Abstract
Purpose
Mutations in the SOX2 and CHX10 genes have been reported in patients with anophthalmia and/or microphthalmia. In this study, we evaluated 34 anophthalmic/microphthalmic patient DNA samples (two sets of siblings included) for mutations and sequence variants in SOX2 and CHX10.
Methods
Conformational sensitive gel electrophoresis (CSGE) was used for the initial SOX2 and CHX10 screening of 34 affected individuals (two sets of siblings), five unaffected family members, and 80 healthy controls. Patient samples containing heteroduplexes were selected for sequence analysis. Base pair changes in SOX2 and CHX10 were confirmed by sequencing bidirectionally in patient samples.
Results
Two novel heterozygous mutations and two sequence variants (one known) in SOX2 were identified in this cohort. Mutation c.310 G>T (p. Glu104X), found in one patient, was in the region encoding the high mobility group (HMG) DNA-binding domain and resulted in a change from glutamic acid to a stop codon. The second mutation, noted in two affected siblings, was a single nucleotide deletion c.549delC (p. Pro184ArgfsX19) in the region encoding the activation domain, resulting in a frameshift and premature termination of the coding sequence. The shortened protein products may result in the loss of function. In addition, a novel nucleotide substitution c.*557G>A was identified in the 3′-untranslated region in one patient. The relationship between the nucleotide change and the protein function is indeterminate. A known single nucleotide polymorphism (c. *469 C>A, SNP rs11915160) was also detected in 2 of the 34 patients. Screening of CHX10 identified two synonymous sequence variants, c.471 C>T (p.Ser157Ser, rs35435463) and c.579 G>A (p. Gln193Gln, novel SNP), and one non-synonymous sequence variant, c.871 G>A (p. Asp291Asn, novel SNP). The non-synonymous polymorphism was also present in healthy controls, suggesting non-causality.
Conclusions
These results support the role of SOX2 in ocular development. Loss of SOX2 function results in severe eye malformation. CHX10 was not implicated with microphthalmia/anophthalmia in our patient cohort.
Introduction
Anophthalmia (an absence of eye structures) and/or microphthalmia (an abnormally small eye) are rare disorders with a prevalence of 0.2–0.4 per 10,000 births in developed countries [1-3]. One or both eyes are affected in isolation or as part of other birth defects [4-9]. The disease exhibits diverse patterns of genetic inheritance, and the severity is variable due to the genetic heterogeneity of the ocular malformation.
Mutations in several human genes are associated with anophthalmia and microphthalmia. Among them, mutations in SOX2 appear to account for most cases [4,10-14]. SOX2, which is located at chromosome 3q26.3-q27, encodes 317 amino acids that belong to the high-mobility-group (HMG) DNA-binding protein family. Expressed in embryonic stem cells and a wide variety of tissues during early development, SOX2 plays an important role in cell differentiation and early organogenesis [15,16]. In ocular tissues, the inhibition of SOX2 expression in the developing Xenopus retina has been shown to reduce cell proliferation and allows cells to develop into non-neuronal cell types [17]. Working in concert with PAX6, SOX2 regulates downstream target genes such as the crystalline genes to guide early lens development [18].
Mutations in SOX2 account for approximately 10% of anophthalmia and microphthalmia cases [4,10,19]. Previously reported cases of anophthalmia and microphthalmia were associated with SOX2 whole-gene deletions or coding sequence mutations [4,10-13,19,20]. In this study, a cohort of 34 anophthalmia/microphthalmia patients was screened for mutations in SOX2. Two novel heterozygous mutations and two sequence variants (one novel, one known SNP) were identified in 6 (two siblings) out of 34 patients. This confirms the importance of SOX2 mutation screening in patients with anophthalmia/microphthalmia. In screening unaffected family members, we also found an individual with a pathogenic allele suggesting the novel finding that autosomal dominant SOX2 mutations can be non-penetrant.
As part of our large scale screening campaign, mutation analysis of CHX10 was also performed. The homeodomain protein CHX10 has been shown to be essential for ocular development [21], and autosomal recessive mutations of the gene have been implicated in microphthalmia in both humans and mice [22-24]. Screening of the same cohort of anophthalmia/microphthalmia patients identified two synonymous sequence variants in exon 3 and one non-synonymous sequence variant in exon 5. This non-synonymous polymorphism was also present in normal controls. We did not find sequence variants in other exons of CHX10, suggesting that CHX10 mutations are less frequent in patients with anophthalmia.
Methods
Subjects
To identify existing and potentially novel mutations of SOX2 and CHX10 in a cohort of anophthalmia and microphthalmia patients, we undertook a candidate gene screening approach. We screened 34 affected individuals (two sets of siblings), five unaffected family members, and 80 healthy controls. Patient demographics are summarized in Appendix 1. The study was approved by the Institutional Review Board for Human Subject Research at the Children’s Hospital of Philadelphia and at the Albert Einstein Medical Center. The study also conformed to the tenets of the Declaration of Helsinki. Informed consent was obtained from all subjects whose blood and DNA samples were used in the analysis. Peripheral blood samples (10–20 ml) were collected, and genomic DNA was extracted using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN).
Conformation-sensitive-gel-electrophoresis
Conformation-sensitive-gel-electrophoresis (CSGE) was used for initial SOX2 and CHX10 gene screenings. Standard polymerase chain reaction (PCR) was performed on genomic DNA of affected patients, unaffected relatives, and healthy controls to amplify SOX2 and CHX10 exonic sequences (primer sets are available upon request). An additional 120 base pairs of intronic sequences at both the 5′-end and 3′-end were also included. PCR amplification was performed in a reaction volume of 25 μl containing 50 ng of genomic DNA, 200 μM dNTP, 0.4 μM of each primer pair, 1.5 mM MgCl2, 1X PCR buffer, 1X Q-solution, and 1.25 units of Taq polymerase (Qiagen, Valencia, CA). A single annealing temperature of 55 °C was used for all primers. PCR products were separated by electrophoresis in an 8% polyacrylamide gel for detection of heteroduplexes and homoduplexes. Patient samples containing heteroduplexes were selected for sequence analysis. Base pair changes in SOX2 and CHX10 were confirmed by sequencing bidirectionally in patient samples.
Results
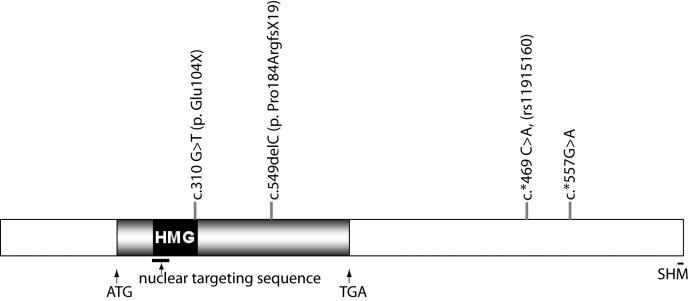
Of the 34 patients (32 independent probands) analyzed, three (two siblings) showed novel mutations in coding sequence and three had two sequence variants in the 3′-untranslated region (UTR) of SOX2 (Figure 1 and Table 1). Mutation, c.310 G>T (p. Glu104X), found in one patient, was in the region encoding the HMG DNA-binding domain and resulted in a change from glutamic acid to a stop codon. The second mutation, noted in two affected siblings, was a single nucleotide deletion c.549delC (p. Pro184ArgfsX19) in the region encoding the activation domain, resulting in a frameshift and premature termination of the coding sequence. In addition, a novel nucleotide substitution, c.*557G>A, was identified in the 3′-untranslated region in one patient, and a known single nucleotide polymorphism (c. *469 C>A, SNP rs11915160) was also detected in 2 out of 34 patients (Appendix 1).
Figure 1.
SOX2 gene structure and mutations in anophthalmia/microphthalmia. The 5′- and 3′- untranslated regions are shown as blank boxes, and the coding region is shown as a filled box. The start (ATG) and the stop (TGA) codons are indicated. Mutations detected in this study are indicated. The nuclear targeting sequence is underlined. HMG=high mobility group, G=guanosine, T=thymidine, C=cytidine, A=adenosine, Glu=glutamic acid, Pro=proline, Arg=arginine, X=stop, FS=frameshift.
Table 1. Sequence variations in SOX2.
| ID | Fragment 4 | Sequence results | Fragment 5 | Sequence results | Fragment 8 | Sequence results | Fragment 9 | Sequence results |
| 012A | homoduplex | homoduplex | heteroduplex | *469C>A1 | homoduplex | |||
| 013A | homoduplex | homoduplex | heteroduplex | *469C>A1 | homoduplex | |||
| 018A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 023A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 023I | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 025A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 027A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 028A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 029A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 030A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 031A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 033A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 034A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 035A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 037A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 038A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 040A | heteroduplex | 310G>T | homoduplex | homoduplex | homoduplex | |||
| 041A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 042A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 043A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 044A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 045A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 046A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 047A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 048A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 049A | homoduplex | homoduplex | homoduplex | heteroduplex | *557G>A | |||
| 050A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 051A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 052A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 053D | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 053A | homoduplex | homoduplex | homoduplex | homoduplex | ||||
| 054A | homoduplex | quadruplet | 549delC | homoduplex | homoduplex | |||
| 054H | homoduplex | triplet | 549delC | homoduplex | homoduplex | |||
| 055A | homoduplex | homoduplex | homoduplex | homoduplex |
The entire genomic sequence of SOX2 was screened by CSGE. Samples which show heteroduplex, triplet, and quadruplet on CSGE are sequenced bi-directionally. Homoduplex indicates two identical alleles. Heteroduplex, triplet, and quadruplet indicate different alleles. One allele is the wild type sequence and the other one is a sequence variant differing from the wild type one. Novel mutation at c.310 G>T results in a nonsense amino acid change at p. Glu104X. The novel single nucleotide deletion at c.549delC results in a frame shift mutation and premature termination at 19 amino acids downstream of the deletion site (p.Pro184ArgfsX19). The novel single nucleotide substitution at c.*557G>A is in the 3′-UTR and does not predictably change the SOX2 protein. Another single nucleotide substitution at *469 C>A is a known SNP (rs11915160) located in the 3′-UTR. G=guanosine, T=thymidine, C=cytidine, A=adenosine, Glu=glutamic acid, Pro=proline, Arg=arginine, X=Stop, FS=frameshift; 1SNP rs11915160
Patient 40A is an eight-year-old child with bilateral anophthalmia. Clinical features included dilated ventricles with ventriculo-peritoneal shunt placement for hydrocephalus, microcephaly, atrophic optic nerves, chiasm, posterior corpus callosum and splenium, and abnormal electroencephalograms with no overt seizures. The patient has short stature and delayed motor and mental development. He has an unusual ataxic gait but is independently mobile with a walker. He has begun to speak in three to four word sentences.
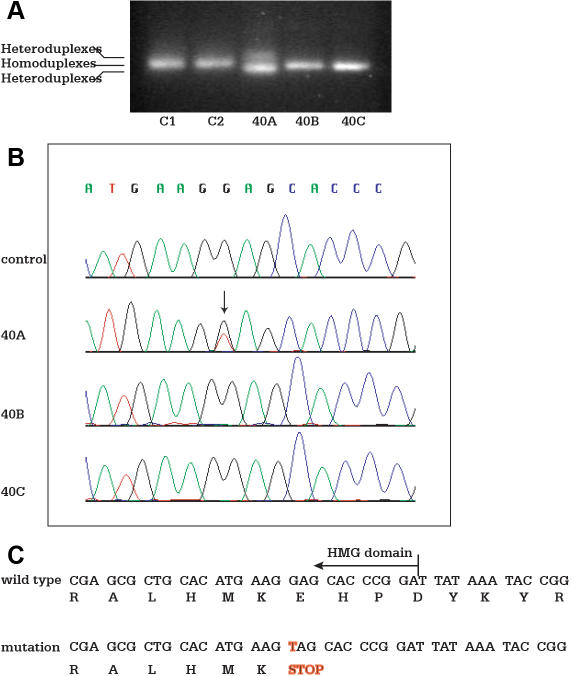
The amplified genomic fragments from the patient (40A) showed faster and slower migrating products comparing to that of the wild type observed in samples from the parents (40B and 40C) and normal controls (C1 and C2) with CSGE (Figure 2A). The heteroduplexes suggest a mutation in the fragment. The genomic DNA from this patient was sequenced bidirectionally. Two alleles were identified (Figure 2B), one with a wild type allele and one with a nucleotide substitution at c.310 G>T of the coding sequence, resulting in a nonsense amino acid change (p. Glu104X). The predicted shorter peptide is truncated at the end of the HMG domain, which results in the deletion of the entire COOH-terminal activation domain (Figure 2C). The mutation, located in the HMG domain, is not directly involved in DNA binding based on NMR-spectroscopy structure from the Protein Data Bank (PDB). Both parents were homozygous for the wild type allele.
Figure 2.
Mutation in the SOX2 HMG domain. Genomic DNA from affected patients, unaffected relatives, and healthy controls are amplified by PCR. PCR products are separated by electrophoresis in an 8% polyacrylamide gel for detection of heteroduplexes and homoduplexes. Genomic DNA samples are then sequenced for detection of sequence variants. A: Heteroduplexes detected by CSGE is shown. Lanes C1 and C2 represent the healthy controls; Lane 40A represents the proband; Lane 40B represents the mother of the proband; and Lane 40C is the father of the proband. Sample from patient 40A shows heteroduplex comparing to homoduplex observed in samples from the parents (40B and 40C) and normal controls (C1 and C2). B: Single nucleotide substitution at c.310 G>T of the coding sequence is identified in patient 40A, but not in the parents (40B and 40C) and normal controls (C1 and C2). C: Mutation at c.310 G>T results in a nonsense amino acid change at p. Glu104X as illustrated. HMG=high mobility group.
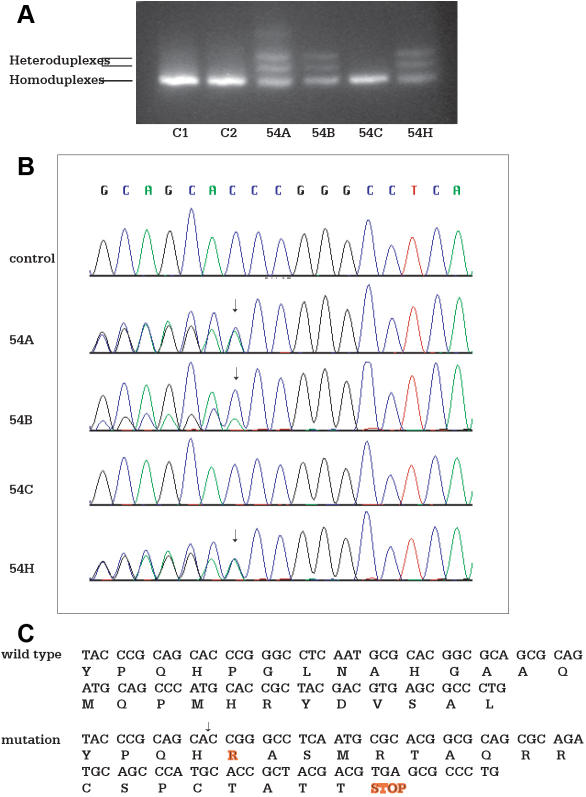
Patient 54A is a nine-year-old child with right anophthalmia and left microphthalmia. Her clinical features included a right middle fossa arachnoid cyst and partial absence of the posterior aspect of the corpus callosum including the splenium. She had mild hydrocephalus and partial complex seizures. Other abnormalities were growth and thyroid hormone deficiency, developmental delays, and a wide-based ataxic gait. She also presented with facial asymmetry with a beaked nose, small widely-spaced teeth, and diastasis recti. The parents were unaffected. CSGE analysis showed three slower migrating product bands in addition to the wild type, indicative of heterozygosity (Figure 3A). The affected sibling of the proband (54H), who was diagnosed with bilateral anophthalmia by ultrasound in utero, had two slower migrating bands and the wild type product. An amniocentesis was performed, and a chromosome study was that of a normal female with a 46XX karyotype. Her birthweight of 8 lbs 15 ounces and length of 52 cm are appropriate for a term infant. Her newborn hearing screening test was normal. An orbital ultrasound shortly after birth revealed the absence of globe structures bilaterally. A head MRI at age of four months showed partial agenesis of the corpus callosum and hypoplasia of the orbits. Minimal ocular tissue was noted in the orbit with visualization of the extraocular muscles and lacrimal glands. The optic nerves were not visualized. The maxillae were hypoplastic.
Figure 3.
Mutation in the SOX2 activation domain. Genomic DNA from affected patients, unaffected relatives, and healthy controls are amplified by PCR. PCR products are separated by electrophoresis in an 8% polyacrylamide gel for detection of heteroduplexes and homoduplexes. Genomic DNA samples are then sequenced for detection of sequence variants. A: Heteroduplexes detected by CSGE is shown. Lanes C1 and C2 represent the healthy controls; Lane 54A represents the proband; Lane 54B represents the mother of the proband; Lane 54C represents the father of the proband; and Lane 54H represents the affected sibling of the proband. The additional slower band in patient 54A may represent a conformational intermediate formed during gel electrophoresis, resulting in a different mobility compared to homoduplexes and the triplet. B: Sequence analysis shows two alleles in the proband 54A, the affected sibling 54H, and the clinically normal mother 54B. One allele is the wild type sequence and the other one is single nucleotide deletion at c.549delC. The father 54C is homozygous for the wild type allele. C: The single nucleotide deletion at c.549delC results in a frame shift mutation and premature termination at 19 amino acids downstream of the deletion site (p.Pro184ArgfsX19). The mutation is predicted to delete part of the C-terminal activation domain and to produce a truncated peptide.
At approximately nine months, she began treatment with thyroid supplementation for hypothyroidism. She is of a low growth percentile with normal growth hormone levels. She had urticaria pigmentosa at 14 months of age. At the age of 16 months, she sat without support, rolled, and babbled with consonants and vowels. She finger feeds and has no feeding difficulties. Her development is delayed but is more advanced than that of her sister, the proband, at the equivalent age. The mother has normal vision and an unremarkable ophthalmologic examination. She has normal motor abilities and intellect. A head MRI scan of the mother was unremarkable.
Sequence analysis detected two alleles in the proband, the affected sibling, and the clinically normal mother (54B). One allele was the wild type sequence and the other was a c.549delC (p.Pro184ArgfsX19) in the coding sequence (Figure 3B). The deletion changes the reading frame and results in a premature stop codon 19 amino acids downstream of the deletion site. The mutation is predicted to delete part of the COOH-terminal activation domain and to produce a truncated peptide (Figure 3C). The father (54C) of the proband was homozygous for the wild type allele.
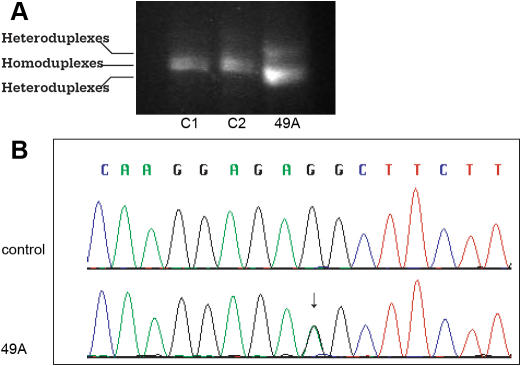
Patient 49A is a 17-year-old female with bilateral anophthalmia. Her clinical features include a ventricular-septal defect, which resolved without surgical intervention, and fusion of two primary teeth of the left lower jaw. She is developmentally normal and is currently applying to four-year colleges. The parents were unaffected. A heterozygous sequence variant was identified in this individual in the 3′-UTR by CSGE (Figure 4A). Sequence analysis detected two alleles, one with a wild type sequence and one with a nucleotide substitution, c.*557G>A (Figure 4B). The sequence alteration does not predictably change the SOX2 protein and is not a previously described SNP (dbSNP Build 128). A review of transcription factor databases did not uncover known regulatory elements associated with the nucleotide substitution. DNA was unavailable from additional unaffected family members of the proband.
Figure 4.
SOX2 mutation in the 3′- untranslated region (3′-UTR). Genomic DNA from the affected patient and healthy controls are amplified by PCR. PCR products are separated by electrophoresis in an 8% polyacrylamide gel for detection of heteroduplexes and homoduplexes. Genomic DNA samples are then sequenced for detection of sequence variants. A: Heteroduplexes detected by CSGE is shown. Lanes C1 and C2 represent the healthy controls and Lane 49A represents the proband. Sample from patient 49A shows heteroduplex comparing to homoduplex observed in samples from normal controls (C1 and C2). B: A nucleotide substitution at c.*557G>A of the 3′-UTR is identified in patient 49A, but not in normal controls (C). The sequence variant does not predictably change the SOX2 protein and is not a previously described SNP.
To determine that the above sequence alterations detected in anophthalmia and microphthalmia patients were not single nucleotide polymorphisms (SNPs), 80 healthy control individuals were examined by direct sequencing analysis. None of the above sequence alterations were found in the normal control subjects. During this screening, we also detected SNP c.*469 C>A in 2 out of 34 patients. The SNP is a known variant and located at 3′ UTR (SNP rs11915160).
To evaluate sequence variations in CHX10, the same cohort of patients were screened by CSGE and direct sequencing. The majority of sequence variations were detected in exon 3 (Table 2), and the mutations were all synonymous, c.471 C>T (p.Ser157Ser, rs35435463) and c.579 G>A (p. Gln193Gln, novel SNP). Another sequence variation was found in exon 5, c.871 G>A (p. Asp291Asn, novel SNP), in one affected individual resulting in a non-synonymous polymorphism. Using healthy controls, the same sequence variant was found, suggesting non-causality.
Table 2. Sequence variations in CHX10.
| Person ID | Exon 3 | Sequence results | Exon 5 | Sequence results |
| 012A | homoduplex | homoduplex | ||
| 013A | homoduplex | homoduplex | ||
| 018A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 023A | homoduplex | homoduplex | ||
| 023I | homoduplex | homoduplex | ||
| 025A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 027A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 029A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 030A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 031A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 033A | homoduplex | homoduplex | ||
| 034A | homoduplex | homoduplex | ||
| 035A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 037A | homoduplex | homoduplex | ||
| 038A | homoduplex | homoduplex | ||
| 040A | homoduplex | homoduplex | ||
| 041A | homoduplex | homoduplex | ||
| 042A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 043A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 044A | homoduplex | homoduplex | ||
| 045A | homoduplex | homoduplex | ||
| 046A | homoduplex | homoduplex | ||
| 047A | homoduplex | homoduplex | ||
| 048A | homoduplex | homoduplex | ||
| 049A | heteroduplex | c.579 G>A (Q193Q) | homoduplex | |
| 050A | homoduplex | homoduplex | ||
| 051A | homoduplex | homoduplex | ||
| 052A | homoduplex | homoduplex | ||
| 053A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 054A | homoduplex | homoduplex | ||
| 054H | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 055A | homoduplex | heteroduplex | c.871 G>A (p.D291N)* | |
| 056A | homoduplex | homoduplex | ||
| 058A | homoduplex | homoduplex | ||
| 059A | heteroduplex | c.471 C>T (p.S157S) | homoduplex | |
| 061A | homoduplex | homoduplex |
All sequence variations detected in CHX10 were in exons 3 and 5. Homoduplex indicates two identical alleles. Heteroduplex indicates different alleles. One allele is the wild type sequence and the other one is a sequence variant differing from the wild type one. The single nucleotide substitution at c.471 C>T is a known SNP (rs35435463) and results in a synonymous change in the protein sequence (p.Ser157Ser). The single nucleotide substitution at c.579 G>A is a novel SNP and results in a synonymous change in the protein sequence (p. Gln193Gln). Another novel sequence variation, c.871 G>A, is identified in exon 5 in one affected individual and results in a non-synonymous change in the protein sequence (p. Asp291Asn). This sequence variation is also identified in healthy controls. G=guanosine, T=thymidine, C=cytidine, A=adenosine, S=Ser=serine, Q=Gln=glutamine, D=Asp=aspartic Acid, N=Asn=asparagine. The asterisk indicates that the heteroduplex is detected in 4 of 50 healthy control samples.
Discussion
The present study identified two novel mutations and two sequence variants (one novel, one known SNP) in SOX2 in 6 (two siblings) out of 34 anophthalmia/microphthalmia/ coloboma patients. The frequency of mutations in SOX2 in anophthalmia and microphthalmia patients is estimated at 10% for this study, which is consistent with previous reports [4,14,19]. A mutational analysis of CHX10 of the same cohort of anophthalmia/microphthalmia patients identified two synonymous sequence variants in exon 3 and one non-synonymous sequence variant in exon 5. The non-synonymous polymorphism was also present in normal controls, suggesting it is not causative. Further sequence analysis of other genes associated with anophthalmia and microphthalmia such as PAX6 [7,25], BCOR [26], OTX2 [27], SIX6 [28,29], and CHD7 [30,31] is ongoing in this study cohort to determine mutation associations.
Two mutations, a nonsense mutation and a frameshift mutation, were located in the coding region of SOX2, and both resulted in truncation of the SOX2 protein. The shortened protein product may result in loss of function. In animal studies, homozygous mice for SOX2 null allele are embryonic lethal [16] and heterozygous mice with one null allele are normal compared to that of the wild type [32]. Further reduction of the level of SOX2 expression by deleting a neural cell-specific enhancer on a heterozygous background resulted in reduced viability and neurodegeneration [32]. These observations suggest that the abnormal phenotype may become apparent when SOX2 expression falls below a certain threshold, or loss of function in one allele reduces SOX2 expression below that level. One of the mutations detected in this study was at the 3′ UTR. Although the relationship between the mutation at the 3′-UTR and anophthalmia/ microphthalmia is indeterminate, one possibility is that the nucleotide substitution at c.*557G>A affects SOX2 expression. This is supported by the observation that the 3′UTR of SOX2 contains regulatory elements that enhance its transcriptional activity in embryonic stem cells [33].
Of interest is the transmission of a SOX2 mutation from an unaffected mother to her two daughters in the example of sibling patients 54A and 54H. Gonosomal mosaicism, evidenced by the presence of a mutation in buccal cells but not in white cell-derived DNA, has been reported for SOX2 in an unaffected parent [34]. We were able to identify this mutation in genomic DNA derived from white blood cells in the unaffected mother. This suggests that this mother may not be mosaic in this circumstance. We propose that the anophthalmia phenotype appears to be non-penetrant. Non-penetrant mutations have been reported previously in sonic hedgehog (SHH) [34] and in fibroblast growth factor receptor 1 (FGFR) [35]. This example underscores the importance of testing seemingly unaffected parents once a mutation is discovered in their offspring regardless of their phenotype and points out that some individuals with a SOX2 mutation may be non-penetrant.
Our patients with coding region mutations had SOX2 phenotypes consistent with previous studies [4,10-12]. All patients with mutations had bilateral anophthalmia and unilateral severe microphthalmia (40A, 49A, 54H) with the exception of patient 54A who had unilateral anophthalmia instead of bilateral anophthalmia. This suggests that SOX2 mutations cause a more severe disruption of normal eye development and that microphthalmia/anophthalmia are of a spectrum of the same disease. The patients with coding sequence mutations (40A, 54A, and 54H) had significant brain abnormalities. Patient 40A showed dilated ventricles and microcephaly. Patient 54A was diagnosed with a right middle fossa arachnoid cyst and partial absence of posterior aspect of corpus callosum including the splenium. Patient 54H, the sibling of patient 54A, had partial agenesis of the corpus callosum. Brain structure changes are also consistent with other studies and are supported by murine models of SOX2 deficiency with neurodegeneration and impaired brain neurogenesis [32]. Hormonal deficiencies and growth retardation are also important phenotypic associations with SOX2 coding sequence changes (patients 40A, 54A, and 54H). Ragge et al. [10] have reported a case of anophthalmia/microphthalmia caused by a frameshift mutation in SOX2 (case 9, c.628delA, p.Met210fs211X). The genetic location of that mutation is similar to the proband 54A and her sibling 54H (c.549delC, p.Pro184ArgfsX19) in this study. The neurological phenotypes in this study, including brain malformations and seizure activity, were also present in their case [10]. Patient 49A with a 3′-UTR mutation had a cardiac ventricular-septal defect with dental anomalies. Different forms of craniofacial and dental defects were also noted in some patients with coding region mutations of SOX2 gene. Patient 54A (Appendix 1) had facial asymmetry and small, widely spaced teeth. One of the patients (case #1) reported by Ragge et al. [10] also showed craniofacial dysmorphisms and widely spaced teeth. The SOX2 UTR mutations appear to be associated with severe eye development but are perhaps less likely to manifest with neurological manifestations.
In conclusion, this study confirms that heterozygous loss of function mutations in SOX2 cause anophthalmia and microphthalmia and represent approximately 10% of patients ascertained for anophthalmia and microphthalmia but may also be non-penetrant. Mutations in CHX10 are less frequently found. Genetic screening of other candidate genes and the identification of genetic factors influencing SOX2 expression may help to determine the susceptibility of various disease alleles in the occurrence of this severe form of eye malformation and also help to identify factors that may influence SOX2 expression. Further characterization of genetic interactions among causative genes may also help to further our understanding of the formation of the eye.
Acknowledgments
The authors would like to thank all families for their participation in the study. We wish to thank the Children’s Hospital General Clinical Research Center for their support for this study. This work was supported by NIH Grants RO1 EY014685 and 2PEY01583, Research To Prevent Blindness Inc., The Mabel E. Leslie Endowment Funds, NIH/NCRR Mo1-RR-000240 (all T.L.Y.), and the Albert Einstein Society of the Albert Einstein Healthcare Network (A.S.).
Appendix 1. Patient demographics and mutations and sequence variants in SOX2 and CHX10 genes.
To access the data, click or select the words “Appendix 1”. This will initiate the download of a compressed (.zip) archive that contains the file. This file should be uncompressed with an appropriate program (the particular program will depend on your operating system).
References
- 1.Shaw GM, Carmichael SL, Yang W, Harris JA, Finnell RH, Lammer EJ. Epidemiologic characteristics of anophthalmia and bilateral microphthalmia among 2.5 million births in California, 1989–1997. Am J Med Genet A. 2005;137:36–40. doi: 10.1002/ajmg.a.30840. [DOI] [PubMed] [Google Scholar]
- 2.Kallen B, Tornqvist K. The epidemiology of anophthalmia and microphthalmia in Sweden. Eur J Epidemiol. 2005;20:345–50. doi: 10.1007/s10654-004-6880-1. [DOI] [PubMed] [Google Scholar]
- 3.Lowry RB, Kohut R, Sibbald B, Rouleau J. Anophthalmia and microphthalmia in the Alberta congenital anomalies surveillance system. Can J Ophthalmol. 2005;40:38–44. doi: 10.1016/S0008-4182(05)80115-2. [DOI] [PubMed] [Google Scholar]
- 4.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–3. doi: 10.1038/ng1120. see comment.12612584. [DOI] [PubMed] [Google Scholar]
- 5.Sandler D, Mancuso A, Becker T, Zori R, Hellrung J, Silverstein J, Burton V. Hamosh a, Williams C. Association of anophthalmia and esophageal atresia. Am J Med Genet. 1995;59:484–91. doi: 10.1002/ajmg.1320590415. [DOI] [PubMed] [Google Scholar]
- 6.Seller MJ, Davis TB, Fear CN, Flinter FA, Ellis I, Gibson AG. Two sibs with anophthalmia and pulmonary hypoplasia (the Matthew-Wood syndrome). Am J Med Genet. 1996;62:227–9. doi: 10.1002/(SICI)1096-8628(19960329)62:3<227::AID-AJMG5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 7.Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. Pax6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–71. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 8.Bonneau D, Guichet A, Boussion F, Lepinard C, Biquard F, Descamps P. Absence of deletion at the SOX2 locus in a case of microphthalmia and esophageal atresia. Am J Med Genet A. 2004;131:204. doi: 10.1002/ajmg.a.30180. [DOI] [PubMed] [Google Scholar]
- 9.Forrester S, Kovach MJ, Reynolds NM, Urban R, Kimonis V. Manifestations in four males and an obligate carrier of the Lenz microphthalmia syndrome. Am J Med Genet. 2001;98:92–100. [PubMed] [Google Scholar]
- 10.Ragge NK, Lorenz B, Schneider A, Bushby K, de Sanctis L, de Sanctis U, Salt A, Collin JR, Vivian AJ, Free SL, Thompson P, Williamson KA, Sisodiya SM, van Heyningen V, Fitzpatrick DR. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005;135:1–7. doi: 10.1002/ajmg.a.30642. discussion 8. [DOI] [PubMed] [Google Scholar]
- 11.Hagstrom SA, Pauer GJ, Reid J, Simpson E, Crowe S, Maumenee IH, Traboulsi EI. SOX2 mutation causes anophthalmia, hearing loss, and brain anomalies. Am J Med Genet A. 2005;138:95–8. doi: 10.1002/ajmg.a.30803. [DOI] [PubMed] [Google Scholar]
- 12.Zenteno JC, Gascon-Guzman G, Tovilla-Canales JL. Bilateral anophthalmia and brain malformations caused by a 20-bp deletion in the SOX2 gene. Clin Genet. 2005;68:564–6. doi: 10.1111/j.1399-0004.2005.00518.x. [DOI] [PubMed] [Google Scholar]
- 13.Guichet A, Triau S, Lepinard C, Esculapavit C, Biquard F, Descamps P, Encha-Razavi F, Bonneau D. Prenatal diagnosis of primary anophthalmia with a 3q27 interstitial deletion involving SOX2. Prenat Diagn. 2004;24:828–32. doi: 10.1002/pd.997. [DOI] [PubMed] [Google Scholar]
- 14.Verma AS, Fitzpatrick DR. Anophthalmia and microphthalmia. Orphanet J Rare Dis. 2007;2:47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–84. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 16.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Raay TJ, Moore KB, Iordanova I, Steele M, Jamrich M, Harris WA, Vetter ML. Frizzled 5 signaling governs the neural potential of progenitors in the developing xenopus retina. Neuron. 2005;46:23–36. doi: 10.1016/j.neuron.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–86. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakrania P, Robinson DO, Bunyan DJ, Salt A, Martin A, Crolla JA, Wyatt A, Fielder A, Ainsworth J, Moore A, Read S, Uddin J, Laws D, Pascuel-Salcedo D, Ayusa C, Allen L, Collin JR, Ragge NK. SOX2 anophthalmia syndrome: 12 new cases demonstrating broader phenotype and high frequency of large gene deletions. Br J Ophthalmol. 2007;91:1471–6. doi: 10.1136/bjo.2007.117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driggers RW, Macri CJ, Greenwald J, Carpenter D, Avallone J, Howard-Peebles PN, Levin SW. Isolated bilateral anophthalmia in a girl with an apparently balanced de novo translocation: 46,xx,t(3;11)(q27;p11.2). Am J Med Genet. 1999;87:201–2. doi: 10.1002/(sici)1096-8628(19991126)87:3<201::aid-ajmg1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 21.Liu IS, Chen JD, Ploder L, Vidgen D, van der Kooy D, Kalnins VI, McInnes RR. Developmental expression of a novel murine homeobox gene (ChHX10): Evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–93. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 22.Ferda Percin E, Ploder LA, Yu JJ, Arici K, Horsford DJ, Rutherford A, Bapat B, Cox DW, Duncan AM, Kalnins VI, Kocak-Altintas A, Sowden JC, Traboulsi E, Sarfarazi M, McInnes RR. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 23.Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL. Vidgen d, Hoover F, Goldman D, Kalnins VI, Roderick TH, Taylor BA, Hankin MH, McInnes RR. Ocular retardation mouse caused by CHX10 homeobox null allele: Impaired retinal progenitor proliferation and bipolar cell differentiation. Nat Genet. 1996;12:376–84. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- 24.Bar-Yosef U, Abuelaish I, Harel T, Hendler N, Ofir R, Birk OS. CHX10 mutations cause non-syndromic microphthalmia/ anophthalmia in Arab and Jewish kindreds. Hum Genet. 2004;115:302–9. doi: 10.1007/s00439-004-1154-2. [DOI] [PubMed] [Google Scholar]
- 25.Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–5. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 26.Ng D, Thakker N, Corcoran CM, Donnai D, Perveen R, Schneider A, Hadley DW, Tifft C, Zhang L, Wilkie AO, van der Smagt JJ, Gorlin RJ, Burgess SM, Bardwell VJ, Black GC, Biesecker LG. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–6. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 27.Ragge NK, Brown AG, Poloschek CM, Lorenz B, Henderson RA, Clarke MP, Russell-Eggitt I, Fielder A, Gerrelli D, Martinez-Barbera JP, Ruddle P, Hurst J, Collin JR, Salt A, Cooper ST, Thompson PJ, Sisodiya SM, Williamson KA, Fitzpatrick DR, van Heyningen V, Hanson IM. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005;76:1008–22. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, Rodriguez de Cordoba S. Genomic cloning and characterization of the human homeobox gene six6 reveals a cluster of six genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61:82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- 29.Gallardo ME, Rodriguez de Cordoba S, Schneider A, Dwyer MA, Ayusa C, Bovolenta P. Analysis of the developmental six6 homeobox gene in patients with anophthalmia/microphthalmia. Am J Med Genet A. 2004;129:92–4. doi: 10.1002/ajmg.a.30126. [DOI] [PubMed] [Google Scholar]
- 30.Vissers LELM, van Ravenswaaij CMA, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, Hamel BC, Schoenmakers EF, Brunner HG, Veltman JA, van Kessel AG. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–7. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 31.Lalani SR, Safiullah AM, Fernbach SD, Harutyunyan KG, Thaller C, Peterson LE, McPherson JD, Gibbs RA, White LD, Hefner M, Davenport SL, Graham JM, Bacino CA, Glass NL, Towbin JA, Craigen WJ, Neish SR, Lin AE, Belmont JW. Spectrum of CHD7 mutations in 110 individuals with charge syndrome and genotype-phenotype correlation. Am J Hum Genet. 2006;78:303–14. doi: 10.1086/500273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, deBiasi S, Nicolis SK. SOX2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–19. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 33.Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of SOX-2 regulatory region which is under the control of Oct-3/4–SOX-2 complex. Nucleic Acids Res. 2002;30:3202–13. doi: 10.1093/nar/gkf435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schimmenti LA, de la Cruz J, Lewis RA, Karkera JD, Manligas GS, Roessler E, Muenke M. Novel mutation in Sonic hedgehog in non-syndromic colobomatous microphthalmia. Am J Med Genet A. 2003;116:215–21. doi: 10.1002/ajmg.a.10884. [DOI] [PubMed] [Google Scholar]
- 35.Muenke M, Schell U, Hehr A, Robin NH, Losken HW, Schinzel A, Pulleyn LJ, Rutland P, Reardon W, Malcolm S, Winter RM. A common mutation in the fibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat Genet. 1994;8:269–74. doi: 10.1038/ng1194-269. [DOI] [PubMed] [Google Scholar]