Abstract
Gene flow within populations can occur by sexual and/or parasexual means. Analyses of experimental and in silico work are presented relevant to possible gene flow within the aspergilli. First, the discovery of mating-type (MAT) genes within certain species of Aspergillus is described. The implications for self-fertility, sexuality in supposedly asexual species and possible uses as phylogenetic markers are discussed. Second, the results of data mining for heterokaryon incompatibility (het) and programmed cell death (PCD) related genes in the genomes of two heterokaryon incompatible isolates of the asexual species Aspergillus niger are reported. Het-genes regulate the formation of anastomoses and heterokaryons, may protect resources and prevent the spread of infectious genetic elements. Depending on the het locus involved, hetero-allelism is not tolerated and fusion of genetically different individuals leads to growth inhibition or cell death. The high natural level of heterokaryon incompatibility in A. niger blocks parasexual analysis of the het-genes involved, but in silico experiments in the sequenced genomes allow us to identify putative het-genes. Homologous sequences to known het- and PCD-genes were compared between different sexual and asexual species including different Aspergillus species, Sordariales and the yeast Saccharomyces cerevisiae. Both het- and PCD-genes were well conserved in A. niger. However some point mutations and other small differences between the het-genes in the two A. niger isolates examined may hint to functions in heterokaryon incompatibility reactions.
Keywords: apoptosis, ascomycete, Aspergillus fumigatus, Aspergillus nidulans, Aspergillus niger, heterokaryon incompatibility, MAT, mating type, Neurospora crassa, Podospora anserina, Saccharomyces cerevisiae, self/non-self recognition
INTRODUCTION
Self and non-self recognition is a common requirement for all living organisms. Most taxa have developed specific systems to identify self and non-self. Fungi exhibit two types of compatibility systems based on self and non-self recognition, namely sexual compatibility and somatic or heterothallic (in)compatibility (Dyer et al. 1992, Leslie 1993). Such systems are of great significance as they govern the degree to which gene flow can occur between members of a species, with consequences for evolution and recognition of species (Taylor et al. 1999a). Maintaining gene flow within a species is advantageous as this may lead to increased genotypic variation allowing adaptation to changing environments (Milgroom 1996). However, unrestricted gene flow may lead to resource plundering by other genotypes (Debets & Griffiths 1998).
Many filamentous fungi are able to reproduce both sexually and asexually, depending on environmental conditions (Dyer & Paoletti 2005). In a nutritionally rich environment fungi generally produce mitotic spores (the anamorphic state), but when conditions become unfavourable for vegetative growth, they may initiate sexual reproduction (teleomorphic state). There are two main types of sexual breeding system evident in fungi: homothallism and heterothallism (Dyer et al. 1992). Homothallic strains are self-fertile, though may also be capable of outcrossing, whereas heterothallic strains are self-incompatible and require the presence of a compatible mating partner for the sexual cycle to occur. In addition there are numerous species which are apparently restricted to propagation by asexual means with no known sexual cycle (Taylor et al. 1999). Sexual compatibility in heterothallic fungi is governed by so called “mating-type” (MAT) genes, with two mating types MAT1-1 and MAT1-2 present in heterothallic filamentous ascomycetes (Turgeon & Yoder 2000). These differ according to DNA present at a single MAT locus, with highly divergent DNA sequence (termed an “idiomorph”) present in isolates of opposite mating type. By convention MAT1-1 isolates contain an idiomorph including a MAT gene encoding a protein with an alpha-box domain. In contrast, MAT1-2 isolates contain an idiomorph including a MAT gene encoding a regulatory protein with a specific high mobility group (HMG) type DNA-binding domain (Turgeon & Yoder 2000). Mating-type genes regulate initial stages of the mating process such as pheromone signalling and plasmogamy leading to the production of ascogenous hyphae in ascomycetes, and may also have roles in later internuclear recognition (Lengeler et al. 2000, Coppin et al. 2005, Debuchy & Turgeon 2006). Intriguingly, MAT genes have also been shown to be required for sexual reproduction in homothallic ascomycetes (Paoletti et al. 2007). Mating-type genes also have other functions in some species, for example affecting vegetative incompatibility in Neurospora crassa (Glass et al. 1988).
The lack of a known sexual cycle does not mean automatically the lack of recombination. Indeed, there is evidence of genotypic diversity and gene flow in some supposedly asexual fungal species (Geiser et al. 1998, Paoletti et al. 2005). One possible explanation for such observations is that during the vegetative state of its life a fungus is able to undergo hyphal fusion, karyogamy and mitotic recombination, in the so-called “parasexual cycle” (Pontecorvo 1956). Finding an appropriate partner for mitotic recombination is a crucial aspect of the parasexual cycle, similarly to the sexual mating process. A complex set of heterokaryon-incompatibility genes and associated network of cellular machinery are responsible for the acceptance or rejection of partners in parasexuality (Glass et al. 2000, Saupe 2000).
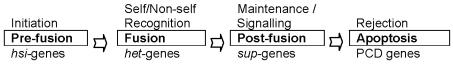
There are four steps in this parasexual cycle, which can be distinguished by the gene sets governing the steps (Fig. 1) (Leslie and Zeller 1996). The initial step, named pre-fusion, is controlled by genes involved in pheromone production and receptors, and in heterokaryon self-incompatibility (hsi genes). The fusion step, in which the interhyphal anastomoses formed and results in a heteroplasmon, is governed by self/non-self recognition genes, like heterokaryon incompatibility (het) (Glass and Kuldau 1992) or vegetative incompatibility (vic) genes (Leslie 1993). Effects of het-genes can be influenced by modifier (mod) genes. After fusion the biochemical pathways leading to non-self recognition and cell death may be influenced by genes including suppressor (sup) genes, which can modify the signal. In the final step programmed cell death (PCD) genes initiate processes leading to apoptosis.
Fig. 1.
Gene families involved in steps of vegetative incompatibility.
The precise function(s) of heterokaron incompatibility in fungi is not yet clear. There are three main theories to explain the existence of these vegetative self/non-self recognition systems in fungi. One theory, the so called allorecognition hypothesis, proposes that it is a bona fide reaction between genetically different individuals, which may limit the spread of harmful cytoplasmic or nuclear elements (Caten 1972) or prevent resource plundering (Debets and Griffiths 1998). In these examples the role of het-genes is to preserve genetic individuality. The second, alternative, theory suggests that het-genes simply arose accidentally during evolution. The existence of genes with dual function, like the mat-a/mat-A loci in N. crassa (Saupe 2000), supports the second theory. These genes sometimes behave as het-genes, whilst fulfilling other cellular functions at other times. Finally it has been suggested that vegetative incompatibility may promote the initiation of sexual reproduction in some species as a result of non-self recognition (Dyer et al. 1992).
There have been a limited number of studies attempting to test these hypotheses. For example, if het-genes are present to prevent formation of heterokaryons between genetically different individuals [i.e. to protect a local population from “invaders”] then an evolutionary trend towards generation of new alleles might be predicted, to avoid “pollution” of the isolates belonging to the same incompatibility groups (Saupe et al. 1994). However, population genetic modelling suggests that selection favouring more than a few alleles is expected to be extremely weak (Nauta and Hoekstra 1993). In contrast to positive selection in non-allelic systems, certain het loci are under maintaining/balancing selection. A well studied example is the N. crassa het-c with three known alleles. These alleles show trans-species polymorphism and balancing selection (Wu et al. 1998).
In this article, we describe experimental work assessing the occurrence of genes relating to mating and heterokaryon incompatibility processes in the aspergilli, and their possible roles in gene flow within species.
Occurrence of mating-type genes in the aspergilli
Although mating-type genes were first reported from filamentous fungi by Glass et al. in 1988, it has proved difficult to clone such genes by traditional molecular biology methods due to high sequence divergence between major ascomycete lineages. Indeed, the only regions conserved across divergent taxa are the alpha-domain in MAT1-1 family proteins, and the HMG-domain in MAT1-2 proteins, and even these show relatively poor sequence conservation [see examples in Debuchy & Turgeon (2006)]. This prevented initial attempts to identify MAT genes from Aspergillus species (e.g. Arie et al. 1997). However, two major developments in recent years have allowed mating-type genes to be identified from the aspergilli,. The first was the use of a degenerate-PCR approach, using primers designed to anneal to conserved sequence present at the HMG MAT1-2 locus, to amplify MAT1-2 sequence from species of Aspergillus (Singh et al. 1999, Paoletti et al. 2005, 2007). The second was the public release of genome sequence data from certain species of Aspergillus, meaning that whole genomes could be screened by bioinformatic approaches (e.g. BLAST searches) to determine the presence of MAT genes (Archer & Dyer 2004, Galagan et al. 2005, Pel et al. 2007). The use of these two techniques has lead to a series of major discoveries concerning sexuality and gene flow in Aspergillus species, with implications for species identity, as will be described.
Mating-type genes in sexual aspergilli
The first MAT gene to be reported from the aspergilli was an HMG-domain family gene from the homothallic sexual species A. nidulans (teleomorph Emericella nidulans), which was identified by a degenerate PCR approach (Dyer 2002; Paoletti et al. 2007). Subsequent BLAST searching of the A. nidulans genome revealed the presence of an alpha-domain family gene within the same genome (Dyer et al. 2003). The genes were found to be on different chromosomes and were named MAT1 (alpha-domain on chromosome 6) and MAT2 (HMG-domain on chromosome 3) to recognise the fact that they occupied different genetic loci (Turgeon & Yoder 2000, Paoletti et al. 2007). This compares to other homothallic ascomycetes in which alpha- and HMG-domain MAT genes, if both present, have most often been found linked at the same single MAT locus (DeBuchy & Turgeon 2006). It was suggested by Galagan et al. (2005) that the arrangement of MAT loci in A. nidulans may have arisen as a result of a chromosome break and translocation event(s) from an ancestral single Aspergillus MAT locus. The role of the MAT genes in A. nidulans was subsequently investigated by Paoletti et al. (2007) who used gene deletion/replacement, overexpression and RNA interference approaches to demonstrate that both MAT1 and MAT2 genes are required for completion of the sexual cycle in A. nidulans. ΔMAT1 and ΔMAT2 gene deletant mutants were unable to form ascospores, although sterile cleistothecia were produced. This was a significant discovery, as it showed that MAT genes, normally associated with control of sexual compatibility in heterothallic species, are also required for sexual development in this model homothallic species. The only other MAT genes to be identified from a known sexual species of Aspergillus has been the identification of both alpha- and HMG-domain genes again together within the genome of the homothallic species Neosartorya fischeri (Rydholm et al. 2007). The genes were present at unlinked loci, and where therefore termed MAT1 and MAT2 respectively. However, the arrangement and synteny of MAT loci differed from that seen in A. nidulans, and it was suggested that homothallism in this species had arisen by a segmental chromosome duplication and translocation event (Rydholm et al. 2007). There have so far been no reports of MAT gene isolation from heterothallic Aspergillus species, reflecting the fact that the vast majority of sexual aspergilli are homothallic, with only four heterothallic species so far identified (Dyer 2007).
Mating-type genes in asexual aspergilli
Ironically, all the other MAT genes reported from the aspergilli have come from asexual species, which supposedly lack a sexual cycle. The detection of MAT genes in asexual species might appear surprising, but is thought to reflect the fact that asexual species have evolved from sexual ancestors by loss of sexuality (Geiser et al. 1996), therefore “sex-related” genes may be retained in the genome though prone to mutation and loss by genetic drift. Both genomic and experimental work was first used to identify apparently functional (i.e. lacking any frameshift or stop codon mutation) MAT genes from the opportunistic pathogen A. fumigatus (Pöggeler 2002, Varga 2003, Dyer and Paoletti 2005, Paoletti et al. 2005). The species was later shown to contain a complement of other genes required for sexual reproduction (Galagan et al. 2005; Nierman et al. 2005). Intriguingly a survey of 290 worldwide clinical and environmental isolates of A. fumigatus, using a newly-developed multiplex mating-type PCR diagnostic, revealed that all isolates contained either a MAT1-1 alpha-domain gene or a MAT1-2 HMG-domain gene and that MAT1-1 and MAT1-2 genotypes were present in a near 1:1 ratio (Paoletti et al. 2005). This resembled the pattern that might be expected to be seen in a heterothallic sexual species. Furthermore, Paoletti et al. (2005) showed that mating-type, pheromone-precursor and pheromone-receptor genes were expressed, again consistent with heterothallism. In parallel work using either degenerate PCR or genomic screening, MAT genes have also been identified from other “asexual” species including A. oryzae (Galagan et al. 2005), A. niger (Pel et al. 2007), A. clavatus, A. sojae, A. flavus and A. parasiticus (Dyer 2007). Significantly isolates of both MAT1-1 and MAT1-2 genotype have been detected in near equal number for many of these species, the only exception being from the A. niger “black aspergilli” group which shows a strong bias towards isolates containing an alpha-domain MAT1-1 family gene (Dyer 2007; Varga, Kocsubé, Pál, Debets, Eyres, Baker, Samson & Dyer unpubl. data). A. niger was also shown to contain a complement of genes required for sexual reproduction, although possible mutation was evident in at least one gene (Pel et al. 2007).
Taken as a whole, these results are highly significant because they suggest that certain “asexual” aspergilli might retain a latent ability for sexual reproduction. Indeed, there is evidence from population genetic studies of A. flavus (Geiser et al. 1998) and A. fumigatus (Dyer & Paoletti 2005, Paoletti et al. 2005, Pringle et al. 2005) of high genetic diversity and genetic recombination within populations of these species. It is possible that this is a result of meiotic exchanges in the near past. However, there is also the tantalising possibility that these, and perhaps other aspergilli categorised as “asexual”, might posses an extant cryptic sexual state which has yet to be identified. This perhaps might be a result of a slow decline in sexual fertility in the majority of isolates due to selection for asexuality, but the retention of fertile isolates as a subset of wild populations (Dyer & Paoletti 2005).
Mating-type genes and species identity
Because MAT genes evolve at a relatively fast rate it has been suggested that they might be particularly suited to phylogenetic analysis to resolve species identity and inter-species taxonomic relationships (Turgeon 1998). At present there is insufficient MAT sequence data available to allow meaningful phylogenies to be constructed in the aspergilli. A further obstacle is that it would be necessary to obtain homologous MAT sequence from all species under examination (e.g. MAT1-2 gene sequence from all species) and for some species the necessary MAT1-1 or MAT1-2 data may be lacking as the majority of isolates might be of the opposite mating type. However, there remains the prospect that accumulating MAT data may provide a means to resolve closely related aspergilli taxa, complementing the use of other genes and markers presently used in phylogenetic studies.
Genetic control of heterokaryon incompatibility in ascomycete fungi
For filamentous fungi the establishment of hyphal anastomoses, both within and between individuals of the same species, is considered to be of high importance. A limitation to intermycelial fusions is heterokaryon incompatibility which is widespread among fungi and prevents the coexistence of genetically dissimilar nuclei within a common cytoplasm. The adaptive significance of heterokaryon incompatibility is unclear but it may serve to limit the spread of detrimental cytoplasmic or nuclear elements (Caten 1972) or prevent resource plundering (Debets & Griffiths 1998). A het locus can be any locus, at which heteroallelism is not tolerated in a heterokaryon (Saupe 2000). When heterokaryon-incompatible strains fuse, the resulting heterokaryotic hyphae are rapidly compartmentalised and destroyed (often with surrounding cells) or seriously inhibited in their growth, depending on the involved het locus. Heterokaryotic cells are often destroyed within 30 min after hyphal fusion. The process of destruction of hyphal compartments shows similarity at the microscopic level in different fungi, and some steps have common features even with multicellular metazoan programmed cell death (PCD) (Glass & Kaneko 2003).
Since heterokaryon incompatible strains can be sexually compatible, the number of het-genes that segregate from a cross can be deduced from the progeny. Genetic analysis of heterokaryon incompatibility was performed in a few sexual fungi. The number of identified het-genes varied with species. There are at least eight and maximum 18 het loci in A. nidulans (Anwar et al. 1993), six in Cryphonectria parasitica (Cortesi & Milgroom 1998), at least 11 in N. crassa (Glass et al. 2000) and nine in P. anserina (Saupe 2000). The majority of the incompatibility reactions is regulated by allelic systems, where two (e.g. mat-a/mat-A in N. crassa, Saupe 2000) or more alleles (e.g. het-c Groveland, Oakridge and Panama alleles in N. crassa, Sarkar et al. 2002) of the same locus interact. In other cases, two distinct loci trigger non-allelic incompatibility (e.g. het-c/het-e in P. anserina, Saupe 2000). In non-allelic systems, incompatible alleles can be present in the same haploid nucleus in the progeny and thus vegetative incompatibily may occur also in homokaryotic cells, like in the het-r/het-v incompatibility in P. anserina. Such homokaryotic strains can be obtained for each non-allelic system, and are named self-incompatible (SI) strains (Bourges et al. 1998).
Identification of heterokaryon (in)compatibility related genes in the aspergilli
Unlike ascomycete species which have a sexual and usually also an asexual reproduction cycle, for many asexual aspergilli the only way to achieve (mitotic) recombination is via the parasexual cycle. There is evidence from studies of field isolates of species including A. niger, A. terreus, A. versicolor, A. glaucus and E. nidulans that heterokaryon incompatibility is widespread in the aspergilli (Croft & Jinks 1977). Mitotic recombination has been used for genetic analysis of mutants in an isogenic background of A. niger and the construction of a genetic map (Debets et al. 1993). But, natural isolates of black aspergilli are highly incompatible with each other, efficiently blocking virus transfer as well as the formation of heterokaryons (van Diepeningen et al. 1997). As a result, mitotic recombination between genetically dissimilar isolates is also blocked, so genetic analysis cannot reveal the genetic basis of heterokaryon incompatibility in A. niger.
In this study we present the results of data mining in the genomes of two heterokaryon incompatible isolates of the asexual A. niger species. We compared incompatibility and cell death related proteins of the two A. niger isolates with each other and similar proteins of related sexual (A. nidulans) and asexual aspergilli (A. fumigatus, A. oryzae, A. terreus), two members of the Sordariales (P. anserina, N. crassa) and the yeast Saccharomyces cerevisiae. Our analyses identified the major putative het-genes in the genome of A. niger and related aspergilli. These findings can be used for further functional analysis of candidate het-genes.
In silico comparison of yeasts and filamentous fungi
A list of known genes involved in either programmed cell death from S. cerevisiae or involved in heterokayon incompatibility and/or programmed cell death in N. crassa or P. anserina was constructed based on the literature (Table 1). Protein forms of genes were blasted against genomic databases: A. fumigatus preliminary sequence data was obtained from The Institute for Genomic Research website. The A. nidulans, N. crassa (release 7) and A. terreus sequence data were from the Aspergillus nidulans, Neurospora crassa and Aspergillus terreus Sequencing Projects, Broad Institute of MIT and Harvard. The A. oryzae sequences were available on the server of National Institute of Technology and Evaluation (NITE). The P. anserina genome was published by the Institut de Génétique et Microbiologie (Université de Paris-Sud XI/CNRS). A. niger ATCC1015 sequence data were produced by the US Department of Energy Joint Genome Institute and A. niger CBS513.88 sequence data by the DSM Research BV (Table 2).
Table 1.
Genes used in this study involved in heterokaryon incompatibility in N. crassa and P. anserina and in Programmed Cell Death (PCD) in S. cerevisiae. All genes are given with their presumed function and references. ID numbers for N. crassa and S. cerevisiae proteins refer to the numbers given in their respective sequencing projects, the ID numbers for the P. anserina proteins were taken from GenBank. The table is an expanded v. of the table in Glass and Kaneko (2003).
| Species | Class | Gene | Function | References | |
|---|---|---|---|---|---|
| N. crassa | |||||
| Heterokaryon incompatibility genes | |||||
| het-6OR (NCU03533.2) | allelic het-gene, TOL/HET-6/HET-E domain | Saupe 2000 | |||
| het-c (NCU3125.2) | allelic het-gene, signal peptide | Sarkar et al. 2002 | |||
| Saupe et al. 2000 | |||||
| un-24 (NCU03539.2) | allelic het-gene, ribonucleotide reductase large subunit | Saupe 2000 | |||
| Smith et al. 2000 | |||||
| Suppressor genes | |||||
| tol (NCU04453.2) | TOL/HET-6/HET-E domain | Shiu & Glass 1999 | |||
| vib-1 (NCU03725.2) | regulation of conidiation and maybe of nrAPase | Xiang & Glass 2002 | |||
| Incompatibility related genes | |||||
| ham-2 (NCU03727.2) | hyphal fusion, putative transmembrane protein | Xiang et al. 2002 | |||
| pin-c (NCU03494.2) | allelic gene with HET-domain, linked to het-c | Kaneko et al. 2006 | |||
| rnr-a (NCU07887.2) | suppresses un-24 temperature sensitive mutation, ribonucleotide reductase small subunit | Kotierk & Smith 2001 | |||
| P. anserina | |||||
| Heterokaryon incompatibility genes | |||||
| het-c2 (AAA20542) | nonallelic het-gene interacts with het-d and het-e, glycolipid transfer protein | Saupe et al. 1994 | |||
| het-d2y (AAL37301) | nonallelic het-gene against het-c2, GTP-binding, WD repeat, TOL/HET-6/HET-E domain | Espagne et al. 2002 | |||
| het-e4s (AAL37297) | nonallelic het-gene against het-c2, GTP-binding, WD repeat, TOL/HET-6/HET-E domain | Espagne et al. 2002 | |||
| het-S (AAB88771) | allelic het-gene, prion analog | Coustou et al. 1997 | |||
| Incompatibility related genes | |||||
| idi-1 (AAC24119) | induced by het-c/e and r/v incompatibility, signal peptide | Dementhon et al. 2003 | |||
| Bourges et al. 1998 | |||||
| idi-2 (AAC24120) | induced by het-r/v incompatibility, signal peptide | Bourges et al. 1998 | |||
| idi-3 (AAC24121) | induced by het-c/e and r/v incompatibility, signal peptide | Bourges et al. 1998 | |||
| idi-4 (j/b-a) | bZIP motif, putative trans-activation domain | Dementhon et al. 2004, 2005 | |||
| (AAT40415) | |||||
| idi-6 (pspA) | induced by het-c/e and r/v incompatibility, subtilisin-like serine protease | Paoletti et al. 2001 | |||
| (AAC03564) | Reichard et al. 2000 | ||||
| idi-7 (AAN41258) | ortholog of the S. cerevisiae aut7p | Pinan-Lucarre et al. 2003 | |||
| Modifier genes | |||||
| mod-A (AAC25496) | modifier of het-c/e, c/d and r/v incompatibility, SH3-binding motif | Barreau et al. 1998 | |||
| Bourges et al. 1998 | |||||
| mod-D (AAC24766) | modifier of het-c/e incompatibility, G protein α subunit | Loubradou et al. 1999 | |||
| mod-E (AAB97626) | modifier of het-r/v incompatibility, HSP90 | Loubradou et al. 1997 | |||
| S. cerevisiae | |||||
| Programmed Cell Death genes | |||||
| atp4 (YPL078C) | F0F1-ATPase | Matsuyama et al. 1998 | |||
| cdc48 (YDL126C) | cell division cycle, AAA ATPase, fusion of ER-derived vesicles | Madeo et al. 1997 | |||
| hel10 (YNL208W) | unknown | Ligr et al. 2001 | |||
| hel13 (YOR309C) | unknown | Ligr et al. 2001 | |||
| mca1/yca1 (YOR197W) | metacaspase | Madeo et al. 2002 | |||
| nsr1 (YGR159C) | rRNA processing | Ligr et al. 2001 | |||
| ppa1 (YHR026W) | vacuolar H+-ATPase | Ligr et al. 2001 | |||
| rsm23 (YGL129C) | mitochondrial small ribosomal unit | Madeo et al. 2002 | |||
| sar1 (YPL218W) | ER to Golgi transport | Ligr et al. 2001 | |||
| stm1 (YLR150W) | suppressor of pop2 and tom2 | Ligr et al. 2001 | |||
| tor1 (YJR066W) | regulation of cell death, phosphatidylinositol 3-kinase | Rohde et al. 2001 | |||
| Dementhon et al. 2003 | |||||
| Fitzgibbon et al. 2005 | |||||
Table 2.
Genome databases and their websites used in this research.
| Species | Strain | Website | References |
|---|---|---|---|
| A. fumigatus | Af293 | http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1 | Nierman et al. 2005 |
| A. nidulans | FGSC A4 | http://www.broad.mit.edu/annotation/genome/aspergillus_nidulans/Blast.html | Galagan et al. 2005 |
| A. niger | CBS513.88 | http://www.dsm.com/en_US/html/dfs/genomics_aniger.htm | Pel et al. 2007 |
| A. niger | ATCC1015 | http://genome.jgi-psf.org/cgi-bin/runAlignment?db=Aspni1&advanced=1 | DOE -JGI |
| A. oryzae | RIB40 | http://www.bio.nite.go.jp/dogan/MicroTop?GENOME_ID=ao | Machida et al. 2005 |
| A. terreus | NIH2624 | http://www.broad.mit.edu/annotation/genome/aspergillus_terreus/Blast.html | Broad Institute of Harvard and MIT |
| N. crassa | OR74A | http://www.broad.mit.edu/annotation/genome/neurospora/Blast.html | Galagan et al. 2003 |
| P. anserina | S | http://podospora.igmors.u-psud.fr/blast_ol.html | P. anserina genome project |
| S. cerevisiae | S288C | http://seq.yeastgenome.org/cgi-bin/nph-blast2sgd | Saccharomyces Genome Database |
The in silico experiments performed with the different sequenced and available genomes show that the majority of the PCD genes from S. cerevisiae have homologs in the filamentous fungi, only hel10 and hel 13, whose functions are unknown, are mostly missing (Table 3). In contrast the majority of vegetative incompatibility and cell death related genes from N. crassa and P. anserina can be found in the other filamentous fungi, but many of them, like het-6, het-c, tol, vib-1, pin-c, het-c2, het-s, idi-1, idi-2, idi-3, idi-4 and mod-A, are missing in the baker”s yeast. Yeast and filamentous fungi diverged approximately 1.1 billion years ago (Cai et al. 2006) and yeast have a different, mainly unicellular, life style. Only the HET-D and HET-E proteins which have GTP binding capacity and the so-called WD (tryptophan-aspartate) repeats (Espagne et al. 2002) yielded many homologous sequences in S. cerevisiae. Of course these genes may have pleiotropic functions and may have a different function in S. cerevisiae than controlling anastomoses formation. The idi-7 gene of P. anserina is classified as being an ortholog of the S. cerevisiae aut7p gene coding for a protein whose binding to the membrane represents an early step in vesicle formation (Lang et al. 1998). Not surprisingly this protein is quite well preserved between both filamentous fungi and yeast as are the genes coding for ribonucleotide reductases and serine proteases.
Table 3.
HET and incompatibility related proteins in filamentous fungi and yeast. ID numbers for N. crassa and S. cerevisiae proteins refer to the numbers given in their respective sequencing projects, the ID numbers for the P. anserina proteins were taken from GenBank.
| Species | Class | Protein | Species (Strains) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. fumigatus (Af 293) | A. nidulans (FGSC A4) | A. niger (CBS513.88) | A. niger (ATCC1015) | A. oryzae (RIB40) | A. terreus (NIH 2624) | N. crassa (OR74A) | P. anserina (S) | S. cerevisiae (S288C) | ||||
| N. crassa | ||||||||||||
| Heterokaryon incompatibility genes | ||||||||||||
| HET-6 (NCU03533.2) | • | • | • | • | • | • | • | • | ||||
| HET-C (NCU03125.2) | • | • | • | • | • | • | • | • | ||||
| UN-24 (NCU03539.2) | • | • | • | • | • | • | • | • | • | |||
| Suppressor genes | ||||||||||||
| TOL (NCU04453.2) | • | • | • | • | • | • | • | |||||
| VIB-1 (NCU03725.2) | • | • | • | • | • | • | • | • | ||||
| Incompatibility related genes | ||||||||||||
| HAM-2 (NCU03727.2) | • | • | • | • | • | • | • | • | • | |||
| PIN-C (NCU03494.2) | • | • | • | • | • | • | • | |||||
| RNR-A (NCU07887.2) | • | • | • | • | • | • | • | • | • | |||
| P. anserina | ||||||||||||
| Heterokaryon incompatibility genes | ||||||||||||
| HET-C2 (AAA20542) | • | • | • | • | • | • | • | • | ||||
| HET-D2Y (AAL37301) | • | • | • | • | • | • | • | • | • | |||
| HET-E4S (AAL37297) | • | • | • | • | • | • | • | • | • | |||
| HET-S (AAB88771) | • | • | • | |||||||||
| Incompatibility related genes | ||||||||||||
| IDI-1 (AAC24119) | • | |||||||||||
| IDI-2 (AAC24120) | • | |||||||||||
| IDI-3 (AAC24121) | • | |||||||||||
| IDI-4/ JLB-A (AAT40415) | • | • | • | • | ||||||||
| IDI-6/ PSP (AAC03564) | • | • | • | • | • | • | • | • | • | |||
| IDI-7 (AAN41258) | • | • | • | • | • | • | • | • | • | |||
| Modifier genes | ||||||||||||
| MOD-A (AAC25496) | • | • | • | |||||||||
| MOD-D (AAC24766) | • | • | • | • | • | • | • | • | • | |||
| MOD-E (AAB97626) | • | • | • | • | • | • | • | • | • | |||
| S. cerevisiae | ||||||||||||
| Programmed Cell Death genes | ||||||||||||
| ATP4 (YPL078C) | • | • | • | • | • | • | • | • | • | |||
| CDC48 (YDL126C) | • | • | • | • | • | • | • | • | • | |||
| HEL10 (YNL208W) | • | • | • | |||||||||
| HEL13 (YOR309C) | • | • | ||||||||||
| MCA1/YCA1 (YOR197W) | • | • | • | • | • | • | • | • | • | |||
| NSR1 (YGR159C) | • | • | • | • | • | • | • | • | ||||
| PPA1 (YHR026W) | • | • | • | • | • | • | • | • | • | |||
| RSM23 (YGL129C) | • | • | • | |||||||||
| SAR1 (YPL218W) | • | • | • | • | • | • | • | • | • | |||
| STM1 (YLR150W) | • | • | • | • | • | • | • | • | ||||
| TOR1 (YJR066W) | • | • | • | • | • | • | • | • | • | |||
Some genes like the un-24 gene, known from N. crassa to be involved in heterokaryon incompatibility and coding for the ribonucleotide reductase large subunit, are well conserved in the filamentous fungi and have a highly similar homolog in S. cerevisiae (Supplementary Table 1). Whereas the putative transmembrane protein HAM-2 is very well conserved within the filamentous fungi, it differs considerably from the S. cerevisiae homolog.
These results show that most of the genes involved in programmed cell death are well conserved among both the filamentous fungi and the yeast S. cerevisiae, but that many genes involved in heterokaryon incompatibility are not.
In silico comparison of Sordariales and Eurotiales
N. crassa and P. anserina belong to the Sordariales, whereas the aspergilli belong to the Eurotiales. Comparing the incompatibility/apoptosis gene sets between these two groups of filamentous fungi (see Table 3 and Suppl. Table 1), the most remarkable difference is shown by P. anserina, bearing many more idi (induced during incompatibility) gene homologs than other fungi and missing the suppressor gene stm1. Among these fungi, P. anserina bears the most different gene set and the largest set of genes with het-domains: HET-6 has 35, HET-D 94, PIN-C 51 and TOL 48 homologs below the threshold of e-10. Some of these homologous sequences overlap between the different genes due to their conserved het-domains. A possible explanation for this can be in its life cycle. P. anserina is a saprophytic fungus, which feeds on partially digested materials in the dung of herbivorous animals. As a coprophilic fungus it grows in synchrony and under rather high density with competitors for the same ephemeral and limited substrate. Therefore, the risk of exploitation or genetic infection by others may be relatively high in comparison to most other fungi. An efficient way to limit exchange of genetic materials is heterokaryon incompatibility, which process is governed by the so called het-genes. In P. anserina the majority of the incompatibility reactions is due to non-allelic interactions (exception is het het-s/het-S reaction). Het-d and het-e trigger incompatibility with the het-c2. Both het-d and het-e encode HET domain proteins, and due to the presence of this domain, these proteins seem to be responsible for nonself recognition in filamentous fungi, including P. anserina (Kaneko et al. 2006). Whether the huge number of HET domain protein homologs thus reflects the relative importance of fusion-rejection systems in the life history of P. anserina is unclear, though this has been suggested for coprophiles (Buss 1982). Of course, het-domain containing genes also may have other functions than just heterokaryon incompatibility reactions and there certainly seems to be a large family of het-domain genes.
Comparing the phylogenies of different proteins in the different species (see e.g. Fig. 2 with the HET-6 gene homologs from A. niger and N. crassa) one can see that the homologs of these genes show old polymorphisms. The sequences found in the A. niger strains are often very similar. The most similar homologs in N. crassa however, can be quite different from the A. niger sequences. Old duplications of the ancestor genes with possible new functions are also visible in the phylogenies.
Fig. 2.
Neighbour-joining tree of HET-6 proteins from N. crassa and two A. nigers. Strains on the tree: A1: A. niger CBS513.88; A2: A. niger ATCC1015; N: N. crassa OR74A. Accession numbers of the different homologs are given from the respective databases. Sequence alignment and bootstrapping was performed with ClustalX (Thompson et al. 1997). Trees were visualised by Treeview (Page, 1996).
Within the tested set of proteins, there are no exclusive proteins for members of the Sordariales. Only the MOD-A protein has but one hit in the aspergilli: the protein blast in A. oryzae resulted in one hit with only low similarity (e-11) (Suppl. Table 1).
In silico comparison of the different aspergilli
S. cerevisiae PCD- and N. crassa and P. anserina HET-, modifier and suppressor protein sequences were used to search the genomes of A. fumigatus Af 293, A. nidulans FGSC A4, A. oryzae RIB40, and A. terreus NIH2624 for homologs (Table 2). Bi-directional best hit analyses were performed with as criterion for homologs an accepted E-value of <e-10.
Comparing the different Aspergillus species for their putative heterokaryon incompatibility and cell death related proteins, there are no large differences between the species in the presence of certain proteins, in agreement with previous findings (Fedorova et al. 2005; Table 3). But, in the number of homologs of HET-6, TOL and TOL-related PIN-C proteins there is a surprisingly big difference between the asexual and (supposedly) sexual lines. Whereas there are at least 10 HET-6, 9 TOL and 11 PIN-C homologs in the asexual strains, in A. nidulans and A. fumigatus we found only 2 and 3 HET-6, 0 and 1 TOL, and 0 and 1 PIN-C homologs respectively (Suppl. Table 1). This finding could be explained by the capability of sex: A. nidulans is able to reproduce sexually, and there are indirect proofs for the presence of a sexual life cycle in A. fumigatus (Varga 2003, Paoletti et al. 2005). The other aspergilli (A. niger, A. oryzae, A. terreus) are known as asexual species. The question in this case is why do asexual species bear much more HET domain genes? As it was mentioned above these genes are the main components of non-allelic incompatibility, therefore there could be a disadvantage for fungi with a sexual cycle to have such genes.
Another reason could be the proposed function for heterokaryon incompatibility in limiting the spread of detrimental cytoplasmic or nuclear elements (Caten 1972). In A. niger and related black Aspergillus species dsRNA mycoviruses occur in approximately 10 % of the natural isolates. These mycoviruses are effectively transferred to all asexual spores (van Diepeningen et al. 2006). Tests showed that heterokaryon incompatibility indeed efficiently blocks the transfer of mycoviruses in these black aspergilli (van Diepeningen et al. 1997). In sexual A. nidulans no dsRNA viruses were found in nature and also here artificially introduced viruses efficiently find their way to the asexual spores. However, when sexual spores are produced the mycoviruses are excluded from the offspring (Coenen et al. 1997). Therefore A. nidulans has an extra option to get rid of parasitic elements through its sexual cycle and thus heterokaryon incompatibility may be less important between A. nidulans strains.
In silico comparison of the two A. niger genomes
Using strains with different spore colors and different auxotrophic mutations or dominant resistances, one can test for the formation of heterokaryotic mycelium on media on which the single partners are unable to grow. Different mutant lines were isolated from DSM Research BV”s Strain CBS 513.88, DOE Joint Genome Institute”s culture collection strain ATCC1015 and our lab strain N400 (ATCC 9029; CBS 120.49), making it possible to test for heterokaryon incompatibility between these strains. Strains ATCC1015 and our laboratory strain N400 (ATCC 9029; CBS 120.49) proved heterokaryon compatible with one another. That they thus belong to the same vegetative compatibility group suggests that they share a common clonal ancestor. However, DSM Research BV”s Strain CBS 513.88 proved incompatible with the two other strains. Thus the two genomes sequenced by DSM and DOE Joint Genome Institute respectively are from heterokaryon incompatible strains. We searched the genomic databases of these two A. niger strains for incompatibility/apoptosis related genes (Table 2). For blastp searches we used the apoptosis-like PCD proteins of S. cerevisiae and the HET, modifier and suppressor protein sequences of N. crassa and P. anserina (Table 1). For validation of the identified A. niger sequences, a bi-directional best hit analysis was performed, using the polypeptide sequence of the identified A. niger ORFs as a query for a blastp search at the N. crassa, P. anserina, S. cerevisiae and GenBank database (http://ncbi.nih.gov/BLAST; Altschul et al. 1990). As criterion for homologs we used an accepted E-value of <e-10.
Similarly to the other asexual aspergilli, we found a high number of HET domain proteins (PIN-C, HET-6, HET-D and TOL homologs). Nearly all het-genes were highly similar between both A. niger strains, differences were limited to a few substitutions but are potentially crucial for incompatibility reactions. However, the two sequenced A. niger strains differ in their sets of heterokaryon incompatibility and apoptosis related genes. Although they largely possess the same gene set, strain CBS513.88 lacks a HET-S homologue and strain ATCC1015 lacks the IDI-4/JLB-A and HEL10 homologs (Table 3). In nearly all of their putative HET proteins the two A. niger isolates show little to no variation in sequence. The two strains are heterokaryon incompatible and the differences -sometimes only simple substitutions, sometimes small stretches of amino acids - in the known indel regions of some of the putative het-genes may explain the observed incompatibility reaction between the two strains (Table 4). For the idi-6 (psp) genes two alleles are present, both very similar with very few substitutions. However, for the MCA1/YCA1 (a meta-caspase) one pair is completely identical, whereas the second pair shows more differences and is only 77 % identical.
Table 4.
A comparison between the different heterokaryon incompatibility and programmed cell death related genes in the two sequenced A. niger genomes (CBS513.88 and ATCC1015). If two proteins differ in size, the longer one is the basis for counting percentage of identities, similarities and gaps. Gaps are counted only in the homologous region.
| Function | Protein | Type and size of difference | Identities | Gaps | ||
|---|---|---|---|---|---|---|
| Heterokaryon incompatibility genes (N. crassa & P. anserina) | ||||||
| HET-6 | only HET domain motifs are slightly conserved | - | - | |||
| HET-C | indel: 622 | 791/793 (99 %) | 1/793 | |||
| substitution: 196 | (0 %) | |||||
| UN-24 | no difference | (100 %) | (0 %) | |||
| HET-C2 | no difference | (100 %) | ||||
| HET-D / HET-E | diverse proteins with WD40 repeats, but no remarkable similarity | - | ||||
| Suppressor genes (N. crassa) | ||||||
| TOL | very diverse proteins, with conserved HET domain motifs | - | - | |||
| VIB-1 | substitution: 195 | 585/586 (99 %) | (0 %) | |||
| Incompatibility related genes (N. crassa & P. anserina) | ||||||
| HAM-2 | substitution: 756 | 1066/1067 (99 %) | (0 %) | |||
| PIN-C | very diverse proteins, with conserved HET domain motifs | - | - | |||
| RNR-A | no difference | (100 %) | (0 %) | |||
| IDI-6 / PSP | 1st pair: indel: 534 | 531/535 (99 %) | ||||
| (2 alleles) | substitutions: 398, 525 | |||||
| 2nd pair: substitutions: 398, 436-443, 450-459, 471-514 | 413/416 (99 %) | |||||
| IDI-7 | no difference | (100 %) | ||||
| Modifier genes (P. anserina) | ||||||
| MOD-D (2 alleles) | Members of the two allele pairs are 100 % identical, between the pairs there are some difference: 1-167 variable part, 1-60 and 168-360 more conserved region. | |||||
| MOD-E | substitution: 244 | 672/702 (96 %) | (0 %) | |||
| Programmed Cell Death genes (S. cerevisiae) | ||||||
| ATP4 | no difference | 100 % | 0 % | |||
| CDC48 | no difference | 100 % | 0 % | |||
| HEL 13 | present only in CBS 513.88 | - | - | |||
| MCA1 / YCA1 | 1st pair 438/438 (100 %) | (0 %) | ||||
| (2 alleles) | ||||||
| 2nd allele: indel region: 1-56, 75-104, 237, 443-447 | ||||||
| subtitutions: 57-75, 105-106. 236, 440-442 | 2nd pair 341/441 (77 %) | 1/333 (0 %) | ||||
| NSR1 | diverse proteins, with short conserved motifs | - | - | |||
| PPA1 | no difference | 100 % | 0 % | |||
| SAR1 | no difference | 100 % | 0 % | |||
| STM1 | indel region: 8-13 | 297/303 (98 %) | 6/303 | |||
| (1 %) | ||||||
| TOR1 | substitution: 16 | 2389/2390 (99 %) | (0 %) | |||
The regions of these genes involved in self/non-self recognition may be under positive selection and single-amino-acid differences can be sufficient to trigger incompatibility (Saupe 2000). Thus, the observed small differences may be an explanation for the observed heterokaryon incompatibility between the two A. niger strains. However, as both P. anserina and N. crassa seem to have selected different sets of heterokaryon incompatibility genes to block intermycelial transfer, aspergilli may use a completely different set of genes as well.
DISCUSSION
Little is known about the nature of gene flow in natural populations of aspergilli. However, there is clear evidence of recombination within populations (e.g. Geiser et al. 1994, Geiser et al. 1998, Paoletti et al. 2005). This may have arisen through sexual and/or parasexual means. The presence or absence of gene flow in populations has significant implications for speciation within the aspergilli, which may proceed at different rates depending on the presence of recombination or clonality (Taylor et al. 1999a).
In the particular case of the presumed asexual A. niger we used an in silico study to assess the genetic basis of heterokaryon incompatibility. Comparisons were made of genome sequences of two different A. niger strains that are heterokaryon incompatible, together with genome sequences of four closely related sexual and asexual species. We searched these databases with genes known to be involved in heterokaryon incompatibility or apoptosis in P. anserina, N. crassa and S. cerevisiae. Our aim was to find out whether the same genes may be involved in the incompatibility reactions between different A. niger isolates as the ones found to interact in N. crassa and P. anserina, fungi that have different sets of active het-genes. Few differences were found between the two sequenced A. niger genomes, but many of the known heterokaryon incompatibility genes were indeed present in the A. niger genomes. Some of the examined het-genes were even found to have many homologs.
Further practical research is needed to find a satisfying explanation for the high level of incompatibility in the natural populations of black aspergilli and to pinpoint functional het-genes in the species. As a result of our data mining, the sequences of the known incompatibility genes are available for functional analysis, to uncover the secrets of incompatibility between the natural isolates of black aspergilli.
Acknowledgments
We want to thank all institutes, companies and individuals involved in the genome projects of the species used in this study: A. fumigatus preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. A. niger CBS513.88 sequence data were obtained from the DSM Research B.V., the A. niger ATCC1015 sequence data from the DOE Joint Genome Institute. The A. nidulans genome data are from the Aspergillus Sequencing Project, the Aspergillus terreus genome data from the Aspergillus terreus Sequencing Project, and the release 7 data of the N. crassa genome were all from the Broad Institute of Harvard and MIT (http://www.broad.mit.edu). A. oryzae data were obtained from the NITE Biological Resource Center. Data from the complete sequence of P. anserina were obtained from the P. anserina genome project and the S. cerevisiae genome data from the Saccharomyces Genome Database.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic Local Alignment Search Tool. Journal of Molecular Biology 215: 403-410. [DOI] [PubMed] [Google Scholar]
- Archer DB, Dyer PS (2004). From genomics to post-genomics in Aspergillus. Current Opinion in Microbiology 7: 499-504. [DOI] [PubMed] [Google Scholar]
- Arie T, Christiansen SK, Yoder OC, Turgeon BG (1997). Efficient cloning of ascomycete mating-type genes by PCR amplification of the conserved MAT HMG box. Fungal Genetics and Biology 21: 118-130 [PubMed] [Google Scholar]
- Barreau C, Iskandar M, Loubradou G, Levallois V, Bégueret J (1998). The mod-A suppressor of nonallelic heterokaryon incompatibility in Podospora anserina encodes a proline-rich polypeptide involved in female organ formation. Genetics 149: 915-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourges N, Groppi A, Barreau C, Clavé C, Bégueret J (1998). Regulation of gene expression during the vegetative incompatibility reaction in Podospora anserina: characterization of three induced genes. Genetics 150: 633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss LW (1982). Somatic-cell parasitism and the evolution of somatic tissue compatibility. Proceedings of the National Academy of Sciences of the U.S.A. 79: 5337-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai JJ, Woo PCY, Lau SKP, Smith DK, Yuen KY (2006). Accelerated evolutionary rate may be responsible for the emergence of lineage-specific genes in Ascomycota. Journal of Molecular Evolution 63: 1-11. [DOI] [PubMed] [Google Scholar]
- Caten CE (1972). Vegetative incompatibility and cytoplasmic infection in fungi. Journal of General Microbiology 72: 221-229. [DOI] [PubMed] [Google Scholar]
- Coenen A, Kevei F, Hoekstra RF (1997). Factors affecting the spread of double stranded RNA mycoviruses in Aspergillus nidulans. Genetical Research 69: 1-10. [DOI] [PubMed] [Google Scholar]
- Coppin E, de Renty C, Debuchy R (2005). The function of the coding sequences for the putative pheromone precursor in Podospora anserina is restricted to fertilization. Eukaryotic Cell 4: 407-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou V, Deleu C, Saupe S, Bégueret J (1997). The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proceedings of the National Academy of Sciences of the U.S.A. 94: 9773-9778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft JH, Jinks JL (1977). Aspects of the population genetics of Aspergillus nidulans. In: Genetics and Physiology of Aspergillus. pp. 349-360 (eds. JE Smith & JA Pateman). Academic Press, London.
- Debets AJM, Griffiths AJF (1998). Polymorphism of het-genes prevents resource plundering in Neurospora crassa. Mycological Research 102: 1343-1349. [Google Scholar]
- Debuchy R, Turgeon BG (2006). Mating-type structure, evolution, and function in Euascomycetes. In; The Mycota I: Growth, Differentiation and Sexuality, U. Kües and R. Fischer, ed. (Berlin: Springer-Verlag), pp. 293-323.
- Dementhon K, Paoletti M, Pinan-Lucarré B, Loubradou-Bourges N, Sabourin M, Saupe SJ, Clavé C (2003). Rapamycin mimics the incompatibility reaction in the fungus Podospora anserina. Eukaryotic Cell 2: 238-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dementhon K, Saupe SJ, Clavé C (2004). Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus Podospora anserina. Molecular Microbiology 53: 1625-1640. [DOI] [PubMed] [Google Scholar]
- Dementhon K, Saupe SJ (2005). DNA-Binding specificity of the IDI-4 basic leucine zipper factor of Podospora anserina defined by systematic evolution of ligands by exponential enrichment (SELEX). Eukaryotic Cell 4: 476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepeningen AD van, Debets AJM, Hoekstra RF (1997). Heterokaryon incompatibility blocks virus transfer among natural isolates of black aspergilli. Current Genetics 32: 209-17. [DOI] [PubMed] [Google Scholar]
- Diepeningen AD van, Debets AJM, Hoekstra RF (2006). Dynamics of dsRNA mycoviruses in the black Aspergillus population. Fungal Genetics and Biology 43: 446-452. [DOI] [PubMed] [Google Scholar]
- Dyer PS (2002). A MAT-2 mating-type gene in the homothallic fungus Aspergillus nidulans. In; 7th International Mycological Congress Book of Abstracts, 331. University of Oslo.
- Dyer PS (2007). Sexual Reproduction and Significance of MAT in the aspergilli. In: Sex in Fungi: Molecular Determination and Evolutionary Principles (eds J Heitman, JW Kronstad, JW Taylor & LA Casselton). ASM Press (in press).
- Dyer PS, Ingram DS, Johnstone K (1992). The control of sexual morphogenesis in the Ascomycotina. Biological Reviews 67: 421-458. [Google Scholar]
- Dyer PS, Paoletti M (2005). Reproduction in Aspergillus fumigatus: sexuality in a supposedly asexual species? Medical Mycology 43 (Supplement1): S7-S14. [DOI] [PubMed] [Google Scholar]
- Dyer PS, Paoletti M, Archer DB (2003). Genomics reveals sexual secrets of Aspergillus. Microbiology 149: 2301-2303. [DOI] [PubMed] [Google Scholar]
- Espagne E, Balhadère P, Penin M-L, Barreau C, Turcq B (2002). HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics 161: 71-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova ND, Badger JH, Robson GD, Wortman JR, Nierman WC (2005). Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics 6: 177-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon GJ, Morozov IY, Jones MG, Caddick MX (2005). Genetic analysis of the TOR pathway in Aspergillus nidulans. Eukaryotic Cell 4: 1595-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CPC, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 438: 859-868. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Bastürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D”Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005). Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105-1115. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Arnold ML, Timberlake WE (1994). Sexual origins of British Aspergillus nidulans. Proceedings of the National Academy of Sciences U.S.A. 91: 2349-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser DM, Pitt JI, Taylor JW (1998). Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proceedings of the National Academy of Sciences U.S.A. 95: 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser DM, Timberlake WE, Arnold ML (1996). Loss of meiosis in Aspergillus. Molecular Biology and Evolution 13: 809-817. [DOI] [PubMed] [Google Scholar]
- Glass NL, Kuldau GA (1992). Mating type and vegetative incompatibility in filamentous ascomycetes. Annual Review of Phytopathology 30: 201-224. [DOI] [PubMed] [Google Scholar]
- Glass NL, Jacobson DJ, Shiu PKT (2000). The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annual Review of Genetics 34: 165-186. [DOI] [PubMed] [Google Scholar]
- Glass NL, Kaneko I (2003). Fatal attraction: nonself recognition and heterokaryon incompatibility in filamentous fungi. Eukaryotic Cell 2: 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Vollmer SJ, Staben C, Grotelueschen J, Metzenberg RL, Yanofsky C (1988). DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 241: 570-573. [DOI] [PubMed] [Google Scholar]
- Kaneko I, Dementhon K, Xiang Q, Glass NL (2006). Nonallelic interactions between het-c and a polymorphic locus, pin-c, are essential for nonself recognition and programmed cell death in Neurospora crassa. Genetics 172: 1545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotierk M, Smith ML (2001). Unpublished sequence, direct submission Submitted (22-FEB-2001) Biology, Carleton University, 1125 Colonel By Drive, Ottawa, Ont K1S 5B6, Canada.
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M (1998). Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO Journal 17: 3597-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D”Souza C, Harashima T, Shen WC, Wang GP, Pan X, Waugh M, Heitman J (2000). Signal transduction cascades regulating fungal development and virulence. Microbiology and Molecular Biology Reviews 64: 746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie JF (1993). Fungal vegetative incompatibility. Annual Review of Phytopathology 31: 127-150. [DOI] [PubMed] [Google Scholar]
- Leslie JF, Zeller KA (1996). Heterokaryon incompatibility in fungi - more than just another way to die. Journal of Genetics 75: 415-424. [Google Scholar]
- Ligr M, Velten I, Froehlich E, Madeo F, Ledig M, Froehlich K-U, Wolf DH, Hilt W (2001). The proteosomal substrate Stm1 participates in apoptosis-like cell death in yeast. Molecular Biology of the Cell 12: 2422-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubradou G, Bégueret J, Turcq B (1997). A mutation in an HSP90 gene affects the sexual cycle and suppresses vegetative incompatibility in the fungus Podospora anserina. Genetics 147: 581-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubradou G, Bégueret J, Turcq B (1999). MOD-D, a G-alpha subunit of the fungus Podospora anserina, is involved in both regulation of development and vegetative incompatibility. Genetics 152: 519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto KI, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Rao JP, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005). Genome sequencing and analysis of Aspergillus oryzae. Nature 438: 1157-1161. [DOI] [PubMed] [Google Scholar]
- Madeo F, Froehlich E, Froehlich KU (1997). A yeast mutant showing diagnostic markers of early and late apoptosis. Journal of Cell Biology 139: 729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F, Herker E, Maldener C, Wissing S, Laechelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Froehlich KU (2002). A caspase-related protease regulates apoptosis in yeast. Molecular Cell 9: 911-917. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Xu Q, Velours J, Reed JC (1998). The mitochondrial F0F1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Molecular Cell 1: 327-336. [DOI] [PubMed] [Google Scholar]
- Milgroom MG (1996). Recombination and the multilocus structure of fungal populations. Annual Review of Phytopathology 34: 457-477. [DOI] [PubMed] [Google Scholar]
- Nauta MJ, Hoekstra RF (1993). Evolutionary dynamics of spore killers. Genetics 135: 923-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latge JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O”Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail M, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Cordoba S, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW (2005). Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438: 1151-1156. [DOI] [PubMed] [Google Scholar]
- Page RDM (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357-358. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Castroviejo M, Bégueret J, Clavé C (2001). Identification and characterization of a gene encoding a subtilisin-like serine protease induced during the vegetative incompatibility reaction in Podospora anserina. Current Genetics 39: 244-252. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latgé JP, Denning DW, Dyer PS (2005). Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Current Biology 15: 1242-1248. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Seymour FA, Alcocer MJC, Kaur N, Calvo AM, Archer DB, Dyer PS (2007). Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Current Biology 17 (in press). [DOI] [PubMed]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JA, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EG, Debets AJ, Dekker P, van Dijck PW, van Dijk A, Dijkhuizen L, Driessen AJ, d”Enfert C, Geysens S, Goosen C, Groot GS, de Groot PW, Guillemette T, Henrissat B, Herweijer M, van den Hombergh JP, van den Hondel CA, van der Heijden RT, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel MJ, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij NN, Ram AF, Rinas U, Roubos JA, Sagt CM, Schmoll M, Sun J, Ussery D, Varga J, Vervecken W, van de Vondervoort PJ, Wedler H, Wosten HA, Zeng AP, van Ooyen AJ, Visser J, Stam H. (2007). Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology 25: 221-231. [DOI] [PubMed] [Google Scholar]
- Pinan-Lucarre B, Paoletti M, Dementhon K, Coulary-Salin B, Clavé C (2003). Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Molecular Microbiology 47: 321-333. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G (1956). The parasexual cycle in fungi. Annual Review of Microbiology 10: 393-400. [DOI] [PubMed] [Google Scholar]
- Pöggeler S (2002). Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Current Genetics 42: 153-160. [DOI] [PubMed] [Google Scholar]
- Pringle A, Baker DM, Platt JL, Wares JP, Latgé JP, Taylor JW (2005). Cryptic speciation in the kosmopolieten and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59: 1886-1899. [PubMed] [Google Scholar]
- Reichard U, Cole GT, Hill TW, Ruchel R, Monod M (2000). Molecular characterization and influence on fungal development of ALP2, a novel serine proteinase from Aspergillus fumigatus. International Journal of Medical Microbiology 290: 549-558. [DOI] [PubMed] [Google Scholar]
- Rohde J, Heitman J, Cardenas ME (2001). The TOR kinases link nutrient sensing to cell growth. Journal of Biological Chemistry 276: 9583-9586. [DOI] [PubMed] [Google Scholar]
- Rydholm, C, Dyer PS, Lutzoni F (2007). DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryotic Cell 6: 868-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Iyer G, Wu J, Glass NL (2002). Nonself recognition is mediated by HET-C heterocomplex formation during vegetative incompatibility. EMBO Journal 21: 4841-4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe SJ, Descamps C, Turcq B, Bégueret J (1994). Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proceedings of the National Academy of Sciences of the U.S.A. 91: 5927-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe SJ, Clavé C, Sabourin M, Bégueret J (2000). Characterization of hch, the Podospora anserina homolog of the het-c heterokaryon incompatibility gene of Neurospora crassa. Current Genetics 38: 39-47. [DOI] [PubMed] [Google Scholar]
- Saupe SJ (2000). Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiology and Molecular Biology Reviews 64: 489-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu PK, Glass NL (1999). Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics 151: 545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Dyer PS, Ashby AM (1999). Intra-specific and inter-specific conservation of mating-type genes from the discomycete plant-pathogenic fungi Pyrenopeziza brassicae and Tapesia yallundae. Current Genetics 36: 290-300. [DOI] [PubMed] [Google Scholar]
- Smith ML, Micali OC, Hubbard SP, Mir-Rashed N, Jacobson DJ, Glass NL (2000). Vegetative incompatibility in the het-6 region of Neurospora crassa is mediated by two linked genes. Genetics 155: 1095-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Geiser DM, Burt A, Koufopanou V (1999a). The evolutionary biology and population genetics underlying fungal strain typing. Clinical Microbiology Reviews 12: 126-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Fisher MC (1999b). The evolution of asexual fungi: reproduction, speciation and classification. Annual Review of Phytopathology 37: 197-246. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon BG (1998). Application of mating type gene technology to problems in fungal biology. Annual Review of Phytopathology 36: 115-137. [DOI] [PubMed] [Google Scholar]
- Turgeon BG, Yoder OC, 2000. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genetics and Biology 31: 1-5. [DOI] [PubMed] [Google Scholar]
- Varga J (2003). Mating type gene homologues in Aspergillus fumigatus. Microbiology 149: 816-819. [DOI] [PubMed] [Google Scholar]
- Varga J, Rinyu E, Kevei F, Toth B, Kozakiewicz Z (1998). Double stranded RNA mycoviruses in species of Aspergillus sections Circumdati and Fumigati. Canadian Journal of Microbiology 44: 569-574. [DOI] [PubMed] [Google Scholar]
- Warn PA, Sharp A, Anderson MJ, Denning DW (2006). Single-spore cultures of the sequences strain of Aspergillus fumigatus (Af293) have significantly different mortality rates in a murine model of IPA. 2nd Advances Against Aspergillosis Meeting Syllabus P031.
- Wu J, Saupe SJ, Glass NL (1998). Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in a group of filamentous fungi. Proceedings of the National Academy of Sciences of the U.S.A. 95: 12398-12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Glass NL (2002). Identification of vib-1, a locus involved in vegetative incompatibility mediated by het-c in Neurospora crassa. Genetics 162: 89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Q, Rasmussen C, Glass NL (2002). The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160: 169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]