Abstract
The DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and Artemis are classical nonhomologous DNA end-joining (C-NHEJ) factors required for joining a subset of DNA double-strand breaks (DSB), particularly those requiring end processing. In mature B cells, activation-induced cytidine deaminase (AID) initiates class switch recombination (CSR) by introducing lesions into S regions upstream of two recombining CH exons, which are processed into DSBs and rejoined by C-NHEJ to complete CSR. The function of DNA-PKcs in CSR has been controversial with some reports but not others showing that DNA-PKcs–deficient mice are significantly impaired for CSR. Artemis-deficient B cells reportedly undergo CSR at normal levels. Overall, it is still not known whether there are any CSR-associated DSBs that require DNA-PKcs and/or Artemis to be joined. Here, we have used an immunoglobulin (Ig)H locus-specific fluorescent in situ hybridization assay to unequivocally demonstrate that both DNA-PKcs and, unexpectedly, Artemis are necessary for joining a subset of AID-dependent DSBs. In the absence of either factor, B cells activated for CSR frequently generate AID-dependent IgH locus chromosomal breaks and translocations. We also find that under specific activation conditions, DNA-PKcs−/− B cells with chromosomal breaks are eliminated or at least prevented from progressing to metaphase via a p53-dependent response.
Classical nonhomologous DNA end joining (C-NHEJ), which ligates DNA ends with no or short stretches of homology, is a major DNA double-strand break (DSB) repair pathway in somatic mammalian cells (for review see reference 1). Known NHEJ factors include Ku70, Ku80, the DNA–protein kinase catalytic subunit (DNA-PKcs), Artemis, XRCC4, ligase IV (Lig4), and the recently described XLF/Cernunnos protein (1–3). Ku70/Ku80 heterodimers bind DSBs and recruit and activate DNA-PKcs to form the DNA-PK holoenzyme. With respect to NHEJ, DNA-PKcs autophosphorylates and activates the endo/exonuclease activity of Artemis, a factor capable of processing complex DNA ends, such as hairpins, before ligation by the XRCC4–Lig4 complex (1–3). Ku70, Ku80, XRCC4, and Lig4 are considered “core” C-NHEJ factors, as they are conserved in evolution and required to join all types of DNA ends. DNA-PKcs and Artemis appear to have evolved in higher eukaryotes and are not required for all types of C-NHEJ (see below), but rather have a particular role in joining ends that need processing (1).
In developing lymphocytes, the Ig and T cell receptor variable region exons are assembled from germline variable (V), diversity (D), and joining (J) gene segments (for review see reference 4). To initiate V(D)J recombination, the RAG endonuclease introduces DSBs between V, D, and J coding sequences and flanking recombination signal sequences (RSs) to generate blunt, 5′ phosphorylated RS ends and covalently sealed hairpin coding ends. Coding and RS ends, respectively, are joined via C-NHEJ to form coding and RS joins (4). Both coding and RS joining require the core C-NHEJ factors. However, although DNA-PKcs and Artemis are required for coding joins, substantial RS joining occurs without DNA-PKcs, and RS joining is essentially normal without Artemis (5, 6). In this context, coding join formation requires the opening and processing of hairpins, which is achieved via DNA-PKcs–activated Artemis endonuclease activity (7, 8), whereas RS joining only involves direct ligation of the blunt 5′ phosphorylated RS ends. Given that RS joining is variably decreased and frequently imprecise in DNA-PKcs-deficient but not in Artemis-deficient cells, the role of DNA-PKcs in NHEJ likely extends beyond Artemis-associated functions. In the latter context, DNA-PKcs activates other factors in the context of the general DSB response (9) and possibly during class switch recombination (CSR) (10).
In activated B cells, DSBs are introduced into the IgH locus during IgH CSR, a process that allows the V(D)J exon first expressed with Cμ constant region exons to be expressed with one of a set of downstream CH exons (e.g., Cγ, Cα, and Cε) (11). CH exons are preceded in the genome by long repetitive switch (S) regions. Transcription-dependent deamination of cytidines in S regions by activation-induced cytidine deaminase (AID) leads to S region DSBs (11), which are joined to complete CSR. S region junctions lack long stretches of homology, leading to the suggestion that they are joined primarily by end joining (12). CSR has been studied in different C-NHEJ–deficient backgrounds by complementing the V(D)J recombination deficiency with preassembled knock-in variable region exons at the IgH and IgL loci (13–16). Consistent with a role for C-NHEJ, Ku70 and Ku80 deficiencies completely abrogate CSR, although proliferation defects of Ku-deficient B cells raise the possibility that, at least in part, the CSR defect of Ku-deficient B cells may reflect the role of Ku in processes besides C-NHEJ (13, 15). In this regard, XRCC4 or Lig4 deficiency reduces CSR, but only to levels ranging from 20 to 50% of control levels (17, 18). This defect results from defective end joining, as a large proportion of IgH loci in XRCC4- or Lig4-deficient B cells suffer chromosomal breaks that often are involved in translocations (18). Residual CSR in XRCC4- or Lig4-deficient B cells is performed by a clearly distinct, alternative end-joining pathway that strongly prefers to join ends with micro-homologies (18).
Mice completely deficient for DNA-PKcs (DNA-PKcs−/− mice) have been generated by gene-targeted mutation (5). DNA-PKcs−/− B cells proliferate normally but have a severe CSR defect to most IgH classes except IgG1, for which CSR ranges from moderately reduced to essentially normal in both outbred and inbred backgrounds (14, 16). Mouse B cells homozygous for the scid mutation (SCID B cells), which leads to generation of a mutant DNA-PKcs protein that lacks kinase activity (19), were reported to show reduced IgH class switching to all IgH constant regions, but not as severely reduced as observed for DNA-PKcs−/− mice (20–22). One potential explanation for the more severe defect in IgH CSR in DNA-PKcs-deficient versus SCID B cells is that the latter might produce a relatively intact DNA-PKcs protein that harbors putative noncatalytic functions. However, the precise mechanism of the CSR defects in DNA-PKcs–deficient and SCID B cells was not characterized; in particular, whether or not it involved an end-joining defect or some other DNA-PKcs function. Notably, a recent report, based on analyses of SCID B cells or a line of DNA-PKcs mutant mice that harbored a targeted mutation similar to the scid mutation, led to the conclusion that CSR can occur essentially normally in the absence of DNA-PKcs (23). In this regard, Artemis-deficient B cells have no obvious CSR defect as assayed by standard methods (16), suggesting that potential DNA-PKcs functions implicated by the CSR defect in DNA-PKcs-deficient mice are not mediated via Artemis. The lack of a detectable CSR defect in Artemis-deficient B cells suggested that most DSBs generated during CSR, like RS ends during V(D)J recombination, do not require processing for joining or that they are processed via an Artemis-independent pathway (16).
To unequivocally elucidate potential roles of DNA-PKcs and Artemis in CSR, we now use a highly sensitive IgH locus-specific fluorescent in situ hybridization (FISH) assay to investigate unequivocally whether DNA-PKcs and Artemis have roles in end joining and in suppression of chromosomal translocations in the context of CSR.
RESULTS
Genomic instability in DNA-PKcs– and Artemis-deficient B cells activated for CSR
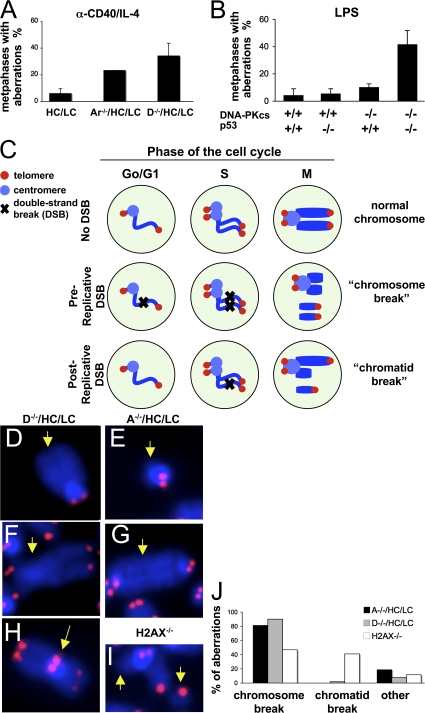
DNA-PKcs– or Artemis-deficient B cells were generated via introduction of preassembled Ig heavy chain (HC) and light chain (LC) knock-in alleles to rescue B cell development in the absence of V(D)J recombination, as described previously (14, 16). To assess whether defects in either of these C-NHEJ factors results in genomic instability in mature B cells, we used telomere FISH (T-FISH) to analyze metaphase spreads of DNA-PKcs–deficient (D−/−/HC/LC) or Artemis-deficient (A−/−HC/LC) B cells that were activated for 4 d with α-CD40/IL-4 to stimulate CSR to IgG1 and IgE (24). T-FISH analyses revealed that 34.2 ± 9.6% of D−/−/HC/LC B cells contained at least one chromosomal aberration, compared with 6.0 ± 3.7% of HC/LC control B cells (n = 4 independent stimulations; Fig. 1 A and Table S1, which is available at http://www.jem.org/cgi/content/full/jem.20080044/DC1). Likewise, A−/−HC/LC B cells stimulated in parallel similarly displayed increased chromosomal instability, with 23.3% of metaphases harboring at least one chromosomal aberration (Fig. 1 A and Table S1). Similarly, the total number of chromosomal aberrations was increased from 6.0 ± 3.7% in HC/LC control B cells to 42.5 ± 13.2% and 26.7 ± 4.7% in D−/−/HC/LC and A−/−HC/LC cultures, respectively (Table S1). Therefore, DNA-PKcs and Artemis function in the suppression of chromosomal instability in α-CD40/IL-4–activated mature B cells with D−/−/HC/LC B cells harboring approximately double the number of chromosomal breaks found in A−/−HC/LC B cells (Fig. 1 A and Table S1), consistent with Artemis-independent roles for DNA-PKcs in general DSB repair, similar to what has been found for V(D)J recombination and CSR (6, 16).
Figure 1.
DNA-PKcs and Artemis maintain chromosomal stability in activated B cells. (A) Quantification of genomic instability in D−/−/HC/LC and A−/−/HC/LC B cells by T-FISH. Metaphase spreads from D−/−/HC/LC, A−/−/HC/LC B, and control HC/LC B cells activated with anti-CD40/IL-4 for 4 d were hybridized with a telomere-specific PNA probe, and chromosomal aberrations were quantified in 30 metaphases per culture. Bars represent average and standard deviation of three mice per genotype in three independent stimulations. (B) Quantification of genomic instability in D−/−/HC/LC and D−/−/p53−/−/HC/LC B cells activated with LPS for 4 d and stained with a telomere probe. Bars represent average and standard deviation of five mice per genotype in five independent stimulations. (C–J) Distribution of chromosomal aberrations in B cells deficient for DNA-PKcs, Artemis, or H2AX. As diagrammed in C, DSBs introduced before replication typically result in chromosome-type aberrations at metaphases, whereas DSBs introduced after DNA replication result in chromatid-type aberrations. Mainly chromosome-type aberrations are observed in B cells deficient for DNA-PKcs (D, chromosome break; F, chromosomal translocation) or Artemis (E, chromosome break; G, dicentric), whereas H2AX deficiency results in both chromosome-type and chromatid-type breaks (an example of a chromatid break is shown in I). End-to-end chromosomal fusions were also observed in DNA-PKcs–deficient B cells (H). Quantification is shown in J.
We also used T-FISH to analyzed metaphase spreads from D−/−/HC/LC or A−/−HC/LC B cells that were activated for 4 d with LPS to stimulate CSR to IgG2b and IgG3. Surprisingly, we found very low levels of genomic instability in LPS-stimulated D−/−/HC/LC or A−/−HC/LC B cells (Fig. 1 B and Table S2, which is available at http://www.jem.org/cgi/content/full/jem.20080044/DC1). To further elucidate the basis of this unexpected result, we generated and analyzed D−/−/HC/LC B cells that were also p53 deficient (D−/−/HC/LC/P). T-FISH analyses of LPS-activated D−/−/HC/LC/P B cells revealed substantial genomic instability as compared with p53-deficient control B cells (Fig. 1 B and Table S2). Thus, these findings suggest that LPS-activated DNA-PKcs–deficient cells also harbor substantial levels of genomic instability but apparently are eliminated via a p53-dependent checkpoint.
To assess whether the chromosomal aberrations found in α-CD40/IL-4–activated DNA-PKcs– or Artemis-deficient B cells resulted from pre- or postreplicative lesions, we classified all chromosomal aberrations observed in metaphases from α-CD40/IL-4–activated D−/−/HC/LC and A−/−/HC/LC B cells as involving one chromatid (chromatid-type) or the two sister chromatids (chromosome-type; see Fig. 1 C for schematic). Although chromosome-type aberrations could potentially result from independent breakage of the two sister chromatids after replication, they typically reflect a DSB introduced before DNA replication, which is then replicated. In contrast, chromatid-type breaks typically originate from unrepaired DSBs introduced after replication (25). Deficiency for either DNA-PKcs or Artemis resulted nearly exclusively in chromosome breaks, or chromosomal rearrangements derived from them (dicentrics, robertsonian translocations), with chromatid breaks comprising ≤1% of the observed aberrations (Fig. 1, D–G, for examples, Fig. 1 J for quantification, and Table S1), similar to what has been found for Lig4-deficient mouse embryonic fibroblasts or XRCC-4–deficient B cells (18, 26). As expected (27), B cells deficient for H2AX, which facilitates DSB repair by both NHEJ and homologous recombination, contained similar frequency of chromosome and chromatid breaks (Fig. 1 I for example and Fig. 1 J for quantification), suggesting that DSB repair mediated by both DNA-PKcs and Artemis largely involves pre-replicative DSBs. Finally, B cells deficient for DNA-PKcs showed a low frequency of end-to-end chromosomal fusions with telomere signals at the fusion point (Fig. 1 H), reflecting the role of DNA-PKcs in maintaining genomic stability by capping chromosome ends (28).
DNA-PKcs is required for end joining of IgH locus DSBs during CSR
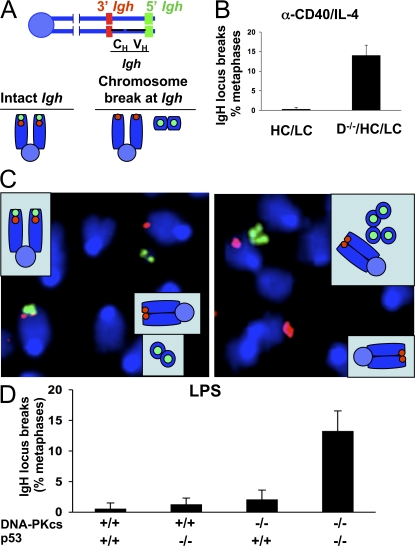
We have used a sensitive IgH locus FISH assay (27) to determine directly whether DNA-PKcs has any function in the end-joining phase of CSR. After treatment of D−/−/HC/LC B cells with α-CD40/IL-4 for 4 d to induce class switching to IgG1 and IgE, we observed normal proliferation levels and surface IgG1 expression that ranged from somewhat decreased to approximately equal to that of HC/LC controls (Figs. S1 and S2, available at http://www.jem.org/cgi/content/full/jem.20080044/DC1). As expected (14), CSR to IgE was abolished in the absence of DNA-PKcs (not depicted). IgH locus FISH analysis revealed IgH-specific chromosomal breaks in 14.0 ± 2.6% D−/−/HC/LC B metaphases (n = 4 independent stimulations; range, 11–16%) compared with 0.25 ± 0.5% (range, 0–1%) in four control HC/LC cultures (Fig. 2, A and B, and C for examples), indicating that a substantial subset of IgH locus DSBs generated after α-CD40/IL-4 treatment fail to rejoin in the absence of DNA-PKcs. LPS-treated D−/−/HC/LC B cells failed to undergo substantial CSR to IgG2b or IgG2 (not depicted), but, as for general DSBs, also failed to show substantially increased IgH locus breaks relative to control cells (Fig. 2 D and Table S2). Notably, LPS-treated D−/−/HC/LC B cells that were also p53 deficient also failed to undergo significant switching to IgG2b or IgG3 but, similar to our findings with general DSBs, did show substantial levels of IgH locus breaks (13.2 ± 3.3% of metaphases; range, 10–18%; n = 5 independent stimulations), confirming that DSBs introduced in the context of CSR to IgG2b or IgG3 are also dependent on DNA-PKcs (Fig. 2 D and Table S2). Finally, as previously observed in DSB response factor–deficient backgrounds and XRCC4-deficient activated B cells (18, 27), all observed IgH locus breaks involved two sister chromatids (chromosome breaks; not depicted), consistent with the notion that DNA-PKcs is required for rejoining S region lesions that are initiated in pre-replicative stages of the cell cycle (29).
Figure 2.
DNA-PKcs and Artemis are required for end joining of a subset of DSBs during CSR. (A) Schematic of the IgH locus in the subtelomeric region of mouse chromosome 12 and location of 3′ and 5′ IgH flanking probes. A chromosome break at IgH is defined as dissociation of the 3′ and 5′ IgH signals, which appear in two distinct DAPI fragments. (B) Frequency of IgH locus breaks determined by IgH FISH on metaphase spreads of DNA-PKcs–deficient B cells activated with α-CD40/IL-4. Bars represent average and standard deviation of three mice in three independent stimulations. 100 metaphases per mice were analyzed. (C) Partial metaphase spreads of activated DNA-PKcs–deficient B cells hybridized with 3′ (red) and 5′ (green) IgH probes. Chromosomal breaks at one IgH allele (left) or at both alleles (right) were observed. (D) Frequency of IgH locus breaks determined by IgH FISH on metaphase spreads of DNA-PKcs–deficient B cells (± p53-deficient) activated with LPS. Bars represent the average and standard deviation of five independent experiments.
ATM deficiency and its associated checkpoint deficiencies can lead to RAG-dependent IgH locus chromosomal breaks introduced in the context of V(D)J recombination in pro–B cells that persist through development and are retained in mature B cells (30). To rigorously demonstrate that the DSBs we observe in DNA-PKcs–deficient B cells are introduced in the context of attempted CSR, we quantified the frequency of IgH locus breaks in α-CD40/IL-4–activated B cells doubly deficient for AID and DNA-PKcs (D−/−/AID−/−/HC/LC), as well as in AID−/−/ATM−/− B cells. Neither AID−/−/ATM−/− or D−/−/AID−/−/HC/LC B cells undergo CSR upon activation (Fig. S2). Notably, although AID−/−/ATM−/− B cells were found to still harbor IgH locus breaks in 8.0 ± 2.0 of metaphases (range, 6–10%; Table I and Table S3 and Fig. S3, which are available at http://www.jem.org/cgi/content/full/jem.20080044/DC1) similarly to what has been described (30), AID deficiency completely eliminated the occurrence of IgH locus breaks in α-CD40/IL-4–activated DNA-PKcs–deficient B cells (Table I, Table S3, and Fig. S3), indicating that the vast majority, if not all, IgH locus breaks observed in the absence of DNA-PKcs result from failure to repair a subset of CSR-specific DNA lesions.
Table I.
Effect of AID deficiency of IgH locus stability in activated B cells deficient for DNA-PKcs, Artemis, or ATM
| Genotype | No. mice (no. metaphases) |
% Metaphases with IgH locus breaks (ave ± SD) |
|---|---|---|
| DNA-PKcs/AID | ||
| HL | 3 (150) | 2.0 ± 2.0 |
| AID−/−/HC/LC | 2 (100) | 0.0 ± 0.0 |
| DNA-PKcs−/−/HC/LC | 3 (150) | 15.3 ± 3.1 |
| DNA-PKcs−/−/ AID−/−/HC/LC |
2 (100) | 0.0 ± 0.0 |
| Artemis/AID | ||
| HL | 2 (100) | 1.0 ± 1.4 |
| AID−/−/HC/LC | 2 (100) | 0.0 ± 0.0 |
| Artemis−/−/HC/LC | 6 (300) | 9.7 ± 2.7 |
| Artemis−/−/ AID−/−/HC/LC |
3 (150) | 0.0 ± 0.0 |
| ATM/AID | ||
| wild-type | 2 (100) | 0.0 ± 0.0 |
| AID−/− | 1 (50) | 0.0 ± 0.0 |
| ATM−/− | 3 (150) | 18.0 ± 4.0 |
| ATM−/−/ AID−/− | 3 (150) | 8.0 ± 2.0 |
B cells of the indicated genotypes were activated with α-CD40/IL-4 for 4 d, and metaphase spreads were hybridized with BAC probes flanking IgH (3′ and 5′ IgH). IgH locus-specific breaks (end-dissociated probes) were scored in 50 metaphases per culture. See Table S3 for data on individual cultures.
Artemis is required for the repair of a subset of IgH locus DSBs during CSR
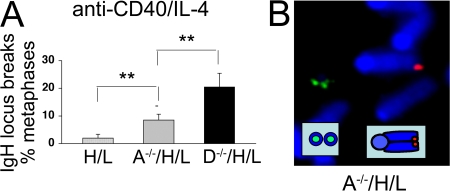
Cellular assays for class switching generally rely on finding a decrease in the level of expression of a particular IgH isotype. Given the general variability in IgH CSR, such “negative” assays are only useful to detect large defects in CSR. On the other hand, the FISH assay for chromosomal IgH locus breaks is a “positive” assay and can detect defects in end joining during CSR in only a few percent of the activated B cells (27). Therefore, we took advantage of the high sensitivity of our IgH FISH assay to investigate a potential role for Artemis in the end joining of a small fraction of the DSBs generated during CSR. As reported previously (16), α-CD40/IL-4–stimulated A−/−/HC/LC B cells proliferated normally and expressed comparable levels of sIgG1 as their HC/LC counterparts (Figs. S1 and S2). However, IgH FISH analyses revealed chromosomal breaks at the IgH locus in 8.8 ± 2.8% of A−/−/HC/LC metaphases (range, 6% to 12%; n = 4 independent stimulations) as compared with ≤1% in all four control HL cultures (Fig. 3). As reported above in the context of general breaks, experiments where DNA-PKcs and Artemis-deficient B cells were stimulated in parallel invariably showed a greater frequency of IgH locus breaks in the absence of DNA-PKcs than in the absence of Artemis (Fig. 3 A and Table S1), consistent with Artemis-independent roles for DNA-PKcs in CSR (16). As Artemis is also required for the rejoining of RAG-dependent DSBs at coding ends, as well as for repairing a subset of general DSBs that require end processing (1), we also assayed activated B cells doubly deficient for Artemis and AID to confirm that the IgH locus breaks observed in activated Artemis-deficient B cells resulted from defective CSR. AID deficiency abolished both CSR (Fig. S2) and IgH locus-specific chromosomal breaks in Artemis-deficient B cells (Table I, Table S3, and Fig. S3), conclusively demonstrating that Artemis is required for the joining of a small subset of IgH CSR-related DSBs.
Figure 3.
IgH locus instability in activated Artemis-deficient B cells. (A) Quantification of the frequency of IgH locus breaks in D−/−/HC/LC, A−/−/HC/LC, and control HC/LC B cells activated in parallel. The frequency of split 3′5′ IgH signals for each genotype is shown. Bars represent the average and standard deviation of three independent stimulations. A representative example of a chromosomal break at the IgH locus in an A−/−/HC/LC metaphase is shown in B.
DNA-PKcs and Artemis suppress AID-dependent chromosomal translocation in mature B cells undergoing CSR
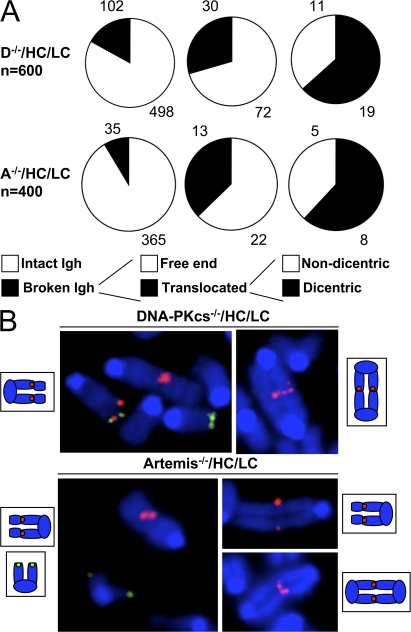
In the absence of DSB response factors or the Ku or XRCC4 C-NHEJ factors, AID-dependent IgH locus chromosomal breaks introduced in B cells activated for CSR are fused to other breaks at high frequency to generate translocations (18, 27, 31). Moreover, DNA-PKcs/p53 double-deficient or Artemis/p53 double-deficient mice succumb to pro–B cell lymphomas with clonal translocations that arise from aberrant repair of RAG-dependent chromosomal breaks (32, 33). To test whether deficiency for DNA-PKcs or Artemis similarly promotes chromosomal translocations with breakpoints within the IgH locus, we analyzed 102 CD40/IL-4–activated D−/−/HC/LC and 35 A−/−/HC/LC B cells metaphases that contained a chromosomal break at the IgH locus (from a total pool of 600 D−/−/HC/LC and 400 A−/−/HC/LC metaphases, respectively) for participation in translocations. The IgH locus was translocated in 30/102 (29.4%) D−/−/HC/LC HL metaphases and in 13/35 (37.1%) A−/−/HC/LC metaphases (Fig. 4 A, and B for examples). Moreover, 19/30 (63.3%) and 8/13 (61.5%) of these D−/−/HC/LC HL and A−/−/HC/LC translocations, respectively, were dicentrics (Fig. 4 A), highly unstable cytogenetic aberrations that trigger breakage-fusion-bridge cycles and thereby can amplify genomic instability (1).
Figure 4.
Frequent chromosomal translocations involving the IgH locus in B cells deficient for DNA-PKcs or Artemis. (A) Analysis of metaphase spreads of α-CD40/IL-4–activated B cells deficient for DNA-PKcs or Artemis for genomic instability at the IgH locus. Chromosomes 12 with a break at IgH were further classified as free ends or translocated. For those involved in translocations, formation of dicentric was noted. Examples are shown in B. Chromosome 12 translocates to other centric or acentric chromosomes, as diagrammed. Only the 3′ (red) or the 5′ (green) IgH probe is detected at the translocation junction, indicating that the breakpoint for chromosome 12 lies within IgH.
DISCUSSION
Previous work demonstrated a requirement for XRCC4 and Lig4, core components of C-NHEJ, in the end-joining phase of CSR (18). In contrast, a potential role for DNA-PKcs in CSR has been controversial (14, 23), whereas Artemis was thought to be dispensable for CSR (16). We now demonstrate AID-dependent IgH locus-specific chromosomal breaks in 10–20% of activated DNA-PKcs−/− B cell metaphases obtained from B cells stimulated with cytokine combinations that induce CSR to different IgH isotypes in vitro (i.e., IgG1, IgE, IgG2b, or IgG3). These findings provide unequivocal evidence that DNA-PKcs, in fact, does play a significant role in the end-joining phase of CSR, regardless of the type of stimulation. In the context of activation with LPS, our observed frequency of IgH locus breaks (∼15% of all B cell metaphases) may well account for the severe CSR defect to both IgG2b and IgG3. In the context of activation with α-CD40/IL-4, where the fraction of cultured cells that effect CSR is typically two- or threefold higher than in LPS-activated cultures, our observed frequency of IgH locus breaks (15–20% of all B cell metaphases) likely reflects both abolished CSR to IgE combined with the modest defect in CSR to IgG1. Unexpectedly, we also detected AID-dependent IgH locus breaks in a significant fraction of activated Artemis-deficient B cells, albeit at a lower frequency than observed in DNA-PKcs–deficient B cells.
The requirement for Artemis for joining a smaller fraction of DSBs during IgH CSR as compared with DNA-PKcs most likely reflects the ends of DSBs in S regions generated in the context of in vitro B cell activation being heterogeneous (11). DNA-PKcs–activated Artemis can process a variety of complex DNA structures in vitro (34) and is specifically required in vivo for the repair of a subset of complex DNA ends that requires extensive processing, such as hairpins at coding ends during V(D)J recombination (6) or a subset of ionizing radiation–induced DSBs (35). However, an Artemis-independent role for DNA-PKcs has been clearly established both for ligation of blunt signal ends during V(D)J recombination (5, 6) and for the repair of ionizing radiation–induced DSBs (35). Thus, in the context of CSR, a subset of AID-dependent lesions with complex ends might similarly require DNA-PKcs–mediated Artemis activation for repair, whereas others only require DNA-PKcs, perhaps because they do not need extensive processing or they are processed via an Artemis-independent mechanism. Artemis-independent roles for DNA-PKcs may include an intrinsic scaffolding function in the alignment/synapses of DNA ends (36, 37) and/or activation of DNA damage response (DDR) factors required for CSR. In the latter context, DNA-PKcs phosphorylates H2AX to form γ-H2AX, a chromatin modification that correlates with CSR (29), at DSBs (38). Moreover, deficiency for H2AX also impairs end joining of S regions during CSR (27, 31). In addition, MDC1, another DSB response factor that promotes end joining of S regions during CSR (27), binds to DNA-PKcs and promotes its activation by autophosphorylation (39).
Even though DNA-PKcs likely plays a role in regulating the DDR, our cytogenetic analysis points to clear differences in the requirement for some DDR factors and C-NHEJ factors in the context of the cell cycle. Both chromosome and chromatid breaks are readily observed in metaphase spreads of B cells (and other cell types) deficient for H2AX or, to a lesser extent, ATM (27), consistent with a role for these factors in the repair of both pre- and postreplicative DNA lesions. In contrast, chromatid breaks are rarely observed in B cells deficient for DNA-PKcs or Artemis (this study), XRCC4 (18), or in mouse embryonic fibroblasts deficient for Lig4 (26), suggesting that the action of the NHEJ is dominant in pre-replicative phases of the cell cycle. In contrast, IgH locus breaks observed in the absence of either DDR or NHEJ components are invariably of the chromosome type (18, 27, and this study), supporting the notion that the initiating lesion in CSR is introduced before DNA replication (29).
Like DNA-PKcs, ATM is a PI3-like kinase required for normal rejoining of DNA ends across both RAG- and AID-dependent DSBs (27, 40). However, major differences exist as to the fate of unrepaired DNA ends in the absence of one or the other of these two kinases. First, we document here that, unlike persistent RAG-initiated DSB in ATM-deficient mice (30), unrepaired RAG-dependent DSBs do not persist to the mature B cell stage in DNA-PKcs–deficient mice. Second, although general and IgH locus-specific chromosomal breaks were abundant in ATM−/− B cells activated either with a-CD40 plus IL-4 or with LPS, such breaks are observed in α-CD40 plus IL-4–activated DNA-PKcs−/− B cells, but not in LPS-activated DNA-PKcs−/− B cells. However, the breaks are observed in LPS-activated DNA-PKcs−/− B cells that are also p53 deficient, indicating that DNAPKcs−/− B cells with DSBs are eliminated, or at least prevented from progressing to metaphase, via a p53-dependent response after LPS stimulation. Although there are other conceivable interpretations, these findings suggest that α-CD40 plus IL-4 activation conditions obviate the p53-dependent response to DSBs in activated DNA-PKcs−/− B cells, and that ATM deficiency also eliminates this response even under LPS stimulation conditions. Of note, α-CD40 plus IL-4 stimulation is thought to mimic in vitro the T cell–dependent germinal center reaction, where the normal p53-dependent cellular response to DSBs is normally blunted in a transient manner to prevent inappropriate growth arrest or apoptosis during CSR (41). Finally, as H2AX−/−, MDC1−/−, and 53BP1−/− B cells also show varying degrees of general and IgH locus breaks under α-CD40 plus IL-4 or LPS activation conditions (27), it would appear that deficiencies for other DDR factors can obviate this p53-dependent response under LPS activation conditions.
Our findings that IgH locus breaks frequently participate in chromosomal translocations in DNA-PKcs−/− and Artemis−/− cells activated for CSR are in accord with findings of increased IgH-myc translocations in activated B cells from SCID mice (31) and end-to-end chromosomal fusions in DNA-PKcs– and Artemis-deficient cells (28, 42) (Fig. 1). Thus, in the context of CSR, DNA-PKcs and Artemis prevent chromosomal translocations by promoting end joining of IgH locus DSBs to other IgH locus DSBs on the same chromosome. In their absence, IgH locus breaks can be frequently joined to breaks on other chromosomes via DNA-PKcs– and Artemis-independent pathways of end joining, a process that may be enhanced due to the increased frequency of unrepaired breaks on other chromosomes. DNA-PKcs– or Artemis-deficient pro–B cells that are also deficient for the p53 cell cycle checkpoint factor are predisposed to pro–B lymphomas that harbor oncogenic translocations that appear to be mediated by aberrant V(D)J recombination (32, 33). By analogy, our current findings suggest that mutations of DNA-PKcs or Artemis in combination with cell cycle checkpoint defects might synergize in the generation of oncogenic IgH locus translocations that occur during attempted CSR, a process that contributes to B cell lymphomas or multiple myeloma (31, 43).
MATERIALS AND METHODS
Mice.
129Sve mice deficient for DNA-PKcs or Artemis and harboring preassembled IgH (B1-8-HC; H) inserted (knocked-in) into the endogenous JH locus and IgL (3–83k-LC; L) knocked-in to the J region of the Igκ locus were described previously (14, 16, 44, 45). Mice deficient for ATM, p53, and AID were described previously (46–48). All mouse protocols were approved by the Institutional Animal Care and Use Committee of Children's Hospital.
Mature B cell purification and cultures.
CD43− populations were obtained from splenocyte single cell suspensions by incubation with CD43 beads (Miltenyi Biotec) and negative selection and activated by incubation with 1 μg/ml α-CD40 antibody (BD Biociences) and 20 ng/ml IL-4 (R&D Systems).
Assessment of CSR by flow cytometry (FACs).
Stimulated B cells (0.5 million) were washed with 2% FCS/PBS, stained with antibodies labeled with CyChrome (B220), fluorescein (IgG2b and IgG1), or phycoerithrin (IgE and IgG3), and analyzed in a FACScan using CELLQuest (Becton Dickinson) or FloJo (Tree Star) software. At least 10,000 events of live lymphoid cells were recorded.
FISH probes.
We detected the 3′ end of the IgH locus using BAC199 and the 5′ end using BAC207, as described previously (27). For telomere staining, a Cy3-conjugated (TTAGGG)3 peptide nucleic acid (PNA) probe was purchased from Applied Biosystems.
PNA FISH of TTAGGG sequences.
Telomere staining was performed as described previously (27). In brief, slides were denatured at 80°C for 3 min, hybridized with a Cy3-labeled PNA telomeric probe (Cy3-(TTAGGG)3) in 70% formamide at room temperature for 2 h, washed, dehydrated, and mounted in Vectashield with DAPI (Vector Laboratories). Metaphase images were captured using a Nikon Eclipse microscope equipped with a CCD camera (Applied Spectral Imaging) and a 63× objective lens.
Two-color FISH.
BACs were labeled with either biotine (Biotin-Nick Translation Mix; Roche) or digoxigenin (Dig-Nick Translation Mix; Roche), as per the manufacturer's instructions, and FISH was performed as described previously (27). In brief, 200 ng of each probe DNA was precipitated with mouse Cot1 DNA (Invitrogen), resuspended, and co-denatured in hybridization solution (50% formamide, 2X SSC, 10% dextran sulfate, and 0.15% SDS) for 5 min at 76°C. After incubation at 37°C for 16 h, slides were washed, incubated with avidin-Cy3 and antidigoxigenin-FITC, and mounted in Vectashield with DAPI (Vector Laboratories). Metaphase images were captured using a Nikon Eclipse microscope equipped with a CCD camera (Applied Spectral Imaging) and a 63× objective lens.
Online supplemental material.
Fig. S1 shows the efficiency of CSR by FACS analysis of surface Ig expression in activated B cells deficient for DNA-PKcs or Artemis. Fig. S2 shows the effect of AID deficiency on CSR measured by FACS analysis of surface Ig expression in activated B cells deficient for DNA-PKcs, Artemis, or ATM. Fig. S3 shows the effect of AID deficiency on IgH locus stability in activated B cells deficient for DNA-PKcs, Artemis, or ATM. Table S1 provides an analysis of genomic stability in α-CD40/IL-4–activated B cells deficient for DNA-PKcs or Artemis. Table S2 provides data on genomic stability in LPS-activated B cells deficient for DNA-PKcs and the effect of p53 inactivation. Table S3 shows the effect of AID deficiency on genomic stability in activated B cells deficient for DNA-PKcs, Artemis, or ATM. The online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20080044/DC1.
Supplemental Material
Acknowledgments
We thank members of the Alt laboratory for helpful discussions.
S. Franco was supported by a Long-Term Fellowship of the European Molecular Biology Organization (EMBO) and by the National Institutes of Health (NIH) under Ruth L. Kirschstein National Research Service Award T32 CA09382. F.W. Alt is supported by NIH grants AI31541, AI35714, and CA92625. F.W. Alt is an Investigator, and G. Li was an Associate of the Howard Hughes Institute.
The authors have no conflicting financial interests.
Abbreviations used: AID, activation-induced cytidine deaminase; C-NHEJ, classical nonhomologous DNA end joining; CSR, class switch recombination; DDR, DNA damage response; DNA-PKcs, DNA–protein kinase catalytic subunit; DSB, double-strand break; Lig4, ligase IV; FISH, fluorescent in situ hybridization; HC, heavy chain; LC, light chain; PNA, peptide nucleic acid; RS, recombination signal sequence; T-FISH, telomere FISH.
References
- 1.Mills, K.D., D.O. Ferguson, and F.W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77–95. [DOI] [PubMed] [Google Scholar]
- 2.Buck, D., L. Malivert, R. de Chasseval, A. Barraud, M.C. Fondaneche, O. Sanal, A. Plebani, J.L. Stephan, M. Hufnagel, F. le Deist, et al. 2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 124:287–299. [DOI] [PubMed] [Google Scholar]
- 3.Ahnesorg, P., P. Smith, and S.P. Jackson. 2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 124:301–313. [DOI] [PubMed] [Google Scholar]
- 4.Jung, D., C. Giallourakis, R. Mostoslavsky, and F.W. Alt. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. [DOI] [PubMed] [Google Scholar]
- 5.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D.T. Weaver, and F.W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 9:367–376. [DOI] [PubMed] [Google Scholar]
- 6.Rooney, S., J. Sekiguchi, C. Zhu, H.L. Cheng, J. Manis, S. Whitlow, J. DeVido, D. Foy, J. Chaudhuri, D. Lombard, and F.W. Alt. 2002. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 10:1379–1390. [DOI] [PubMed] [Google Scholar]
- 7.Ma, Y., U. Pannicke, K. Schwarz, and M.R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 108:781–794. [DOI] [PubMed] [Google Scholar]
- 8.Goodarzi, A.A., Y. Yu, E. Riballo, P. Douglas, S.A. Walker, R. Ye, C. Harer, C. Marchetti, N. Morrice, P.A. Jeggo, and S.P. Lees-Miller. 2006. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 25:3880–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiff, T., M. O'Driscoll, N. Rief, K. Iwabuchi, M. Lobrich, and P.A. Jeggo. 2004. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 64:2390–2396. [DOI] [PubMed] [Google Scholar]
- 10.Reina-San-Martin, B., H.T. Chen, A. Nussenzweig, and M.C. Nussenzweig. 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J. Exp. Med. 200:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri, J., and F.W. Alt. 2004. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 4:541–552. [DOI] [PubMed] [Google Scholar]
- 12.Dunnick, W., G.Z. Hertz, L. Scappino, and C. Gritzmacher. 1993. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 21:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manis, J.P., Y. Gu, R. Lansford, E. Sonoda, R. Ferrini, L. Davidson, K. Rajewsky, and F.W. Alt. 1998. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 187:2081–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manis, J.P., D. Dudley, L. Kaylor, and F.W. Alt. 2002. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 16:607–617. [DOI] [PubMed] [Google Scholar]
- 15.Casellas, R., A. Nussenzweig, R. Wuerffel, R. Pelanda, A. Reichlin, H. Suh, X.F. Qin, E. Besmer, A. Kenter, K. Rajewsky, and M.C. Nussenzweig. 1998. Ku80 is required for immunoglobulin isotype switching. EMBO J. 17:2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rooney, S., F.W. Alt, J. Sekiguchi, and J.P. Manis. 2005. Artemis-independent functions of DNA-dependent protein kinase in Ig heavy chain class switch recombination and development. Proc. Natl. Acad. Sci. USA. 102:2471–2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soulas-Sprauel, P., G. Le Guyader, P. Rivera-Munoz, V. Abramowski, C. Olivier-Martin, C. Goujet-Zalc, P. Charneau, and J.P. de Villartay. 2007. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 204:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan, C.T., C. Boboila, E.K. Souza, S. Franco, T.R. Hickernell, M. Murphy, S. Gumaste, M. Geyer, A.A. Zarrin, J.P. Manis, et al. 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 449:478–482. [DOI] [PubMed] [Google Scholar]
- 19.Danska, J.S., D.P. Holland, S. Mariathasan, K.M. Williams, and C.J. Guidos. 1996. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol. Cell. Biol. 16:5507–5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook, A.J., L. Oganesian, P. Harumal, A. Basten, R. Brink, and C.J. Jolly. 2003. Reduced switching in SCID B cells is associated with altered somatic mutation of recombined S regions. J. Immunol. 171:6556–6564. [DOI] [PubMed] [Google Scholar]
- 21.Rolink, A., F. Melchers, and J. Andersson. 1996. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 5:319–330. [DOI] [PubMed] [Google Scholar]
- 22.Bosma, G.C., J. Kim, T. Urich, D.M. Fath, M.G. Cotticelli, N.R. Ruetsch, M.Z. Radic, and M.J. Bosma. 2002. DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J. Exp. Med. 196:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiefer, K., J. Oshinsky, J. Kim, P.B. Nakajima, G.C. Bosma, and M.J. Bosma. 2007. The catalytic subunit of DNA-protein kinase (DNA-PKcs) is not required for Ig class-switch recombination. Proc. Natl. Acad. Sci. USA. 104:2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkelman, F.D., J. Holmes, I.M. Katona, J.F. Urban Jr., M.P. Beckmann, L.S. Park, K.A. Schooley, R.L. Coffman, T.R. Mosmann, and W.E. Paul. 1990. Lymphokine control of in vivo immunoglobulin isotype selection. Annu. Rev. Immunol. 8:303–333. [DOI] [PubMed] [Google Scholar]
- 25.McClintock, B. 1939. The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc. Natl. Acad. Sci. USA. 25:405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mills, K.D., D.O. Ferguson, J. Essers, M. Eckersdorff, R. Kanaar, and F.W. Alt. 2004. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 18:1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franco, S., M. Gostissa, S. Zha, D.B. Lombard, M.M. Murphy, A.A. Zarrin, C. Yan, S. Tepsuporn, J.C. Morales, M.M. Adams, et al. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 21:201–214. [DOI] [PubMed] [Google Scholar]
- 28.Goytisolo, F.A., E. Samper, S. Edmonson, G.E. Taccioli, and M.A. Blasco. 2001. The absence of the dna-dependent protein kinase catalytic subunit in mice results in anaphase bridges and in increased telomeric fusions with normal telomere length and G-strand overhang. Mol. Cell. Biol. 21:3642–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen, S., R. Casellas, B. Reina-San-Martin, H.T. Chen, M.J. Difilippantonio, P.C. Wilson, L. Hanitsch, A. Celeste, M. Muramatsu, D.R. Pilch, et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callen, E., M. Jankovic, S. Difilippantonio, J.A. Daniel, H.T. Chen, A. Celeste, M. Pellegrini, K. McBride, D. Wangsa, A.L. Bredemeyer, et al. 2007. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 130:63–75. [DOI] [PubMed] [Google Scholar]
- 31.Ramiro, A.R., M. Jankovic, E. Callen, S. Difilippantonio, H.T. Chen, K.M. McBride, T.R. Eisenreich, J. Chen, R.A. Dickins, S.W. Lowe, et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rooney, S., J. Sekiguchi, S. Whitlow, M. Eckersdorff, J.P. Manis, C. Lee, D.O. Ferguson, and F.W. Alt. 2004. Artemis and p53 cooperate to suppress oncogenic N-myc amplification in progenitor B cells. Proc. Natl. Acad. Sci. USA. 101:2410–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo, Y., S.M. Wright, S.A. Maas, T.L. Alley, L.B. Caddle, S. Kamdar, J. Affourtit, O. Foreman, E.C. Akeson, D. Shaffer, et al. 2007. The nonhomologous end joining factor Artemis suppresses multi-tissue tumor formation and prevents loss of heterozygosity. Oncogene. 26:6010–6020. [DOI] [PubMed] [Google Scholar]
- 34.Ma, Y., K. Schwarz, and M.R. Lieber. 2005. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst.). 4:845–851. [DOI] [PubMed] [Google Scholar]
- 35.Riballo, E., M. Kuhne, N. Rief, A. Doherty, G.C. Smith, M.J. Recio, C. Reis, K. Dahm, A. Fricke, A. Krempler, et al. 2004. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 16:715–724. [DOI] [PubMed] [Google Scholar]
- 36.DeFazio, L.G., R.M. Stansel, J.D. Griffith, and G. Chu. 2002. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 21:3192–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spagnolo, L., A. Rivera-Calzada, L.H. Pearl, and O. Llorca. 2006. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 22:511–519. [DOI] [PubMed] [Google Scholar]
- 38.Park, E.J., D.W. Chan, J.H. Park, M.A. Oettinger, and J. Kwon. 2003. DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner. Nucleic Acids Res. 31:6819–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lou, Z., B.P. Chen, A. Asaithamby, K. Minter-Dykhouse, D.J. Chen, and J. Chen. 2004. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 279:46359–46362. [DOI] [PubMed] [Google Scholar]
- 40.Bredemeyer, A.L., G.G. Sharma, C.Y. Huang, B.A. Helmink, L.M. Walker, K.C. Khor, B. Nuskey, K.E. Sullivan, T.K. Pandita, C.H. Bassing, and B.P. Sleckman. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470. [DOI] [PubMed] [Google Scholar]
- 41.Phan, R.T., and R. Dalla-Favera. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 432:635–639. [DOI] [PubMed] [Google Scholar]
- 42.Rooney, S., F.W. Alt, D. Lombard, S. Whitlow, M. Eckersdorff, J. Fleming, S. Fugmann, D.O. Ferguson, D.G. Schatz, and J. Sekiguchi. 2003. Defective DNA repair and increased genomic instability in Artemis-deficient murine cells. J. Exp. Med. 197:553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuppers, R. 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat. Rev. Cancer. 5:251–262. [DOI] [PubMed] [Google Scholar]
- 44.Pelanda, R., S. Schwers, E. Sonoda, R.M. Torres, D. Nemazee, and K. Rajewsky. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 7:765–775. [DOI] [PubMed] [Google Scholar]
- 45.Sonoda, E., Y. Pewzner-Jung, S. Schwers, S. Taki, S. Jung, D. Eilat, and K. Rajewsky. 1997. B cell development under the condition of allelic inclusion. Immunity. 6:225–233. [DOI] [PubMed] [Google Scholar]
- 46.Donehower, L.A., M. Harvey, B.L. Slagle, M.J. McArthur, C.A. Montgomery Jr., J.S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 356:215–221. [DOI] [PubMed] [Google Scholar]
- 47.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 48.Borghesani, P.R., F.W. Alt, A. Bottaro, L. Davidson, S. Aksoy, G.A. Rathbun, T.M. Roberts, W. Swat, R.A. Segal, and Y. Gu. 2000. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. USA. 97:3336–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.