Abstract
The effects of insulin on the mammalian target of rapamycin, mTOR, were investigated in 3T3-L1 adipocytes. mTOR protein kinase activity was measured in immune complex assays with recombinant PHAS-I as substrate. Insulin-stimulated kinase activity was clearly observed when immunoprecipitations were conducted with the mTOR antibody, mTAb2. Insulin also increased by severalfold the 32P content of mTOR that was determined after purifying the protein from 32P-labeled adipocytes with rapamycin⋅FKBP12 agarose beads. Insulin affected neither the amount of mTOR immunoprecipitated nor the amount of mTOR detected by immunoblotting with mTAb2. However, the hormone markedly decreased the reactivity of mTOR with mTAb1, an antibody that activates the mTOR protein kinase. The effects of insulin on increasing mTOR protein kinase activity and on decreasing mTAb1 reactivity were abolished by incubating mTOR with protein phosphatase 1. Interestingly, the epitope for mTAb1 is located near the COOH terminus of mTOR in a 20-amino acid region that includes consensus sites for phosphorylation by protein kinase B (PKB). Experiments were performed in MER-Akt cells to investigate the role of PKB in controlling mTOR. These cells express a PKB-mutant estrogen receptor fusion protein that is activated when the cells are exposed to 4-hydroxytamoxifen. Activating PKB with 4-hydroxytamoxifen mimicked insulin by decreasing mTOR reactivity with mTAb1 and by increasing the PHAS-I kinase activity of mTOR. Our findings support the conclusion that insulin activates mTOR by promoting phosphorylation of the protein via a signaling pathway that contains PKB.
The mammalian target of rapamycin, mTOR (1), also known as FRAP (2) or RAFT (3), is a key element of a signaling pathway involved in the control of protein synthesis and mitogenesis. This large protein (predicted Mr = 289,000) contains several structural motifs, including a high-affinity binding site for rapamycin-FKBP12 (FKBP12, FK506-binding protein of Mr = 12,000) and a catalytic domain homologous to those domains found in phosphatidylinositol 3-kinases (PI 3-kinases) (4). The function of mTOR in cells is potently inhibited by rapamycin, a characteristic that has facilitated the identification of mTOR-mediated processes, such as the insulin-stimulated phosphorylation of the translational regulators PHAS-I (5, 6) (also known as 4E-BP1) and p70S6K (7). Phosphorylation of PHAS-I results in the release of eukaryotic initiation factor 4E (eIF4E), thereby increasing the translation of capped mRNAs (8). Phosphorylation in the appropriate sites activates p70S6K, which then phosphorylates ribosomal protein S6 (7). Although much has been learned about the cellular processes regulated by mTOR, very little is known either about how mTOR itself signals or about the mechanisms involved in the control of mTOR by hormones and growth factors.
PI 3-kinase is a point of convergence in the pathways by which several hormones and growth factors stimulate the phosphorylation of PHAS-I and p70S6K (7, 8). Activation of PI 3-kinase leads to activation of protein kinase B (PKB) (9–11), which is believed to bind by its pleckstrin homology domain to products of the PI 3-kinase reaction (12). The membrane localization appears to be sufficient for PKB to be phosphorylated and activated by PDK1 (13, 14), although full activation requires an uncharacterized second kinase, referred to as PDK2 (13). Activating PKB in cells mimics insulin by increasing the phosphorylation of both PHAS-I (15, 16) and p70S6K (11, 17), although there is no evidence that PKB directly phosphorylates either protein. The pathway downstream of PKB has not been defined, and whether mTOR is a downstream element in the PI 3-kinase/PKB pathway or in a separate signaling pathway is a point of debate (7). Results obtained with rapamycin and the PI 3-kinase inhibitor wortmannin have provided arguments for both sides. For example, incubating cells with either rapamycin or wortmannin abolished activation of p70S6K by insulin (18) and promoted dephosphorylation of the same sites in the kinase (19), suggesting that mTOR and PI 3-kinase are in the same pathway. The finding that deleting residues 2–46 from the NH2 terminus of the p70S6K isoform, p85S6K, resulted in a truncated kinase whose activation was insensitive to rapamycin but still inhibited by wortmannin (20) supports the argument that mTOR and PI 3-kinase are found in separate signaling pathways.
A major problem in investigating mechanisms involved in the control of mTOR function has been the lack of a biochemical measure of mTOR activity. The recent discovery that mTOR is able to phosphorylate PHAS-I in vitro demonstrates that mTOR has the potential to signal as a protein kinase and provides a means to investigate mechanisms involved in the regulation of the protein (21, 22). In this report we present evidence that insulin stimulates the phosphorylation and activation of mTOR via the PI 3-kinase/PKB signaling pathway.
MATERIALS AND METHODS
Culture and Incubation of Cells.
3T3-L1 murine adipocytes were cultured in 10-cm-diameter dishes and used in experiments 8–10 days after removal of the differentiation medium (6). Cells were incubated at 37°C for 2.5 hr in Hepes-buffered saline (HBS) (145 mM NaCl/5.4 mM KCl/1.4 mM CaCl2/1.4 mM MgCl2/0.1 mM NaPi/5 mM glucose/0.5% BSA/10 mM NaHepes, pH 7.4), then incubated without or with insulin as indicated. Extracts were prepared as described previously (6) after homogenizing the cells in 1 ml of homogenization buffer, which contained 50 mM NaF, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM sodium vanadate, 0.1 μM microcystin-LR, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, 10 mM KPi, and 50 mM β-glycerophosphate, pH 7.4.
32P-labeling experiments were performed essentially as described previously (23). Briefly, the HBS was supplemented with Na32Pi (2.5 mCi/5 ml; 1 mCi = 37 MBq), and cells were incubated at 37°C for 3 hr. Additions of insulin were made at the appropriate time before the end of this incubation period to give the desired treatment time. To terminate the incubation, the medium was aspirated and the cells were rinsed and homogenized (1 ml of homogenization buffer per dish) in a glass homogenization tube with a Teflon pestle driven at 1,000 rpm. Homogenates were centrifuged at 10,000 × g for 20 min, and the supernatants were retained for analyses. The protein content was determined by using bicinchoninic acid (24).
NIH 3T3 cells or a transfected line of NIH 3T3 cells (MER-Akt) stably expressing myrAkt Δ4–129-ER, a conditionally active, hemagglutinin (HA)-tagged form of PKB (15, 25), were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. Confluent cultures were incubated at 37°C for approximately 15 hr in medium without serum. The medium was then replaced with fresh medium containing 1 mg/ml BSA, and the cells were incubated with insulin or 4-hydroxytamoxifen. Extracts for assessing mTOR were prepared as described for preparing extracts of 3T3-L1 adipocytes, except that the homogenization buffer was supplemented with 0.1% Tween 20. For measuring PKB activity, extracts were prepared as described previously (9), except that the cell lysis solution contained 0.5% Triton X-100, 10 μg/ml each of aprotinin and leupeptin, and no bacitracin.
Immunoprecipitation of mTOR and HA-Tagged PKB Fusion Protein.
Antibodies, designated mTAb1 and mTAb2, to peptides having sequences identical to positions 2335–2450 (DTNAKGNKRSRTRTDSYS) and 1272–1290 (ARRVSKDDWKEWKRRKSKE), respectively, in mTOR were generated in rabbits and affinity-purified (22). Immunoprecipitations were performed with either nonimmune IgG or the appropriate mTOR antibody coupled to staphylococcal protein A-agarose beads as described previously (22). The HA-tagged PKB fusion protein was immunoprecipitated by using 12CA5 anti-HA monoclonal antibody as described previously (9).
Affinity Purification of mTOR.
Extract samples (1 ml) were incubated for 2 hr at 4°C with 20 μl of rapamycin⋅GST-FKBP12 gel, which was then washed as described previously (21). The gel was generated by incubating 20 μl of glutathione-agarose beads with 100 μg of glutathione-S-transferase (GST)-FKBP12 fusion protein and 5 μM rapamycin (21).
Measurements of Protein Kinase Activity.
To measure the mTOR protein kinase, immune complexes were suspended in 50 μl of reaction mix, which contained 50 mM NaCl, 1 mM DTT, 10 μM inhibitory peptide of cAMP-dependent protein kinase (26), 0.2 μM microcystin-LR, 20 μg/ml [His6-PHAS-I] (27), 0.2 mM [γ-32P]ATP (1 μCi/nmol), 10 mM MnCl2, 10 mM NaHepes, and 50 mM β-glycerophosphate (pH 7.4). The samples were incubated at 30°C for 30 min before the reactions were terminated by adding SDS sample buffer. The activity of the HA-PKB fusion protein was measured with myelin basic protein as substrate as described previously (9).
Electrophoretic Analyses.
Samples were subjected to SDS/PAGE by using the method of Laemmli (28). 32P-labeled proteins were detected by autoradiography after drying the gels. mTOR (22) and PHAS-I (29) were detected by immunoblotting.
Other Materials.
His6-PHAS-I (27) and GST-FKBP12 (1) were expressed in bacteria and purified as described previously. Rapamycin and FK506 were obtained from Calbiochem– Novabiochem and Fujisawa Pharmaceuticals, respectively. [γ-32P]ATP was from New England Nuclear. Recombinant catalytic subunit of type 1 protein phosphatase (PP1C, α isoform) was provided by Anna DePaoli Roach, Indiana University School of Medicine.
RESULTS
Activation of mTOR by Insulin.
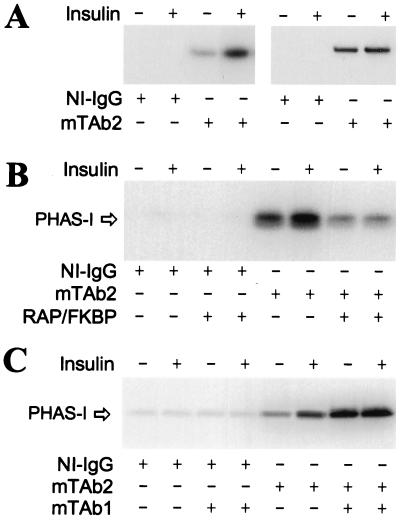
To investigate the regulation of mTOR, the protein was immunoprecipitated from extracts of 3T3-L1 adipocytes by using the antibody mTAb2. The PHAS-I kinase activity was approximately 4 times as high in the mTOR immune complexes derived from insulin-treated cells as in the complexes from control cells (Fig. 1A). Very little kinase activity was detected when immunoprecipitations were conducted with nonimmune IgG (Fig. 1A), supporting the conclusion that the insulin-stimulated kinase activity was due to mTOR. Insulin treatment did not change the amount of mTOR immunoprecipitated (Fig. 1A). Thus, the increase in PHAS-I kinase activity was not because of an increase in the amount of mTOR in the assay.
Figure 1.
Insulin-stimulated mTOR protein kinase activity. 3T3-L1 adipocytes were incubated without insulin or with 10 milliunits/ml insulin for 30 min. Extracts were prepared and immunoprecipitations were performed with either nonimmune IgG (NI-IgG) or mTAb2. Beads were suspended in reaction mix containing [γ-32P]ATP and His6-PHAS-I, and then incubated for 30 min. After the reactions had been terminated, samples were subjected to SDS/PAGE and autoradiograms of the dried gels were prepared. (A) An autoradiogram showing 32P-labeled His6-PHAS-I (Left) and an immunoblot prepared with mTAb2 (Right) are presented. Other immunoprecipitations were performed as described above. (B) Prior to incubation in reaction mix, beads were incubated for 30 min at 22°C in buffer A alone or buffer A supplemented with 200 μg/ml GST-FKBP12 (FKBP) and 5 μM rapamycin. Buffer A contained 50 mM NaCl, 10 mM NaHepes, and 50 mM β-glycerophosphate (pH 7.4). The beads were then washed twice before incubation in reaction mix for 30 min. Proteins were eluted and subjected to SDS/PAGE. An autoradiogram showing 32P-labeled His6-PHAS-I is presented. (C) Before incubation in reaction mix, beads were suspended in 25 μl of buffer A and incubated for 30 min at 22°C without or with 5 μg of mTAb1. The beads were washed twice with buffer A before incubation in reaction mix. The autoradiogram shows 32P-labeled His6-PHAS-I.
As previously observed with the rat brain and recombinant mTOR proteins (21), mTOR immunoprecipitated from extracts of 3T3-L1 adipocytes exhibited a marked preference for Mn2+ over Mg2+ (P.H.S., G.J.B., and J.C.L., unpublished observations). The insulin-stimulated PHAS-I kinase was inhibited by rapamycin-FKBP12 (Fig. 1B), but not by FK506–FKBP12 (P.H.S., G.J.B., and J.C.L., unpublished observations). The preferential inhibition by rapamycin-FKBP12 is a hallmark of an mTOR-mediated process (4).
The antibody mTAb1 has been shown to markedly increase the protein kinase activity of rat brain mTOR, suggesting that it binds to a regulatory domain in mTOR (22). Incubating immunoprecipitated adipocyte mTOR with mTAb1 also increased the PHAS-I kinase activity (Fig. 1C). No effect of insulin on mTOR activity over the increase produced by mTAb1 was observed. The finding that an mTOR antibody is capable of increasing the activity of the mTAb2-immunoprecipitated protein provides additional evidence that the PHAS-I kinase activity observed is due to mTOR rather than a copurifying kinase.
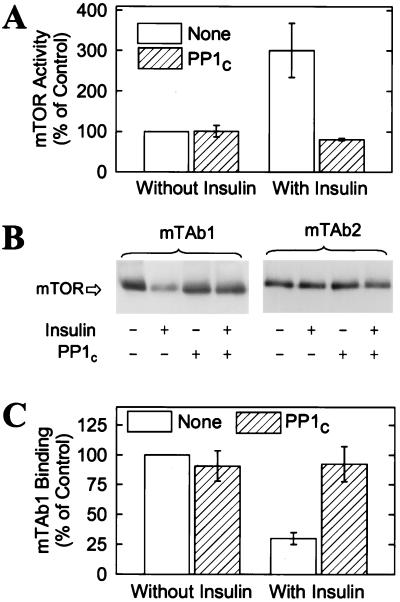
Insulin-Stimulated Phosphorylation of mTOR.
The stable activation of mTOR produced by insulin is indicative of a mechanism involving covalent modification. Incubating mTOR immune complexes with PP1C abolished the effect of insulin on the mTOR protein kinase (Fig. 2A). The effect of the phosphatase was markedly inhibited when the incubation was conducted in the presence of microcystin-LR (P.H.S., G.J.B., and J.C.L., unpublished observations), which inhibits PP1C (30). These findings strongly suggest that phosphorylation is responsible for the activation of mTOR by insulin. Results obtained with mTAb1 provided additional evidence that insulin stimulates the phosphorylation of mTOR. Incubating adipocytes with the hormone markedly decreased the binding of mTOR and mTAb1 (Fig. 2 B and C). In contrast, the reactivity of mTOR toward mTAb2 was not affected by insulin (Fig. 2B). Incubating affinity-purified mTOR preparations with PP1C increased mTAb1 reactivity and abolished the effect of insulin (Fig. 2C). Thus, it seems reasonable to conclude that the insulin-stimulated decrease in binding of mTAb1 is due to phosphorylation of mTOR. The marked reduction in mTAb1 binding is indicative of nearly stoichiometric modification of mTOR.
Figure 2.
Reversal by PP1C of the effects of insulin on mTOR. (A) 3T3-L1 adipocytes were incubated at 37°C for 30 min in HBS without or with 10 milliunits/ml insulin before extracts were prepared and mTOR was immunoprecipitated. The beads were then suspended in 50 μl of buffer A (see legend to Fig. 1) containing 1 mM MnCl2 and incubated at 22°C for 30 min without or with 0.5 μg of recombinant PP1C. The beads were washed three times in buffer A (1 ml per wash) containing 500 nM microcystin-LR, then incubated for 30 min at 30°C in 50 μl of reaction mix before 32P incorporation into PHAS-I was determined. The results are expressed as percentages of the 32P incorporation observed with mTOR immunoprecipitated from control cells and are mean values ± SE of three experiments. (B) Other adipocytes were incubated without or with 10 milliunits/ml insulin for 30 min before mTOR was affinity purified by using the rapamycin⋅GST-FKBP12 resin. The beads were incubated at 22°C for 2 hr without or with PP1C, then washed as described above. Proteins were eluted, subjected to SDS/PAGE, and transferred to Immobilon membranes. After an immunoblot was prepared with mTAb1, the membrane was stripped and reprobed with mTAb2. (C) The relative amounts of mTAb1 bound to mTOR were determined by laser densitometry. Mean values ± SE of three experiments are presented.
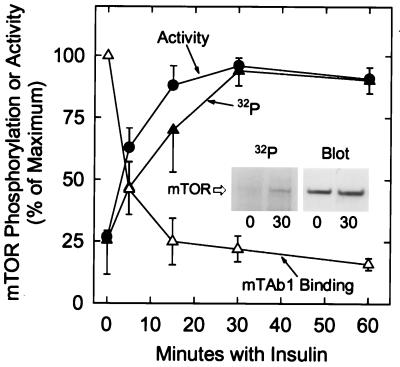
To investigate further the phosphorylation of mTOR, adipocytes were incubated in medium containing 32Pi. The cells were then incubated for increasing times before mTOR was immunoprecipitated and subjected to SDS/PAGE. 32P-labeled mTOR was detected by autoradiography after transfer to Immobilon P. Insulin rapidly increased the 32P content of mTOR (Fig. 3). The maximal hormonal effect, which represented an increase of approximately 4-fold, was observed after 30 min with insulin, and it was maintained for at least 30 more min. The increase in mTOR activity produced by insulin also reached a stable plateau (Fig. 3). The time course of mTOR activation by insulin closely resembled that of mTOR phosphorylation, particularly when assessed by mTAb1 immunoblotting (Fig. 3).
Figure 3.
Time courses of mTOR phosphorylation and activation in response to insulin. 3T3-L1 adipocytes were incubated in HBS or in HBS supplemented with 32Pi (0.5 mCi/ml) for 3 hr, then homogenized. Insulin (10 milliunits/ml) was added at the appropriate time prior to homogenization to give the treatment times indicated. mTOR activity was measured after immunoprecipitation with mTAb2. [32P]mTOR was affinity purified from extracts by using rapamycin⋅GST-FKBP12 beads, and samples were subjected to SDS/PAGE before proteins were electrophoretically transferred to Immobilon P (Millipore) membranes. An autoradiogram was prepared to allow measurements of the relative amounts of 32P-labeled mTOR (see Inset for example). An immunoblot was then prepared with mTAb2 to estimate the amounts of the mTOR protein present. mTOR activity was measured after immunoprecipitation with mTAb2 from extracts of cells incubated without 32P. Samples of these extracts were subjected to SDS/PAGE and blotted with mTAb1. The relative amounts of mTAb1 reactivity (▵) were determined by laser densitometry and are expressed as a percentage of control. mTOR activities (•) and 32P contents (▴) are expressed as percentages of the respective maximal levels. The results presented are mean values ± SE from three experiments.
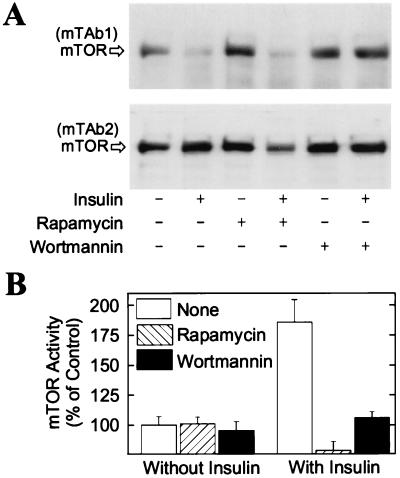
Activation of mTOR by Insulin Is Potently Inhibited by Wortmannin.
There is strong evidence that both mTOR and PI 3-kinase are involved in the control of PHAS-I phosphorylation by insulin (8, 31). To address the question of whether mTOR is positioned downstream of PI 3-kinase or in a separate signaling pathway, 3T3-L1 adipocytes were incubated with wortmannin at a concentration (100 nM) that abolishes activation of PI 3-kinase but has a minimal effect on the kinase activity of mTOR (21). The effect of insulin on decreasing mTAb1 binding to mTOR was abolished by wortmannin but not by rapamycin (Fig. 4A), consistent with the interpretation that PI 3-kinase is upstream of mTOR.
Figure 4.
Wortmannin-sensitive modification of the COOH-terminal region of mTOR in response to insulin. (A) 3T3-L1 adipocytes were incubated without additions or with either 25 nM rapamycin or 100 nM wortmannin for 1 hr. After addition of 10 milliunits/ml insulin as indicated, the incubations were continued for 30 min. mTOR was affinity purified and subjected to SDS/PAGE. Immunoblots prepared with mTAb1 and mTAb2 are presented. (B) In other experiments, mTOR was immunoprecipitated with mTAb2, then incubated in reaction mix for 30 min. The results represent the relative amounts of 32P incorporated into His6-PHAS-I, expressed as percentages of control, and are mean values ± SE from three experiments.
The increase in the PHAS-I kinase activity of mTOR produced by insulin was inhibited by both wortmannin and rapamycin (Fig. 4B). Again, the inhibition by wortmannin is consistent with the placement of PI 3-kinase upstream of mTOR. That rapamycin treatment of cells inhibited mTOR activity is consistent with the very high binding affinity between mTOR and rapamycin–FKBP12 (4), as the inactive ternary complex formed in cells would be expected to persist in cell extracts.
Activation of PKB Leads to the Phosphorylation and Activation of mTOR.
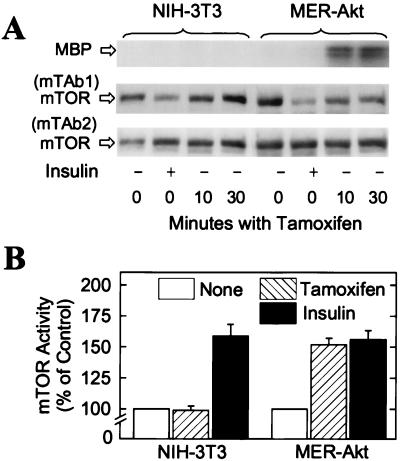
To investigate the possible regulation of mTOR by PKB, we utilized a transfected line of NIH 3T3 cells, designated MER-Akt, that express a recombinant PKB fusion protein engineered to be conditionally activated in response to tamoxifen (15). The protein consists of myristoylated PKB (25) fused in-frame with a mutant form of the murine estrogen receptor that no longer binds 17β-estradiol but is still responsive to the synthetic steroid 4-hydroxytamoxifen (32). Incubating MER-Akt cells with 4-hydroxytamoxifen for 10 min resulted in a dramatic increase in the activity of the PKB fusion protein (Fig. 5A). This rapid activation is consistent with the previously described model in which binding of hormone (in this case 4-hydroxytamoxifen) stimulates the release of the 90-kDa heat shock protein from the estrogen-binding domain, thereby allowing access of other proteins (presumably PDK-1 or PDK-2) to the chimeric molecule (33). 4-Hydroxytamoxifen rapidly decreased mTAb1 binding to mTOR in the MER-Akt cells, but not in the parental NIH 3T3 cells (Fig. 5A). Insulin, which is able to activate endogenous PKB, decreased mTAb1 binding in both the parental NIH 3T3 cells and the MER-Akt cells (Fig. 5A).
Figure 5.
Effect of PKB activation on mTOR phosphorylation and activity. (A) NIH 3T3 cells or MER-Akt cells were incubated without or with either 10 milliunits/ml insulin for 30 min or with 1 μM 4-hydroxytamoxifen (Tamoxifen) for 10 min or 30 min. PKB activity was assessed with myelin basic protein (MBP) as substrate after the PKB fusion protein had been immunoprecipitated from extracts with the anti-HA monoclonal antibody. Samples of extracts were subjected to SDS/PAGE, and mTAb1 and mTAb2 immunoblots were prepared. An autoradiogram showing 32P-labeled MBP and mTOR immunoblots are presented. (B) In separate experiments, cells were incubated for 30 min without additions (None), with 10 milliunits/ml of insulin, or with 1 μM 4-hydroxytamoxifen. mTOR was immunoprecipitated by using mTAb2, then incubated in reaction mix for 30 min. The results represent the relative amounts of 32P incorporated into His6-PHAS-I, expressed as percentages of control, and are mean values ± SE from three experiments.
The effects of insulin and 4-hydroxytamoxifen on mTOR activity were consistent with the effects of mTOR phosphorylation as assessed by mTAb1 reactivity. Thus, insulin activated mTOR in both the parental cells and the MER-Akt cells (Fig. 5B), although the increase in kinase activity was less than observed in 3T3-L1 adipocytes. 4-Hydroxytamoxifen increased the PHAS-I kinase activity of mTOR in the MER-Akt cells but not in the parental cells (Fig. 5B). 4-Hydroxytamoxifen also increased the phosphorylation of endogenous PHAS-I in the MER-Akt cells, but not in the control cells (15).
DISCUSSION
The kinase domain near the COOH terminus is essential for mTOR to effect increases in the phosphorylation of PHAS-I (21) and p70S6K (34) in cells. Although homologous to the catalytic domain of PI 3-kinase, the mTOR kinase transfers phosphate from ATP to proteins. mTOR autophosphorylates when incubated with ATP (34, 35), and it directly phosphorylates PHAS-I in vitro, as initially shown by Brunn et al. (21) and recently confirmed (36). By using PHAS-I as a substrate, we have obtained direct evidence that the protein kinase activity of mTOR may be increased in response to insulin and growth factors that activate PKB. We have also demonstrated that mTOR is phosphorylated in response to insulin, and that phosphorylation of mTOR in the appropriate site(s) increases its activity. The loss of mTAb1 binding indicates that insulin-stimulated phosphorylation occurs in a site(s) located near the COOH terminus of mTOR. We propose that phosphorylation in the COOH-terminal region activates mTOR, because at least one phosphorylation site appears to be located in the epitope for mTAb1, whose binding markedly increases mTOR activity. It should be stressed that phosphorylation of multiple sites might contribute to the regulation of mTOR activity. Because of the relatively small amounts of 32P-labeled mTOR that were recovered, it was not feasible to identify the sites of phosphorylation.
On the basis of the antigenic peptide, mTAb1 is presumed to bind in the region of mTOR having the following sequence: DTNAKGNKRSRTRTDS*YS, where S* corresponds to Ser-2448. This residue is located in a very good consensus motif for phosphorylation by PKB (Arg-Xaa-Arg-Yaa-Zaa-Ser/Thr-Hyd, where Xaa is any amino acid, Yaa is preferably a Ser/Thr, Zaa is any small amino acid except Gly, and Hyd is a bulky hydrophobic residue) (37). Thr-2446 is another potential site for phosphorylation by PKB. The antigenic peptide was phosphorylated in vitro by PKB at 25% of the rate at which the kinase phosphorylated Crosstide (GRPRTSSFAEG) (P.H.S., G.J.B., and J.C.L., unpublished observations), which is a very good peptide substrate for PKB. However, we have been unable to phosphorylate either affinity-purified or immunoprecipitated mTOR with purified PKB. Although it seems possible that the in vitro kinase reactions lack factors found in the intracellular milieu that are required to support phosphorylation of mTOR by PKB, we cannot conclude that PKB directly phosphorylates mTOR. Nevertheless, the finding that tamoxifen rapidly activates mTOR in MER-Akt cells provides strong evidence that the PKB pathway regulates mTOR.
As PKB activation is known to depend on activation of PI 3-kinase (9–11), our results challenge the widely held view that PI 3-kinase is not an upstream element in the mTOR signaling pathway. This model was based on studies of the activation of p70S6K in which mTOR was not directly assessed. The major argument for placing PI 3-kinase and mTOR in separate pathways is the finding that removing the NH2-terminal region of p70S6K results in a truncated kinase that when expressed in cells is resistant to inhibition by rapamycin but still inhibited by wortmannin (20). However, these results are also consistent with a model in which PI 3-kinase is an upstream element in two pathways, only one of which contains mTOR.
Multiple pathways are clearly involved in the control of p70S6K. The activity of the kinase depends on a complex pattern of phosphorylation catalyzed by three or more protein kinases which phosphorylate at least 10 distinct sites (7, 12, 38, 39). Insulin promotes the phosphorylation of several sites, including four S/T-P sites located in the COOH-terminal region of p70S6K. Although the S/T-P sites are not essential for activation, their phosphorylation facilitates phosphorylation of other sites, including Thr-389. This important site functions as a gatekeeper, as its phosphorylation induces a conformational change that allows phosphorylation of Thr-229 by PDK1 (38, 39). Thr-229 is located in the activation loop and must be phosphorylated for kinase activity. Rapamycin treatment of cells inhibits phosphorylation of Thr-229, Thr-389, Ser-404, and Ser-411 (19). Except for Ser-411, which is one of the S/T-P sites, these phosphorylated residues are flanked by aromatic amino acids.
Interestingly, immunoprecipitated mTOR was recently reported by Burnett et al. (36) to phosphorylate Thr-389 in vitro. This finding would seem to provide an explanation of the rapamycin sensitivity, not only of this site but also of Thr-229, by means of the gatekeeper function of Thr-389. A difficulty with this interpretation is the earlier report that phosphorylation of Thr-389 was inhibited by wortmannin, but not by rapamycin, in a mutant p70S6K lacking NH2- and COOH-terminal domains (40). The lack of inhibition of Thr-389 by rapamycin formed the basis of the argument of Dennis et al. (40) that mTOR does not mediate the phosphorylation of this site. On the other hand, an NH2-terminal truncated p70S6K containing Thr-389 appeared to be a much better in vitro substrate for mTOR than was full-length p70S6K (36). Moreover, with the concentration of wortmannin (250 nM) used in the study of Dennis et al. (40) significant inhibition not only of PI 3-kinase but also of mTOR would have been expected (35). Thus, it is conceivable that when expressed in cells, a mutant p70S6K with the appropriate NH2-terminal truncation could appear resistant to rapamycin, which does not fully inhibit mTOR, and such a mutant might still be sensitive to wortmannin, which binds in the catalytic domain and is able to abolish mTOR kinase activity. However, it is a concern that rapamycin⋅FKBP12 had little if any effect on the in vitro phosphorylation of full-length p70S6K attributed to mTOR (36). Additional work will be needed to reconcile this finding with the marked inhibition of Thr-389 phosphorylation by rapamycin treatment of cells, and to explain the rapamycin sensitivities of Ser-404 and Ser-411.
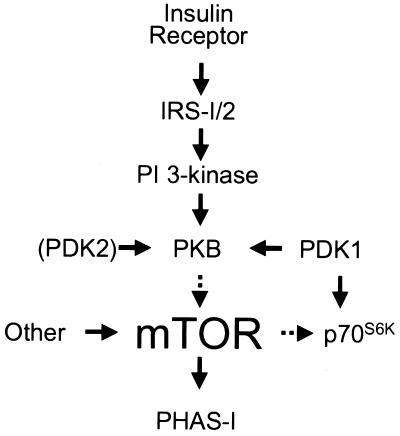
The control of PHAS-I appears less complicated than that of p70S6K. Although five sites in PHAS-I are phosphorylated in adipocytes, all five are sensitive to rapamycin and are found in a S/T-P motif (41). Moreover, the same five sites were phosphorylated in vitro by mTOR (22), and their phosphorylation by mTOR was markedly inhibited by rapamycin⋅FKBP12, but by neither FK506⋅FKBP12 nor rapamycin alone (21, 22). Thus, there is strong evidence that mTOR phosphorylates PHAS-I in vivo. With the present discovery that PKB mediates the activation of mTOR, an almost complete picture of the steps leading to PHAS-I phosphorylation has emerged (Fig. 6). This signal transduction pathway is one of the few that can be traced from the receptor to an important metabolic effect of insulin, in this case the stimulation of protein synthesis.
Figure 6.
A signal transduction pathway leading to the phosphorylation and activation of mTOR in response to insulin. Binding of insulin to its receptor activates the receptor tyrosine kinase, which phosphorylates insulin receptor substrate (IRS)-1 and/or –2 (42). Phosphotyrosyl residues in these proteins serve as docking sites for PI 3-kinase, whose activation leads to activation of PKB. As described in the text, activation of PKB results in phosphorylation and activation of mTOR, although we stress that other kinases may be involved. Once activated, mTOR phosphorylates the translational repressor, PHAS-I, which dissociates from eIF-4E, thereby increasing the amount of eIF-4E available to mediate translation initiation (8).
Acknowledgments
We thank Annette Farris for technical assistance. This work was supported by National Institutes of Health Grants DK52753, DK28312, and AR41180 (to J.C.L.) and DK34926 (to R.A.R.). G.J.B. is supported by a postdoctoral fellowship from the Juvenile Diabetes Foundation.
ABBREVIATIONS
- eIF4E
eukaryotic initiation factor 4E
- FKBP12
FK506-binding protein of Mr = 12,000
- HA
hemagglutinin
- GST
glutathione-S-transferase
- mTOR
mammalian target of rapamycin
- PI 3-kinase
phosphatidylinositol 3-kinase
- PKB
protein kinase B
- PP1C
the catalytic subunit of type 1 protein phosphatase
References
- 1.Sabers C J, Martin M M, Brunn G J, Williams J M, Dumont F J, Wiederrecht G, Abraham R T. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 2.Brown E J, Albers M W, Shin T B, Ichikawa K, Keith C T, Lane W S, Schreiber S L. Nature (London) 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 3.Sabatini D M, Erdjument-Bromage H, Lui M, Tempst P, Snyder S H. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 4.Abraham R T, Wiederrecht G. Annu Rev Immunol. 1996;14:483–510. doi: 10.1146/annurev.immunol.14.1.483. [DOI] [PubMed] [Google Scholar]
- 5.Graves L M, Bornfeldt K E, Argast G M, Krebs E G, Kong X, Lin T-A, Lawrence J C., Jr Proc Natl Acad Sci USA. 1995;92:7222–7226. doi: 10.1073/pnas.92.16.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin T-A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C., Jr J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 7.Proud C G. Trends Biochem Sci. 1996;21:181–185. [PubMed] [Google Scholar]
- 8.Lawrence J C, Jr, Abraham R T. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 9.Kohn A D, Kovacina K S, Roth R A. EMBO J. 1995;14:4288–4295. doi: 10.1002/j.1460-2075.1995.tb00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke T F, Yang S I, Chan T O, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 11.Burgering B M T, Coffer P J. Nature (London) 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 12.Downward J. Science. 1998;279:673–674. doi: 10.1126/science.279.5351.673. [DOI] [PubMed] [Google Scholar]
- 13.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R J, Reese C B, Cohen P. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 14.Stokoe D, Stephens L R, Copeland T, Gaffney P R J, Reese C B, Painter G F, Holmes A B, McCormick F H, Hawkins P T. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 15.Kohn A, Barthel A, Kovacina K S, Boge A, Wallach B, Summers S A, Birnbaum M, Scott P H, Lawrence J C, Jr, Roth R A. J Biol Chem. 1998;273:11937–11943. doi: 10.1074/jbc.273.19.11937. [DOI] [PubMed] [Google Scholar]
- 16.Ueki K, Yamamoto-Honda R, Kaburagi Y, Yamauchi T, Tobe K, Burgering B M T, Coffer P J, Komuro I, Akanuma Y, Yazaki Y, Kadowaki T. J Biol Chem. 1998;273:5315–5322. doi: 10.1074/jbc.273.9.5315. [DOI] [PubMed] [Google Scholar]
- 17.Kohn A D, Takeuchi F, Roth R A. J Biol Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 18.Chung J, Grammer T C, Lemon K P, Kazlauskas A, Blenis J. Nature (London) 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 19.Han J-W, Pearson R B, Dennis P B, Thomas G. J Biol Chem. 1998;270:21396–21403. doi: 10.1074/jbc.270.36.21396. [DOI] [PubMed] [Google Scholar]
- 20.Weng Q-P, Andrabi K, Kozlowski M T, Grove J R, Avruch J. Mol Cell Biol. 1995;15:2333–2340. doi: 10.1128/mcb.15.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J C, Jr, Abraham R T. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 22.Brunn G J, Fadden P, Haystead T A J, Lawrence J C., Jr J Biol Chem. 1997;272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- 23.Lin T-A, Lawrence J C., Jr J Biol Chem. 1996;271:30199–30204. doi: 10.1074/jbc.271.47.30199. [DOI] [PubMed] [Google Scholar]
- 24.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 25.Kohn A D, Summers S A, Birnbaum M J, Roth R A. J Biol Chem. 1996;271:31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 26.Cheng H-C, Kemp B E, Pearson R B, Smith A J, Misconi L, Van Patten S M, Walsh D A. J Biol Chem. 1986;261:989–992. [PubMed] [Google Scholar]
- 27.Haystead T A J, Haystead C M M, Hu C, Lin T-A, Lawrence J C., Jr J Biol Chem. 1994;269:23185–23191. [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lin T-A, Kong X, Haystead T A J, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 30.Cohen P. Ann Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- 31.Brown E J, Schreiber S L. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 32.Littlewood T D, Hancock D C, Danielian P S, Parker M G, Evan G I. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherrer L C, Picard D, Massa E, Harmon J M, Simons S S, Jr, Yamamoto K R, Pratt W B. Biochemistry. 1993;32:5381–5386. doi: 10.1021/bi00071a013. [DOI] [PubMed] [Google Scholar]
- 34.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Nature (London) 1997;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 35.Brunn G J, Williams J, Sabers C, Wiederrecht G, Lawrence J C, Jr, Abraham R T. EMBO J. 1996;15:5256–5267. [PMC free article] [PubMed] [Google Scholar]
- 36.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alessi D R, Caudwell F B, Andjelkovic M, Hemmings B A, Cohen P. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 38.Alessi D R, Kozlowski M T, Weng Q-P, Morrice N, Avruch J. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 39.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 40.Dennis P B, Pullen N, Kozma S C, Thomas G. Mol Cell Biol. 1996;16:6242–6251. doi: 10.1128/mcb.16.11.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fadden P, Haystead T A J, Lawrence J C., Jr J Biol Chem. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- 42.White M F, Kahn C R. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]