Abstract
Only a limited number of target molecules have been shown to be recognized by colon tumor-reactive T cells, limiting the options for the development of immunotherapies for patients with colon cancer. The current studies were undertaken in an attempt to generate tumor-reactive T cells that could be used to identify and characterize novel colon tumor-associated antigens. Multiple CD4+ T-cell clones isolated either from tumor-infiltrating lymphocytes or peripheral blood mononuclear cells that were sensitized in vitro with autologous tumor cells from a colon cancer patient, 1869, recognized autologous tumor cells in a class II HLA-DR-restricted manner. One of the peripheral blood mononuclear cell clones, clone C111, was used to screen pools of clones that were generated from an autologous colon tumor cell line cDNA library. A cDNA clone that was isolated encoded a protein that was termed colorectal tumor-associated antigen-1 (COA-1). This product was recognized in the context of the two autologous HLA-DRβ1 alleles, HLA-DRβ1*0402 and DRβ1*1301. The nucleotide sequence of the COA-1 transcript was nearly identical to multiple expressed sequence tag sequences that encode variants of Socius, a protein that was found recently to bind to members of the Rnd family of GTPases. The COA-1 gene was expressed at relatively comparable levels in colorectal and melanoma tumor cells, EBV-infected B cells, normal B cells, and cultured fibroblast cell lines. However, the gene that was isolated from normal cell types contained a single nucleotide substitution, resulting in an amino acid change near the COOH terminus of the protein. Although the minimal epitope recognized by CD4+ cells was encoded by sequences that were upstream from this substitution, C111 T cells did not appear to recognize the normal gene product. Therefore, this alteration may either affect the localization or the processing of this gene product, which may at least in part be responsible for the differential recognition of tumor and normal cells.
INTRODUCTION
Colon cancer is a leading cause of mortality in Western countries. Despite the improvement of surgery and chemotherapy treatments, the 5-year survival rate has not altered significantly over several decades (1, 2). Immunological therapies have been intensively investigated in patients with melanoma, where treatment with IL-2,2 as well as the adoptive transfer of in vitro cultured TIL, has been found to result in cancer regression in a significant percentage of patients (3, 4). In contrast, immunotherapy has not provided a benefit to colorectal cancer patients, which may be because of the poor immunological characterization of this cancer, limiting the treatment options for patients with this disease (5, 6). The presence of a CD8+ T-cell infiltrate in colon cancer has prognostic value (7); nevertheless, the presence of an inflammatory infiltrate was not linked to systemic immunity against cancer in this report. The loss of HLA class I expression both in vitro and in vivo has been described frequently in colorectal cancers, and appears to be associated with tumor progression (8–10). The limited availability of in vitro established tumor lines and specific T lymphocytes has, in addition, hindered analysis of the role of the immune system in colorectal cancer. Although many tumor-associated antigens have been identified, the majority of these are either limited in their expression to melanoma or are expressed in melanoma as well as in a number of other histologies, including breast, ovarian, lung, and prostate tumors (11). Candidate antigens that appear to be overexpressed in colon cancer such as CEA, the epithelial cell adhesion molecule EP-CAM, HER-2/neu, and cyclophilin B, have been evaluated as potential targets for colorectal cancer therapy by carrying out in vitro sensitizations of PBMCs with candidate peptides from these molecules that bind to particular HLA alleles. However, a relatively few potential epitopes have been identified using this approach, and the T cells that have been generated using many of these peptides did not efficiently recognize native, unmanipulated tumor cells (12–15). This report details the results of studies that resulted in the identification of a gene that encodes an antigen recognized by colon tumor-reactive T cells.
MATERIALS AND METHODS
Cell Lines and Antibodies
Colon cancer lines were generated from tumor liver metastases of five patients admitted to the Surgery Branch, National Cancer Institute, NIH. The cell lines were generated from the tumor samples by cutting the tissue into small fragments, followed by filtration through sterile gauze. The tumor cells were cultured in collagen-coated six-well plates (Becton Dickinson, Franklin Lakes, NJ) in ACL-4 medium (Invitrogen, Carlsbad, CA) containing 10% FBS plus MEGM SingleQuots (Clonetics, Walkersville, MD) that contained epidermal growth factor (10 ng/ml), insulin (5 μg/ml), hydrocortisone (0.5 μg/ml), gentamicin (50 μg/ml), and amphotericin-B (50 ng/ml). Fresh medium was added to the cells every 5 days, and fibroblasts were depleted from the cultures by carrying out a short-term treatment with trypsin. Immunofluorescent staining assays to assess cell surface HLA gene expression were carried out using the anticlass I mAb W6/32 and the anti-DR mAb L243 (Becton Dickinson). The cell lines were stained using the mAb BerEP4 (DAKO, Cupertino, CA) that is directed against a cell surface molecule appears to be limited in its expression to epithelial tissues, and intracellular staining was carried out using the cytokeratin reactive mAbs CD18, LP34, and MNF116 (DAKO). Analysis of the expression of CEA, a molecule that is overexpressed frequently in colon tumor, was carried out using the mAb Col-1 (Zymed, South San Francisco, CA). The presence of fibroblasts in the cultured colon tumor cell lines was assessed using the mAb 5B5 (DAKO) that was directed against the β subunit of prolyl-4-hydroxylase, a protein involved with the synthesis of collagen. Flow cytometry was carried out using a FACScan (Becton Dickinson). The established colon cancer lines SW1463, SW480, and Colo205 were obtain from American Type Culture Collection (Manassas, Virginia). The melanoma cell line 1681, the fibroblast cell line 1519, and the EBV-transformed B cell lines 1869 and 1519 were established in the Surgery Branch and were cultured in RPMI 1640 plus 10% FBS. The normal B cell lines 1847, 1681, 1872, and 1869 were generated, as described previously (16), by culturing PBL in Iscove’s medium (Invitrogen) plus 10% human serum in the presence of 100 IU/ml of CD40L (Immunex, Seattle, WA) and 100 IU/ml of recombinant human IL-4 (PharMingen, San Diego, CA). The MHC class I and class II typing of the PBL and of the tumor lines used in this study was determined by single-stranded oligonucleotide probe-PCR typing carried out in the NIH HLA typing laboratory, and is summarized in Table 1. Antibodies used to carry out TCR analysis were obtained from Beckman/Coulter (Miami, FL) or Pierce/Endogen (Rockford, IL).
Table 1.
MHC haplotype of cell lines
| A | B | C | DRβ1 | DRβ3–5 | DQ | |
|---|---|---|---|---|---|---|
| 1869 | 3, 24 | 35, 38 | 0401, 1203 | 0402, 1301 | 3*01, 4*01 | 03, 06 |
| 1870 | 24 | 35 | 04 | 1202 | 3*03 | 03 |
| 1872 | 02, 03 | 07, 4402 | 0501, 0702 | 0401, 1501 | 4*01, 5*01 | 03, 06 |
| 1681 | 01, 0201 | 08, 44 | N.D. | 0301, 0402 | 3*0101, 4*01 | 0301, 0402 |
| 1847 | 02 | 18, 44 | 05, 0701 | 0401, 1301 | 3*01, 4*01 | 03, 06 |
| 1519 | 24, 32 | 1401, 4402 | 05, 08 | 0701, 1301 | 3*01, 4*01 | 02, 06 |
Identification and Characterization of Tumor-Reactive T Cells
Tumor-reactive T lymphocytes were generated from PBMC and TILs derived from colon cancer patients. Incubation of PBMCs with autologous tumor cells that had been irradiated with 150 Gy was carried out at a tumor cell:lymphocyte ratio of 1:5 in RPMI 1640 containing 300 IU/ml of recombinant human IL-2 plus 10% HS. The cultures were stimulated weekly for a period of 5–6 weeks with autologous irradiated tumor cells. Cultures of TIL were established by initially plating fresh uncultured tumors at 5 × 105 cells/well in 24-well plates in RPMI 1640 containing 10% HS and 1000 IU/ml of IL-2. Tumor cells used for T-cell stimulation were cultured for at least 10 days in RPMI 1640 containing 10% HS to avoid the generation of T cells with reactivity against FBS. In addition, to optimize or up-regulate the expression of MHC molecules by tumor cells, these cells were incubated with IFN-γ (500 IU/ml) for 48 h. The reactivity of the T-cell lines against colon cancer lines was examined by incubation of 2 × 104 or, for some of the assays, 5 × 104 T cells in flat-bottomed 96-well plate in the presence of 5 × 104 autologous or allogeneic tumor cells. After overnight incubation at 37°C in 5% CO2, the supernatants were collected, and T-cell responses were evaluated using anti-IFN-γ antibodies (Endogen, Rockford, IL) in a sandwich ELISA assay. After 3 weeks of culture the T-cell lines were cloned by limiting dilution in the presence of allogeneic PBMCs that had been irradiated with 50 Gy in RPMI 1640 containing 30 ng/ml of OKT3 mAb in RPMI 1640 plus 10% HS. The following day, fresh medium plus recombinant human-IL-2 (300 IU/ml) was added to the cultures. After 2 weeks of culture, growth-positive wells were screened for their ability to release IFN-γ in response to tumor stimulation. The T lymphocytes from sensitized PBMCs that were chosen for additional analysis, C4, C49, and C111, were isolated from cultures that were plated at 5 cells/well, but only 27% of the wells were positive for growth under these conditions, indicating that some or all of these cells may represent T-cell clones. Analysis carried out with antibodies directed against TCR families indicated that >95% of clone C4 T cells expressed a TCR reactive with an anti-Vβ5 reactive antibody, whereas C49 failed to express TCRs detected by any of the commercial antibodies. Amplification of the clone C111 TCR Vβ region product carried out using RT-PCR indicated that this clone expressed a single sequence derived from the Vβ18 germ-line gene. Flow cytofluorometric analysis indicated that ~80% of C111 T cells expressed Vβ18, but contaminating feeder cells used to expand the T-cell clone may be responsible for the discrepancy between these results. Two CD4+ tumor-reactive T-cell cultures, C5 and C15, were also identified from 1869 TIL. These cultures were isolated from cells that were plated at 1 cell/well, and as only 3% of the wells that were plated were positive for growth, these are likely to represent T-cell clones. In addition, these cultures appeared to stain homogeneously with an antibody directed against Vβ2, additionally indicating that these represented T-cell clones. Tumor-reactive cultures were then expanded in the presence of allogeneic PBL that were irradiated with 50 Gy in RPMI 1640 containing phytohemagglutinin (1 μg/ml) and IL-2 (300 IU/ml). Immunofluorescent analysis of positive cultures was carried out using mAb directed against CD3, CD4, CD8, CD16, and CD56 (Becton Dickinson). Antibody blocking assays were carried out by preincubating target cells for 1 h with W6/32, an antibody directed against a pan-MHC class I epitope, or L243, a mAb directed against a pan-HLA class II DR epitope. The T cells were then added to target cells and IFN-γ release measured after an overnight incubation.
CIITA Transduction of Tumor Lines
To induce stable expression of cell surface MHC class II molecules, the tumor lines 1869 col, SW480, and Colo205 were transduced with a recombinant retrovirus that was generated by cloning the gene that encoded the human CIITA into the retroviral expression vector pCLRCX (17). The transduced 1869 tumor cells were then sorted using a FACSvantage cell sorter to obtain cells that homogeneously expressed relatively high levels of cell surface HLA class II expression.
Isolation of MHC Class II DRβ1 Molecules
The DRβ1*0402 gene was isolated by carrying out an RT-PCR with RNA derived from the tumor line 1869 col, and the DRβ1*1301 gene was obtained by carrying out an RT-PCR with RNA derived from an autologous T-cell line. Primers that were used to amplify HLA-DR were: 5′-TCCAGCATGGTGTGTCTGA-3′ and 5′-CCTTGAATGTGGTCATCT-3′. Two additional primers were designed to specifically amplify the HLA-DR13 gene product: 5′CGTTTCTTGGAG-TACTCTACGTC-3′ and 5′-CCACCGCGGCCCGCTCGTCT-3′. The isolated products were cloned in the plasmid vector pCR-Blunt (Invitrogen) and sequenced using an ABI Prism 310 Genetic analyzer (Perkin-Elmer, Shelton, CT). The genes were then cloned in the eukaryotic expression vectors pCDNA3.1 (Invitrogen) and the retroviral expression vector CLRCX4. Constructs encoding either of the HLA-DRβ1 genes were cotransfected along with a construct encoding the HLA-DRβ gene into 293 cells. Stable transfectants were stained with the FITC-labeled anti-HLA-DR mAb L243, and cells that were strongly positive for the expression of the cell surface HLA-DR molecules were isolated using a FACSvantage cell sorter (Becton Dickinson). To induce the expression of molecules involved with HLA class II antigen processing, such as the class II Ii, DMA, and DMB genes, the 293 cells that had been transfected with the HLA-DR constructs were then transduced with recombinant retroviral supernatants generated using the CLRC-CIITA construct, as described previously (17).
cDNA Library Construction and Screening
Total RNA was extracted from 1869 col tumor line using TRIzol (Life Technologies, Inc.) and poly(A) RNA was then isolated using poly(A) Tract (Promega, Madison, WI). The poly(A) RNA was then converted to cDNA using the SuperScript cDNA Synthesis kit (Invitrogen) and cloned in the episomal mammalian expression vector pEAK8 (Edge BioSystems, Gaithersburg, MD). The pEAK8 vector had been modified by cloning a fragment encoding amino acids 1–80 of the human Ii downstream of the EF1-α promoter to express the cDNA inserts as fusion constructs and target the gene products to the HLA class II antigen presentation pathway. The recombinant cDNA was then electroporated into DH10B electrocompetent cells (Invitrogen), and plasmid pools containing ~50 cDNA recombinants prepared as described previously (18). The 293 cell lines that were transfected with HLA-DRβ1*0402 (293-DR0402) or HLA-DRβ1*1301 (293-DR13) were transiently transfected with DNA prepared from the cDNA pools (200 ng) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s directions. To conserve C111 T cells, screening assays were initially carried out by transfecting a mixture of 5 × 104 293-DR*0402 and 5 × 104 293-DR*1301 cells with cDNA library pools in 96 well flat-bottomed plates. The following day the cells were washed and 1 × 105 cells T cells in AIM-V medium plus 2% HS were added each well. After 18 h of incubation at 37°C and 5% CO2, 100 μl of supernatant was collected, and the IFN-γ release was evaluated by ELISA. For subsequent assays, cDNA pools and clones were transfected into 293 cells that expressed only a single HLA DR allele, and these cells were tested for their ability to stimulate C111 T cells.
5′ RACE
Total RNA was extracted from the 1869 col tumor cell line and a 5′ RACE was performed using the Smart RACE cDNA amplification kit according the manufacturer’s instructions (Clontech, Franklin Lakes, NJ). The RT-PCR products were cloned into the pCDNA 3.1 Topo cloning vector (Invitrogen), and recombinant DNA was prepared for sequence analysis. In addition, amplification of the full-length COA-1 gene products was carried out using the Advantage 2 PCR kit (Clontech). The amplification was carried out by incubation at 95°C for 1 min, followed by 35 amplification cycles consisting of a 30-s incubation at 95°C, a 30-s annealing step at 62°C, and a 2-min extension step at 68°C.
Identification of T-Cell Epitopes
Peptides of 20 or 21 amino acids in length that overlapped by 15 amino acids that were encoded by the long open reading frame of the original cDNA clone that was isolated were synthesized by solid-phase method using a peptide synthesizer (AMS 422; Gilson Co., Inc. Middleton, WI). The purity of the peptides was verified by mass spectrometry (Tuft’s Core Facility, Boston, MA). Allogeneic B cells (1 × 105 cells/well) that expressed either the DRβ1*0402 or the DRβ1*1301 molecules were incubated with 50 μg/ml in 100 μl/well of Iscove’s medium plus 10% HS in flat-bottomed-96-well plates. After 3 h, 1–5 × 104 T cells were added to the wells in 150 μl/well of medium and incubated for 18 h at 37°C and 5% CO2, followed by measurement of IFN-γ release by ELISA.
RESULTS
Generation and Characterization of Colon Cancer Lines
Cultured colon cancer lines were initially established from liver metastasis specimens obtained from five colorectal cancer patients. Analysis of one of the most rapidly proliferating cell lines that was obtained, 1869 col, demonstrated that these cells expressed a common epithelial marker, expressed cytokeratins associated with epithelial cells (Fig. 1), and maintained a morphology in tissue culture that was typical of epithelial cells (data not shown). In contrast, the cell lines did not stain with an antibody directed against the β subunit of prolyl-4-hydroxylase, a cell surface marker expressed in fibroblasts. Taken together, these results indicated that these cells were of epithelial origin and represented colon cancer cell lines, and did not contain significant numbers of normal cells. The 1869 col cell line expressed uniform levels of MHC class I molecules, whereas low or undetectable levels of cell surface MHC class II molecules were found on the same cells (Fig. 1), but treatment of the 1869 col cells with IFN-γ resulted in strong up-regulation of HLA class II expression (data not shown). The CEA molecule represents a marker that is expressed at high levels in vivo on colon tumor cells as well as on many colon tumor cell lines, but is not expressed by fibroblasts or hepatic cells. Analysis of 1869 col cells indicated that they expressed CEA (Fig. 1), and the additional colon tumor cell lines that were generated appeared to express similar levels of this gene product (data not shown). An early passage of the 1869 col cell line demonstrated high level expression of CEA, and lower but still significant levels of CEA expression were observed at later passages of 1869 col cells (Fig. 1). These observations are consistent with previous studies in which heterogeneous expression of CEA was observed on a variety of colon tumor cell lines (19).
Fig. 1.
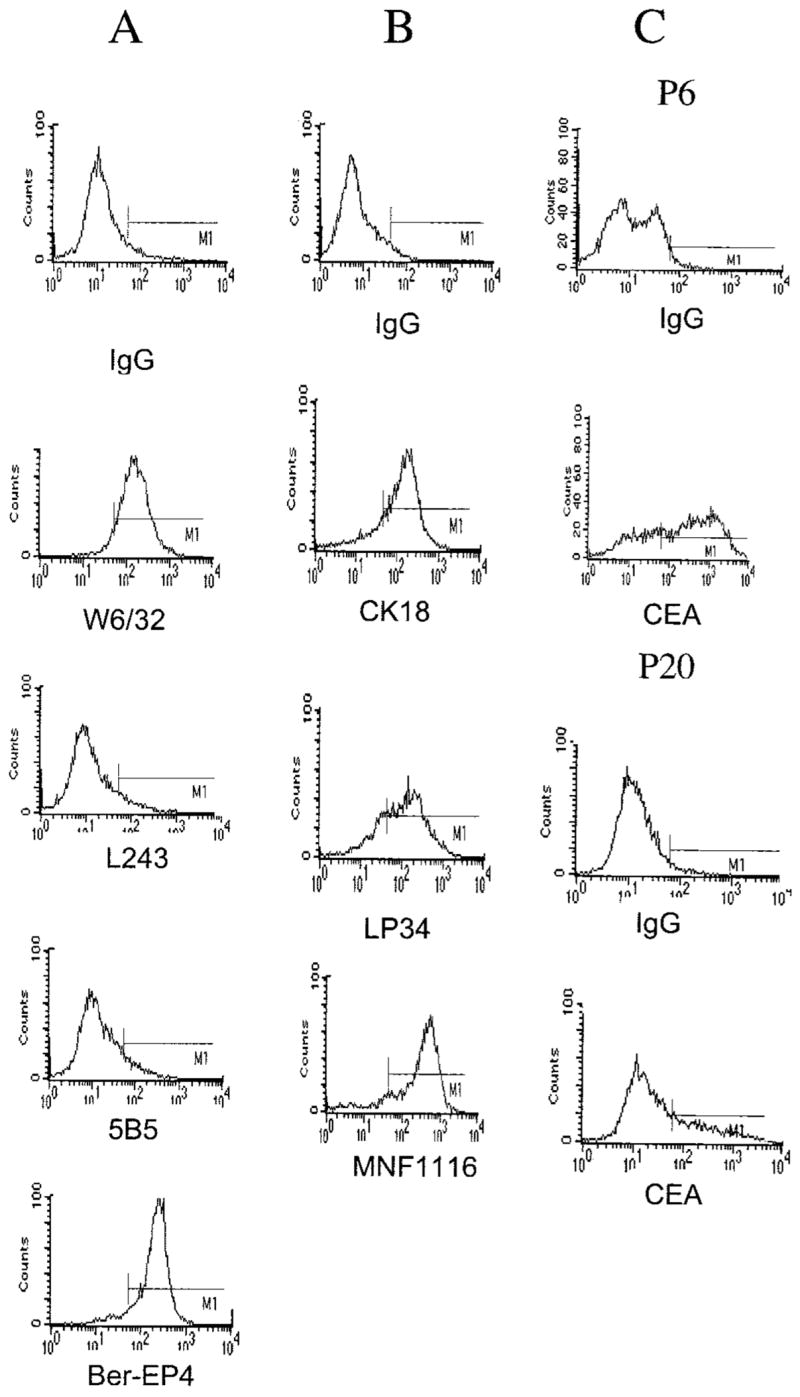
Phenotypic characterization of the colorectal cancer line 1869 col. A, antibodies directed against MHC class I (W6/32) and class II (L243) molecules, an epithelium marker (Ber-EP4), and the β subunit of prolyl-4-hydroxylase (5B5), a protein expressed exclusively in fibroblasts, were used to stain the 1869 col cell line. B, intracellular staining was carried out using three cytokeratin reactive mAbs, CK18, which reacts with cytokeratin 18, LP34, which reacts with multiple cytokeratins, and MNF116, which reacts with cytokeratins 5, 6, 8, 17, and probably 19. C, staining of 1869 col cells at passage 6 (P6) and passage 20 (P20) was carried out with the anti-CEA mAb Col-1.
Isolation and Characterization of Colon Cancer Rreactive T Lymphocytes
In the initial attempts to derive colon tumor reactive T cells, TIL from patient 1869 were cultured in high-dose IL-2. In addition, autologous tumor cells that had been treated with IFN-γ to up-regulate HLA class II gene expression were used to carry out in vitro mixed lymphocyte tumor cultures with PBMCs from patient 1869. Three CD4+ tumor-reactive T-cell clones, C4, C49, and C111, were initially selected for additional analysis on the basis of their high degree of reactivity with the autologous tumor cell line. The three clones derived from PBMCs released IFN-γ in response to autologous tumor cells that had been treated with IFN-γ, and these clones released significantly higher levels of IFN-γ in response to 1869 tumor cells that had been treated with the CIITA and sorted for cells that constitutively expressed high levels of cell surface HLA class II molecules (Table 2). Relatively low levels of IFN-γ were released after stimulation with the autologous 1869 EBV B cell line from the three T-cell clones. All of the T-cell clones released IFN-γ and granulocyte macrophage colony-stimulating factor but not IL-4 after stimulation with HLA class II-positive tumor cells (data not shown), indicating that they represent cells of the T-helper 1 cell phenotype, but were not tested for their cytotoxic activity because of limitations in the numbers of cells that were available.
Table 2.
Specific recognition of colon cancer lines by CD4+ clones from patient 1869
| T cellb |
|||||
|---|---|---|---|---|---|
| Target cells | Antibodya | HLA-DRβ1 | C4 | C49 | C111 |
| None | None | <8c | <8 | <8 | |
| 1869 col + IFN-γd | None | *0402, | 234 | 1213 | 536 |
| W6/32 | 212 | 1100 | 442 | ||
| L243 | 107 | 97 | 17 | ||
| 1869 col CIITA | None | *0402, | 536 | 5178 | 5005 |
| W6/32 | 527 | 4987 | 4249 | ||
| L243 | 47 | 254 | 305 | ||
| 1870 col + IFN-γ | None | *1202 | <8 | <8 | <8 |
| W6/32 | <8 | <8 | <8 | ||
| L243 | <8 | <7.8 | <8 | ||
| 1872 col + IFN-γ | None | *0401, | <8 | <8 | <8 |
| W6/32 | <8 | <8 | <8 | ||
| L243 | <8 | <8 | <8 | ||
| SW 480 CIITA | None | *0103, | 879 | 968 | 963 |
| W6/32 | 780 | 902 | 996 | ||
| L243 | 571 | 129 | 127 | ||
| Colo 205 CIITA | None | *0401, | 68 | 942 | 686 |
| W6/32 | 76 | 951 | 669 | ||
| L243 | 78 | 170 | 489 | ||
| 1869 EBV-B | *0402, | 52 | 126 | 322 | |
Target cells were preincubated for 1 h with either the anti-MHC class I mAb W6/32 or the anti-HLA DR mAb L243 before addition to T cells.
2 × 104 T cells were incubated with 5 × 104 target cells in flat bottomed 96-well plate in 250 μl of AIMV 2% HS. After 18 h, the supernatants IFN-γ secretion was evaluated by ELISA.
pg/ml of IFN-γ.
Where indicated, target cells were preincubated for 48 h with 500 IU of IFN-γ.
To test whether the clones isolated from the PBMCs recognized tumor cells in an MHC-restricted manner, cytokine release assays were carried out in the presence of anti-HLA class I and class II-specific antibodies using stimulator cells bearing a variety of MHC haplotypes (Table 1). The results indicated that the C4, C49, and C111 T-cell clones recognized the autologous tumor cells in the context of the HLA DR class II restriction element (Table 2). The C49 and C111 T-cell clones also recognized the CIITA transduced allogeneic MHC class II+ colon cancer lines SW480 and Colo 205 that shared expression of HLA-DRβ1*1301 with the autologous tumor, and this recognition was blocked by preincubation of the tumor cell lines with the anti-HLA-DR mAb. Generally the responses were inhibited between 50 and 90% by preincubation with the anti-HLA DR antibody, whereas <20% inhibition was observed with the anti-HLA class I antibody. The response of the C4 line to the SW480 CIITA-treated tumor cell lines, as well as the response of C111 to the Colo205 CIITA, were only partially inhibited by anti-HLA DR antibody, which might reflect the fact that these T cells can recognize additional ligands other than the classical TCR. The C4, C49, and C111 clones released low but significant levels of IFN-γ in response to autologous EBV-B cells, as well as an allogeneic EBV-B cell line that shared expression of HLA DRβ1*1301 with autologous cells. Normal B cells that were generated by stimulating autologous PBMCs with CD40 ligand plus IL-4, as well as an allogeneic fibroblast cell line that shared expression of HLA DRβ1*1301 with the 1869 col tumor and that was treated with IFN-γ to up-regulate HLA class II gene expression, stimulated little or no cytokine release from these T cells (Table 3). Two CD4+ T cell clones from TIL 1869 that responded in preliminary assays to autologous HLA class II-positive tumor cells were also tested for their ability to recognize autologous as well as allogeneic colon tumor cell lines. Clones C4, C49, and C111, as well as two clones derived from 1869 TIL, C5 and C15, responded to the allogeneic colon tumor cell line 1847 col that shared expression of the HLA-DRβ1*1301 gene product with the autologous tumor (Table 3). In contrast, the allogeneic 1872 col cell line that did not share expression of any HLA DR gene products with the 1869 col tumor failed to stimulate significant cytokine release from the T-cell clones.
Table 3.
CD4+ clones recognized colon cancer lines but not normal B or fibroblast cells sharing MHC class II molecules
| T cellb |
|||||||
|---|---|---|---|---|---|---|---|
| TIL
|
PBL
|
||||||
| Stimulator | Antibodya | HLA-DRβ1 genotype | C5 | C15 | C4 | C49 | C111 |
| None | None | <8c | <8 | <8 | <8 | <8 | |
| 1869 col CIITA | None | *0402, *1301 | 8695 | 1259 | 12328 | 12749 | 15269 |
| L243c | 279 | 162 | 511 | 524 | 790 | ||
| 1847 col + IFN-γd | None | *0401, *1301 | 2008 | 457 | 598 | 9758 | 11576 |
| L243 | 2055 | 327 | 585 | 790 | 2938 | ||
| 1872 col + IFN-γ | None | *0401, *1501 | 72 | <8 | 61 | <8 | 66 |
| L243 | 75 | <8 | 60 | <8 | 41 | ||
| 1869 EBV-B | None | *0402, *1301 | 79 | 116 | 122 | 232 | 209 |
| 1519 EBV-B | None | *0701, *1301 | 112 | 24 | 99 | 106 | 220 |
| 1519 Fibroblast + IFN-γ | None | *0701, *1301 | <8 | <8 | <8 | 55 | 62 |
| 1869 CD40LBe | None | *0402, *1301 | <8 | <8 | <8 | <8 | 45 |
| Stimulator | Antibodya | T cellb | |||||
|
|
|||||||
| C111 | |||||||
| None | 23c | ||||||
| 1869 col CIITA | 15269 | ||||||
| 1869 col CIITA | L243 | 790 | |||||
| 1681 mel + IFN-γd | 10298 | ||||||
| 1681 mel + IFN-γ | L243 | 253 | |||||
| 1869 CD40L-Be | 65 | ||||||
| 1681 CD40L-Be | 22 | ||||||
Where indicated, target cells were preincubated for 1 h with the anti-HLA DR mAb L243.
2 × 104 of the indicated T cells were incubated with 5 × 104 target cells in flat bottomed 96-well plate in 250 μl of AIMV 2% HS. After 18 h the supernatants IFN-γ secretion was evaluated by ELISA.
pg/ml of IFN-γ.
Where indicated, target cells were preincubated for 48 h with 500 IU of IFN-γ.
B cells from patients 1869 and 1681 were in vitro cultured with CD40L (100 IU/ml) and IL-4 (100 IU/ml).
Identification of the Antigen Recognized by C111 T Cells
Additional studies aimed at identifying tumor antigens expressed on 1869 col cells focused on C111 T cells, which was the only T-cell clone that expanded sufficiently to allow the cDNA library to be screened. The results of studies carried out with additional tumor histologies indicated that C111 T cells did not recognize two allogeneic renal cell lines, as well as a prostate tumor cell line that shared expression of HLA-DRβ1*1301 with the 1869 col cell line (data not shown). A single allogeneic melanoma cell line that expressed HLA-DRβ1*0402 was identified, 1681 mel. Cell surface HLA class II expression was up-regulated after treatment of the 1681 mel cell line with IFN-γ, and the treated cells were recognized by C111 T cells, indicating that certain tumor types shared expression of the antigen recognized by these T cells (Table 3).
Stable transfectants of the 293 cell line that expressed either the autologous MHC class II DRβ1*0402 or 1301 gene product molecules were then mixed in equal numbers and transiently transfected with DNA pools generated from the autologous tumor cell cDNA library. The positive pool that was initially identified after the screening of ~3 × 104 clones, 4G3, appeared to sensitize either 293-DRβ1*0402 or 1301 target cells for recognition by C111 T cells, and a single cDNA clone that could sensitize target cells for recognition by C111 T cells, 1D8, was identified (Fig. 2). An assay carried out by transfection of the 293-DRβ1*0402+ or 1301+ cell lines individually with the 1D8 cDNA indicated that either of these HLA class II restriction elements could present the T-cell epitope to C111 T cells. In contrast, 293 cell lines that expressed the HLA-DRβ1*0101, 0401, 0701, or 1601 class II alleles failed to stimulate these T cells after transfection of the 1D8 cDNA clone (data not shown), indicating that presentation of this epitope to C111 T cells may be limited to the two autologous HLA-DR alleles expressed by 1869 col cells. Additional screening of the cDNA library resulted in the isolation of a second cDNA clone that was nearly identical to the 1D8 clone. The isolation of a second clone with a nearly identical sequence provides evidence supporting the contention that this represents the natural transcript encoding the antigen recognized by C111 T cells.
Fig. 2.
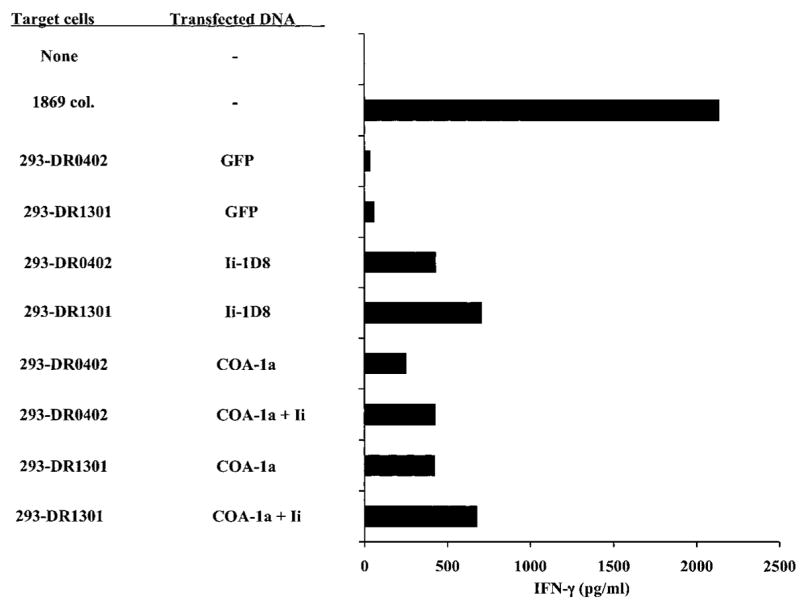
A cDNA clone isolated from the 1869 cDNA library encodes an antigen recognized by C111 T cells. The 293 cells expressing the MHC DRβ1*0402 or 1301 molecules were transfected with the 1D8 cDNA clone, or COA-1a, which corresponds to nucleotides 209-1318 of the COA-1 gene (Fig. 3). Target cells were either transfected with the COA-1a product alone or were cotransfected with a mixture of COA-1a and the full-length HLA class II Ii. Additional targets were transfected with a control plasmid encoding green fluorescent protein. Eighteen h after the addition of 5 × 104 C111 T cells to the transfectants, supernatants were collected and IFN-γ release was measured by ELISA.
Characterization of Colorectal Tumor-Associated Antigen COA-1
The 1D8 insert contained a 44-bp poly(A) tail at the 3′ end, but appeared to represent a partial cDNA clone, because it was only 291 bp in length. The 5′ end of the gene product that was expressed in the 1869 col cell line was then isolated by carrying out a 5′ RACE reaction using nested internal primers complementary to the sequence of the 1D8 clone. Sequencing of products that were cloned from this reaction indicated that a 1412-bp product represented the predominant transcript of the gene in the 1869 col cell line that encoded the antigen recognized by C111 T cells, which was designated COA-1 (Fig. 3). Comparison of the COA-1 sequence with the genomic DNA sequence database indicated that this product was derived from 13 exons, but at least two additional alternatively spliced products of this gene were isolated from the RACE reaction. An alignment of the COA-1 transcript with the human EST database indicated that this was identical or nearly identical to several sequences obtained from normal human brain, placenta, ovary, and testis, as well as sequences obtained from a variety of adenocarcinomas. The 5′ end of the transcript cloned from the RACE reaction corresponded to the 5′ end of several EST sequences found in the database, and the 3′ end of the original cDNA clone corresponded to the 3′ end of the EST transcripts derived from several cell lines, indicating that these may represent the authentic 5′ and 3′ ends of the predominant COA-1 colon tumor cell transcript. The COA-1 sequence was also nearly identical to that of a transcript encoding the human homologue of the rat Socius protein, a molecule that was cloned recently on the basis of its ability to bind to a member of the Rnd family of GTPases (20).
Fig. 3.
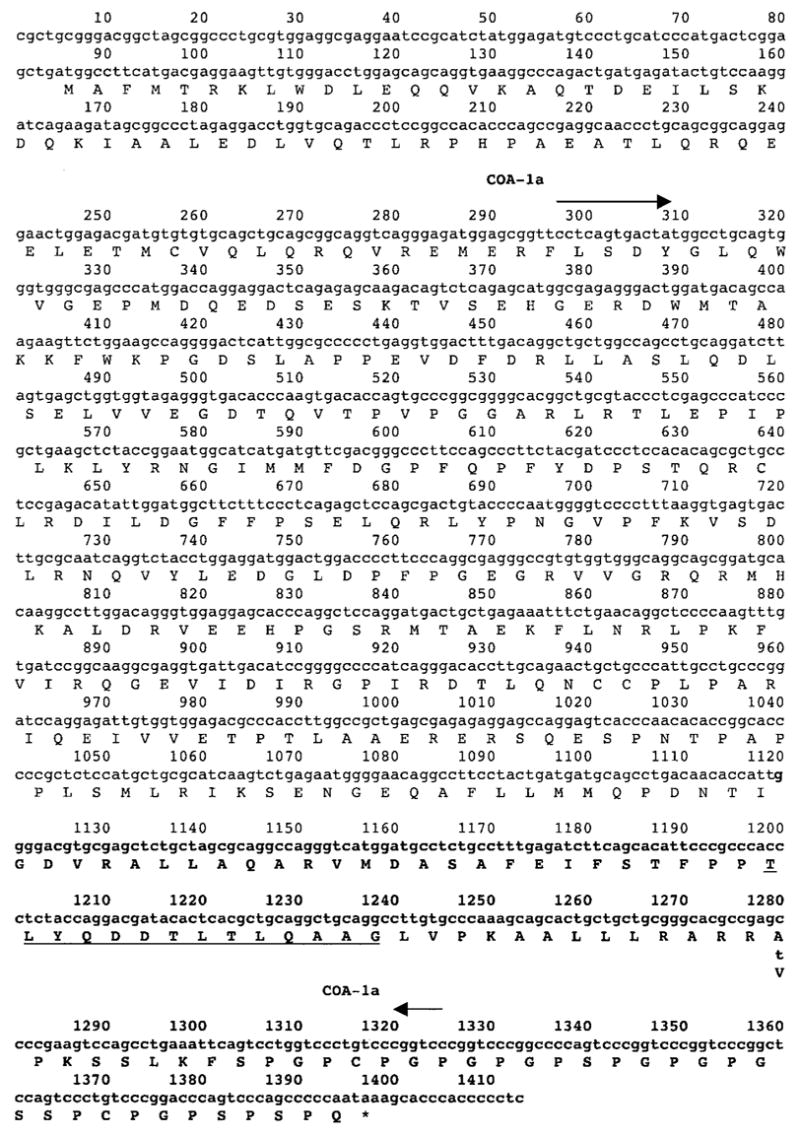
Sequence of the COA-1 gene isolated from the mRNA of the tumor line 1869 col. The COA-1 gene was isolated by RT-PCR from the 1869 col tumor cell line. The sequence of the 1D8 cDNA clone is indicated by bold letters, the amino acid sequence corresponding to the T-cell epitope is underlined, and the single nucleotide difference between the normal and tumor transcripts at position 1280 is noted. The position of the primers used to generate the COA-1a product is indicated by the arrows, and the amino acid sequence of the longest open reading frame in this transcript, which is similar to the Socius gene product (20), is noted beneath the nucleotide sequence.
Forward and reverse primers located at or near the 5′ and 3′ ends of the putative COA-1 gene product were then used to carry out an RT-PCR from 1869 RNA, because the RACE products that had been cloned only comprised a portion of the normal transcript. When RT-PCR was carried out with several primers that were proximal to the putative 5′ end of the transcript in combination with primers that were complementary to the highly repetitive G/C rich sequence near to the 3′ end of the COA-1 transcript, a variety of nonspecific transcripts were generated (data not shown). A product that was designated COA-1a was, however, amplified from 1869 col RNA using two primers that encompassed the region between nucleotides 290 and 1318 of the putative full-length COA-1 transcript. Transfectants that coexpressed the COA-1a gene along with either HLA-DRβ1*0402 or 1301 appeared to stimulate comparable levels of cytokine release from C111 T cells to those transfected with the truncated 1D8 cDNA clone, indicating that the full-length gene can be processed relatively efficiently (Fig. 2). Cotransfection of the COA-1a gene with a construct encoding the full-length human Ii had little or no effect on the recognition of target cells transfected with the COA-1a product by C111 T cells. Thus, either the levels of Ii expression in 293 cells that were also transfected with a construct encoding the CIITA gene product was adequate for recognition of this epitope, or Ii expression does not have a significant impact on the processing of the COA-1 epitope. In addition, the COA-1a product was not fused with amino acids 1–80 of the human Ii molecule, which had been shown previously to enhance the recognition of some HLA class II antigens (21). The observation that the fusion of the cDNA clone with the Ii did not appear to enhance recognition by the CD4+ T cells indicates that the COA-1 antigen may naturally target the endogenous HLA class II processing pathway in colon tumor cells.
The expression pattern of the COA-1 gene was then examined in several colorectal, melanoma, and EBV-B cell lines, as well as in several normal cell lines, which included CD40L stimulated B cell and fibroblast cell lines. The results of Northern blot analysis indicated that this gene was expressed at relatively low levels in colon and melanoma tumor cell lines, EBV B cells, normal B cells, and fibro-blasts, and quantitative TaqMan RT-PCR indicated that the levels of expression did not differ significantly between these cells (data not shown).
The observation that the level of expression of the COA-1 gene did not differ significantly between cell lines that were or were not recognized by C111 T cells indicated that these cells might express similar but nonidentical products. Therefore, transcripts of the COA-1 gene that were expressed in the autologous and allogeneic CD40L-stimulated B cells, as well as allogeneic fibroblast cell lines, were isolated using RT-PCR and sequenced. The results of sequencing carried out with the bulk RT-PCR products indicated that CD40L-stimulated B cells and fibroblast cell lines predominantly expressed products that appeared to be identical to the COA-1 transcript derived from 1869 col cells with the exception of a single substitution of a T for a C residue at nucleotide position 1280, resulting in a change at amino acid 399. The COA-1 transcripts that were expressed in CD40L B cells were isolated by carrying out RT-PCR and cloning the resultant products. Ten of 10 clones from the CD40L-B cells that were sequenced contained a T at position 1280 but were otherwise identical to the 1869 col COA-1 transcript. Amplification of the COA-1 gene product from allogeneic colorectal tumor lines SW1463, SW480, and 1847 col, as well as the 1681 mel line, indicated that these cells predominantly expressed products containing a C residue at position 1280, as determined by sequencing the bulk, uncloned RT-PCR products that were amplified from these cells (data not shown). Two peaks of comparable heights that corresponded to C and T residues at position 1280 of the COA-1 transcript were derived by sequencing the uncloned RT-PCR product from autologous EBV B cells, indicating that these products may be expressed at similar levels in these cells. The results obtained using RNA from autologous CD40L-stimulated B cells, EBV B cells, and the colon tumor cell lines were confirmed by repeated analysis carried out on products obtained from four independent RT-PCR reactions, indicating that the residue found at nucleotide 1280 of the COA-1 transcripts did not represent a PCR mutation (data not shown).
In an attempt to evaluate the significance of the single bp change at position 1280 in the COA-1a sequence, the RT-PCR products obtained from autologous CD40L-stimulated B cells were cloned in a eukaryotic expression vector. A plasmid containing the COA-1a transcript that was amplified from normal B cells was then compared with products cloned from 1869 col cells for its ability to sensitize 293-DR*0402 or 293-DR*1301 cells for recognition by C111 T cells. Target cells expressing either of the autologous HLA-DR genes that were transfected with the COA-1a or 1D8 gene products, but not the product that was isolated from CD40L activated B cells, stimulated cytokine release from C111 T cells (Fig. 4). These results indicated that there was a correlation between the recognition of normal B cells and tumor cells, and the ability of the COA-1 gene products that were expressed by these cells to sensitize targets for recognition by C111 T cells.
Fig. 4.
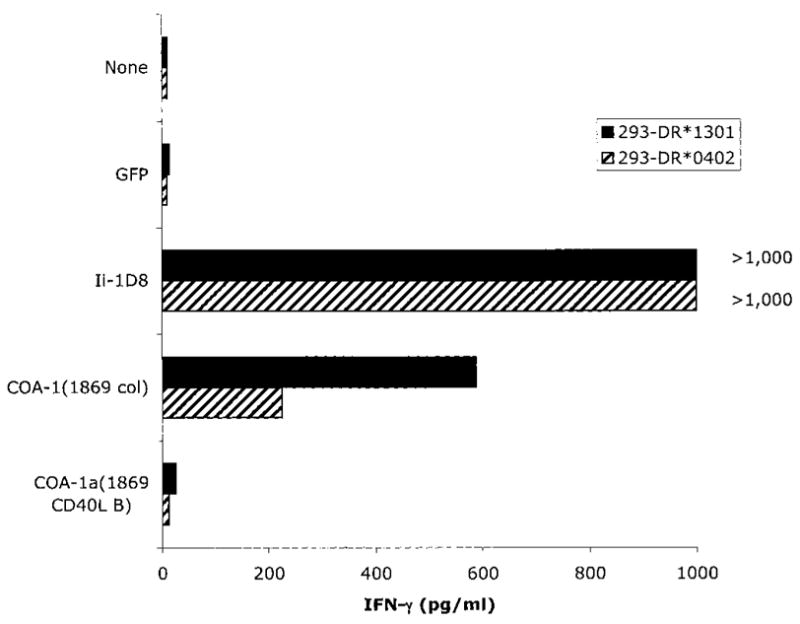
The COA-1 transcript derived from normal B cells is not recognized by the clone C111 T cells. 293 cells expressing the indicated MHC DRβ1 molecules were transfected with COA-1a cDNAs isolated by RT-PCR from either the 1869 col cell line or from 1869 CD40L-stimulated B cells. The green fluorescent protein and Ii-1D8 constructs were used as negative and positive controls, respectively. Eighteen h after the addition of 5 × 104 C111 T cells to the transfectants, supernatants were collected, and IFN-γ release was measured by ELISA.
Identification of the Epitope Recognized by the CD4+ Clone C111
The results of transfection studies carried out using truncated COA-1 gene products indicated that the C111 T-cell epitope was encoded by a region located between nucleotides 1121 and 1288 of the COA-1 transcript. The longest open reading frame in the COA-1 transcript, which overlapped with the Socius gene product (20), was used as the basis for the synthesis of peptides that were used in an attempt to identify the T-cell epitope recognized by C111 T cells. Peptides that were 20 or 21 amino acids in length and that overlapped by either 14 or 16 amino acids were than synthesized and tested for their ability to sensitize target cells for recognition by C111 T cells. Because autologous normal B cells could not be efficiently expanded, allogeneic normal B cells expressing either DRβ1*0402 or DRβ1*1301 were used to carry out these assays. The 1681 and 1847 CD40L-stimulated normal B cell lines shared expression of HLA-DRβ1*0402 and HLA-DRβ1*1301 molecules, respectively, with the autologous tumor cell line. These cells were incubated with the panel of peptides and then tested for their ability to stimulate cytokine release from C111 T cells. The results indicated that 1681 and 1847 CD40LB cells that were pulsed with either of the two overlapping peptides FSTFPPTLYQDDTLTLQAAG and TLYQDDTLTLQAA-GLVPKAA stimulated significant cytokine release from C111 T cells. Thus, these T cells appear to recognize the peptide TLYQD-DTLTLQAAG, which represents the overlapping region in these peptides. The L at position two, the T at the position 7, and L at position 10 in this sequence appeared to conform to an HLA binding motif that has been identified for the HLA-DRβ1*0402 class II allele (22); however, it was not possible to identify the potential anchor residues in this sequence that were involved in binding to the HLA-DRβ1*1301 allele. Nevertheless, these observations provide evidence that C111 T cells recognize a single peptide epitope in the context of either the HLA-DRβ1*0402 or 1301 class II gene products.
DISCUSSION
The identification of antigens that are naturally processed and presented on tumor cells represents an important goal of cancer immunotherapy. A wide variety of antigens have been identified using T cells that were initially generated after tumor stimulation; however, many of the antigens that have been identified are expressed in melanoma exclusively or are expressed on only a relatively small percentage of other tumors from other histologies. Although “reverse immunology” approaches that use the use of candidate antigens or epitopes to stimulate tumor reactive T cells has had some success, only a limited array of antigens that are expressed on common tumor types such as colon carcinomas have been identified using this approach.
The results of initial attempts to identify antigens recognized by colon tumor-reactive T cells are presented in this report. Several tumor-reactive CD4+ T lymphocytes were isolated from PBMCs and TILs that were obtained after the establishment of autologous cultured colon tumor cell lines. These studies focused on a single clone of CD4+ T cells, C111, that responded strongly to autologous tumor cells, and demonstrated low but significant reactivity with autologous EBV B cells, but failed to respond to autologous CD40L-stimulated B cells. The gene encoding this antigen, termed COA-1, was isolated by screening an autologous cDNA library with clone C111 T cells. This gene appeared to be nearly identical to the gene encoding the human homologue of the rat Socius protein that was cloned recently using a yeast two-hybrid screening assay in which a member of the Rnd family of GTPases was used as bait (20). The Socius product was expressed at high levels in rat testis, but was expressed at significantly lower levels in rat lung, thymus, and brain.
The longest open reading frame in the COA-1 transcript encodes a 437 amino acid product that corresponds to a portion of the human Socius gene product, and two overlapping peptides derived from this open reading frame were identified that could sensitize target cells expressing either HLA-DRβ1*0402 or 1301. The stimulation observed with peptide-pulsed targets was weak relative to that seen with the tumor cell lines that were recognized, and a minimum concentration of ~10 μM was needed to stimulate significant cytokine release from C111 T cells (Table 4). Peptides derived from nonmutated tumor antigens such as tyrosinase (23) and TRP-1 or TRP-2 (17) have also been found to stimulate only relatively low levels of cytokine release from HLA class II-restricted, tumor-reactive T cells, and minimal concentrations of between 1 and 10 μM of the peptides identified in these studies were required to sensitize target cells for T-cell recognition. This may reflect that fact that these represent nonmutated self-antigens, and that self-tolerance results in the deletion of T cells that recognize peptides that bind to class II molecules with high affinity. In addition, the autologous tumor cell line should present the COA-1 peptide in the context of both the HLA-DRβ1*0402 and 1301 restriction elements, leading to enhanced stimulation of T cells reactive with this epitope. Transfectants expressing the COA-1 product also stimulated significantly less cytokine release from C111 T cells than the autologous tumor cell line that had been induced to express high levels of HLA class II molecules. However, one potential explanation for this observation is that the HLA class II-positive 293 cells used as targets for transfection of the COA-1 gene products fail to express optimal levels of accessory molecules associated with the processing of this epitope.
Table 4.
Identification of the COA-1-derived epitopes recognized by the CD4+ clone C111
| Stimulator | HLA-DRβ1 | No peptide | |||||
|---|---|---|---|---|---|---|---|
| None | <8a | ||||||
| 1869 col | 0402, 1301 | 2186 | |||||
| 1681 CD40LB | 0301, 0402 | <8 | |||||
| 1847 CD40LB | 0401, 1301 | <8 | |||||
| Peptide concentration (μg/mM)
|
|||||||
| Peptideb | 100 | 50 | 25 | 12.5 | 6.25 | ||
|
| |||||||
| 1681 CD40LB | 0301, 0402 | FSTFPPTLYQDDTLTLQAAG | 105 | 236 | 69 | <7.8 | <7.8 |
| 1681 CD40LB | TLYQDDTLTLQAAGLVPKAA | 51 | 159 | <7.8 | <7.8 | <7.8 | |
| 1681 CD40LB | DDTLTLQAAGLVPKAALLLRA | 11 | 16 | <7.8 | <7.8 | <7.8 | |
| 1681 CD40LB | LQAAGLVPKAALLLRARRAP | 21 | 12 | <7.8 | <7.8 | <7.8 | |
| 1847 CD40LB | 0401, 1301 | ASAFEIFSTFPPTLYQDDTL | < | <7.8 | <7.8 | <7.8 | <7.8 |
| 1847 CD40LB | FSTFPPTLYQDDTLTLQAAG | 226 | 397 | 296 | 79 | <7.8 | |
| 1847 CD40LB | TLYQDDTLTLQAAGLVPKAA | 79 | 326 | <7.8 | <7.8 | <7.8 | |
| 1847 CD40LB | DDTLTLQAAGLVPKAALLLRA | 22 | 33 | <7.8 | <7.8 | <7.8 | |
| 1847 CD40LB | LQAAGLVPKAALLLRARRAP | 52 | 32 | <7.8 | <7.8 | <7.8 | |
The CD4+ T-cell clone C111 was added at 2 × 104 cells/well at the final volume of 250 μl/well of ISCOVE’s plus 10% HS, and after 18 h of incubation, the supernatants were collected and the IFN-γ release was evaluated by ELISA.
Peptides of 20 or 21 amino acids overlapping by 15 amino acids were synthesized using the putative COA-1 protein, in the 1D8 region (1012–1318 bp). 4 × 105/ml of B cells sharing one of the DRβ1 molecules (*0402 or *1301) with the autologous tumor 1869, were incubated for 3 h at 37°C and 5% CO2 in the presence or not (−) of the peptides at the final volume of 100 μl/well in ISCOVE’s plus 10% HS.
The COA-1 transcript is nearly identical to sequences derived from a variety of tissues and tumor cell lines. However, these transcripts comprise a large array of >20 alternatively spliced products that are derived from at least 15 exons residing at the chromosome 1p36.1-p35 locus. The COA-1 product expressed in colon tumor cell lines appeared to contain a unique splicing pattern that did not correspond to any of the transcripts identified in the EST and GenBank databases, which may not encode products recognized by C111 T cells. Two nearly identical COA-1 gene products were amplified from EBV B cells, one of which was identical to that isolated from the colon tumor cells, and a second that contained a single nucleotide alteration at position 1280 that resulted in a substitution of a valine residue for the alanine residue at position 399 encoded by the dominant colon tumor cell product. It is not clear why C111 T cells only appeared to weakly recognize EBV B cells expressing the appropriate HLA class II gene products, but these observations could result from inherent differences in the antigen processing abilities of colon tumor cells and EBV-transformed B cells. Previous results have suggested that differences in the proteosomal subunits expressed by various cells may significantly influence antigen recognition, which provides one potential explanation for this finding (24). The RT-PCR products that were amplified from normal B cells and fibroblasts also appeared to uniquely encode the COA-1 variant that expressed a valine residue at amino acid 399, and target cells that were transfected with the COA-1 product that was amplified from normal cell lines were not recognized by C111 T cells. Thus, it appears that normal B cells and fibroblasts either fail to express the COA-1 transcript that can be processed and presented to C111 T cells or express this product at only relatively low levels. The mechanisms involved in the preferential expression of these two transcripts are unknown, but these may represent the products of two nearly identical genes of which the expression is differentially regulated. Attempts to amplify and sequence COA-1 genomic sequences derived from these cell lines have yielded ambiguous results, but future studies may help to clarify this issue. Nevertheless, the correlation between expression of these transcripts and the ability of C111 T cells to recognize the epitope encoded by these products provides additional evidence that this represents the natural product recognized by these T cells and not a peptide mimic of the natural epitope.
An additional observation that is unexplained is how the alteration at position 399 affects recognition of the cell epitope comprised of amino acids 371–384 of the COA-1 transcript. Results of a previous study indicated that alteration of a distal residue can influence the ability of tumor-reactive CD4+ T cells to recognize a mutated product of the CDC-27 gene product (21). Preliminary results presented in the prior study indicated that altered intracellular targeting of the mutated CDC-27 gene product may have played an important role in influencing processing of this gene product. Investigation of the cellular localization of the COA-1 protein in normal and tumor cells may help to indicate whether a similar mechanism may be involved with T-cell recognition of this product.
Transfection studies, as well as peptide pulsing experiments indicated that either of the autologous HLA-DRβ1 alleles, DRβ1*0402 or DRβ1*1301, could present the T-cell epitope to clone C111 T cells, which may potentially enhance the immunogenicity of this peptide in patient 1869 as well as other individuals that express these class II alleles. However, this observation is not unique, as examples of promiscuous recognition of class II as well as class I restricted epitopes have been noted in previous studies. In one report, CD4+ T cells were identified that also recognized an epitope of the herpes simplex type 2 virus virion protein VP16 in the context of DRβ1*0402, 1102, or 1301, but not several closely related DR4, 11, or 13 subtypes (25). The sequences of the DRβ1*0402, 1102, and 1301 molecules are identical in a polymorphic region between amino acids 67 and 71, and site-directed mutagenesis studies demonstrated that these residues were critical for the recognition of the viral epitope.
High levels of lymphocyte infiltration into tumors has been shown in some studies to be correlated with a good prognosis (26), but detailed investigations of the reactivity of infiltrating T cells have not been carried out. The expression of HLA class II molecules on colorectal cancer cells is also a favorable prognostic marker (27, 28). Previous studies resulted in the isolation of HLA class I (29) and class II (30, 31) -restricted tumor-reactive T cells from colon cancer patients, but only a limited panel of shared tumor-specific antigens were identified in these studies. This report presents the description of an immunogenic CD4+ T-cell epitope derived from a previously undescribed colorectal cancer antigen that represents a potential target for immunotherapy in patients with this disease. Evaluation of the in vitro immunogenicity of the COA-1 epitope in DRβ1*0402- and *1301-positive colorectal patients may provide support for the use of this antigen in the immunotherapy of patients with this disease.
Footnotes
The abbreviations used are: IL, interleukin; TIL, tumor-infiltrating lymphocyte; CEA, carcinoembryonic antigen; PBMC, peripheral blood mononuclear cell; mAb, mono-clonal antibody; FBS, fetal bovine serum; TCR, T-cell receptor; HS, human serum; RT-PCR, reverse transcription-PCR; PBL, peripheral blood lymphocyte; CIITA, class II transactivator; poly(A), polyadenylic acid; RACE, rapid amplification of cDNA ends; COA, colorectal antigen; EST, expressed sequence tag; Ii, invariant chain.
References
- 1.DeCosse JJ, Tsioulias GJ, Jacobson JS. Colorectal cancer: detection, treatment, and rehabilitation. CA Cancer J Clin. 1994;44:27–42. doi: 10.3322/canjclin.44.1.27. [DOI] [PubMed] [Google Scholar]
- 2.Harrington DP. The tea leaves of small trials. J Clin Oncol. 1999;17:1336–1338. doi: 10.1200/JCO.1999.17.5.1336. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA, Packard BS, Aebersold PM, Solomon D, Topalian SL, Toy ST, Simon P, Lotze MT, Yang JC, Seipp CA, Simpson C, Carter C, Bock S, Schwartzentruber D, Wei JP, White DE. Use of tumor infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. Preliminary report. N Engl J Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 4.Mukherji B, Chakraborty NG. Immunobiology and immunotherapy of melanoma. Curr Opin Oncol. 1995;7:175–184. doi: 10.1097/00001622-199503000-00014. [DOI] [PubMed] [Google Scholar]
- 5.Riethmuller G, Holz E, Schlimok G, Schmiegel W, Raab R, Hoffken K, Gruber R, Funke I, Pichlmaier H, Hirche H, Buggisch P, Witte J, Pichlmayr R. Monoclonal antibody therapy for resected Dukes’ C colorectal cancer: seven-year outcome of a multicenter randomized trial. J Clin Oncol. 1998;16:1788–1794. doi: 10.1200/JCO.1998.16.5.1788. [DOI] [PubMed] [Google Scholar]
- 6.Vermorken JB, Claessen AM, van Tinteren H, Gall HE, Ezinga R, Meijer S, Scheper RJ, Meijer CJ, Bloemena E, Ransom JH, Hanna MG, Jr, Pinedo HM. Active specific immunotherapy for stage II and stage III human colon cancer: a randomised trial. Lancet. 1999;353:345–350. doi: 10.1016/S0140-6736(98)07186-4. [DOI] [PubMed] [Google Scholar]
- 7.Naito Y, Saito K, Shiiba K, Ohuchi A, Saigenji K, Nagura H, Ohtani H. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 1998;58:3491–3494. [PubMed] [Google Scholar]
- 8.Browning M, Petronzelli F, Bicknell D, Krausa P, Rowan A, Tonks S, Murray N, Bodmer J, Bodmer W. Mechanisms of loss of HLA class I expression on colorectal tumor cells. Tissue Antigens. 1996;47:364–371. doi: 10.1111/j.1399-0039.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 9.Garrido F, Cabrera T, Concha A, Glew S, Ruiz-Cabello F, Stern PL. Natural history of HLA expression during tumour development. Immunol Today. 1993;14:491–499. doi: 10.1016/0167-5699(93)90264-L. [DOI] [PubMed] [Google Scholar]
- 10.Coulie PG, Ikeda H, Baurain JF, Chiari R. Antitumor immunity at work in a melanoma patient. Adv Cancer Res. 1999;76:213–242. doi: 10.1016/s0065-230x(08)60778-2. [DOI] [PubMed] [Google Scholar]
- 11.Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsang KY, Zhu M, Nieroda CA, Correale P, Zaremba S, Hamilton JM, Cole D, Lam C, Schlom J. Phenotypic stability of a cytotoxic T-cell line directed against an immunodominant epitope of human carcinoembryonic antigen. Clin Cancer Res. 1997;3:2439–2449. [PubMed] [Google Scholar]
- 13.Akagi J, Nakagawa K, Egami H, Ogawa M. Induction of HLA-unrestricted and HLA-class-II-restricted cytotoxic T lymphocytes against MUC-1 from patients with colorectal carcinomas using recombinant MUC-1 vaccinia virus. Cancer Immunol Immunother. 1998;47:21–31. doi: 10.1007/s002620050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brossart P, Stuhler G, Flad T, Stevanovic S, Rammensee HG, Kanz L, Brugger W. Her-2/neu-derived peptides are tumor-associated antigens expressed by human renal cell and colon carcinoma lines and are recognized by in vitro induced specific cytotoxic T lymphocytes. Cancer Res. 1998;58:732–736. [PubMed] [Google Scholar]
- 15.Ito M, Shichijo S, Tsuda N, Ochi M, Harashima N, Saito N, Itoh K. Molecular basis of T cell-mediated recognition of pancreatic cancer cells. Cancer Res. 2001;61:2038–2046. [PubMed] [Google Scholar]
- 16.Lapointe R, Lemieux R, Olivier M, Darveau A. Tyrosine kinase and cAMP-dependent protein kinase activities in CD40- activated human B lymphocytes. Eur J Immunol. 1996;26:2376–2382. doi: 10.1002/eji.1830261016. [DOI] [PubMed] [Google Scholar]
- 17.Robbins PF, El-Gamil M, Li YF, Zeng G, Dudley M, Rosenberg SA. Multiple HLA class II-restricted melanocyte differentiation antigens are recognized by tumor-infiltrating lymphocytes from a patient with melanoma. J Immunol. 2002;169:6036–6047. doi: 10.4049/jimmunol.169.10.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robbins PF, El-Gamil M, Li YF, Kawakami Y, Loftus D, Appella E, Rosenberg SA. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183:1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Conejo T, Olmo N, Turnay J, Navarro J, Lizarbe A. Characterization of tumorigenic sub-lines from a poorly tumorigenic human colon-adenocarcinoma cell line. Int J Cancer. 1996;67:668–675. doi: 10.1002/(SICI)1097-0215(19960904)67:5<668::AID-IJC13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Katoh H, Harada A, Mori K, Negishi M. Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers. Mol Cell Biol. 2002;22:2952–2964. doi: 10.1128/MCB.22.9.2952-2964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science (Wash DC) 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 22.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 23.Topalian SL, Gonzales MI, Parkhurst M, Li YF, Southwood S, Sette A, Rosenberg SA, Robbins PF. Melanoma-specific CD4+ T cells recognize nonmutated HLA-DR-restricted tyrosinase epitopes. J Exp Med. 1996;183:1965–1971. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morel S, Levy F, Burlet-Schiltz O, Brasseur F, Probst-Kepper M, Peitrequin AL, Monsarrat B, Van Velthoven R, Cerottini JC, Boon T, Gairin JE, Van den Eynde BJ. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12:107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 25.Doherty DG, Penzotti JE, Koelle DM, Kwok WW, Lybrand TP, Masewicz S, Nepom GT. Structural basis of specificity and degeneracy of T cell recognition: pluriallelic restriction of T cell responses to a peptide antigen involves both specific and promiscuous interactions between the T cell receptor, peptide, and HLA-DR. J Immunol. 1998;161:3527–3535. [PubMed] [Google Scholar]
- 26.Di Giorgio A, Botti c, Tocchi A, Mingazzini P, Flammia M. The influence of tumor lymphocyte infiltration on long term survival of surgically treated colrectal cancer patients. Int Surg. 1992;77:256–260. [PubMed] [Google Scholar]
- 27.Kinihiro M, Tanaka S, Haruma K, Yoshihara M, Sumii K, Kajiyama G, Shimamoto F. Combined expression of HLA-DR antigen and proliferating cell nuclear antigen correlate with colorectal cancer prognosis. Oncology. 1998;55:326–333. doi: 10.1159/000011870. [DOI] [PubMed] [Google Scholar]
- 28.Ransom JH, Pelle B, Hanna MG., Jr Expression of class II major histocompatibility complex molecules correlates with human colon tumor vaccine efficacy. Cancer Res. 1992;52:3460–3466. [PubMed] [Google Scholar]
- 29.Yang D, Nakao M, Shichijo S, Sasatomi T, Takasu H, Matsumoto H, Mori K, Hayashi A, Yamana H, Shirouzu K, Itoh K. Identification of a gene coding for a protein possessing shared tumor epitopes capable of inducing HLA-A24-restricted cytotoxic T lymphocytes in cancer patients. Cancer Res. 1999;59:4056–4063. [PubMed] [Google Scholar]
- 30.Bremers AHA, Andreola S, Leo E, Gallino F, Rini F, Lombardo C, Belli F, Kuppen PJK, Parmiani G, Castelli C. T cell responses in colorectal cancer patients: evidence for class-II HLA restricted recognition of shared tumor-associated antigens. Int J Cancer. 2000;88:956–961. doi: 10.1002/1097-0215(20001215)88:6<956::aid-ijc19>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 31.Saeterdal I, Bjorheim J, Lislerud K, Gjertsen MK, Bukholm IK, Olsen OC, Nesland JM, Eriksen JA, Moller M, Lindblom A, Gaudernack G. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc Natl Acad Sci USA. 2001;98:13255–13260. doi: 10.1073/pnas.231326898. [DOI] [PMC free article] [PubMed] [Google Scholar]


