Abstract
Multiple recent studies have demonstrated the limited utility of morphological methods used singly for species identification of clinically relevant aspergilli. It is being increasingly recognised that comparative sequence based methods used in conjunction with traditional phenotype based methods can offer better resolution of species within this genus. Recognising the growing role of molecular methods in species recognition, the recently convened international working group meeting entitled “Aspergillus Systematics in the Genomic Era” has proposed several recommendations that will be useful in such endeavors. Specific recommendations of this working group include the use of the ITS regions for inter section level identification and the β-tubulin locus for identification of individual species within the various Aspergillus sections.
Keywords: Emericella, molecular phylogeny, pathogenic aspergilli, polyphasic taxonomy, section Aspergillus section Terrei, section Usti
INTRODUCTION
aspergilli cause a wide spectrum of infections including cutaneous manifestations, otomycosis, and invasive infections such as pulmonary aspergillosis and endocarditis. Pulmonary aspergillosis may range from invasive pulmonary aspergillosis (IPA) in severely immunocompromised patients to chronic necrotising aspergillosis in mildly immunocompromised populations. The risk of IPA appears to be much higher in hematopoietic stem cell transplant patients and in patients with leukemia, where the attributable mortality rate was 38.5 % according to a recent study (Pagano et al. 2007). Aspergillus fumigatus remains the predominant agent of IPA, followed by either A. terreus or A. flavus depending on the medical center. Recently, IPA due to A. ustus and other rare aspergilli such as A. alliaceus (Balajee et al. 2007), A. lentulus (Balajee et al. 2005a) and A. udagawae (Balajee et al. 2006) have been reported.
The genus Aspergillus was originally divided into subgenera and groups (Raper & Fennell 1965) but the current classification scheme replaces the designation “group” with “section” (Gams et al. 1985) to conform to rules of the International Code of Botanical Nomenclature. Currently, the genus Aspergillus is classified into 7 subgenera that are in turn sub-divided into several sections comprised of related species (Gams et al. 1985). Clinical microbiology laboratories rely heavily on morphology-based identification methods for Aspergillus species wherein diagnostic criteria include the recognition of asexual or sexual structures and their characteristics such as shape, size, color, ornamentation and/or mode of attachment. Unfortunately, numerous difficulties exist in such a phenotype-based scheme largely because these characteristics are unstable, and clinical aspergilli sometimes manifest atypically with slow sporulation and aberrant conidiophore formation. Additionally, members of the section Fumigati have overlapping morphological characteristics, with several genetically distinct species existing within a single morphospecies.
Clinically, identification of unknown Aspergillus clinical isolates to species may be important given that different species have variable susceptibilities to multiple antifungal drugs. Thus, knowledge of the species identity may influence the choice of appropriate antifungal therapy. For example, in vitro and in vivo studies have demonstrated that A. terreus isolates are largely resistant to the antifungal drug amphotericin B, A. ustus isolates appear to be refractory to azoles, and A. lentulus and Petromyces alliaceus have low in vitro susceptibilities to a wide range of antifungals including amphotericin B, azoles, and echinocandins (Balajee et al. 2005a, 2007). Comparative DNA sequence-based identification formats appear to be promising in terms of speed, ease, objectivity and economy for species identification. Multiple genes ranging from the universal ribosomal DNA regions ITS and the large ribosomal subunit D1-D2 to protein encoding genes such as the β-tubulin and calmodulin gene regions have been evaluated to delimit species within aspergilli.
In spite of the shift of fungal identification formats into the molecular arena as evidenced by numerous publications, there is no consensus on the gene/genes that can be used for species identification in the genus Aspergillus. As a first step, a group of experts met at the “International Workshop on Aspergillus Systematics in the Genomic Era” [Utrecht, The Netherlands; April 2007] and presented research data on species identification strategies available to identify aspergilli. Throughout the meeting, research pertaining to the utility of the ITS region for inter section level classification of Aspergillus was presented. The session “Species identification in the clinical setting” was proposed specifically to deliberate on the utility of comparative sequence analyses of protein coding loci for intra section level species identification. This communication is a report of the research findings presented at this session. At the end of the report, we present the recommendations proposed by the Aspergillus working group for inter species level recognition of clinically relevant aspergilli and for identification of species within the sections Fumigati, Terrei, Usti and Emericella nidulans.
Pathogenic species in Aspergillus section Fumigati and species delimitation based on polyphasic taxonomy
The most common causative agent of aspergillosis is A. fumigatus with rare reports of invasive infections caused by species of Neosartorya. However, clinical isolates of A. fumigatus are not necessarily morphologically uniform, and mistaken identification of these taxa by morphological characteristics has occurred in the past. In order to develop diagnostic techniques, it is essential to clarify intra- and interspecies diversity in A. fumigatus and closely related species using robust techniques.
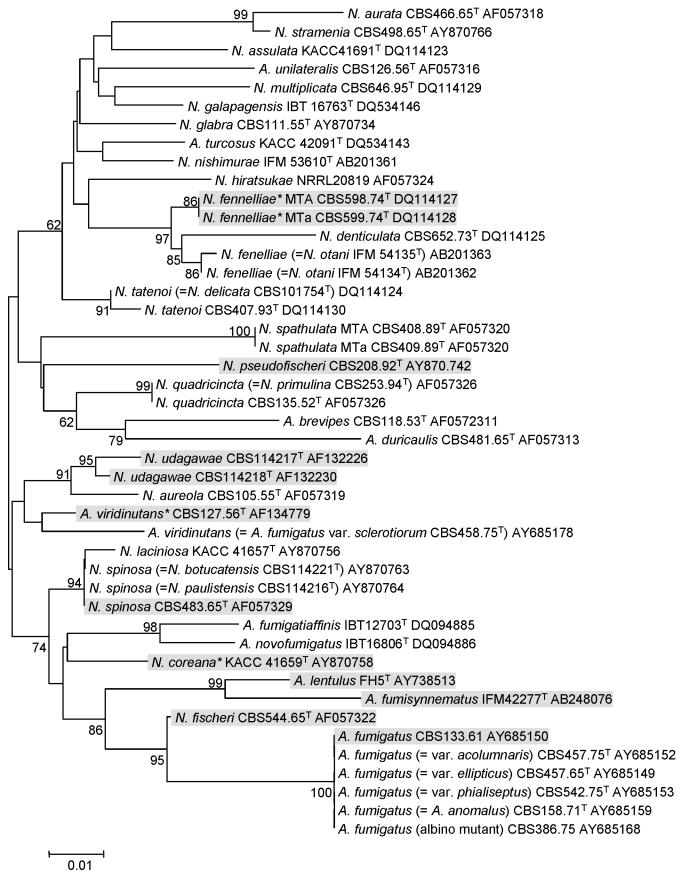
Recently, A. lentulus isolated from clinical specimens in the U.S.A. was described as a new species; members of this species were not able to survive at 48 °C, and this species has high in vitro MICs to several different classes of antifungals (Balajee et al. 2005a). Members of this species were distinct from the other species in this section, which includes the varieties of A. fumigatus. Two additional new species, A. fumigatiaffinis and A. novofumigatus, had also been proposed (Hong et al. 2005) by investigators who analyzed the species within the section Fumigati using a polyphasic approach that included phenotypic characters such as macro- and micro-morphology, growth temperature regimes, and extrolite patterns, and genotypic characters including RAPD-PCR and multi-locus sequence typing (MLST) of partial β-tubulin, calmodulin and actin genes. From these results, 30 species were accepted within the section Fumigati (Hong et al. 2006) and their taxonomic positions are shown in Fig. 1. Although A. fumigatus is the predominant agent of aspergillosis, several species in the section have also been reported from clinical samples: A. fumisynnematus (Yaguchi et al. 2007), A. lentulus (Balajee et al. 2005), A. viridinutans species complex (Hong et al. 2005, 2006; Katz et al. 2005), Neosartorya coreana (Hong et al. 2006; Katz et al. 2005), N fennelliae ((Kwon-Chung & Kim 1974), N. fischeri (Chim et al. 1998; Gori et al. 1998), N. pseudofischeri (often as the anamorph A. thermomutans status) (Balajee et al. 2005b; Coriglione et al. 1990; Padhye et al. 1994), N. spinosa (Summerbell et al. 1992; Gerber et al. 1973), N. hiratsukae (Guarro et al. 2002) and N. udagawae (Balajee et al. 2006). In the case of N. coreana and N. fennelliae, pathogenicity in humans has not been established yet.
Fig. 1.
Phylogenetic tree of Aspergillus section Fumigati species inferred from Neighbour-Joining analysis of partial β-tubulin gene sequence. The shaded species have been reported from clinical environment.
Because of the re-evaluation of section Fumigati based on polyphasic methods, the molecular identification of the A. fumigatus isolates recovered as causative agents of mycosis in humans and animals at the Medical Mycology Research Center, Chiba University, Japan (MMRC) was investigated (Yaguchi et al. 2007). Several other species within the section Fumigati were also included in the analyses. The phylogenetic relationships among A. fumigatus and related species, including Neosartorya species, were analyzed by sequencing partial regions of the β-tubulin, hydrophobin and calmodulin genes. The gene regions were sequenced directly from the PCR products by using primer pairs Bt2a and Bt2b, rodA1 and rodA2, and cmd5 and cmd6, respectively. PCR products were sequenced with the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems) and phylogenetic analyses were performed by the maximum parsimony (MP) and Neighbour-joining (NJ) methods. Comparative analyses of tree topologies from MP and NJ analyses showed no differences, and the three trees based on the three loci were similar.
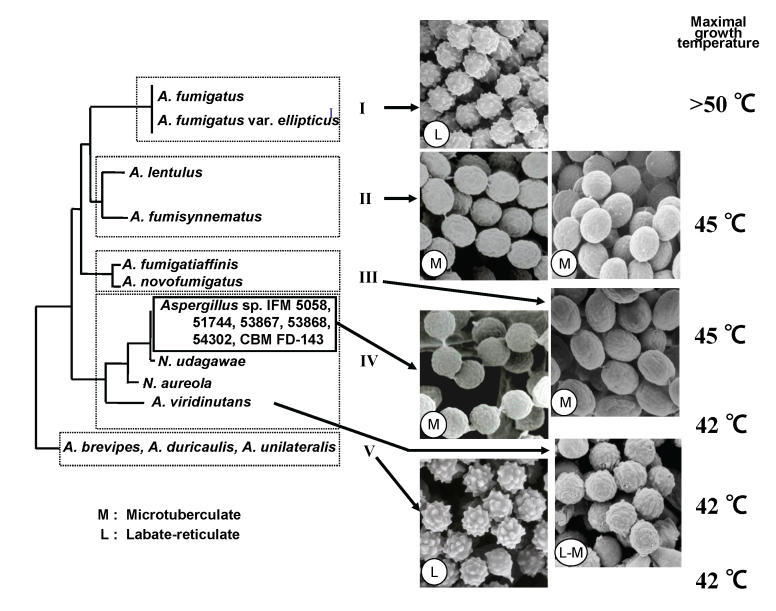
Results of this study showed that the species within the section Fumigati could be divided into five clades: clade I, typical strains of A. fumigatus including A. fumigatus var. ellipticus (Raper & Fennell 1965) and A. arvii (Aho et al. 1994); clade II, species including A. lentulus and A. fumisynnematus (Horie et al. 1993); clade III, species including A. fumigatiaffinis and A. novofumigatus; clade IV, atypical strains of A. fumigatus including A. viridinutans Katz et al. 1998; Varga et al. 2000); and clade V, species including A. brevipes, A. duricaulis and A. unilateralis. Most of the examined strains from clinical specimens in Japan clustered together in clade I. The other strains from clinical specimens fell into clades II and IV, and none of the clinical isolates clustered within clades III and V.
Correlations among morphology, maximal growth temperatures, minimal inhibitory concentrations (MICs) of antifungal agents, and phylogeny of isolates within the section Fumigati were also analyzed (Fig. 2). Scanning electron microscopy examination of these isolates showed that the conidial ornamentations of isolates belonging to clades I and V were lobate-reticulate (Kozakiewicz 1989), while those of A. viridinutans were intermediate between lobate-reticulate and microtuberculate. All strains in clade II and the six variant isolates in clade IV (IFM 5058, 51744, 53867, 53868, 54302 and CBM FD-0143) had conidia with microtuberculate ornamentation. These six strains are very closely related to N. udagawae (Horie et al. 1995), a heterothallic species isolated from soil in Brazil. However, mating between these strains and N. udagawae did not occur. It is often difficult to perform successful mating experiments on clinical isolates and fungi that have been routinely sub-cultured. Therefore these strains need to be investigated further before they are identified as the anamorphic state of N. udagawae.
Fig. 2.
Correlation among phylogeny, detailed morphology and maximal growth temperatures on Aspergillus section Fumigati. The letters L and M refer to the conidial ornamentation as observed by scanning electron microscopy.
The maximal growth temperatures and MICs of antifungal agents were also examined. The maximal growth temperatures of clades I, II, III, IV and V were above 50 °C, 45 °C, 45 °C, 42 °C and 42 °C, respectively. These phenotypic data may be useful for classification of species within Aspergillus section Fumigati. The isolates of A. lentulus demonstrated lower in vitro susceptibilities to amphotericin B than other isolates in section Fumigati (Balajee et al. 2005a). In conclusion, these studies showed that species of Aspergillus section Fumigati were divisible into five distinct clades by molecular analyses. Further, results revealed good correlation between phylogenetic and phenotypic characteristics. Comparative sequence analyses of the benA and cal regions offered good resolution and can be used for species delimitation within the section Fumigati.
Molecular phylogeny in Aspergillus section Terrei
Although A. terreus is a less common cause of invasive pulmonary aspergillosis when compared to A. fumigatus, infections due to these aspergilli appear to be increasing in frequency in certain hospitals worldwide (Baddley et al. 2003; Lass-Florl et al. 2000). Infections due to these organisms are difficult to treat because of both in vitro and in vivo refractoriness of the organism to the antifungal drug amphotericin B. In addition, A. terreus often causes disseminated infection with increased lethality compared with other Aspergillus spp. (Iwen et al. 1998; Steinbach et al. 2004; Walsh et al. 2003). Aspergillus terreus may be nosocomial in origin with potential reservoirs including construction activity, soil of potted plants, and water distribution systems in hospital environments (Lass-Florl et al. 2000; Anaissie et al. 2002; Flynn et al. 1993). In spite of the emerging threat due to this opportunistic pathogen, little is known about the genetic diversity and population structure of A. terreus.
A. terreus grows on potato dextrose agar at 25 °C as beige to buff to cinnamon brown colonies with reverse of the colony appearing yellow. Microscopically, conidial heads are biseriate and columnar with smooth walled conidiophores; conidia are globose and smooth. Globose, sessile, hyaline accessory conidia are frequently produced on submerged hyphae and are also produced in vivo during infection. Based on these phenotypical characteristics, A. terreus has been described as the only member of section Terrei and includes two varieties - A. terreus var. africanus and A. terreus var. aureus. However, molecular studies using the D1-D2 regions of the 28S RNA and the ITS regions (intergenic spacer regions 1 and 2 including the 5.8S rRNA) have indicated that this section should be expanded to include a number of other species (Varga et al. 2005). Recently, a three locus phylogenetic approach using partial regions of the protein coding genes β-tubulin, calmodulin and enolase has been attempted to characterise the genetic variability of a large number of A. terreus isolates [Balajee et al. in prep.]. These results suggest that A. terreus var. aureus should be raised to species status and that several cryptic species probably exist within isolates identified as A. terreus. Interestingly, members of one cryptic species/clade included clinical isolates recovered predominantly as colonising agents in the immunocompetent population [Balajee et al. in prep.]. Although the results of this study demonstrated the usefulness of the three-locus sequence strategy for species recognition in section Terrei, comparative sequence analyses of the benA region alone appeared to be a good marker for species recognition in this section.
Emericella species causing invasive infections
Invasive infections caused by Emericella species are uncommon in humans. Infections due to E. nidulans (anamorph Aspergillus nidulans) appear to occur predominantly in patients with chronic granulomatous disease (CGD), a rare disorder of phagocytes in which absence of superoxide and hydrogen peroxide production in phagocytes predisposes patients to bacterial and fungal infections. Invasive E. nidulans infections in this patient group are associated with a higher mortality than those caused by A. fumigatus (Dotis et al. 2003). E. nidulans is rarely encountered in other patient populations at risk for aspergillosis, such as those with neutropenia from myeloablative chemotherapy or recipients of a hematopoietic stem cell transplant. The lung is the most common site of infection, followed by subcutaneous or liver abscess, suppurative adenitis, osteomyelitis, fungemia, cellulitis and meningitis (van”t Hek et al. 1998; Winkelstein et al. 2000).
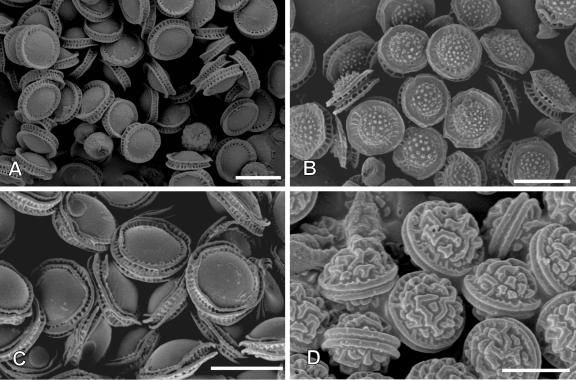
The identification of E. nidulans in clinical microbiology laboratories is commonly based on the characteristic microscopic morphology. The conidiophore typically shows metulae and phialides arranged on the upper part of the flask-shaped vesicle. E. nidulans produces dark green conidia (asexual spores) on brown-tinged conidiophores, and characteristic cleistothecia contain asci with 8 purple-red ascospores (Fig. 3). Hülle cells, thickened large cells associated with cleistothecia, are often abundant in E. nidulans. Another species within the genus Emericella that causes infection in humans is E. quadrilineata, which was reported to cause onychomycosis in one patient and sinusitis in two patients with hematological malignancy (Drakos et al. 1993; Gugnani et al. 2004; Polacheck et al. 1992). The morphologic features that distinguish E. nidulans from E. quadrilineata can be detected only by electron microscopy: the ascospores of E. nidulans have two longitudinal crests, while E. quadrilineata has four short equatorial crests.
Fig. 3.
Scanning electronmicroscopy of ascospores of Emericella species. A. E. nidulans. B. E. echinulata. C. E. quadrilineata. D. E. rugulosa. Scale bars = 5 μm.
A recent cluster of infections caused by Emericella species involving four patients was found to be due to E. quadrilineata based on sequence-based analysis of ITS1 and ITS2 regions [Verweij et al. in prep.]. Morphologically, the isolates were identical to E. nidulans. The identification of the strains involved in the cluster was further confirmed to be E. quadrilineata by sequence analysis of partial regions of the β-tubulin and the calmodulin genes [Verweij et al. in prep.]. In addition, several strains from the collection of the Centraalbureau voor Schimmelcultures were included in the study as well as clinical E. nidulans and E. quadrilineata isolates from published cases or private culture collections (Dotis et al. 2004). Based on the sequence-based analysis, several isolates were reclassified. The availability of molecular techniques in addition to morphological identification was shown to describe a role of E. quadrilineata as opportunistic fungal pathogen. The use of these techniques will help to identify and discriminate more accurately within the current fungal species and give more insight into the pathogenesis of fungal infection.
Pathogenic species in section Usti
A. ustus is a world-wide occurring species commonly found in food, soil and indoor environments (Samson et al. 2002). Aspergillus ustus is also isolated from clinical specimens; however, invasive infections caused by this species are uncommon. In the review by Panackal et. al. (2006), 21 documented cases of human infection were reported in the literature. Since this review, two more studies, a case of fungal endophthalmitis and a cluster of eye infections, have been published (Saracli et al. 2007; Yildiran et al. 2006). The true incidence of infections caused by this species is probably higher than reported, as there are cases where the fungus has not been identified or other instances where the organism was identified but remains unpubl. (Chakrabarti et al. 1998). The mortality associated with invasive aspergillosis caused by A. ustus seems strikingly high - of the 22 reported cases only 6 patients survived. One main reason for this high mortality rate could be the decreased susceptibilities of A. ustus to various antifungal drugs. In vitro susceptibility testing showed that this species has decreased susceptibilities to the antifungal drugs amphotericin B, caspofungin, itraconazole, voriconazole and posaconazole but is susceptible to the allylamine terbinafine (Yildiran et al. 2006; Garcia-Martos et al. 2005; Gene et al. 2001; Pavie et al. 2005).
A ustus (Bainier) Thom and Church (1926), described in 1881 by Bainier as Sterigmatocystis usta, was placed by Thom and Raper together with A. granulosus, in the A. ustus group. This group was revised by Raper and Fennell, who used a broad description of A. ustus and added A. puniceus, A. panamensis, A. conjunctus, and A. deflectus. The use of the name “groups”, a category without nomenclatural standing, was abandoned, and infrageneric taxa were formalised. Subgenera and sections were created and A. ustus became the type species of section Usti, which was placed in the subgenus Nidulantes (Gams et al. 1985). Peterson (2000) compared this phenotype-based classification system with the phylogenetic relationships based on the D1 and D2 regions of the large subunit ribosomal RNA (lsu-DNA). These data show that the type strain of A. ustus (and therefore also the section Usti) was in the subgenus Nidulantes. Because the entire section Usti branched between Emericella species, Usti was deleted and placed in section Nidulantes. However, the invalidation of section Usti by Peterson is rejected here since multiple sections can be linked to one teleomorph, and since the species also form a distinct clade within the subgenus Nidulantes. Based on the study of Peterson (2000), A. ustus, A. puniceus and A. pseudodeflectus are true members of the section Usti, while A. deflectus is tentatively placed in section Nidulantes and A. panamensis and A. conjunctus are in section Sparsi.
Of all the members in the section Usti, A. ustus is most often reported to be a causal agent of invasive infection, whereas A. puniceus and A. pseudodeflectus have never been mentioned in relation with infections. Only one report in available about A. granulosus, where this organism was described as the causal agent of a disseminated infection in a cardiac transplant patient (Fakih et al. 1995). A MLST study using the partial regions of β-tubulin, calmodulin and actin genes showed that A. ustus strains received from multiple centers (Panackal et al. 2006; Yildiran et al. 2006; Verweij et al. 1999) revealed the presence of a new species A. calidoustus (sp. nov.) from predominantly clinical samples [Varga et. al., unpubl. data]. Phenotypic differences between A. ustus and A. calidoustus are easy to recognise, since the latter grows rapidly at 37 °C, while the former does not. Nevertheless, other undescribed species, which are also able to grow at 37 °C, are present in section Usti and the occurrence of these species in clinical samples remains unknown. Thus, identification solely based on morphology appears difficult and unreliable. An additional problem with members of section Usti is that these species rarely cause invasive infections, which makes identification even more difficult. Combining all of above mentioned details, the use of morphology in combination with sequence data is recommended as an approach which will generate a less subjective and more reliable result.
Analyses of the D1-D2 sequences currently deposited in GenBank showed that two main clades are present in the section Usti. In one clade the type cultures of A. ustus (U29791; NRRL 275), A. puniceus (AY216673; CBS 495.65) and A. ustus var. laevis are present (U29788; NRRL 1852); the other clade includes the types of A. granulosus (AF454165; CBS 119.58) and A. pseudodeflectus (AF433123; NRRL 6135). However, the D1-D2 region does not have enough variation for species delimitation. The presence of these two main clades is also confirmed by a phylogenetic analysis of the ITS region of the ribosomal RNA (Varga et. al., unpubl. data). Although identification of medically important aspergilli based on ITS sequence data is more reliable than that based on D1-D2 data (Hinrikson et al. 2005), it is also known that the genetic variability within the ITS region is not sufficient and that some Aspergillus species share identical sequences. This is also the case within the section Usti, where A. pseudodeflectus and A. calidoustus could not be discriminated on ITS data alone [Varga et al. in prep.]. For correct species identification within this section, it is recommended to use the protein coding genes rather than ITS or D1-D2 data. Partial regions of β-tubulin, calmodulin and actin genes were tested and gave good resolution and are therefore excellent identification markers within the section Usti.
RECOMMENDATIONS
The preceding presentations clearly demonstrate that a multi locus sequence identification method where multiple genes (or portions there of) are sequenced and the resultant data are analyzed by phylogenetic methods is a robust strategy for species recognition within the genus Aspergillus. However, this methodology involves significant cost and phylogenetic expertise that are limiting factors in most clinical microbiology laboratories. Additionally, consideration should also be given to the fact that most of these isolates may not be true causative agents of disease and therefore may not warrant species level identification in a diagnostic laboratory. Taken together, a universal single marker that would rapidly and accurately identify Aspergillus isolates to the species level would help support diagnostic microbiology laboratories in their routine identification efforts. Comparative sequence-based methods are finding a place in the clinical microbiology laboratory for fungal species identification, and there is a need for a consensus recommendation for such global markers that can be used with confidence for this purpose.
Recently, the international group of experts that gathered for the workshop entitled “Aspergillus Systematics in the Genomic Era” reviewed research data presented from research groups worldwide on recent genomic investigations, secondary metabolite analyses, multi locus phylogenetic analyses of the genus Aspergillus, and sequence based identification schemes for previously recognised human pathogens within the genus. Deriving from the entire proceedings of the workshop in general and from the session on clinically relevant aspergilli in particular, the following recommendations were proposed as a first step towards formulating a unified sequence-based identification scheme for the genus Aspergillus.
Although not discussed in this session, previous publications and research work presented elsewhere during the meeting revealed the utility of ITS region for identification of Aspergillus isolates to the section level. Thus, as a first recommendation, comparative sequence analyses of the ITS regions, specifically the ITS1 and ITS2 non-coding regions flanking the 5.8S rDNA, was suggested as an appropriate locus for identification of Aspergillus isolates to the level of subgenus/section. Use of the ITS sequence should be sufficient to place most isolates within the appropriate section, but will not provide sufficient sensitivity to discriminate among individual species within the section. Realising the limitations of the ITS regions to identify intrasection species, sequence comparison of the β-tubulin region for species identification within the section (discussed in detail throughout this manuscript) was proposed as the second recommendation.
The ITS region is a convenient universal marker for fungal species identification and most clinical microbiology laboratories still rely on morphology based identification; both these strategies will not identify species within the sections. Considering these two factors and to help support species reporting in clinical microbiology laboratories, the term “complex” was proposed as an alternate to “section”. Thus, whether clinical microbiology laboratories rely on morphological identification methods or an ITS based sequencing strategy, it is advised to report species within the sections Fumigati, Flavi, Nidulantes, Usti and Terrei as “species complex”, for instance, “A. fumigatus complex”. Results reported in this manner should be interpreted as indicating the placement of the isolate within the species complex, but not necessarily indicating a species within that section. The microbiologist and the clinician can then jointly decide whether further DNA sequencing using a protein coding locus is required to identify the individual into a particular species within the section/complex. This information may be necessary when investigating an outbreak, when dealing with infections refractory to antifungal therapy, or when performing applied epidemiologic studies. In all cases, communication between the clinician and the microbiologist is critical to provide results that benefit patient care with the highest value and the least cost.
It must be remembered that these are tentative recommendations that are based on research data available at this time. It should also be reiterated that comparative sequence analyses should be used in tandem with morphological examination for an identification scheme to be successful. We believe that these recommendations will help stimulate further discussion, encourage validations of appropriate loci for comparative sequence strategies, and focus research studies on the clinical relevance of recovering these species from our patient populations.
The contributors to this session were: A. Balajee, Pathogenic species in Aspergillus section Terrei; T. Yaguchi, Pathogenic species in Aspergillus subgenus Fumigati; B. Hong, Speciation in Aspergillus subgenus Fumigati; P. Verweij, Emericella pathogens; J. Houbraken, Pathogenic species in section Usti.
DISCLAIMER
The findings and conclusions in this article are those of the author(s) and do not necessarily represent the views of the CDC.
References
- Aho R, Horie Y, Nishimura K, Miyaji M (1994). Aspergillus arvii spec. nov., a new animal pathogen? Mycoses 37: 389-392. [DOI] [PubMed] [Google Scholar]
- Anaissie EJ, Stratton SL, Dignani MC, Summerbell RC, Rex JH, Monson TP, Spencer T, Kasai M, Francesconi A, Walsh TJ (2002). Pathogenic Aspergillus species recovered from a hospital water system: a 3-year prospective study. Clinical Infectious Diseases 34: 780-789. [DOI] [PubMed] [Google Scholar]
- Baddley JW, Pappas PG, Smith AC, Moser SA (2003). Epidemiology of Aspergillus terreus at a university hospital. Journal of Clinical Microbiology 41: 5525-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Gribskov J, Brandt M, Ito J, Fothergill A, Marr KA (2005b). Mistaken identity: Neosartorya pseudofischeri and its anamorph masquerading as Aspergillus fumigatus. Journal of Clinical Microbiology 43: 5996-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA (2005a). Aspergillus lentulus sp nov., a new sibling species of A. fumigatus. Eukaryotic Cell 4: 625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Nickle D, Varga J, Marr KA (2006). Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryotic Cell 5: 1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee SA, Lindsley MD, Iqbal N, Ito J, Pappas PG, Brandt ME (2007). A non-sporulating clinical isolate identified as Petromyces alliaceus (anamorph Aspergillus alliaceus) by morphological and sequence based methods. Journal of Clinical Microbiology 45: 2701-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Gupta V, Biswas G, Kumar B, Sakhuja VK (1998). Primary cutaneous aspergillosis: our experience in 10 years. Journal of Infection 37: 24-27. [DOI] [PubMed] [Google Scholar]
- Chim CS, Ho PL, Yuen KY (1998). Simultaneous Aspergillus fischeri and herpes simplex pneumonia in a patient with multiple myeloma. Scandinavian Journal of Infectious Diseases 30: 190-191. [DOI] [PubMed] [Google Scholar]
- Coriglione G, Stella G, Gafa L, Spata G, Oliveri S, Padhye AA, Ajello L (1990). Neosartorya fischeri var fischeri (Wehmer) Malloch and Cain 1972 (anamorph: Aspergillus fischerianus Samson and Gams 1985) as a cause of mycotic keratitis. European Journal of Epidemiology 6: 382-385. [DOI] [PubMed] [Google Scholar]
- Dotis J, Panagopoulou P, Filioti J, Winn R, Toptsis C, Panteliadis C, Roilides E (2003). Femoral osteomyelitis due to Aspergillus nidulans in a patient with chronic granulomatous disease. Infection 31: 121-124. [DOI] [PubMed] [Google Scholar]
- Dotis J, Roilides E (2004). Osteomyelitis due to Aspergillus spp. in patients with chronic granulomatous disease: comparison of Aspergillus nidulans and Aspergillus fumigatus. International Journal of Infectious Diseases 8: 103-110. [DOI] [PubMed] [Google Scholar]
- Drakos PE, Nagler A, Or R, Naparstek E, Kapelushnik J, Engelhard D, Rahav G, Ne”emean D, Slavin S (1993). Invasive fungal sinusitis in patients undergoing bone marrow transplantation. Bone Marrow Transplantation 12: 203-208. [PubMed] [Google Scholar]
- Fakih MG, Barden GE, Oakes CA, Berenson CS (1995). First reported case of Aspergillus granulosus infection in a cardiac transplant patient. Journal of Clinical Microbiology 33: 471-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn PM, Williams BG, Hetherington SV, Williams BF, Giannini MA, Pearson TA (1003). Aspergillus terreus during hospital renovation. Infection Control and Hospital Epidemiology 14: 363-365. [DOI] [PubMed] [Google Scholar]
- Gams W, Christensen M, Onions AH, Pitt JI, Samson RA (1985). Infrageneric taxa of Aspergillus. In: Advances in Penicillium and Aspergillus systematics. (Samson RA, Pitt JI, eds.) New York: Plenum Press: 55-62.
- Garcia-Martos P, Garcia-Agudo L, Gutierrez-Calzada J, Ruiz-Aragon J, Saldarreaga A, Marin P (2005). In vitro activity of amphotericin B, itraconazole and voriconazole against 20 species of Aspergillus using the Sensititre microdilution method. Enfermedades Infecciosas y Microbiología Clínica 23: 15-18. [DOI] [PubMed] [Google Scholar]
- Gene J, Azon-Masoliver A, Guarro J, De Febrer G, Martinez A, Grau C, Ortoneda M, Ballester F (2001). Cutaneous infection caused by Aspergillus ustus, an emerging opportunistic fungus in immunosuppressed patients. Journal of Clinical Microbiology 39: 1134-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J, Chomicki J, Brandsberg JW, Jones R, Hammerman KJ (1973). Pulmonary aspergillosis caused by Aspergillus fischeri var. spinosus: report of a case and value of serologic studies. American Jorunal of Clinical Pathology 60: 861-866. [DOI] [PubMed] [Google Scholar]
- Gori S, Pellegrini G, Filipponi F, Capanna SD, Biancofiore G, Mosca F, Lofaro A (1998). Pulmonary aspergillosis caused by Neosartorya fischeri (Aspergillus fischerians) in a liver transplant recipient. Journal de Mycologie Medicale 8: 105-107. [Google Scholar]
- Guarro J, Kallas EG, Godoy P, Karenina A, Gene J, Stchigel A, Colombo AL (2002). Cerebral aspergillosis caused by Neosartorya hiratsukae, Brazil. Emerging Infectious Diseases 8: 989-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gugnani HC, Vijayan VK, Tyagi P, Sharma S, Stchigel AM, Guarro J (2004). Onychomycosis due to Emericella quadrilineata. Journal of Clinical Microbiology 42: 914-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hek LG van ”t, Verweij PE, Weemaes CM, van Dalen R, Yntema JB, Meis JF (1998). Successful treatment with voriconazole of invasive aspergillosis in chronic granulomatous disease. American Journal of Respiratory anod Critical Care Medicine 157: 1694-1696. [DOI] [PubMed] [Google Scholar]
- Hinrikson HP, Hurst SF, Lott TJ, Warnock DW, Morrison CJ (2005). Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. Journal of Clinical Microbiology 43: 2092-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Cho HS, Shin HD, Frisvad JC, Samson RA (2006). Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology 56: 477-486. [DOI] [PubMed] [Google Scholar]
- Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA (2005). Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia 97: 1316-1329. [DOI] [PubMed] [Google Scholar]
- Horie Y, Miyaji M, Nishimura K, Franco MF, Coelho KIR (1995). New and interesting species of Neosartorya from Brazilian soil Mycoscience 36: 199-204. [Google Scholar]
- Horie Y, Miyaji M, Nishimura K, Taguchi H, Udagawa S (1993). Aspergillus fumisynnematus, a new species from Venezuelan soil. Transactions of the Mycological Society Japan 34: 33-37. [Google Scholar]
- Iwen PC, Rupp ME, Langnas AN, Reed EC, Hinrichs SH (1998). Invasive pulmonary aspergillosis due to Aspergillus terreus: 12-year experience and review of the literature. Clinical Infectious Diseases 26: 1092-1097. [DOI] [PubMed] [Google Scholar]
- Katz ME, Dougall AM, Weeks K, Cheetham BF (2005). Multiple genetically distinct groups revealed among clinical isolates identified as atypical Aspergillus fumigatus. Journal of Clinical Microbiology 43: 551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ME, McLoon M, Burrows S, Cheetham BF (1998). Extreme DNA sequence variation in isolates of Aspergillus fumigatus. FEMS Immunolology and Medical Microbiology 20: 283-288. [DOI] [PubMed] [Google Scholar]
- Kozakakiewicz Z (1989). Aspergillus species in stored products. Mycological Papers 161: 1-188. [Google Scholar]
- Kwon-Chung KJ, Kim SJ (1974). A second heterothallic Aspergillus. Mycologia 66: 628-638. [PubMed] [Google Scholar]
- Lass-Florl C, Rath P, Niederwieser D, Kofler G, Wurzner R, Krezy A, Dierich MP (2000). Aspergillus terreus infections in haematological malignancies: molecular epidemiology suggests association with in-hospital plants. Journal of Hospital Infection 46: 31-35. [DOI] [PubMed] [Google Scholar]
- Padhye AA, Godfrey JH, Chandler FW, Peterson SW (1994). Osteomyelitis caused by Neosartorya pseudofischeri. Journal of Clinical Microbiology 32: 2832-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano L, Caira M, Picardi M, Candoni A, Melillo L, Fianchi L, Offidani M, Nosari A (2007). Invasive Aspergillosis in patients with acute leukemia: update on morbidity and mortality-SEIFEM-C Report. Clinical Infectious Diseases 44: 1524-1525. [DOI] [PubMed] [Google Scholar]
- Panackal AA, Imhof A, Hanley EW, Marr KA (2006). Aspergillus ustus infections among transplant recipients. Emerging Infectious Diseases 12: 403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavie J, Lacroix C, Hermoso DG, Robin M, Ferry C, Bergeron A, Feuilhade M, Dromer F, Gluckman E, Molina JM, Ribaud P (2005). Breakthrough disseminated Aspergillus ustus infection in allogeneic hematopoietic stem cell transplant recipients receiving voriconazole or caspofungin prophylaxis. Journal of Clinical Microbiology 43: 4902-4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SW (2000). Phylogenetic relationships in Aspergillus based upon rDNA sequence analysis. In: Integration of modern taxonomic methods for Penicillium and Aspergillus classification. (Samson RA, Pitt JI, eds.) Amsterdam: Harwood Academic Publishers: 323-355.
- Polacheck I, Nagler A, Okon E, Drakos P, Plaskowitz J, Kwon-Chung KJ (1992). Aspergillus quadrilineatus, a new causative agent of fungal sinusitis. Journal of Clinical Microbiology 30: 3290-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper BK, Fennell DI (1965). The Genus Aspergillus. Baltimore: Williams and Wilkins.
- Samson RA, Hoekstra ES, Frisvad JC, Filtenborg O (eds) (2002). Introduction to food and airborne fungi edn 6. Utrecht: Centraal Bureau voor Schimmelcultures.
- Saracli MA, Mutlu FM, Yildiran ST, Kurekci AE, Gonlum A, Uysal Y, Erdem U, Basustaoglu AC, Sutton DA (2007). Clustering of invasive Aspergillus ustus eye infections in a tertiary care hospital: A molecular epidemiologic study of an uncommon species. Medical Mycology 45: 377-384. [DOI] [PubMed] [Google Scholar]
- Steinbach WJ, Benjamin DK, Jr., Kontoyiannis DP, Perfect JR, Lutsar I, Marr KA, Lionakis MS, Torres HA, Jafri H, Walsh TJ (2004). Infections due to Aspergillus terreus: a multicenter retrospective analysis of 83 cases. Clinical Infectious Diseases 39: 192-198. [DOI] [PubMed] [Google Scholar]
- Summerbell RC, de Repentigny L, Chartrand C, St Germain G (1992). Graft-related endocarditis caused by Neosartorya fischeri var. spinosa. Journal of Clinical Microbiology 30: 1580-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Tóth B, Kocsubé S, Farkas B, Szakács G, Téren J, Kozakiewicz Z (2005). Evolutionary relationships among Aspergillus terreus isolates and their relatives. Antonie Van Leeuwenhoek 88: 141-150. [DOI] [PubMed] [Google Scholar]
- Varga J, Tóth B, Rigó K, Debets F, Kozakiewicz Z (2000). Genetic variability within the Aspergillus viridinutans species. Folia Microbiologica 45: 423-428. [DOI] [PubMed] [Google Scholar]
- Verweij PE, van den Bergh MF, Rath PM, de Pauw BE, Voss A, Meis JF (1999). Invasive aspergillosis caused by Aspergillus ustus: case report and review. Journal of Clinical Microbiology 37: 1606-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TJ, Petraitis V, Petraitiene R, Field-Ridley A, Sutton D, Ghannoum M, Sein T, Schaufele R, Peter J, Bacher J, Casler H, Armstrong D, Espinel-Ingroff A, Rinaldi MG, Lyman CA (2003). Experimental pulmonary aspergillosis due to Aspergillus terreus: pathogenesis and treatment of an emerging fungal pathogen resistant to amphotericin B. Journal of Infectious Diseases 188: 305-319. [DOI] [PubMed] [Google Scholar]
- Winkelstein JA, Marino MC, Johnston RB, Jr., Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, Buckley RH, Foster CB, Chanock SJ, Dickler H (2000). Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine 79: 155-169. [DOI] [PubMed] [Google Scholar]
- Yaguchi T, Horie Y, Tanaka R, Matsuzawa T, Ito J, Nishimura K (2007). Molecular phylogenetics of multiple genes on Aspergillus section Fumigati isolated from clinical specimens in Japan. Nippon Ishinkin Gakkai Zasshi 48: 37-46. [DOI] [PubMed] [Google Scholar]
- Yildiran ST, Mutlu FM, Saracli MA, Uysal Y, Gonlum A, Sobaci G, Sutton DA (2006). Fungal endophthalmitis caused by Aspergillus ustus in a patient following cataract surgery. Medical Mycology 44: 665-669. [DOI] [PubMed] [Google Scholar]