Abstract
Canonical Wnt signaling is critically involved in normal hematopoietic development and the self-renewal process of hematopoietic stem cells (HSCs). Deregulation of this pathway has been linked to a large variety of cancers, including different subtypes of leukemia. Lef-1 is a major transcription factor of this pathway and plays a pivotal role in lymphoid differentiation as well as in granulopoiesis. Here, we demonstrate Lef-1 expression in murine HSCs as well as its expression in human leukemia. Mice transplanted with bone marrow retrovirally transduced to express Lef-1 or a constitutive active Lef-1 mutant showed a severe disturbance of normal hematopoietic differentiation and finally developed B lymphoblastic and acute myeloid leukemia (AML). Lef-1–induced AMLs were characterized by immunoglobulin (Ig) DH-JH rearrangements and a promiscuous expression of lymphoid and myeloid regulatory factors. Furthermore, single cell experiments and limiting dilution transplantation assays demonstrated that Lef-1–induced AML was propagated by a leukemic stem cell with lymphoid characteristics displaying Ig DH-JH rearrangements and a B220+ myeloid marker− immunophenotype. These data indicate a thus far unknown role of Lef-1 in the biology of acute leukemia, pointing to the necessity of balanced Lef-1 expression for an ordered hematopoietic development.
The canonical Wnt/β-catenin signaling is an evolutionary highly conserved cascade with multiple functions in proliferation, differentiation, and apoptosis, among others (1). In the hematopoietic system, Wnt signaling has been tightly linked to maintenance, activation, and proliferation of hematopoietic stem cells (HSCs) (2–5), and aberrant expression of key players of this pathway is implicated in various leukemias (6).
LEF-1 belongs to the family of LEF-1/T cell factor transcription factors that share homology with high mobility group proteins. LEF-1 acts as a central transcription mediator of Wnt signaling, regulating cell cycle– and growth-relevant genes like Cyclin D1 and c-myc (6–8). Studies on Lef-1 in hematopoietic development have been mainly restricted to the lymphoid lineages, where Lef-1 has functions in T cell development and affects proliferation and apoptosis in pro–B cells (9, 10). Recent reports, however, also discuss specific functions of Lef-1 independent of Wnt signaling (11, 12), suggesting a more complex role of Lef-1 in the development of hematopoietic tissues.
In this report, we demonstrate that Lef-1 is expressed in murine HSCs as well as in different human leukemias. Aberrant expression of Lef-1 in murine BM perturbed normal hematopoietic development and induced B lymphoblastic leukemias and Ig DH-JH–rearranged acute myeloid leukemias (AML), which were propagated by a leukemic stem cell with lymphoid characteristics. Collectively, our data suggest a previously unknown role of LEF-1 in early hematopoiesis and in the biology of acute leukemias.
RESULTS AND DISCUSSION
Expression of Lef-1 in murine hematopoiesis
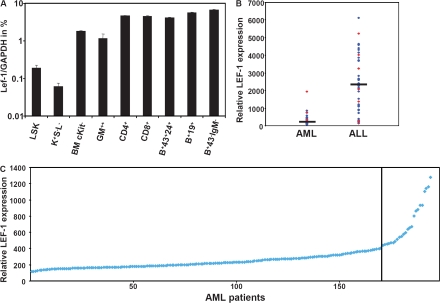
In a first step, we determined Lef-1 mRNA expression levels in early murine hematopoietic development by real-time TaqMan PCR. Lef-1 transcripts were detected at different stages of myeloid and lymphoid development. Of note, murine HSCs expressing cKit and Sca1, but lacking lineage markers (Fig. 1 A, LSK), were also positive for Lef-1 transcription and showed a 2.6-fold higher expression of Lef-1 as compared with more differentiated Sca−/cKit+/Lin− progenitor cells.Lef-1 expression was higher in more mature Gr1/Mac1+ myeloid cells and showed highest expression levels in lymphoid cells (Fig. 1 A). These data show for the first time expression of Lef-1 in murine HSCs and are consistent with data in the human system documenting expression of LEF1 in early human myeloid cells (12).
Figure 1.
Lef-1 expression in murine hematopoietic tissue and human leukemia. (A) Lef-1 mRNA expression in FACS-sorted murine hematopoietic BM subpopulations. Data represent means plus SD of triplicates (n = 3). K+S−L−, cKit+Sca−Lin−; GM++, Gr1+Mac1+; B+43+24+, B220+CD43+CD24+; B+19+, B220+CD19+; B+43−IgM−, B220+CD43−IgM−. (B and C) Expression levels of LEF-1 were determined in two microarray datasets: 120 samples from 13 different leukemia subtypes (B) and a series of 194 expression profiles from AML samples with normal karyotype (C). (B) Comparison of relative LEF-1 mRNA expression in AML and ALL patients. Bars indicate the median. Red dots indicate CALM/AF10+ AML/ALL cases. (C) Relative expression of LEF-1 in 194 AML patient samples with normal karyotype (NK).
Expression of LEF-1 in human leukemias
We extended our analyses to human tissue and analyzed LEF-1 expression in normal BM and in human leukemias by microarray analysis in >300 patients with acute leukemia. Leukemic samples as well as normal BM samples were positive for LEF-1 expression. However, there was an ∼13-fold higher relative expression of LEF-1 in acute lymphoblastic leukemia (ALL) samples (n = 47) compared with AML (n = 73), possibly reflecting the higher expression of the transcription factor in lymphoid tissue. The levels of LEF-1 expression, however, did not correlate with particular subtypes of AML or ALL (not depicted). Although most of the AML cases were characterized by low LEF-1 expression, intriguingly, two of the AML cases showing highest LEF-1 levels were diagnosed as CALM/AF10+ AML (Fig. 1 B), which was recently shown to harbor DJH rearrangements of the IgH chain in a majority of patients. Moreover, it was shown that this leukemia was initiated by a leukemic stem cell with lymphoid characteristics in a murine transplantation model (13). When we examined LEF-1 expression in a larger cohort of AML patients with normal karyotype (n = 194), there was a small subgroup (n = 24) of AML patients with increased LEF-1 expression (range 443 to 1,277 relative units) (Fig. 1 C). Collectively, these results demonstrated that LEF-1 is widely expressed in human lymphoid leukemias, but also in human AML.
Constitutive expression of Lef-1 perturbs normal hematopoietic development
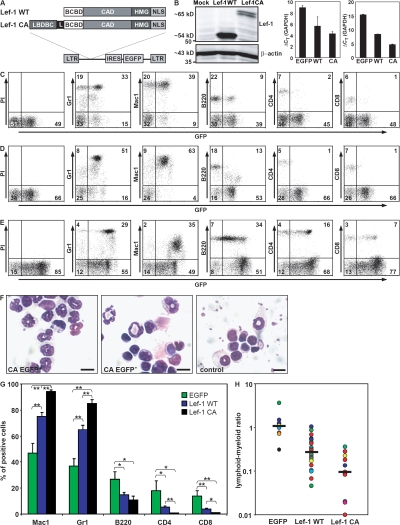
We next sought to determine the functional relevance of Lef-1 in early hematopoiesis. For this, murine BM cells enriched for primitive progenitors were retrovirally engineered to express wild-type Lef-1 (Lef-1 WT) or a constitutive active mutant of Lef-1 (Lef-1 CA) using a murine stem cell virus–based retroviral construct (Fig. 2, A and B) and transplanted into lethally irradiated recipient mice. Mice engrafted with Lef-1 WT– or Lef-1 CA–expressing BM cells showed a profound disturbance of normal hematopoietic development (n = 12). Analysis of peripheral blood (PB) in nondiseased animals showed a twofold reduction of the proportion of B220+ lymphoid cells in the transduced compartment compared with empty vector control mice. Furthermore, both CD4+ T cells and CD8+ T cells were greatly reduced in the enhanced GFP (EGFP)+ compartment in Lef-1–transplanted mice. Simultaneously, PB analysis showed a substantial increase in the proportion of Gr1+ and Mac1+ myeloid cells. In contrast, the nontransduced compartment of the PB of mice transplanted with Lef-1 WT or Lef-1 CA did not show any abnormalities compared with the control (not depicted). This accumulation of mature myeloid cells in the EGFP+ compartment of the PB of transplanted animals was confirmed by morphological analyses. These changes led to an inversion of the lymphoid/myeloid ratio in these animals (Fig. 2, C–H).
Figure 2.
Lef-1 severely perturbs normal hematopoietic differentiation. (A) Retroviral constructs for expression of WT and mutant constitutive active (CA) Lef-1 proteins. As control, the empty EGFP vector was used. BCBD, β-catenin binding domain; CAD, context-dependent activation domain; HMG, high mobility group DNA binding domain; NLS, nuclear localization signal; LBDBC, Lef-1 binding domain of β-catenin; L, linker; LTR, long-terminal repeat; IRES, internal ribosomal entry site; EGFP, enhanced GFP. (B) Expression of Lef-1. Western blot analysis of cellular extracts from NIH 3T3 cells transfected with the different constructs (left image, the molecular mass is indicated); TaqMan real-time RQ-PCR with cDNA (triplicates) of BM cells from Lef-1 WT (n = 2) and Lef-1 CA leukemic mice (n = 2) compared with the endogenous Lef-1 mRNA expression level in a healthy EGFP control animal (middle); and quantification of Lef-1 expression level by RQ-PCR in BM cells freshly infected with Lef-1 WT, Lef-1 CA, and empty vector control using highly purified EGFP+ populations (right). (C–E) Flow cytometry analysis of PB samples of (C) a Lef-1 WT (no. 8) and (D) a Lef-1 CA (no. 10) –engrafted mouse, respectively, compared with a control EGFP animal (E). Representative dot plots with the proportion of positive stained cells within the EGFP+ compartment compared with the EGFP− compartment are shown. (F) Accumulation of mature myeloid cells in Lef1-transplanted animals. Wright-Giemsa–stained cytospins from PB of a Lef-1 CA–transduced mouse (no. 10; left image, GFP+ compartment; middle image, GFP− compartment) and a control animal (right image). Bar, 10 μm. (G) Percentage of lymphoid and myeloid cells in PB in transduced animals. Mean values ± SEM are indicated (n [WT] = 20, n [CA] = 12, n [EGFP] = 8; *, P < 0.02; **, P < 0.001). Significant differences between Lef-1 WT and Lef-1 CA animals are indicated when present. (H) Lymphoid/myeloid ratio in PB of transduced mice. Lymphoid cells included B220+, CD4+, and CD8+ cells, and myeloid cells included Mac1+/Gr1+, Mac1+/Gr1−, and Mac1−/Gr1+ cells. Individual mice are indicated by different colors and were analyzed at one or multiple time points after transplantation. The median is presented as a black bar (*, P < 0.02; **, P < 0.002).
In a comparative analysis, Lef-1 CA seemed to induce a more pronounced perturbation of hematopoietic development than the Lef-1 WT gene. Furthermore, the skewing of the lymphoid/myeloid ratio toward the myeloid lineage was more pronounced in the Lef-1 CA group compared with Lef-1 WT (Fig. 2, C–H).
Collectively, these data demonstrated that constitutive expression of Lef-1 results in a severe perturbation of hematopoiesis in transplanted animals with an increase in the proportion of myeloid cells and a simultaneous proportional decrease of lymphoid cells in the PB. We observed that the alteration of the myeloid/lymphoid ratio was not accompanied by a significant change of the absolute cell numbers of circulating WBCs (not depicted), suggesting that the drop in the lymphoid ratio could possibly result from the skewing of lineage fate decisions toward the myeloid lineage in early progenitors ectopically expressing the Lef-1 transcript. Furthermore, our observation that enforced expression of Lef-1 did not impair maintenance of normal murine B220+/Lin− B cells in vitro compared with nontransduced controls argued against a direct anti-proliferative effect of Lef-1 (not depicted). Of note, a pivotal role of LEF-1 in myeloid differentiation was recently described in the human system (12). The same report also demonstrated that C/EBPα is a direct target gene of LEF-1. Therefore, constitutive expression of Lef-1 in early hematopoietic progenitor cells might result in a shift toward the myeloid lineage. This important role of Lef-1 in myeloid development is supported by our data showing expression in early and myeloid hematopoietic cells in mice.
Previous reports have documented that deregulated expression of LEF-1 can influence both lymphoid and myeloid development. Earlier reports have demonstrated that Lef-1 deficiency is associated with defects in pro–B cell proliferation and survival, but not B cell differentiation (10). The fact that in our model rather similar effects were observed in mice transplanted with WT or constitutive active Lef-1 might suggest that Lef-1 can function independently of Wnt signaling in hematopoietic tissue. In human CD34+ progenitor cells, inhibition of LEF-1 but not of β-catenin affected proliferation and apoptosis of this cell population, supporting a model of a β-catenin–independent function of LEF-1 in early human myelopoiesis (12).
Constitutive Lef-1 induces acute leukemia in transplanted mice
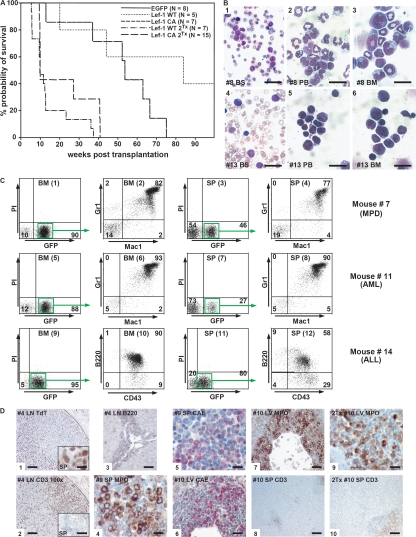
After a median latency of 12 mo, 10 of 12 animals transplanted with BM-expressing Lef-1 WT or Lef-1 CA developed a lethal hematological disease. Median survival of mice in the Lef-1 WT group was 310 d (n = 3, range 141–589 d), whereas 7 of 7 Lef-1 CA–transduced mice died after an average of 377 d after transplant (n = 7, range 91–527 d) (Fig. 3 A). 5 out of 10 evaluable diseased animals showed highly elevated levels of WBCs (P < 0.02), whereas 7 of the mice were characterized by mild to severe anemia (P < 0.001). 8 out of 11 evaluable mice suffered from splenomegaly (P < 0.02) (Table S1, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1). All evaluable diseased mice showed increased numbers of blast cells in the PB, BM (Fig. 3 B), or spleen (SP). Histological sections demonstrated infiltration of blasts in multiple nonhematopoietic organs (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1). According to the Bethesda proposal for classification of nonlymyphoid hematopoietic neoplasms in mice, seven animals suffered from acute leukemia, which was retransplantable, and had >20% blasts in PB, BM, or SP (Fig. 3, A and B, and Table S1) (14). In two mice, a myeloproliferative disease was diagnosed with a blast percentage of ≤20%, accumulation of mature neutrophils in the PB and BM, or lack of transplantability into secondary recipients (Table S1).
Figure 3.
Lef-1 overexpression induces acute leukemia. (A) Survival curve of mice transplanted with BM cells expressing Lef-1 WT, Lef-1 CA, and EGFP as well as survival of secondary recipient mice transplanted with BM from diseased primary mice. (B) Blood smears (BS) and cytospin preparations from PB and BM of a representative animal with AML (no. 8, 1–3) and ALL (no. 13, 4–6). Slides were stained with Wright-Giemsa. Bars: 1, 60 μm; 4, 40 μm; 2 and 5, 30 μm; 3 and 6, 20 μm. (C) Immunophenotype analysis of diseased mice. Representative dot plots from the BM and SP of a mouse with myeloproliferative disease (MPD, 1–4, no. 7), with AML (5–8, no. 11) and ALL (9–12, no. 14). The Mac1/Gr1 and CD43/B220 costainings were gated on GFP+ cells. The percentage of positive-stained cells is indicated. (D) Histochemical and immunohistological analysis of diseased mice. Panels 1–3, B-ALL (mouse no. 4); panels 1 and 2, tumor cells infiltrating lymph nodes staining positive for TdT and negative for CD3; residual normal T cells in the lymph node are CD3+ (2, right side), whereas infiltrated leukemic blast cells are TdT+, but CD3− (1; left side and insets 1 and 2, SP). Panels 4–8: primary AML (4 and 5, mouse no. 9; 6–8, mouse no. 10). Panels 9 and 10: diseased secondary recipient of mouse number 10. H&E, hematoxylin and eosin; MPO, myeloperoxidase; CAE, N-acetyl-chloroacetate esterase; TdT, terminal deoxynucleotidyl transferase; LN, lymph node; LV, liver. Bars: 8, 200 μm; 1, 2, and 10, 100 μm; 3, 6, and 7, 50 μm; 4, 5, 9, and insets 1 and 2, 25 μm.
More detailed analysis demonstrated that four mice died of an AML with maturation (14). The diagnosis of AML was supported by flow cytometry demonstrating a high proportion of Gr1+ and Mac1+ cells in the PB, BM, and SP (Fig. 3 C). In the immunohistochemistry, the blasts were positive for myeloperoxidase and chloracetate esterase and negative for CD3 (Fig. 3 D). Three mice succumbed to acute B lymphoblastic leukemia displaying lymphoid-like blast cells (Fig. 3 B), a high percentage of B220+/CD43+ cells in the PB, BM, and SP (Fig. 3 C) as well as positivity of the blast cells for terminal deoxynucleotidyl transferase (TdT) and B220 in histological sections (Fig. 3 D). In mice suffering from myeloproliferation, a high proportion of BM and splenic cells stained positive for Gr1 and Mac1 (Fig. 3 C). Clonal analysis by Southern blotting of leukemic mice revealed a monoclonal disease in all examined primary and secondary animals (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1). A comparative analysis between mice with Lef-1 WT versus Lef-1 CA positive leukemia showed a trend toward more pronounced disease manifestations in the Lef-1 CA experimental group, although the differences were not significant (Table S1).
Lef-1–induced AMLs are propagated by a leukemic stem cell with lymphoid characteristics
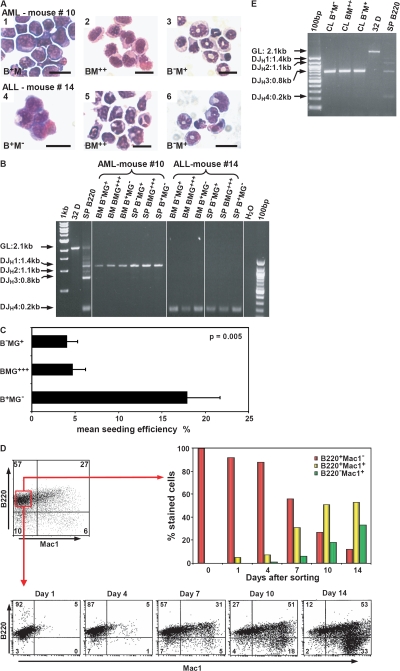
When the ALLs and AMLs were examined in more detail, ALL and AML cases shared key biological characteristics: all leukemias independent of their phenotype were characterized by a distinct B220+Mac1+ cell population in the PB, BM, and SP. In addition, all the diseased mice and, of note, also mice suffering from AML revealed a B220+Mac1− population. In the AML cases, this B220+Mac1− population comprised 6.3 and 1.6% in BM and PB cells, respectively (Table S2, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1). Morphological analysis of the different highly purified BM populations isolated from mice with ALL or AML revealed an immature, blast-like appearance of the B220+Mac1− cells, whereas the B220−Mac1+ cells displayed a more differentiated phenotype with granulocytic characteristics (Fig. 4 A). The immature nature of the B220+ population was further confirmed by FACS analysis showing expression of CD43 and AA.4.1, but negativity for CD19 (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1). We then tested AML and ALL samples for IgH DJH rearrangements. Strikingly, clonal DJH but not VDJH rearrangements were detected in all three subpopulations in acute leukemias independent of the phenotype (Fig. 4 B, Table S2, and not depicted).
Figure 4.
Lef-1–induced acute leukemias are propagated by a LSC with lymphoid characteristics. (A) Morphology of sorted BM populations of a representative AML (1–3, no. 10) and an ALL case (3–6, no. 14). B+M−, B220+/Mac1−; BM++, B220+Mac1+; B−M+, B220−Mac1+. Bars, 20 μm. (B) Multiplex PCR for DJH loci rearrangement in diseased mice. Line 2, germline control (GL, myeloid cell line 32D); line 3, splenic B220 cells. (C) Seeding efficiency of B+MG−, BMG+++, and B−MG+ populations from BM cell cultures of leukemic mice (n = 3; mouse nos. 4, 10, and 11; Table S1). Shown are the mean values ± SEM. (D) B+M− cells from a cell population generated from a single B+M− cell (mouse no. 4) developed into BM2+ and B−M+ populations. (E) Multiplex PCR for DJH rearrangement for highly purified B+M−, BM2+, and B−M+ cells generated from a single B+M− cell in vitro. CL, cell line.
To further explore the characteristics of the B220+ myeloid marker− cell population in these leukemias, B220+Mac1−Gr1− (B+MG−), B220−Mac1+Gr1+ (B−MG+), and B220+Mac1+Gr1+ (BMG+++) cells were highly purified from BM cell cultures of three leukemic mice (AML, n = 2; ALL, n = 1). Of note, the B+MG− cell population displayed the highest proliferative potential in vitro (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1). In addition, the B+MG− cells had the highest proliferative potential at the single cell level compared with the BMG+++ and B−MG+ populations (Fig. 4 C). These data demonstrated that ALL and AML cases induced by Lef-1 shared striking similarities in the hierarchical organization of the malignant clone. B+MG− cells were then isolated from a cell population derived from a single B+MG− cell of a mouse suffering from ALL and propagated in vitro. Importantly, these myeloid marker− DJH-rearranged cells were able to generate the BMG+++ and the B−MG+ cell populations harboring the identical clonal DJH rearrangement (Fig. 4, D and E). These data indicated that in this model, B+MG− cells have the highest proliferative potential at the single cell level and can give rise to DJ-rearranged myeloid cells lacking B220 expression.
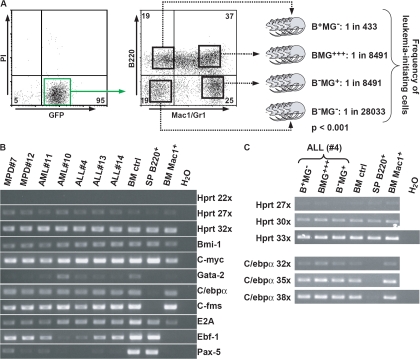
As these observations suggested that the leukemia-propagating cell in Lef-1–induced AML resides in the B+MG− population, we performed a limiting dilution transplantation assay using leukemic BM from a representative AML mouse (Table S1, no. 10) to calculate the frequency of leukemic stem cells (LSCs) in the different subpopulations. In these experiments, the B+MG− cell population was highly enriched for LSCs with a 20- and 60-fold increase of the LSC frequency compared with the BMG+++/B−MG+ and the B−MG− cell compartments, respectively (Fig. 5 A). In summary, these observations indicate that deregulated expression of Lef-1 induces acute B lymphoblastic leukemias as well as DJH-rearranged AMLs, which are propagated by an LSC with lymphoid characteristics. These observations that constitutive expression of Lef-1 induces the development of a leukemic B+M− cell population, which exhibits DH-JH rearrangements and is negative for Pax5, are consistent with reports on the impact of constitutive β-catenin expression in hematopoietic progenitors (15, 16): retroviral expression of β-catenin into murine HSCs induced the outgrowth of c-KitlowSca-1low CD19− cells, which interestingly were highly positive for B220, whereas enforced expression of β-catenin in normal murine myeloid or lymphoid cells led to uncommitted cells with multilineage differentiation potential. In the latter experiments, enforced expression of β-catenin in lymphoid progenitors rapidly increased C/ebpα, reduced Ebf/Pax5 expression, and induced outgrowth of c-kit+Sca-1low/−B220+CD43+AA4.1+ cells with DH-JH rearrangements. These data demonstrated that perturbations in the Wnt/β-catenin pathway can induce a destabilization of lineage fate decisions in early committed progenitors, which might be an important prerequisite for the development of the B220+CD43+AA4.1+ LSC in our model.
Figure 5.
Frequency of leukemia-propagating cells and transcriptional profile analysis. (A) Highly purified B+MG−, BMG+++, B−MG+, and triple negative B−MG− cell populations from a primary mouse with AML (no. 10) were injected into cohorts of secondary mice. The frequency of leukemia-propagating cells in each of the populations is indicated. For details see Table S3, available at http://www.jem.org/cgi/content/full/jem.20071875/DC1. (B) Analysis of mRNA expression of regulatory factors in BM cells of diseased mice by semiquantitative RT-PCR. BM bulk (BM ctrl), Mac1+ BM cells (BM Mac1+), and B220+ splenic cells (SP B220+) of a nontransplanted mouse served as controls. (C) Analysis of C/ebpα expression in subpopulations generated from a B220+Mac1− single cell originating from an ALL-diseased mouse single cell. HPRT, hypoxanthine guanine phoshoribosyltransferase.
Interestingly, the expression profile of crucial transcription factors and growth factor receptors revealed a striking similarity between the leukemic myeloid and lymphoid bulk population. Both AML and ALL cases showed a promiscuous expression of lymphoid and myeloid regulatory factors with positivity for both the myeloid factors C/ebpα and C-fms and the lymphoid transcription factors E2A and Ebf-1, whereas both types of acute leukemias were low/negative for Pax5, which is in line with our observation that ALL and AML cases displayed DJH but not VDJH rearrangements (Fig. 5, B and C) (17–20).
Our observations indicate that deregulated expression of Lef-1 induces acute B lymphoblastic leukemias, but also DJH-rearranged AMLs, which are propagated by a LSC with characteristics similar to early lymphoid progenitors. We just recently reported that murine CALM/AF10+ AML is initiated by a B220+ myeloid marker− leukemic stem cell and that both murine and human CALM/AF10+ AML cases are DJH rearranged (13). Thus, our observations show that not only an appropriate leukemia-specific fusion gene, but also a transcription factor can generate myeloid leukemias propagated by LSCs with certain lymphoid characteristics. The expression of lymphoid antigens on LSCs not present on normal HSCs would have clear clinical implications for the development of innovative treatment strategies that aim at the LSC eradication without major toxicity to the normal stem cell pool.
In summary, our results demonstrate that ordered expression of Lef-1 is not only crucial for normal lymphopoiesis, but also for normal hematopoietic development. Furthermore, we show for the first time that deregulated expression of this transcription factor is directly linked to the development of hematological neoplasia.
MATERIALS AND METHODS
TaqMan PCR analysis.
Quantification of Lef-1 expression in murine hematopoietic subpopulations was assayed by real-time RQ-PCR with primers from Applied Biosystems. Hematopoietic cells were obtained from 8–12-wk-old C3H×C57 Bl mice. For normalization, GAPDH was used. Reactions were run in triplicates with 2 μl cDNA in a total reaction volume of 20 μl using an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Fold expression was calculated by using the ΔΔCT method. For quantification of Lef-1 expression in leukemic mice and freshly transduced BM, cDNA of equivalent amounts of RNA (50 ng) was used to determine the difference in Lef-1 expression. Mean ΔCT values (±SD) were obtained by normalization to the housekeeping gene GAPDH. ΔCT values are inversely correlated to gene expression levels.
Retroviral transduction of primary BM cells and BM transplantation.
Maintenance and breeding of the mice as well as transduction of primary murine BM were conducted as described previously (21, 22). In brief, murine BM cells were harvested from mice pretreated with 150 mg/kg 5-fluorouracil 5 d before being killed, followed by 48 h of prestimulation and subsequent infection by cocultivation with irradiated viral producer cells (GP+E86). For transplantation into primary recipients, 4.0 × 104 to 6.0 × 105 and 9.0 × 104 to 4.0 × 105 EGFP+ successfully transduced cells per mouse for Lef-1 WT (n = 5) and Lef-1 CA (n = 7), respectively, were injected into lethally irradiated recipients together with 3.2 to 4.0 × 105 nontransduced cells. For re-transplantability of hematological diseases, lethally irradiated secondary recipients were injected with 106 primary BM cells together with an equal number of nontransduced carrier cells for radioprotection. The animal studies were approved by the Ethics Committee of Ludwig Maximilians University and abided by the tenets of the revised World Medical Association Declaration of Helsinki (http://www.wma.net/e/policy/b3.htm).
Multiparameter flow cytometry and cell sorting.
Cells were analyzed with a FACSCalibur and sorted with a FACS Sorter (Becton Dickinson). The following fluorochrome-conjugated antibodies were purchased from BD Biosciences or eBioscience: anti–Sca-1, –Gr-1, -Ter119, -CD3, -CD4, -CD8, -Mac1, -CD117, -B220, -CD19, -NK1.1, -AA4.1, -CD43, -IgM, and -CD24.
Histology.
Sections of organs were prepared and stained at the Department of Pathology, GSF (Munich, Germany), using standard protocols, as described previously (22).
Characterization of leukemic cells.
Proliferation capacity of leukemic cells was analyzed by single cell sorting of highly purified subpopulations from BM liquid cultures of diseased animals into 96-well plates in duplicates supplemented with DMEM, 20% FBS, and 6 ng/ml IL-3. After 25 d, plates were analyzed for colony growth. Proliferation and differentiation capacity of cell lines and cell populations was monitored by cell counting with Trypan blue and subsequent FACS staining. DJ rearrangement in the IgH locus was analyzed by multiplex PCR (25) (sequences and protocol upon request).
The frequency of leukemia-propagating cells was determined as described previously (13). Highly purified B220+Gr1−Mac1−, B220+Gr1+Mac1+, B220−Gr1+Mac1+, and triple negative B220−Gr1−Mac1− cell populations from a primary mouse with AML (no. 10) were injected into cohorts of lethally irradiated secondary mice using five dilutions of sorted cell populations ranging from 10 to 5.0 × 104 cells per donor mouse. 7.5 × 105 carrier cells (nontransduced BM from a syngenic disease-free mouse) were added to each sample for radioprotection. Diseased mice were killed and assessed for leukemia development. The frequency of leukemia-propagating cells was calculated using Poisson statistics using the L-Calc Limiting dilution analysis software (version 1.1; StemSoft Inc.).
Transcriptional profiling of both leukemic BM samples and sorted leukemic cell line subpopulations was performed by semiquantitative PCR using intron-spanning primers when available (sequences upon request).
Western blot.
Protein expression of Lef-1 constructs was demonstrated by Western blotting using standard procedures, applying anti–Lef-1 polyclonal goat antibody and secondary anti–goat peroxidase-conjugated antibody (Santa Cruz Biotechnology, Inc.).
Microarray.
Total mRNA was isolated from patient samples and processed and analyzed as described previously (23). The .CEL file data from the samples used in the comparison were normalized together according to the procedure described by Huber et al. (24). Normalized expression data were then analyzed with the R software package and the “boxplot” function (http://www.r-project.org). Expression signal intensities are given on a logarithmic scale or as relative expression. For the comparison of LEF-1 transcript levels, the following different AML or ALL subtypes were analyzed: AML: M2, M3, M4eo, FLT3-length mutation, AML with MLL translocations, and normal karyotype and complex karyotype; ALL: pro–B ALL, Philadelphia chromosome+ ALL, and ALL with MLL rearrangements.
Statistical analysis.
Data were evaluated by using the t test for dependent or independent samples. Differences with p-values of <0.05 were considered statistically significant.
Online supplemental material.
Fig. S1 shows leukemic blast infiltration into nonhematopoietic tissue. Fig. S2 shows Lef-1–induced leukemias to be of monoclonal origin. Fig. S3 highlights highest proliferative potential of B+MG− cells in vitro, and Fig. S4 displays additional immunophenotypic characterization of leukemic B220+ cells. Table S1 shows the critical characteristics of diseased mice, Table S2 displays percentages of B220+ cells in Lef-1–induced leukemias, and Table S3 highlights incidence and latency of leukemias in the CRU. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20071875/DC1.
Supplemental Material
Acknowledgments
We thank Rudolf Grosschedl for kindly providing the Lef-1 constructs, Dirk Eick for discussions, Bianka Ksienzyk and Nicole Behm for technical assistance, the members of the animal facility for maintenance of animals, and Manuela Hummel and Ulrich Mansmann for support with the evaluation of the microarray data.
V.P.S. Rawat and C. Buske were supported by a grant from the DFG (SFB 684 project A7), M. Feuring-Buske was supported by the Deutsche Krebshilfe (70-2968-Fe I to M.F.B.), and C. Buske, M. Feuring-Buske, and S.K. Bohlander were supported by the BMBF (NGFN2 grant 01GS0448).
The authors have no conflicting financial interests.
References
- 1.Clevers, H. 2006. Wnt/beta-catenin signaling in development and disease. Cell. 127:469–480. [DOI] [PubMed] [Google Scholar]
- 2.Kirstetter, P., K. Anderson, B.T. Porse, S.E. Jacobsen, and C. Nerlov. 2006. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat. Immunol. 7:1048–1056. [DOI] [PubMed] [Google Scholar]
- 3.Reya, T., A.W. Duncan, L. Ailles, J. Domen, D.C. Scherer, K. Willert, L. Hintz, R. Nusse, and I.L. Weissman. 2003. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 423:409–414. [DOI] [PubMed] [Google Scholar]
- 4.Scheller, M., J. Huelsken, F. Rosenbauer, M.M. Taketo, W. Birchmeier, D.G. Tenen, and A. Leutz. 2006. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nat. Immunol. 7:1037–1047. [DOI] [PubMed] [Google Scholar]
- 5.Staal, F.J., and H.C. Clevers. 2005. WNT signalling and haematopoiesis: a WNT-WNT situation. Nat. Rev. Immunol. 5:21–30. [DOI] [PubMed] [Google Scholar]
- 6.Reya, T., and H. Clevers. 2005. Wnt signalling in stem cells and cancer. Nature. 434:843–850. [DOI] [PubMed] [Google Scholar]
- 7.He, T.C., A.B. Sparks, C. Rago, H. Hermeking, L. Zawel, L.T. da Costa, P.J. Morin, B. Vogelstein, and K.W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science. 281:1509–1512. [DOI] [PubMed] [Google Scholar]
- 8.Shtutman, M., J. Zhurinsky, I. Simcha, C. Albanese, M. D'Amico, R. Pestell, and A. Ben-Ze'ev. 1999. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 96:5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamura, R.M., M. Sigvardsson, J. Galceran, S. Verbeek, H. Clevers, and R. Grosschedl. 1998. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 8:11–20. [DOI] [PubMed] [Google Scholar]
- 10.Reya, T., M. O'Riordan, R. Okamura, E. Devaney, K. Willert, R. Nusse, and R. Grosschedl. 2000. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 13:15–24. [DOI] [PubMed] [Google Scholar]
- 11.Ross, D.A., and T. Kadesch. 2001. The notch intracellular domain can function as a coactivator for LEF-1. Mol. Cell. Biol. 21:7537–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skokowa, J., G. Cario, M. Uenalan, A. Schambach, M. Germeshausen, K. Battmer, C. Zeidler, U. Lehmann, M. Eder, C. Baum, et al. 2006. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat. Med. 12:1191–1197. [DOI] [PubMed] [Google Scholar]
- 13.Deshpande, A.J., M. Cusan, V.P. Rawat, H. Reuter, A. Krause, C. Pott, L. Quintanilla-Martinez, P. Kakadia, F. Kuchenbauer, F. Ahmed, et al. 2006. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 10:363–374. [DOI] [PubMed] [Google Scholar]
- 14.Kogan, S.C., J.M. Ward, M.R. Anver, J.J. Berman, C. Brayton, R.D. Cardiff, J.S. Carter, S. de Coronado, J.R. Downing, T.N. Fredrickson, et al. 2002. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 100:238–245. [DOI] [PubMed] [Google Scholar]
- 15.Baba, Y., K.P. Garrett, and P.W. Kincade. 2005. Constitutively active beta-catenin confers multilineage differentiation potential on lymphoid and myeloid progenitors. Immunity. 23:599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba, Y., T. Yokota, H. Spits, K.P. Garrett, S. Hayashi, and P.W. Kincade. 2006. Constitutively active beta-catenin promotes expansion of multipotent hematopoietic progenitors in culture. J. Immunol. 177:2294–2303. [DOI] [PubMed] [Google Scholar]
- 17.Fuxa, M., J. Skok, A. Souabni, G. Salvagiotto, E. Roldan, and M. Busslinger. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I.L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell. 3:137–147. [DOI] [PubMed] [Google Scholar]
- 19.Nutt, S.L., P. Urbanek, A. Rolink, and M. Busslinger. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476–491. [DOI] [PubMed] [Google Scholar]
- 20.Roessler, S., and R. Grosschedl. 2006. Role of transcription factors in commitment and differentiation of early B lymphoid cells. Semin. Immunol. 18:12–19. [DOI] [PubMed] [Google Scholar]
- 21.Rawat, V.P., M. Cusan, A. Deshpande, W. Hiddemann, L. Quintanilla-Martinez, R.K. Humphries, S.K. Bohlander, M. Feuring-Buske, and C. Buske. 2004. Ectopic expression of the homeobox gene Cdx2 is the transforming event in a mouse model of t(12;13)(p13;q12) acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 101:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schessl, C., V.P. Rawat, M. Cusan, A. Deshpande, T.M. Kohl, P.M. Rosten, K. Spiekermann, R.K. Humphries, S. Schnittger, W. Kern, et al. 2005. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J. Clin. Invest. 115:2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoch, C., A. Kohlmann, S. Schnittger, B. Brors, M. Dugas, S. Mergenthaler, W. Kern, W. Hiddemann, R. Eils, and T. Haferlach. 2002. Acute myeloid leukemias with reciprocal rearrangements can be distinguished by specific gene expression profiles. Proc. Natl. Acad. Sci. USA. 99:10008–10013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber, W., A. von Heydebreck, H. Sultmann, A. Poustka, and M. Vingron. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 18:S96–S104. [DOI] [PubMed] [Google Scholar]
- 25.Martensson, I.L., F. Melchers, and T.H. Winkler. 1997. A transgenic marker for mouse B lymphoid precursors. J. Exp. Med. 185:653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.