between A. flavus and A. oryzae. While ESTs for all 25 of the aflatoxin pathway genes were found in A. flavus (Yu et al. 2004), no ESTs of these genes were detected in A. oryzae except for aflJ and norA (Akao et al. 2007). Mutations in some putative binding sites for the known transcription regulators may account for the lower expression levels of some genes (Tominaga et al. 2006). A truncation mutation of the pathway specific regulator aflR is known to prevent expression of the aflatoxin biosynthesis homologs in A. sojae. However, complementation of the mutated aflR with an intact copy did not restore the ability of the fungus to produce aflatoxin (Matsushima et al. 2001; Takahashi et al. 2002).
It is possible that other mutations may have been introduced into the A. oryzae genome preventing the expression of most secondary metabolism genes. Recently, detailed sequence analysis of the aflR genes from A. oryzae and A. flavus revealed phylogenetic differences of sequence between the two species (Chang et al. 2006). These results indicate that consideration of particular phenotypes, which includes productivity of some metabolites, should be a useful means to distinguish species efficiently and accurately.
The availability of both oligo-based and Affymetrix GeneChip DNA microarrays provide a new genomic tool to examine genome wide differences between A. flavus and A. oryzae. Such experiments are currently in progress. The observation that A. oryzae fails to produce many of the secondary metabolites that are present in A. flavus suggests that several genes will be differentially expressed between the two fungi. Affymetrix GeneChip DNA microarrays also provide a powerful tool to examine gene content and polymorphism among isolates of A. flavus and A. oryzae. For example, the high degree of DNA correspondence between A. flavus and A. oryzae allows the use of the A. flavus DNA microarrays for CGH (Comparative Genome Hybridisation) analysis across strains of both A. flavus and A. oryzae. Data obtained from a comparison between the two sequenced strains of A. flavus and A. oryzae show the power of this technique. The number of unique genes predicted for each strain by CGH was very similar to that predicted by sequence annotation. The difference between the two techniques was less that 10 % (+/- 3 %, a = 0.05). Currently three strains of each species are being compared for similarity.
In summary, genomic resources provide a new and powerful tool for distinguishing among closely related fungal species. An available genome sequence for both A. flavus and A. oryzae, and whole genome arrays for both fungi provide a means to carefully interrogate these two fungal species and learn more about their genetic relatedness, their evolution, and possibly the effect of domestication on changes in their genome.
Interspecific case study IV: A. fumigatus Af293 versus N. fischeri
Neosartorya fischeri (the teleomorph of A. fischerianus), a very close homothallic sexual relative to A. fumigatus, is found in soil, and its spores are found in many agricultural products. N. fischeri can cause keratitis and possibly pulmonary aspergillosis in transplant patients, but is an extremely rare invasive pathogen (Gerber et al. 1973; Chim et al. 1998). Its inadequacy as a pathogen is interesting in light of its close evolutionary relationship to A. fumigatus, and so comparison of the N. fischeri and A. fumigatus genomes should yield significant clues regarding A. fumigatus virulence and epidemiology. In addition, N. fischeri has a known sexual cycle, and elucidation of A. fumigatus sexual reproduction by comparative genomics would be of immense value to the Aspergillus research community. This would greatly advance A. fumigatus as an experimental system by facilitating genetic studies.
The J. Craig Venter Institute (JCVI) has sequenced the type culture of N. fischeri NRRL 181 by the whole genome random sequencing method (Nierman et al. 2005), using single spore subculture for genomic DNA extraction. Preliminary analysis of the N. fischeri genome, which was made available by the JCVI, has shown that it (32.6 Mb) is 10 % larger than the A. fumigatus Af293 genome (Table 1). There are currently 10 415 predicted protein-coding genes with a mean gene length of 1 466 bp. Comparisons to the genomes of A. fumigatus Af293 revealed 1 739 genes unique to N. fischeri, including several mycotoxin biosynthesis gene clusters. Other notable findings include a large number of transposable elements, which may have contributed to the genome size expansion observed in this species.
In contrast to the intraspecific comparison between A. fumigatus strains discussed above, the genomes of A. fumigatus and N. fischeri are much more divergent (Tables 1, 2). Only 35 % of the A. fumigatus-specific genes can be assigned any biological function (Fig. 1C). Comparison of the A. fumigatus - N. fischeri pair with the A. oryzae - A. flavus comparison, gives support to the proposal that A. oryzae represents a distinct ecotype of A. flavus and not a distinct species (Geiser et al. 1998). Firstly, the identity between orthologous sequences is ∼5-7 % lower for the A. fumigatus - N. fischeri pair than the A. oryzae - A. flavus pair (Table 2). Secondly, whereas more than 10 % of A. fumigatus and N. fischeri genes are species-specific, only < 3 % of genes are species-specific in the A. oryzae - A. flavus comparison, a value remarkably similar to the number of strain-specific genes exhibited by the intraspecific comparison in A. fumigatus.
CONCLUSIONS
These four comparisons highlight the potential usefulness of genomics for the accurate identification of species boundaries in the genus Aspergillus. Genomics can aid Aspergillus taxonomy by serving as a source of novel and unprecedented amounts of comparative data, as a resource for the development of additional diagnostic tools, and finally as a knowledge database about the biological differences between strains and species. It is unlikely that genomics, or any other discipline for that matter, will come up with a golden rule the application of which will solve all taxonomic problems in the clade. Rather, our hope is that genomics can provide an arsenal of data and molecular tools to aid taxonomists” quests for a more accurate delineation of species boundaries in the genus Aspergillus.
Acknowledgments
We thank Bo Jiang for providing the genomic sequence of the A. fumigatus isolate A1163.
References
- Akao T, Sano M, Yamada O, Akeno T, Fujii K, Goto K, Ohashi-Kunihiro S, Takase K, Yasukawa-Watanabe M, Yamaguchi K, Kurihara Y, Maruyama J, Juvvadi PR, Tanaka A, Hata Y, Koyama Y, Yamaguchi S, Kitamoto N, Gomi K, Abe K, Takeuchi M, Kobayashi T, Horiuchi H, Kitamoto K, Kashiwagi Y, Machida M, Akita O (2007). Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Research 14: 47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SE (2006). Aspergillus niger genomics: past, present and into the future. Medical Mycology 44 (Suppl 1): S17-S21. [DOI] [PubMed] [Google Scholar]
- Balajee SA, Nickle D, Varga J, Marr KA (2006). Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryotic Cell 5: 1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Ehrlich KC, Hua SST (2006). Cladal relatedness among Aspergillus oryzae isolates and Aspergillus flavus S and L morphotype isolates. International Journal of Food Microbiology 108: 172-177. [DOI] [PubMed] [Google Scholar]
- Chim CS, Ho PL, Yuen KY (1998). Simultaneous Aspergillus fischeri and Herpes simplex pneumonia in a patient with multiple myeloma. Scandinavian Journal of Infectious Diseases 30: 190-191. [DOI] [PubMed] [Google Scholar]
- Cruickshank PJ, Pitt JI (1990). Isoenzyme patterns in Aspergillus flavus and closely related species. In: Modern concepts in Penicillium and Aspergillus classification. Samson RA, Pitt JI, eds. New York: Plenum Press: 259-267.
- Currie JN (1917). The citric acid fermentation of Aspergillus niger. Journal of Biological Chemistry 31: 15-37. [Google Scholar]
- d”Enfert C (1996). Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5” decarboxylase gene, pyrG, as a unique transformation marker. Current Genetics 30: 76-82. [DOI] [PubMed] [Google Scholar]
- Denning DW (1998). Invasive aspergillosis. Clinical Infectious Diseases 26: 781-803. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T (1971). Genetics of the evolutionary process. New York: Columbia University Press.
- FAO/WHO (1987). Committee on Food Additives. 31. World Health Organization Technical Report Series, Geneva. [PubMed]
- Fedorova ND, Badger JH, Robson GD, Wortman JR, Nierman WC (2005). Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics 6: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D”Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature) 438: 1105-1115. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Pitt JI, Taylor JW (1998). Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proceedings of the National Academy of Sciences of the United States of America 95: 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Chomicki J, Brandsberg JW, Jones R, Hammerman KJ (1973). Pulmonary aspergillosis caused by Aspergillus fischeri var. spinosus: report of a case and value of serologic studies. American Journal of Clinical Pathology 60: 861-866. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, Salzberg SL, White O (2003). Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Research 31: 5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesseltine CW, Sorenson WG, Smith M (1970). Taxonomic studies of aflatoxin-producing strains in Aspergillus flavus group. Mycologia 62: 123-132. [PubMed] [Google Scholar]
- Kasuga T, Taylor JW, White TJ (1999). Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. Journal of Clinical Microbiology 37: 653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klich MA, Mullaney EJ (1987). DNA restriction enzyme fragment polymorphism as a tool for rapid differentiation of Aspergillus flavus from Aspergillus oryzae. Experimental Mycology 11: 170-175. [Google Scholar]
- Klich MA, Pitt JI (1985). The theory and practice of distinguishing species of the Aspergillus flavus group. In: Advances in Penicillium and Aspergillus systematics. Samson RA, Pitt JI, eds. New York: Plenum Press: 211-220.
- Kumeda Y, Asao T (2001). Heteroduplex panel analysis, a novel method for genetic identification of Aspergillus Section Flavi strains. Applied and Environmental Microbiology 67: 4084-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Smiley MJ, Robnett CJ, Wicklow DT (1986). DNA relatedness among wild and domesticated species in the Aspergillus flavus group. Mycologia 78: 955-959. [Google Scholar]
- Lanen JM van, Smith MB (1968). Process of producing glucoamylase and an alcohol product, US Patent 3418211.
- Latge JP (1999). Aspergillus fumigatus and aspergillosis. Clinical Microbiology Reviews 12: 310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CZ, Liou GY, Yuan GF (2006). Comparison of the aflR gene sequences of strains in Aspergillus section Flavi. Microbiology 152: 161-170. [DOI] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005). Genome sequencing and analysis of Aspergillus oryzae. Nature 438: 1157-1161. [DOI] [PubMed] [Google Scholar]
- Matsushima K, Chang PK, Yu J, Abe K, Bhatnagar D, Cleveland TE (2001). Pre-termination in aflR of Aspergillus sojae inhibits aflatoxin biosynthesis. Applied Microbiology and Biotechnology 55: 585-589. [DOI] [PubMed] [Google Scholar]
- Montiel D, Dickinson MJ, Lee HA, Dyer PS, Jeenes DJ, Roberts IN, James S, Fuller LJ, Matsuchima K, Archer DB (2003). Genetic differentiation of the Aspergillus section Flavi complex using AFLP fingerprints. Mycological Research 107: 1427-1434. [DOI] [PubMed] [Google Scholar]
- Murakami H, Takase S, Ishii T (1967). Non-productivity of aflatoxin by Japanese industrial strains of Aspergillus. I. Production of fluorescent substances in agar slant and shaking cultures. Journal of General and Applied Microbiology 13: 323-334. [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latge JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O”Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Cordoba S, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438: 1151-1156. [DOI] [PubMed] [Google Scholar]
- Paisley D, Robson GD, Denning DW (2005). Correlation between in vitro growth rate and in vivo virulence in Aspergillus fumigatus. Medical Mycology 43: 397-401. [DOI] [PubMed] [Google Scholar]
- Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, Bhatnagar D, Cleveland TE, Machida M, Yu J (2006). Whole genome comparison of Aspergillus flavus and A. oryzae. Medical Mycology 44 (Suppl 1): 9-11. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d”Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology 25: 221-231. [DOI] [PubMed] [Google Scholar]
- Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW (2005). Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59: 1886-1899. [PubMed] [Google Scholar]
- Rokas A, Galagan JE (2007). The Aspergillus nidulans genome and a comparative analysis of genome evolution in Aspergillus. In: The aspergilli: genomics, medical applications, biotechnology, and research methods. Osmani SA, Goldman GH, eds. New York: CRC Press (in press).
- Rydholm C, Szakacs G, Lutzoni F (2006). Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryotic Cell 5: 650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor MJ, Richardson T (1979). Applications of microbial enzymes in food systems and in biotechnology. Advances in Applied Microbiology 25: 7-35. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Chang PK, Matsushima K, Yu JJ, Abe K, Bhatnagar D, Cleveland TE, Koyama Y (2002). Nonfunctionality of Aspergillus sojae aflR in a strain of Aspergillus parasiticus with a disrupted aflR gene. Applied and Environmental Microbiology 68: 3737-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21-32. [DOI] [PubMed] [Google Scholar]
- Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D (2006). Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 361: 1947-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom C, Raper KB (1945). A manual of the aspergilli. William and Wilkins, Baltimore.
- Tominaga M, Lee YH, Hayashi R, Suzuki Y, Yamada O, Sakamoto K, Gotoh K, Akita O (2006). Molecular analysis of an inactive aflatoxin biosynthesis gene cluster in Aspergillus oryzae RIB strains. Applied and Environmental Microbiology 72: 484-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DM, Cohan FM, Bhaya D, Heidelberg JF, Kuhl M, Grossman A (2007). Genomics, environmental genomics and the issue of microbial species. Heredity (in press). [DOI] [PubMed]
- Wicklow DT (1983). Taxonomic features and ecological significance of sclerotia. Opelika, AL: Craftmaster Printers.
- Yu JJ, Whitelaw CA, Nierman WC, Bhatnagar D, Cleveland TE (2004). Aspergillus flavus expressed sequence tags for identification of genes with putative roles in aflatoxin contamination of crops. FEMS Microbiology Letters 237: 333-340. [DOI] [PubMed] [Google Scholar]