Abstract
The zinc finger transcription factor GATA-1 requires direct physical interaction with the cofactor friend of GATA-1 (FOG-1) for its essential role in erythroid and megakaryocytic development. We show that in the mast cell lineage, GATA-1 functions completely independent of FOG proteins. Moreover, we demonstrate that FOG-1 antagonizes the fate choice of multipotential progenitor cells for the mast cell lineage, and that its down-regulation is a prerequisite for mast cell development. Remarkably, ectopic expression of FOG-1 in committed mast cell progenitors redirects them into the erythroid, megakaryocytic, and granulocytic lineages. These lineage switches correlate with transcriptional down-regulation of GATA-2, an essential mast cell GATA factor, via switching of GATA-1 for GATA-2 at a key enhancer element upstream of the GATA-2 gene. These findings illustrate combinatorial control of cell fate identity by a transcription factor and its cofactor, and highlight the role of transcriptional networks in lineage determination. They also provide evidence for lineage instability during early stages of hematopoietic lineage commitment.
The zinc finger transcription factor GATA-1 serves as a useful paradigm for studying the role of lineage-specific factors in cell fate determination. Its expression is restricted to select cell types within the hematopoietic system, including erythroid, megakaryocytic, eosinophilic, and mast cells (1). Its only site of expression outside of the hematopoietic system is in Sertoli cells of the testis (2). Gene targeting studies in mice demonstrate an essential role for GATA-1 in erythroid and megakaryocytic terminal maturation (3–5). This activity requires direct physical interaction between GATA -1 and its cofactor, friend of GATA-1 (FOG-1; also known as zfpm1), a large multitype zinc finger–containing protein (6–10). In humans, germline missense mutations that disrupt the binding of GATA-1 to FOG-1 lead to severe thrombocytopenia and dyserthropoietic anemia, in some cases necessitating bone marrow transplantation (11–14).
Both GATA-1 and FOG-1 belong to families of proteins, whose members are expressed in overlapping patterns in diverse tissues. There are six known GATA factor genes (GATA-1, -2, -3, -4, -5, and -6) and two FOG genes (FOG-1 and -2) in vertebrates. All GATA factor members contain highly conserved amino acid sequences within their amino zinc finger that mediate binding to FOG proteins (15). The requirement for direct interaction between GATA and FOG family members extends beyond GATA-1 and FOG-1. Interaction between GATA-2, another hematopoietic-expressed GATA factor, and FOG-1 is required during early megakaryopoiesis in the absence of GATA-1–FOG-1 binding (9). Interaction between GATA-4 and FOG-2, two nonhematopoietic-expressed family members, is required for normal cardiac and gonadal development (16–19). Thus, GATA–FOG protein interactions are required in broad developmental settings.
Despite the critical role that FOG proteins play in modulating GATA factor activity, their mechanism of action remains incompletely understood (for review see reference 20). To date, no sequence-specific high affinity DNA binding activity of FOG proteins has been identified. This infers that FOG proteins exert their influence via protein–protein interactions. Chromatin immunoprecipitation (ChIP) studies indicate that FOG-1 facilitates occupancy by GATA-1 at selected cis-regulatory chromatin elements in hematopoietic cells via a mechanism involving the switching of GATA-1 for GATA-2 (21, 22). However, the molecular details of this activity are unknown. FOG family proteins also interact with at least two transcriptional corepressor complexes: the C-terminal binding protein (CtBP) and the NuRD chromatin remodeling/histone deacetylase transcriptional complexes (23–27). The physiological significance of CtBP binding is not clear given that homozygous FOG-1 knock-in mice containing mutations that disrupt CtBP binding are viable, fertile, and have normal hematopoiesis (24). Interaction with the NuRD complex has not been fully examined in vivo as of yet.
Work over the past several years has shown that GATA-1 also plays a functional role in the terminal stages of mast cell maturation, particularly on expression of the high affinity IgE receptor (FcεRI) (28–32). A previous study from our group showed that FOG-1 expression is absent in cultured mast cell lines, suggesting a possible FOG-independent GATA-1 function in the mast cell lineage (6). In this study, we extend these findings using primary cells and gene-targeted mice to show that indeed GATA-1 functions independent of FOG protein in terminal mast cell maturation. Moreover, we show that FOG-1 potently represses cell fate choice for the mast cell lineage during early multipotent progenitor cell lineage commitment. Remarkably, ectopic expression of FOG-1 in prospectively isolated mast cell progenitors (MCPs) redirects them into erythroid, megakaryocytic, and granulocytic lineages. Collectively, these observations identify FOG-1 as a key negative regulator of mast cell lineage choice, and demonstrate combinational control of cell fate decisions by a transcription factor and its cofactor.
RESULTS
FOG-independent function of GATA-1 in mast cell development
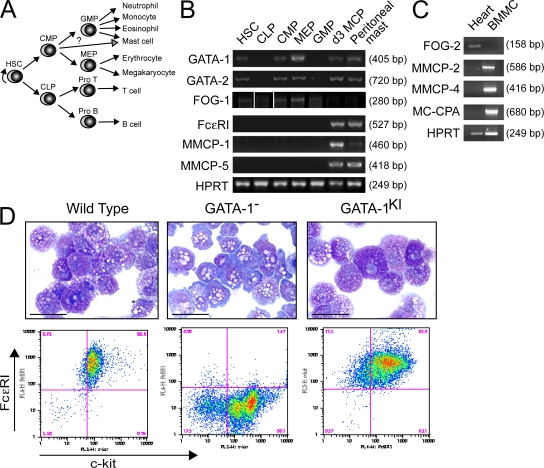
Previously, we reported that FOG-1 expression is absent in the mouse mast cell lines P615 and HC.3 (6). However, its expression in primary mast cells has not been reported. We recently identified a bipotent basophil/MCP cell population that can be immunophenotypically isolated from the mouse spleen (Lin−c-kit+FcγRII/IIIhiβ7hiFcεRIα−/lo) (33). These cells develop exclusively into mast cells and basophils when cultured in a cocktail of cytokines that includes stem cell factor (SCF), IL-3, IL-5, IL-6, IL-7, IL-9, IL-11, GM-CSF, erythropoietin (EPO), and thrombopoietin (TPO). In the present study, we used this system to examine the expression patterns of FOG-1, GATA-1, and GATA-2 as primary cells commit to the mast cell/basophil lineage. As shown in Fig. 1, RT-PCR analysis reveals expression of GATA-1, GATA-2, and FOG-1 in hematopoietic stem cells, common myeloid progenitors, and megakaryocyte/erythroid progenitors (MEPs), and low levels of FOG-1 in common lymphoid progenitors and granulocyte/macrophage progenitors (GMPs; along with low levels of GATA-2). In contrast, MCPs cultured for 3 d in the cytokine cocktail described earlier in this paragraph express relatively high levels of GATA-1 and GATA-2 but no detectable FOG-1. The same is true of primary peritoneal mast cells. Thus, FOG-1 expression is specifically down-regulated as multipotent progenitor cells commit to the mast cell lineage.
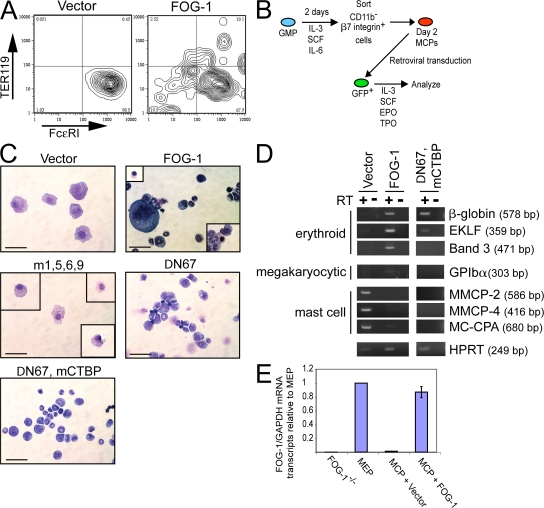
Figure 1.
GATA-1 functions independent of FOG cofactors in mast cell development. (A) Schematic diagram showing hierarchical relationships of the hematopoietic progenitor populations examined in B. (B) RT-PCR analysis of GATA-1, GATA-2, and FOG-1 expression in sorted progenitor cell populations, day 3 MCPs (d3 MCP), and peritoneal mast cells. FcεRI, MMCP-1, and MMCP-5 are included as mast cell–specific marker genes. HPRT serves as the housekeeping gene control. White lines indicate that intervening lanes have been spliced out. (C) RT-PCR analysis of FOG-2 expression in BMMCs (from 6-wk-old cultures) or whole mouse heart tissue. MMCP-2, MMCP-4, and MC-CPA are included as mast cell–specific markers. (D) May-Grunwald-Giemsa stains and FACS analysis for FcεRI and c-kit expression of YSMCs from wild-type, GATA-1−, or GATA-1V205G(KI) male embryos. Bars, 10 μm.
FOG-2, the only other known mammalian FOG gene, is thought to be expressed predominantly outside of the hematopoietic system (15, 34, 35). However, to rule out a possible role in mast cell development, we performed RT-PCR analysis for FOG-2 expression in primary mouse bone marrow–derived mast cells (BMMCs). As shown in Fig. 1 C, we found no detectable FOG-2 mRNA transcripts in these cells despite robust signals for the mast cell genes mouse mast cell protease 2 (MMCP2), MMCP-4, and mast cell carboxypeptidase A (MC-CPA) in the same sample, and amplification of FOG-2 mRNA transcripts from control heart tissue.
It is possible that FOG genes are expressed at levels below our detection limit or that yet additional FOG genes exist. To examine these possibilities, we analyzed yolk sac–derived mast cells (YSMCs) from knock-in mice containing substitution of valine 205 of GATA-1 by glycine (GATA-1V205G). This mutation markedly impairs FOG-1 binding, resulting in lethal anemia and impaired megakaryopoiesis in mice (8, 9). A similar mutation (GATA-1V205M) causes severe X-linked dyserythropoietic anemia and thrombocytopenia in humans, and the homologous mutation in GATA-4 (GATA-4V217G) blocks binding to FOG-2 (11, 19). Because of the embryonic lethality of the mice, it was not possible to examine BMMCs. Instead, we cultured embryonic day 9.5 YSMCs in the presence of IL-3 and SCF for 6 wk, conditions typically used to generate BMMCs. These cells morphologically resemble BMMCs and express equivalent levels of the mast cell genes MC-CPA, MMCP-4, MMCP-5, tryptophan hydroxylase, and microphthalmia-associated transcription factor (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20070544/DC1). As shown in Fig. 1 D, YSMCs cultured from wild-type embryos are highly granular in appearance and nearly all coexpress c-kit and FcεRI on their cell surface. In contrast, YSMCs from male GATA-1− embryos are hypogranular and have markedly impaired FcεRI surface expression, consistent with previous reports of GATA-1low mast cells (28–31). A smaller population of c-kit−FcεRI− cells is also present in these cultures. Importantly, cells cultured from GATA-1V205G male embryo yolk sacs resemble those derived from wild-type, and not GATA-1−, embryos in morphology and FcεRI surface expression. We conclude that, in contrast to the erythroid and megakaryocyte lineages, GATA functions completely independent of FOG proteins in terminal mast cell development.
Inhibition of mast cell development by FOG-1 in vitro and in vivo
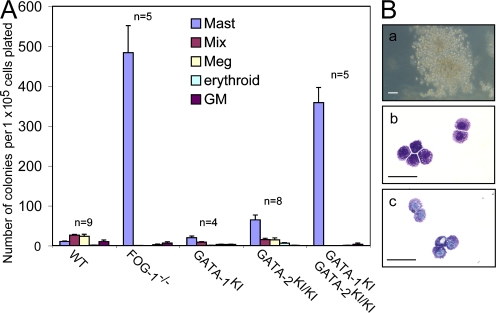
Could FOG-1 actually antagonize mast cell development? This possibility was suggested by the results of yolk sac colony assays from FOG-1−/− mouse embryos. When yolk sac cells from embryonic day 9.5 FOG-1−/− embryos are grown under conditions supporting multilineage growth (EPO, TPO, and SCF), there is a marked expansion of mast cell colonies (∼47-fold) compared with the wild type (Fig. 2 A). Identification of the colonies was based on cell morphology and positive Toluidine blue staining (Fig. 2 B). The absolute increase in mast cell colony numbers (473 ± 69 per 105 cells plated) is significantly greater than the additive loss of mixed, erythroid, and megakaryocytic colonies (48 ± 9 per 105 cells plated), suggesting a true expansion of MCPs rather than a simple defaulting of multipotent progenitors to the mast cell lineage. These results imply that FOG-1 represses mast cell lineage commitment, possibly at the level of common myeloid progenitors or GMPs.
Figure 2.
Loss of FOG-1 results in an increased number of YSMC progenitors in vivo. (A) Yolk sac cultures from WT, FOG-1−/−, GATA-1V205G(KI) male, GATA-2V296G/V296G(KI/KI), and compound GATA-1KI, GATA-2KI/KI male embryonic day 9.5 embryos. Shown are numbers of mast cell, mixed (Mix), megakaryocytic (Meg), erythroid, and GM colonies per 105 cells plated and cultured in 2 IU/ml EPO, 5 ng/ml TPO, and rat 50 ng/ml SCF for 7 d. The number of animals analyzed for each is indicated above the graphs. Error bars represent the SEM. (B) Colony and cell morphology of mast cell colonies from GATA-1KI, GATA-2KI/KI male embryos. (a) Brightfield appearance of colony. Bar, 50 μm. (b) May-Grunwald-Giemsa stain of cytospun cells. Bar, 10 μm. (c) Toluidine blue stain of cytospun cells. Bar, 10 μm.
To determine whether this inhibition requires direct physical interaction between FOG-1 and GATA-1 and/or GATA-2, we performed yolk sac colony assays from embryos containing knock-in mutations that disrupt the binding of FOG-1 to either GATA-1 (GATA-1V205G) and/or GATA-2 (GATA-2V296G) (9). As shown in Fig. 2 A, there is a slight expansion of mast cell colony numbers compared with the wild type in hemizygous GATA-1V205G males (∼1.9-fold) and a moderate expansion in GATA-2V296G/V296G embryos (∼6.2-fold), but a marked expansion in compound GATA-1V205G, GATA-2V296G/V296G embryos (35-fold) to levels similar to those of FOG-1−/− embryos. These results indicate that direct binding of FOG-1 to GATA factors is required for its inhibitory activity on mast cell lineage commitment, and that GATA-1 and GATA-2 play overlapping roles. This mirrors the overlapping FOG-dependent roles of GATA-1 and GATA-2 in early megakaryocytic development (9).
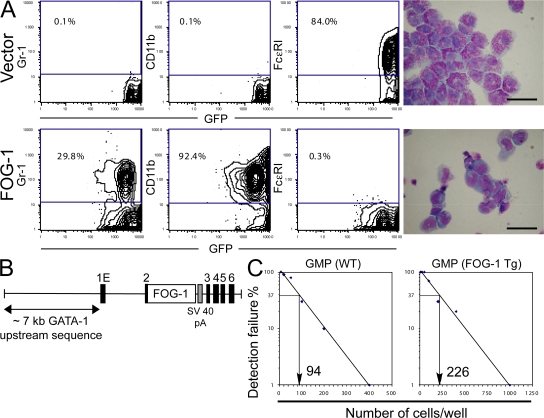
To test directly whether FOG-1 represses mast cell development, we enforced FOG-1 expression in prospectively isolated GMPs using retroviruses and examined the ability of the cells to form mast cells in vitro in a limiting dilution assay (Fig. 3). The retroviral vectors used coexpress enhanced GFP (eGFP) as a bicistronic mRNA, allowing for tracking of the transduced cells. As shown in Fig. 3 A, nearly all of the GFP+ GMPs transduced with the empty control vector and cultured for 3 wk in the presence of IL-3, IL-6, IL-9, and SCF express high levels of FcεRI, contain granular cytoplasm, and lack expression of the granulocytic and monocyte/macrophage markers Gr-1 and CD11b. In contrast, GMPs transduced with FOG-1–expressing retroviruses (GFP+) are almost entirely FcεRI−, lack significant granularity, and express CD11b and, to a lesser extent, Gr-1 when cultured under identical conditions.
Figure 3.
Enforced expression of FOG-1 blocks mast cell development. (A) FACS expression analysis for eGFP, Gr-1, CD11b, or FcεRI in GMPs transduced with retroviruses expressing eGFP alone (vector) or FOG-1–IRES-GFP (FOG-1), and cultured at limiting dilution in the presence of SCF, IL-3, IL-6, and IL-9 for 3 wk. Percentages of total cells that fall within the boxed FACS gates are shown. May-Grunwald-Giemsa stains of the GFP+ cells are shown on the right. Bars, 10 μm. (B) Schematic representation of the transgene used to express FOG-1 in the mast cell lineage. Approximately 7 kb of DNA 5′ to the first erythroid exon (1E) of GATA-1 (noncoding) were used. Full-length FOG-1 cDNA was inserted into exon 2 of GATA-1 upstream of the GATA-1 start codon, followed by an SV40 polyA sequence (gray box). Exons are indicated by black boxes. (C) Limiting dilution analysis of bone marrow GMPs from FOG-1–transgenic mice or wild-type littermates. The percentage of wells that failed to generate mast cells was scored, and the number of cells per well leading to a 37% detection failure rate was calculated.
We also examined the effect of enforced FOG-1 expression on mast cell development in vivo. A transgene containing ∼7 kb of DNA sequence upstream of the first erythroid exon of GATA-1 is sufficient to drive expression in erythroid and megakaryocytic cells and, albeit somewhat weakly, in mast cells and eosinophils (36–38). Because no other well characterized mast cell–specific transgenic promoters have been reported, we chose to use this ∼7-kb GATA-1 upstream sequence to examine the consequences of enforced FOG-1 expression on mast cell lineage commitment (Fig. 3 B). We previously generated FOG-1 transgenic mice using this regulatory region and demonstrated that it rescues the hematopoietic defects of FOG-1−/− mice (39). In this study, we examined the same transgenic line for mast cell production. GMPs were isolated from bone marrow, plated under mast cell growth conditions, and quantitated for mast cell growth by limiting dilution. An ∼2.5-fold greater number of GMPs was required to generate that same number of single cell mast cell clones from these transgenic mice compared with nontransgenic littermates, suggesting a reduction in the number of MCPs in the transgenic mice (Fig. 3 C). The somewhat modest effect, relative to the in vitro studies, is likely caused by relatively weak expression of the transgene in the mast cell lineage. Nonetheless, our data collectively indicate that down-regulation of FOG-1 expression is a prerequisite for mast cell development.
Reprogramming of primary mast cells into alternate lineages by ectopic expression of FOG-1
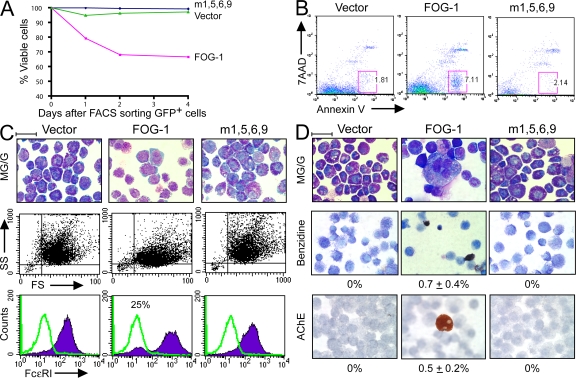
In the preceding experiments, FOG-1 expression was enforced in early multipotent progenitor cells. We next examined the consequences of ectopic FOG-1 expression in cells already committing to the mast cell lineage. In the first set of studies, we generated primary mast cells by culturing whole bone marrow in the presence of IL-3 and SCF for 6 wk. FACS analysis of this starting cell population showed 100% of the cells were c-kit+, Ter119− (erythroid marker), and 96% were FcεRI+ (not depicted). We then infected them with retroviruses packaged with the empty vector, FOG-1, or a missense FOG-1 mutant with impaired GATA factor binding (m1,5,6,9) (40). We previously showed that the m1,5,6,9 FOG-1 protein is stable and properly targeted to the nucleus when expressed in a FOG-1–null hematopoietic cell line (10). All of the constructs coexpress eGFP as a bicistronic mRNA. 2 d after retroviral infection, GFP+ cells were sorted and incubated in the presence of IL-3, SCF, EPO, and TPO for various time points. Cells transduced with the empty vector or the non-GATA–binding FOG-1 mutant retained nearly 100% viability throughout the first 5 d of culture, as measured by vital dye exclusion (Fig. 4 A). In contrast, only ∼70% of the FOG-1–transduced cells survived. Some of the cell death occurs via apoptosis, as indicated by an increased number of Annexin V+/7-amino-actinomycin D− cells (41) in the FOG-1 compared with the empty vector and m1,5,6,9 FOG-1–transduced samples (7.1 vs. 1.8 vs. 2.1%, respectively) when examined 3 d after sorting (Fig. 4 B).
Figure 4.
Ectopic expression of FOG-1 in BMMCs represses mast cell phenotype and produces alternate lineage cells. (A) Percentage of viable GFP+ primary BMMCs retrovirally transduced with eGFP alone (vector), FOG-1–IRES-eGFP (FOG-1), or m1,5,6,9 FOG-1–IRES-GFP (m1,5,6,9). Viability was determined by Trypan blue dye exclusion. Time refers to days after FACS sorting of eGFP+ cells (2 d after retroviral infection). (B) FACS analysis for Annexin V (apoptosis marker) and 7-amino-actinomycin D (necrosis marker) expression of primary BMMCs transduced with each of the retroviral vectors described in A and examined 3 d after FACS sorting of GFP+ cells. Percentages of total cells that fall within the boxed FACS gates are shown. (C) May-Grunwald-Giemsa stains, FACS analysis showing side-scatter (SS) and forward scatter (FS), and FcεRI expression of primary BMMCs transduced with each of the retroviral vectors shown in A, sorted for GFP after 2 d, and cultured for 6 d in 2 U/ml EPO, 5 ng/ml TPO, 10 ng/ml IL-3, and 50 ng/ml SCF. (bottom) The green lines represent negative control staining for FcεRI (no IgE added; see Materials and methods). The purple histograms represent staining for FcεRI (IgE added). The percentage in the middle panel represents the proportion of total stained cells present within the negative staining control peak area. Bar, 10 μm. (D) May-Grunwald-Giemsa, o-dianisidine (benzidine), and acetylcholinesterase (AChE) histochemical stains of the cultures in C incubated for an additional 5 d. Benzidine+ cells stain black/brown; AChE+ cells stain orange. The percentage of positive-staining cells is indicated (±SEM; n = 2; 5,000 cells examined for all samples). Bar, 10 μm.
By 6 d after FACS sorting, cells transduced with either the empty vector or the non-GATA–binding FOG-1 mutant show typical mast cell morphology by May-Grunwald-Giemsa stains. They are highly granular, as indicated by prominent side scatter on FACS analysis, and nearly all express FcεRI on their cell surface (Fig. 4 C). In contrast, many of the FOG-1–transduced cells appear hypogranular, with pale blue cytoplasm and large nuclei with an open chromatin pattern. Loss of granularity is evident by FACS analysis, which shows a marked reduction in cells with high side scattering compared with the control cells (after gating for viable cells). In addition, ∼25% of the viable FOG-1–transduced cells lack surface expression of FcεRI.
Remarkably, after culturing for an additional 5 d in the presence of EPO, TPO, IL-3, and SCF, cells resembling maturing erythrocyte, megakaryocytic, and granulocytic cells became evident in the FOG-1–transduced sample (Fig. 4 D). This was confirmed by staining with benzidine, which turns hemoglobinized cells a dark brown color, and for acetylcholinesterase, a marker of mouse megakaryocytes. Although the frequency of positive-staining cells was quite low (0.7 ± 0.4% benzidine positive and 0.5 ± 0.2% acetycholinesterase positive of 5,000 cells examined each), no positive cells were observed in >5,000 vector or m1,5,6,9-transduced cells examined.
One interpretation of these results is that ectopic FOG-1 expression reprograms “committed” MCPs into the erythroid, megakaryocytic, and granulocytic lineages. However, based on the results so far, we could not exclude the possibility that our cultures contain rare contaminating multilineage progenitors, and that ectopic FOG-1 expression simply selects against cells committing to the mast cell lineage and for those committing to the erythroid, megakaryocytic, and granulocytic pathways. This seems unlikely because the starting cells were cultured for 6 wk in the presence of SCF and IL-3, conditions not expected to sustain multipotent progenitor cells. However, we took two approaches to address this possibility. First, we repeated the experiment using clonally derived primary mast cells. Because primary mast cells can be cultured for extended periods of time, it is possible to clone them at the single cell level. Whole bone marrow was cultured in the presence of IL-3 and SCF for 2 wk, and the cells were cloned by limiting dilution. This was the latest time point possible to begin the cloning and still generate enough cells for retroviral infection and analysis before they senesced. A single cell clone was expanded in IL-3 and SCF for an additional 4 wk. These cells were then infected with the empty vector or FOG-1–expressing retroviruses, sorted for GFP expression after 2 d, and cultured in IL-3, SCF, EPO, and TPO for an additional 6 d. They were then analyzed by FACS analysis for surface expression of FcεRI and Ter-119, an erythroid specific marker. As shown in Fig. 5 A, essentially 100% of the vector-transduced cells were FcεRI+ Ter-119−, which was consistent with the starting population being a clonal mast cell line. In contrast, 22% of the FOG-1–transduced cells were FcεRI−, 12.6% were Ter-119+, and 2.5% were both FcεRI− and Ter-119+. Interestingly, a continuum of FcεRI–Ter-119–coexpressing cells was observed, suggesting an overlapping transition from FcεRI to Ter-119–expressing cells.
Figure 5.
Ectopic expression of FOG-1 in MCPs blocks mast cell maturation and enables alternate lineage development. (A) FACS analysis for TER119 and FcεRI expression of clonal primary BMMCs retrovirally transduced with empty vector or FOG-1–IRES-eGFP–expressing retroviruses. GFP+ cells were sorted 2 d after retroviral infection and cultured in EPO, TPO, IL-3, and SCF for 6 d. (B) Scheme for isolation and retroviral infection of day 2 MCPs. (C) May-Grunwald-Giemsa stains of day 2 MCPs transduced with empty vector, FOG-1, or the FOG-1 mutants m1,5,6,9, ΔN67, and ΔN67, mCTBP expressing retroviruses; cultured in mast cell media containing EPO, TPO, SCF, and IL-3 for 4 d; sorted for GFP expression (GFP+ cells); and cultured for an additional of 5 d in mast cell media containing EPO, TPO, SCF, and IL-3. Boxes indicate cells seen in additional fields. Bars, 10 μm. (D) RT-PCR analysis for erythroid, megakaryocytic, and mast cell marker gene expression in the cells shown in C (harvested 5 d after sorting GFP+ cells). (E) Quantitative RT-PCR analysis for FOG-1 mRNA transcript levels in day 2 MCPs retrovirally transduced with the empty vector or FOG-1, sorted for GFP+ expression after 4 d, and cultured for an additional 5 d in the presence of SCF, IL-3, EPO, and TPO. Levels are displayed relative to those determined in parallel from sorted mouse MEPs and normalized to GAPDH mRNA transcript levels. The relative signal from FOG-1−/− cells (reference 10) is shown as an indicator of experimental background. All measurements were made in triplicate. The error bars represent the SEM.
As a second approach, we prospectively isolated cell populations highly enriched in MCP cells (day 2 MCPs, which were GMPs cultured for 2 d in IL-3, SCF, and IL-6 and sorted for β7hiCD11b− expression) and repeated the FOG-1 transduction experiment (Fig. 5 B). GFP+ cells were sorted after 4 d, and the cells were cultured for an additional 5 d in the presence of SCF, IL-3, EPO, and TPO. As shown in Fig. 5 (C and D), cells transduced with the empty vector have a granular appearance, express the mast cell–specific genes MMCP-2, MMCP-4, and MC-CPA, and lack expression of the erythroid- and megakaryocyte-specific genes β-globin, band 3, erythroid Kruppel-like factor (EKLF), and glycoprotein Ibα (GPIbα) by RT-PCR analysis. Consistent with our earlier observation (Fig. 4), most of the cells transduced with the FOG-1–expressing retroviruses lack typical mast cell morphology, and many resemble cells maturing along the erythroid, megakaryocytic, and granulocytic lineages. RT-PCR analysis shows markedly diminished expression of MMCP-2, MMCP-4, and MC-CPA, and activation of β-globin, band 3, EKLF, and GPIbα. Approximately 4% of the cells were benzidine positive, and 5% expressed acetylcholinesterase based on histochemical staining (not depicted). These higher levels compared with those from 6-wk-old primary BMMCs (Fig. 4) are likely caused by an increased proportion of more immature MCPs, which may be more amenable to lineage reprogramming. Quantitative RT-PCR analysis of the retrovirally transduced MCPs shows FOG-1 expression levels close to those found physiologically in MEPs (Fig. 5 E). As expected, only background levels of the FOG-1 signal were detectable in the MCPs transduced with the empty vector based on comparison to FOG-1−/− cells. Collectively, these data strongly suggest that ectopic expression of FOG-1 redirects committed MCP cells into alternate lineages.
Requirement for interaction with GATA factors but not CtBP and NuRD corepressor complexes
Next, we wanted to explore the mechanism of FOG-1–mediated mast cell repression and lineage reprogramming. First, we tested whether this activity requires direct binding of FOG-1 to GATA factors. As shown in Fig. 5 C, the non-GATA–binding FOG-1 mutant m1,5,6,9 was inactive in day 2 MCP reprogramming, which was consistent with our earlier observations in YSMCs (Fig. 2) and bulk BMMC cultures (Fig. 4). Thus, direct binding to GATA factors appears necessary. We next tested whether interaction between FOG-1 and one or both of its known transcriptional corepressor partners, the NuRD and CtBP complexes, is required. We first examined a truncation FOG-1 mutant molecule (FOG-1 ΔN67) that lacks the N-terminal 67 amino acids, a region that includes the entire NuRD interaction motif (25). This molecule behaved similarly to the full-length protein when expressed in the day 2 MCPs, although megakaryocytic development was not observed, as expected (Fig. 5 C) (10). We then tested a double mutant that also substitutes the CtBP interaction motif PIDLS with the nonbinding sequence PIASS (ΔN67, mCTBP) (24). This construct also blocks mast cell development and leads to activation of some erythroid-specific genes (Fig. 5, C and D). Although expression levels of these mutants were considerably higher than endogenous FOG-1 levels in MEPs (∼18-fold higher for ΔN67 and ∼150–fold higher for ΔN67, mCTBP as measured by quantitative RT-PCR analysis), these results suggest that mechanisms other than or overlapping with transcriptional corepression via the NuRD and CtBP complexes are responsible for FOG-1's antagonistic role in mast cell development.
FOG-1–mediated down-regulation of GATA-2 in mast cells
High level GATA-2 expression is required for mast cell development (28, 42). Grass et al. have studied the transcriptional regulation of GATA-2 and identified a critical enhancer element located ∼2.8 kb upstream of the 1S promoter that contains phylogenetically conserved GATA consensus binding sequences (43). Based on ChIP and transcriptional studies, they proposed a model in which GATA-2 occupies this site in early multipotent progenitor cells and autoactivates GATA-2 gene transcription. As cells mature along the erythroid pathway and GATA-1 levels increase, occupancy of this enhancer element switches from GATA-2 to GATA-1, leading to repression of GATA-2 transcription. In collaborative studies, we showed that this GATA factor switch depends on the presence of FOG-1 (21). Moreover, FOG-1 rapidly and potently represses GATA-2 expression in reconstituted FOG-1−/− cells, and the non-FOG–binding GATA-1 mutant GATA-1V205G fails to repress GATA-2 transcription in reconstituted GATA-1–null erythroid cells (8, 21).
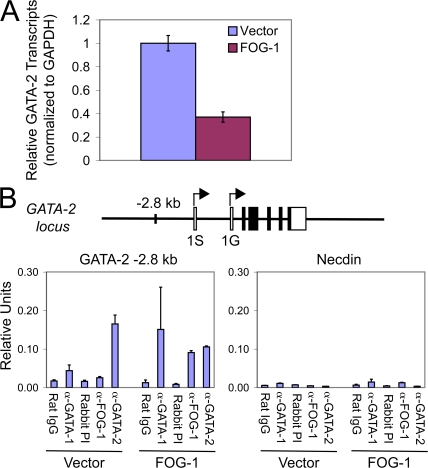
Given GATA-2's essential role in mast cell development, we hypothesized that FOG-1's antagonist activity in mast cell development could be in part caused by GATA factor switching at the GATA-2 −2.8-kb enhancer and subsequent repression of GATA-2 transcription (42, 44). To test this, we examined changes in GATA-2 mRNA transcript levels at the earliest time point possible after FOG-1 retroviral overexpression in GMPs. As shown in Fig. 6 A, we observed a 2.7-fold reduction in GATA-2 transcripts by quantitative RT-PCR in FOG-1– versus empty vector–transduced GMPs sorted 2 d after retroviral infection for GFP expression and analyzed immediately. Because there are insufficient numbers of cells to perform ChIP assays in GMPs, we expressed FOG-1 or the empty vector in BMMCs and examined occupancy of the −2.8-kb region immediately after sorting GFP+ cells 2 d after infection. As shown in Fig. 6 B, control cells (empty vector) contain high levels of GATA-2, low levels of GATA-1, and no detectable FOG-1 (over background) at the −2.8-kb region, as expected. In contrast, significant levels of FOG-1 are detectable at the −2.8-kb site in the FOG-1–transduced cells, and there is a marked increase in GATA-1 and a decrease in GATA-2 occupancy compared with control cells. Only background levels of GATA-1, GATA-2, and FOG-1 were found at the control Necdin promoter, which does not contain GATA binding sites, in both vector- and FOG-1–transduced cells. Thus, FOG-1–mediated GATA factor switching at the −2.8-kb GATA-2 enhancer occurs as an early event after ectopic expression in mast cells. Consequential down-regulation of GATA-2 expression may therefore explain, at least in part, the mechanism of FOG-1's antagonistic activity in mast cell lineage commitment.
Figure 6.
FOG-1–mediated down-regulation of GATA-2 expression correlates with GATA factor switching at the −2.8-kb GATA-2 enhancer element in mast cells. (A) Quantitative RT-PCR analysis of GATA-2 mRNA transcript levels in GFP+ GMPs infected with retroviruses expressing either the empty vector (IRES-GFP) or FOG-1–IRES-GFP, sorted after 2 d, and analyzed immediately. Error bars represent SEM (n = 3). (B) ChIP assay examining occupancy of the GATA-2 −2.8-kb enhancer or the Necdin promoter by GATA-1, GATA-2, and/or FOG-1 in 4-wk-old BMMCs retrovirally transduced with either the empty vector (IRES-GFP) or FOG-1–IRES-GFP and sorted for GFP+ cells after 2 d. A schematic drawing of the mouse GATA-2 locus with the −2.8-kb enhancer element is indicated. 1S and 1G refer to different transcriptional start sites. Normal pooled rat IgG was used as the control for GATA-1 ChIP, and FOG-1 preimmune rabbit serum served as the control for FOG-1 and GATA-2 ChIPs. The level of detected amplicon was normalized to a standard curve of input chromatin and is indicated as relative units. Error bars represent the SEM (n = 2).
DISCUSSION
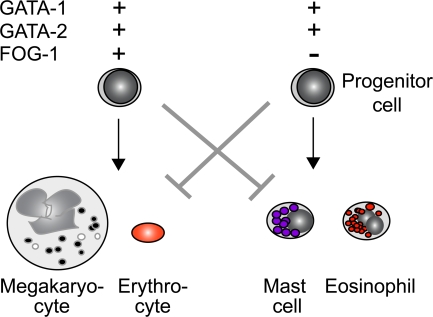
In this study, we use both loss- and gain-of-function approaches to demonstrate that FOG-1 acts in a combinatorial manner with GATA factors to specify the cell fate of multipotent hematopoietic progenitor cells (Fig. 7). Although FOG-1 acts cooperatively with GATA factors in erythroid and megakaryocytic differentiation, it potently antagonizes GATA function in mast cell lineage commitment. This requires direct GATA factor binding and correlates with transcriptional down-regulation of GATA-2 via a GATA factor switching mechanism. Moreover, we show that FOG-1, as a single factor, is capable of redirecting committed primary MCP cells into alternate lineage fates. This provides evidence for lineage plasticity during at least the early stages of hematopoietic lineage commitment.
Figure 7.
Model of combinatorial control of erythrocyte/megakaryocyte versus mast cell/eosinophil lineage commitment by GATA-1 and GATA-2 depending on the presence of FOG-1. + and − refer to the presence or absence, respectively, of expression of each gene. The arrows represent the favored differentiation pathway, and the gray “T” bars represent the blocked differentiation pathway.
Down-regulation of FOG-1 in eosinophil development
Our results are strikingly similar to those reported by Querfurth et al. in their studies of eosinophil development (45). Both eosinophils and mast cells require GATA-1 for complete maturation (46). Like mast cells, FOG-1 expression is extinguished as eosinophils differentiate from multipotent progenitor cells, and this is a prerequisite for their development. Moreover, ectopic expression of FOG-1 in an eosinophilic cell line changes it to a more primitive state. These findings highlight important parallels in the transcriptional control of eosinophil and mast cell development (Fig. 7).
Combinatorial control of hematopoietic cell fate decisions by the Drosophila GATA factor Serpent (Srp) and the FOG orthologue U-shaped (Ush)
The combinatorial control of hematopoietic cell fate determination by GATA and FOG described in this paper is also reminiscent of that reported in studies of Drosophila hematopoiesis. The Drosophila hematopoietic system consists of multipotent hemocyte progenitor cells that give rise to one of three specialized cell types: plasmatocytes, crystal cells, and lamellocytes, all of which function in immune responses (for review see reference 47). Remarkably, most of the same transcription factors required for mammalian hematopoiesis also play essential roles in Drosophila blood development (48). The GATA orthologue Srp is required for hemocyte precursor, crystal cell, and plasmatocyte development. The FOG orthologue Ush is required for hemocyte precursor formation and plasmatocyte differentiation. However, it is selectively down-regulated during crystal cell differentiation (49). This is essential for crystal cell formation, because enforced Ush expression ablates crystal cell development, paralleling our findings of blocked mast cell development by enforced FOG-1 expression (Fig. 3). Conversely, loss-of-function Ush mutants contain marked expansion of crystal cells and a reduced number of plasmatocytes, similar to the enhanced mast cell and impaired erythroid/megakaryocyte production we observe in FOG-1−/− yolk sac cultures (Fig. 2). As in our findings, the effects in Drosophila require direct binding of Ush to Srp, because an isoform of Srp that lacks the equivalent N-terminal zinc finger of vertebrate GATA factors is immune to the inhibitory effects of Ush (50).
Collectively, these studies underscore how combinations of GATA and FOG family members control hematopoietic cell fate decisions. Given the broad developmental requirement for interaction between GATA and FOG family members, such as GATA-4 and FOG-2 in cardiac and gonadal development, it is tempting to speculate that combinatorial GATA–FOG control mechanisms are involved in nonhematopoietic cell fate decisions as well.
Combinatorial control of cell fate decisions by transcription factors and cofactors
Transcription factor combinatorial control mechanisms of cell fate decisions have been appreciated for some time. However, few examples exist involving a transcription factor and a cofactor (i.e., transcription factor protein–protein interactions). One could imagine several possible mechanisms by which such interactions could affect gene programs in a lineage-specific manner: (a) the cofactor could alter the DNA affinity and/or sequence specificity of the transcription factor; (b) the cofactor could enzymatically alter the local chromatin structure, affecting the accessibility of other transcription factors; (c) the cofactor could alter the locus conformation by bridging the transcription factor to another sequence-specific DNA binding protein; and (d) the cofactor could sequester the transcription factor.
Our data are consistent with a novel mechanism in which FOG-1 mediates the switching of two transcription factor family members at specific loci to yield distinct transcriptional outcomes. The molecular details of this switching event remain to be determined. Our data do not exclude the possibility of additional mechanisms operating. Vakoc et al. recently used chromosome conformation capture to show that interaction between GATA-1 and FOG-1 is required for spatial proximity of the distant upstream β-globin locus control region to the β-globin gene proximal promoter during erythroid development (51). This DNA looping event correlates with the onset of β-globin transcription. The absence of FOG-1 in developing mast cells would preclude such a locus reconfiguration and prevent efficient β-globin gene expression.
FOG proteins interact with at least two transcriptional corepressor complexes, NuRD and CtBP. Rodriguez et al. demonstrated promoter cooccupancy of the eosinophilic-specific gene MBP by GATA-1, FOG-1, and the NuRD component MBD3 in induced mouse erythroleukemia (MEL) cells, which was consistent with FOG-1–mediated repression of eosinophil-specific genes in erythroid cells via NuRD (26). We hypothesized that FOG-1's repressive activity in mast cell development involves its recruitment of one or both of these complexes. However, a double mutant molecule incapable of binding either the NuRD or CtBP complexes was as potent as the wild-type molecule in repressing mast cell development from MCPs (Fig. 5). Our results are consistent with a recent report showing that a double ΔN, ΔCtBP FOG-1 mutant represses GATA-1–dependent activation of the FcεRI-β promoter as well as wild-type FOG-1 in transient transfection reporter assays in the PT18 mast cell line (52). These results suggest that mechanisms other than recruitment of the NuRD and/or CtBP complexes are responsible for FOG-1's inhibitory activity on mast cell development.
Regulation of FOG-1 expression
Regardless of the mechanism, our data, in combination with that of Querfurth et al., indicate that repression of FOG-1 expression is critical for mast cell and eosinophil differentiation from multipotent progenitors (45). This begs a question: what turns off FOG-1 during the development of these lineages? The transcriptional regulation of FOG-1 is just beginning to be explored. Welch et al. used bioinformatics to predict critical cis-regulatory elements controlling FOG-1 expression (53). They showed that one such region, predicted cis-regulatory module 1, is occupied by GATA-1 in erythroid cells, which is consistent with their data indicating transcriptional up-regulation of FOG-1 by GATA-1. We examined this same region in primary mast cells and instead found relatively low GATA-1 but high GATA-2 occupancy compared with induced MEL cells (unpublished data). In addition, histone 3 is markedly hypoacetylated in this region, compared with induced MEL cells, consistent with epigenetic silencing of FOG-1 in mast cells. Whether high level GATA-2 occupancy at predicted cis-regulatory module 1 is responsible for FOG-1 down-regulation will require further studies. However, given our findings that FOG-1 represses GATA-2 expression (Fig. 6) (8, 21), it is possible that cross-antagonistic regulatory loops exist between GATA-2 and FOG-1. In this model, high GATA-2 levels silence FOG-1 expression in maturing mast cells, leading to enhanced GATA-2 expression and further FOG-1 repression. Conversely, high FOG-1 levels in maturing erythroid/megakaryocytic cells inhibit GATA-2 expression, leading to further elevation of FOG-1 and reduced GATA-2 expression.
Mast cells in human disease
Mast cells play important roles in human disease, including asthma and type I hypersensitivity reactions. They have also recently been implicated as key intermediaries of regulatory T cell–mediated immune tolerance (54). Our data provide new insights into the molecular control of mast cell development and suggest possible novel approaches to manipulating mast cell–related processes. In theory, derepression of FOG-1 in maturing MCPs should inhibit their production and, possibly, alleviate the secondary effects of mast cell release.
Mastocytosis refers to a heterogeneous group of disorders associated with inappropriate or nonatopic expansion of mast cells. It ranges in clinical severity from localized cutaneous mastocytosis to aggressive systemic mastocytosis. Activating mutations in c-kit or interstitial chromosome 4 deletions that generate a constitutively active FIP1LI–PDGFA fusion receptor tyrosine kinase have been identified in many, but not all, cases of systemic mastocytosis (55–57). Based on our data, it is intriguing to speculate that acquired FOG-1 loss-of-function mutations in early multipotent progenitor or stem cells might also cause or contribute to clonal mast cell expansion in some cases, particularly those with associated eosinophilia and/or myelodysplasia. Examination of such a possibility seems warranted.
Lineage plasticity during the early stages of hematopoietic differentiation
The classical view of hematopoietic development is one in which multipotent progenitor cells irreversibly commit to distinct lineage types during differentiation. However, beginning with the work of Boyd and Schrader (58), the irreversibility of these decisions has been questioned (59–63). Although most of the early studies used transformed cell lines, more recent work using primary cell and in vivo systems support the initial finding (64–69). Our results provide further evidence for lineage plasticity during at least the early stages of hematopoietic cell fate commitment. Although we cannot completely exclude the possibility of selection of rare contaminating multipotent progenitors cells in our system, several aspects of our findings suggest that the FOG-1–mediated block in mast cell development and the appearance of alternate lineage cells represents true “reprogramming.” First, MCPs were prospectively isolated in two rounds of immunophenotypic cell sorting before FOG-1 transduction. Second, FOG-1 reprogramming activity was observed in clonal primary mast cells (and the parent clone was 100% FcεRI positive and Ter119 negative after culture in multilineage-supporting cytokine cocktails). Third, the block in mast cell development and the appearance of abundant alternate lineage cells appears rapidly (over 6–11 d), which seems unlikely if it represents the selection of a very rare contaminating multipotent progenitor cell in the original retrovirally infected cell population.
Although it seems unlikely that FOG-1–induced lineage switching of MCPs occurs physiologically, the findings demonstrate the potential for such plasticity. Whether this occurs via direct transdifferentiation of MCPs to erythroid/megakaryocytic cells or via dedifferentiation followed by redifferentiation along erythroid/megakaryocytic pathways remains to be determined.
Our results, collectively with those of others described in this section, support a model of “lineage instability” during the early stages hematopoietic cell fate determination. In this developmental window, the term “commitment” should perhaps be considered more relative, rather than absolute, reflecting what the cell would do if left unperturbed. As the cell matures, lineage choices may become increasingly stabilized by transcriptional networks involving positive autoregulation and cross-antagonism of alternate lineage transcription factors and cofactors. Is there a point of no return? That is, is there a point in differentiation when reprogramming is not possible? When we attempted to reprogram more mature BMMCs (sorted for FcεRI surface expression), as opposed to MCPs, we observed mostly cell death (unpublished data). Further understanding the mechanisms that mediate the transition from “reversible” to “irreversible” lineage commitment will be of interest and, possibly, clinical utility in the future.
MATERIALS AND METHODS
Materials and constructs.
All cytokines were recombinant preparations purchased from R&D Systems unless otherwise stated. Antibodies for FACS staining were obtained from BD Biosciences unless otherwise stated. The mutant FOG-1 constructs have been previously described (10). The ΔN67, mCTBP FOG-1 double mutant was constructed by recombining the ΔN67 and mCtBP FOG-1 mutants.
Mouse strains.
Previous papers have described the generation of the GATA-1− (3), FOG-1−/− (7), GATA-1V205G (9), GATA-2V296G/V296G (9), and FOG-1 transgenic mice (39) used in this study. All experiments involving mice were approved by the Children's Hospital Boston Animal Care and Use Committee and conform to institutional regulatory standards.
YSMCs.
Collagenase-released yolk sac cells were cultured in methylcellulose (StemCell Technologies Inc.) in the presence of 5 ng/ml mTpo, 2 IU/ml hEpo, and 50 ng/ml of rat SCF (Amgen), as previously described (9). Colonies were scored after 7 d of culture. Cytospins of colonies were performed and analyzed by May-Grunwald-Giemsa and Toluidine blue staining to confirm colony type.
Generation of YSMCs and BMMCs.
For YSMCs, pregnant females were killed at 9.5 d after coitus. Yolk sacs were dissected away from the embryos, digested with 0.1% (wt/vol) collagenase (Worthington Biochemical Corp.) in 20% FCS/calcium and magnesium-free 1× PBS for 90 min at 37°C with agitation, passaged 8× through an 18-gauge needle, and placed in IMDM (Invitrogen) containing 15% FCS. The suspension was centrifuged, and the cell pellets were cultured in IMDM containing 15% FCS, penicillin/streptomycin, gentamicin, 10 ng/ml of mouse IL-3, and 50 ng/ml of mouse SCF (mast cell growth media) for 6 wk (unless stated otherwise in the figure legends), splitting the cell cultures as needed.
For BMMCs, adult male C57/BL6 mice were killed and both femurs were removed. The marrow cavity was flushed with IMDM containing 15% FCS. The samples were vortexed and allowed to sit at room temperature for 5 min to allow large debris to settle. The supernatant was centrifuged, and the cell pellet was resuspended in mast cell growth media. Cells were cultured in mast cell growth media for 6 wk (unless stated otherwise in the text), splitting the cell cultures as needed.
Limiting dilution analysis of MCPs from transgenic mice.
GMPs were purified from wild-type or FOG-1–transgenic mice, as previously described (70), and plated on 96-well culture plates containing mast cell growth media at graded cell numbers using an automatic cell deposition unit system. Cells were cultured for 4 wk and examined in cytospin preparations. Wells positive for mast cell growth were determined morphologically by May-Grunwald-Giemsa and Toluidine blue staining.
Isolation of MCP cells.
For day 2 MCP sorting, purified GMPs were cultured for 48 h in IMDM containing 20% FBS in the presence of 50 ng/ml mSCF, 20 ng/ml mIL-3, and 20 ng/ml mIL-6. Cells were then stained with FITC-conjugated anti–β7 integrin (M293) and PE-conjugated anti-CD11b (M1/70; BD Biosciences). Day 2 MCPs were sorted as CD11b− β7 integrin+ cells.
Retroviral infection of primary mast cells and cell sorting.
Constructs were inserted between the viral ATG and an internal ribosome start site (IRES)–GFP cassette in the mouse myeloproliferative retroviral vector, and retroviral particles were packaged in 293GPG cells and concentrated by ultracentrifugation, as previously described (10, 71). BMMCs, GMPs, or day 2 MCPs were incubated with concentrated retroviral supernatants in mast cell growth media containing 4 μg/ml polybrene, 20 ng/ml mIL-3, 50 ng/ml SCF, 20 ng/ml mIL-6, 5 ng/ml mTPO, and 2 IU/ml hEpo for 12–16 h in a tissue culture incubator at 37°C/5%CO2. After incubation, cells were washed and resuspended in mast cell growth media containing the cytokines listed earlier in this paragraph. After 2 d of additional culture, GFP+ cells were sorted using high speed FACS sorters (Altra or Aria; Becton Dickinson). Reanalysis of sorted cells revealed GFP+ purities of at least 95%. The GFP+ cells were placed in mast cell growth media containing 20 ng/ml mIL-3, 50 ng/ml SCF, 5 ng/ml mTPO, and 2 IU/ml hEPO at a starting concentration of 105 cells per milliliter, and incubated for various amounts of time, as indicated in the figures. Cultures were expanded as necessary.
FACS analysis.
FACS staining for FcεRI surface expression was performed as previously described (72). In brief, all incubations were done at 4°C in DMEM (Invitrogen) containing 2% FCS. Cells were preincubated with monoclonal anti-CD3 (B3B4) and anti-FcγRII/III (clone 2.42G) mAbs for 15 min to block the low affinity IgE and FcγRII/III receptors, respectively. Mouse IgE (anti-DNP) mAb (clone SPE-7; Sigma-Aldrich) was added, and the cells were incubated for an additional 1 h. After washing in DMEM/2%FCS, the cells were incubated with biotinylated anti–mouse IgE (BD Biosciences) for 30 min. The cells were washed again and simultaneously stained with PE–anti–c-kit and allophycocyanin-streptavidin for 30 min. After final washes, the cells were analyzed using a FACSCalibur (Becton Dickinson). Standard FACS staining was used for the analysis of Gr-1, CD11b, and Ter-119 expression.
RT-PCR analysis of gene expression.
Total RNA was extracted from the cells or whole organ (heart) using TRIzol (Invitrogen), according to the manufacturer's instructions. 5 μg of RNase-free glycogen was included as a carrier during RNA precipitation. First-strand cDNA synthesis was performed at 37°C for 1 h using oligodT15 as a primer and MMLV-RT (Roche), according to manufacturer's instructions. PCRs were performed for 22–34 cycles using the Advantage 2 kit (BD Biosciences), according the manufacturer's instructions, and analyzed by electrophoresis in 2% agarose gels containing ethidium bromide. Primer sequences for β-major globin, EKLF, HPRT (73), GPIbα (5), GATA-1, GATA-2, FOG-1, FcεRI, MMCP-1, MMCP-5 (38), and FOG-2 (exons 3/4) (74) have been previously described. Other primer sequences are as follows: MMCP-2, (forward) 5′-GTGATGACTGCTGCACACTG-3′ and (reverse) 5′-CTTGAAGAGTCTGACTCAGG-3′; MMCP-4, (forward) 5′-GTAATTCCTCTGCCTCGTCCT-3′ and (reverse) 5′-CCCAAGGGTTATTAGAAGAGCTC-3′; MC-CPA, (forward) 5′-ACACAGGATCGAATGTGGAG-3′ and (reverse) 5′-TAATGCAGGACTTCATGAGC-3′; and band 3, (forward) 5′-GGCACCTTCCTTCTGGGTCTGGC-3′ and (reverse) 5′-GTCGACATGCGGAGCCTCAGGTC-3′. For quantitative PCR analysis, reactions were performed using SYBR green Supermix (Bio-Rad Laboratories) according to the manufacturer's instructions and analyzed on a real-time PCR machine (MyCycler; Bio-Rad Laboratories). GATA-2 or FOG-1 transcript levels were calculated using the 2ΔΔC(t) method, normalizing to transcript levels of the housekeeping gene GAPDH. Melt curves demonstrated a single product species for all reactions. Sequences of the real-time PCR primers for GATA-2 (exons 3/4), FOG-1 (exons 5/6), and GAPDH are as previously described (21).
ChIP assays.
ChIP assays were performed as previously described (21). In brief, BMMCs cultured for 6 wk in mast cell growth media were treated with 1% formaldehyde (final concentration) for 10 min at room temperature, and the reaction was stopped by adding excess glycine. Nuclei were lysed and the chromatin was sonicated to generate ∼500–1,000-bp fragments. Immunoprecipitation was performed using anti-GATA-1 (N6; Santa Cruz Biotechnologies), followed by rabbit anti–rat IgG (Jackson ImmunoResearch Laboratories), anti-GATA-2 (H-116; Santa Cruz Biotechnologies), or anti–FOG-1 polyclonal antisera (6), or the equivalent protein amount of pooled normal rat IgG (Sigma-Aldrich) or FOG-1 preimmune sera. After washing, the cross-links were reversed and the released DNA fragments were purified by phenol-chloroform extraction and ethanol precipitation. Recovery of specific DNA fragments was assessed by real-time PCR (My-Cycler or iCycler; Bio-Rad Laboratories) and normalized to a serial dilution standard curve of input chromatin for each reaction. PCR primer sets are as follows: Necdin promoter, 5′-GGTCCTGCTCTGATCCGAAG-3′ and 5′-GGGTCGCTCAGGTCCTTACTT-3′; and GATA-2 −2.8 kb, 5′-GCATGGCCCTGGTAATAGCA-3′ and 5′-CAGCCGCACCTTCCCTAA-3′.
Online supplemental material.
Fig. S1 examines the morphology and marker gene expression of YSMCs and BMMCs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20070544/DC1.
Supplemental Material
Acknowledgments
The authors would like to thank Yuko Fujiwara, Sam Katz, Aaron Chang, and Alice Tsang for generation of the mouse lines used in this study. They would also like to acknowledge John Daly and Grigoriy Losyev for assistance with FACS, Emery Bresnick and Soumen Pal for technical advice on ChIP assays, and Jonathan Snow for helpful discussions.
A.B. Cantor is supported by grants from the National Institutes of Health (K08 CA82175 and R01 HL075705). H. Iwasaki is supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (18604006) and the Takeda Science Foundation. S.H. Orkin is an investigator of the Howard Hughes Medical Institute.
The authors have no conflicting financial interests.
Abbreviations used: BMMC, bone marrow–derived mast cell; ChIP, chromatin immunoprecipitation; CtBP, C-terminal binding protein; eGFP, enhanced GFP; EKLF, erythroid Kruppel-like factor; EPO, erythropoietin; FcεRI, high affinity IgE receptor; FOG, friend of GATA; GMP, granulocyte/macrophage progenitor; GPIb, glycoprotein Ib; MC-CPA, mast cell carboxypeptidase A; MCP, mast cell progenitor; MEL, mouse erythroleukemia; MEP, megakaryocyte/erythroid progenitor; MMCP, mouse mast cell protease; SCF, stem cell factor; Srp, Serpent; TPO, thrombopoietin; Ush, U-shaped; YSMC, yolk sac–derived mast cell.
References
- 1.Zon, L.I., Y. Yamaguchi, K. Yee, E.A. Albee, A. Kimura, J.C. Bennett, S.H. Orkin, and S.J. Ackerman. 1993. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood. 81:3234–3241. [PubMed] [Google Scholar]
- 2.Ito, E., T. Toki, H. Ishihara, H. Ohtani, L. Gu, M. Yokoyama, J.D. Engel, and M. Yamamoto. 1993. Erythroid transcription factor GATA-1 is abundantly transcribed in mouse testis. Nature. 362:466–468. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara, Y., C.P. Browne, K. Cunniff, S.C. Goff, and S.H. Orkin. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA. 93:12355–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shivdasani, R.A., Y. Fujiwara, M.A. McDevitt, and S.H. Orkin. 1997. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 16:3965–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vyas, P., K. Ault, C.W. Jackson, S.H. Orkin, and R.A. Shivdasani. 1999. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood. 93:2867–2875. [PubMed] [Google Scholar]
- 6.Tsang, A.P., J.E. Visvader, C.A. Turner, Y. Fujiwara, C. Yu, M.J. Weiss, M. Crossley, and S.H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 90:109–119. [DOI] [PubMed] [Google Scholar]
- 7.Tsang, A.P., Y. Fujiwara, D.B. Hom, and S.H. Orkin. 1998. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 12:1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crispino, J.D., M.B. Lodish, J.P. MacKay, and S.H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell. 3:219–228. [DOI] [PubMed] [Google Scholar]
- 9.Chang, A.N., A.B. Cantor, Y. Fujiwara, M.B. Lodish, S. Droho, J.D. Crispino, and S.H. Orkin. 2002. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl. Acad. Sci. USA. 99:9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantor, A.B., S.G. Katz, and S.H. Orkin. 2002. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol. Cell. Biol. 22:4268–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols, K.E., J.D. Crispino, M. Poncz, J.G. White, S.H. Orkin, J.M. Maris, and M.J. Weiss. 2000. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat. Genet. 24:266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freson, K., K. Devriendt, G. Matthijs, A. Van Hoof, R. De Vos, C. Thys, K. Minner, M.F. Hoylaerts, J. Vermylen, and C. Van Geet. 2001. Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood. 98:85–92. [DOI] [PubMed] [Google Scholar]
- 13.Freson, K., G. Matthijs, C. Thys, P. Marien, M.F. Hoylaerts, J. Vermylen, and C. Van Geet. 2002. Different substitutions at residue D218 of the X-linked transcription factor GATA1 lead to altered clinical severity of macrothrombocytopenia and anemia and are associated with variable skewed X inactivation. Hum. Mol. Genet. 11:147–152. [DOI] [PubMed] [Google Scholar]
- 14.Mehaffey, M.G., A.L. Newton, M.J. Gandhi, M. Crossley, and J.G. Drachman. 2001. X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood. 98:2681–2688. [DOI] [PubMed] [Google Scholar]
- 15.Tevosian, S.G., A.E. Deconinck, A.B. Cantor, H.I. Rieff, Y. Fujiwara, G. Corfas, and S.H. Orkin. 1999. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA. 96:950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tevosian, S.G., A.E. Deconinck, M. Tanaka, M. Schinke, S.H. Litovsky, S. Izumo, Y. Fujiwara, and S.H. Orkin. 2000. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 101:729–739. [DOI] [PubMed] [Google Scholar]
- 17.Svensson, E.C., G.S. Huggins, H. Lin, C. Clendenin, F. Jiang, R. Tufts, F.B. Dardik, and J.M. Leiden. 2000. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat. Genet. 25:353–356. [DOI] [PubMed] [Google Scholar]
- 18.Tevosian, S.G., K.H. Albrecht, J.D. Crispino, Y. Fujiwara, E.M. Eicher, and S.H. Orkin. 2002. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development. 129:4627–4634. [DOI] [PubMed] [Google Scholar]
- 19.Crispino, J.D., M.B. Lodish, B.L. Thurberg, S.H. Litovsky, T. Collins, J.D. Molkentin, and S.H. Orkin. 2001. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 15:839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cantor, A.B., and S.H. Orkin. 2005. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin. Cell Dev. Biol. 16:117–128. [DOI] [PubMed] [Google Scholar]
- 21.Pal, S., A.B. Cantor, K.D. Johnson, T.B. Moran, M.E. Boyer, S.H. Orkin, and E.H. Bresnick. 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. USA. 101:980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letting, D.L., Y.Y. Chen, C. Rakowski, S. Reedy, and G.A. Blobel. 2004. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl. Acad. Sci. USA. 101:476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmes, M., J. Turner, A. Fox, O. Chisholm, M. Crossley, and B. Chong. 1999. hFOG-2, a novel zinc finger protein, binds the co-repressor mCtBP2 and modulates GATA-mediated activation. J. Biol. Chem. 274:23491–23498. [DOI] [PubMed] [Google Scholar]
- 24.Katz, S.G., A.B. Cantor, and S.H. Orkin. 2002. Interaction between FOG-1 and the corepressor C-terminal binding protein is dispensable for normal erythropoiesis in vivo. Mol. Cell. Biol. 22:3121–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, W., M. Nakazawa, Y.Y. Chen, R. Kori, C.R. Vakoc, C. Rakowski, and G.A. Blobel. 2005. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 24:2367–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez, P., E. Bonte, J. Krijgsveld, K.E. Kolodziej, B. Guyot, A.J. Heck, P. Vyas, E. de Boer, F. Grosveld, and J. Strouboulis. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 24:2354–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, A.C., A.E. Roche, J. Wilk, and E.C. Svensson. 2004. The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J. Biol. Chem. 279:55017–55023. [DOI] [PubMed] [Google Scholar]
- 28.Harigae, H., S. Takahashi, N. Suwabe, H. Ohtsu, L. Gu, Z. Yang, F.Y. Tsai, Y. Kitamura, J.D. Engel, and M. Yamamoto. 1998. Differential roles of GATA-1 and GATA-2 in growth and differentiation of mast cells. Genes Cells. 3:39–50. [DOI] [PubMed] [Google Scholar]
- 29.Migliaccio, A.R., R.A. Rana, M. Sanchez, R. Lorenzini, L. Centurione, L. Bianchi, A.M. Vannucchi, G. Migliaccio, and S.H. Orkin. 2003. GATA-1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA-1low mouse mutant. J. Exp. Med. 197:281–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda, K., C. Nishiyama, T. Tokura, Y. Akizawa, M. Nishiyama, H. Ogawa, K. Okumura, and C. Ra. 2003. Regulation of cell type-specific mouse Fc epsilon RI beta-chain gene expression by GATA-1 via four GATA motifs in the promoter. J. Immunol. 170:334–340. [DOI] [PubMed] [Google Scholar]
- 31.Nishiyama, C., T. Ito, M. Nishiyama, S. Masaki, K. Maeda, N. Nakano, W. Ng, K. Fukuyama, M. Yamamoto, K. Okumura, and H. Ogawa. 2005. GATA-1 is required for expression of Fc{epsilon }RI on mast cells: analysis of mast cells derived from GATA-1 knockdown mouse bone marrow. Int. Immunol. 17:847–856. [DOI] [PubMed] [Google Scholar]
- 32.Ghinassi, B., M. Sanchez, F. Martelli, G. Amabile, A.M. Vannucchi, G. Migliaccio, S.H. Orkin, and A.R. Migliaccio. 2007. The hypomorphic Gata1low mutation alters the proliferation/differentiation potential of the common megakaryocytic-erythroid progenitor. Blood. 109:1460–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arinobu, Y., H. Iwasaki, M.F. Gurish, S. Mizuno, H. Shigematsu, H. Ozawa, D.G. Tenen, K.F. Austen, and K. Akashi. 2005. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc. Natl. Acad. Sci. USA. 102:18105–18110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, J.R., T.A. McKinsey, H. Xu, D.Z. Wang, J.A. Richardson, and E.N. Olson. 1999. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 19:4495–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svensson, E.C., R.L. Tufts, C.E. Polk, and J.M. Leiden. 1999. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. USA. 96:956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDevitt, M.A., Y. Fujiwara, R.A. Shivdasani, and S.H. Orkin. 1997. An upstream, DNase I hypersensitive region of the hematopoietic-expressed transcription factor GATA-1 gene confers developmental specificity in transgenic mice. Proc. Natl. Acad. Sci. USA. 94:7976–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jasinski, M., P. Keller, Y. Fujiwara, S.H. Orkin, and M. Bessler. 2001. GATA1-Cre mediates Piga gene inactivation in the erythroid/megakaryocytic lineage and leads to circulating red cells with a partial deficiency in glycosyl phosphatidylinositol-linked proteins (paroxysmal nocturnal hemoglobinuria type II cells). Blood. 98:2248–2255. [DOI] [PubMed] [Google Scholar]
- 38.Iwasaki, H., S. Mizuno, R. Mayfield, H. Shigematsu, Y. Arinobu, B. Seed, M.F. Gurish, K. Takatsu, and K. Akashi. 2005. Identification of eosinophil lineage–committed progenitors in the murine bone marrow. J. Exp. Med. 201:1891–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katz, S.G., A. Williams, J. Yang, Y. Fujiwara, A.P. Tsang, J.A. Epstein, and S.H. Orkin. 2003. Endothelial lineage-mediated loss of the GATA cofactor Friend of GATA 1 impairs cardiac development. Proc. Natl. Acad. Sci. USA. 100:14030–14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox, A.H., C. Liew, M. Holmes, K. Kowalski, J. Mackay, and M. Crossley. 1999. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 18:2812–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vermes, I., C. Haanen, H. Steffens-Nakken, and C. Reutelingsperger. 1995. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J. Immunol. Methods. 184:39–51. [DOI] [PubMed] [Google Scholar]
- 42.Tsai, F.Y., and S.H. Orkin. 1997. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 89:3636–3643. [PubMed] [Google Scholar]
- 43.Grass, J.A., M.E. Boyer, S. Pal, J. Wu, M.J. Weiss, and E.H. Bresnick. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA. 100:8811–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walsh, J.C., R.P. DeKoter, H.J. Lee, E.D. Smith, D.W. Lancki, M.F. Gurish, D.S. Friend, R.L. Stevens, J. Anastasi, and H. Singh. 2002. Cooperative and antagonistic interplay between PU.1 and GATA-2 in the specification of myeloid cell fates. Immunity. 17:665–676. [DOI] [PubMed] [Google Scholar]
- 45.Querfurth, E., M. Schuster, H. Kulessa, J.D. Crispino, G. Doderlein, S.H. Orkin, T. Graf, and C. Nerlov. 2000. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 14:2515–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, C., A.B. Cantor, H. Yang, C. Browne, R.A. Wells, Y. Fujiwara, and S.H. Orkin. 2002. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195:1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans, C.J., V. Hartenstein, and U. Banerjee. 2003. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev. Cell. 5:673–690. [DOI] [PubMed] [Google Scholar]
- 48.Fossett, N., and R.A. Schulz. 2001. Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation. 69:83–90. [DOI] [PubMed] [Google Scholar]
- 49.Fossett, N., S.G. Tevosian, K. Gajewski, Q. Zhang, S.H. Orkin, and R.A. Schulz. 2001. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc. Natl. Acad. Sci. USA. 98:7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fossett, N., K. Hyman, K. Gajewski, S.H. Orkin, and R.A. Schulz. 2003. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc. Natl. Acad. Sci. USA. 100:11451–11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vakoc, C.R., D.L. Letting, N. Gheldof, T. Sawado, M.A. Bender, M. Groudine, M.J. Weiss, J. Dekker, and G.A. Blobel. 2005. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 17:453–462. [DOI] [PubMed] [Google Scholar]
- 52.Maeda, K., C. Nishiyama, T. Tokura, H. Nakano, S. Kanada, M. Nishiyama, K. Okumura, and H. Ogawa. 2006. FOG-1 represses GATA-1-dependent FcepsilonRI beta-chain transcription: transcriptional mechanism of mast-cell-specific gene expression in mice. Blood. 108:262–269. [DOI] [PubMed] [Google Scholar]
- 53.Welch, J.J., J.A. Watts, C.R. Vakoc, Y. Yao, H. Wang, R.C. Hardison, G.A. Blobel, L.A. Chodosh, and M.J. Weiss. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 104:3136–3147. [DOI] [PubMed] [Google Scholar]
- 54.Lu, L.F., E.F. Lind, D.C. Gondek, K.A. Bennett, M.W. Gleeson, K. Pino-Lagos, Z.A. Scott, A.J. Coyle, J.L. Reed, J. Van Snick, et al. 2006. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature. 442:997–1002. [DOI] [PubMed] [Google Scholar]
- 55.Nagata, H., A.S. Worobec, C.K. Oh, B.A. Chowdhury, S. Tannenbaum, Y. Suzuki, and D.D. Metcalfe. 1995. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc. Natl. Acad. Sci. USA. 92:10560–10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Longley, B.J., Jr., D.D. Metcalfe, M. Tharp, X. Wang, L. Tyrrell, S.Z. Lu, D. Heitjan, and Y. Ma. 1999. Activating and dominant inactivating c-KIT catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc. Natl. Acad. Sci. USA. 96:1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pardanani, A., R.P. Ketterling, S.R. Brockman, H.C. Flynn, S.F. Paternoster, B.M. Shearer, T.L. Reeder, C.Y. Li, N.C. Cross, J. Cools, et al. 2003. CHIC2 deletion, a surrogate for FIP1L1-PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood. 102:3093–3096. [DOI] [PubMed] [Google Scholar]
- 58.Boyd, A.W., and J.W. Schrader. 1982. Derivation of macrophage-like lines from the pre-B lymphoma ABLS 8.1 using 5-azacytidine. Nature. 297:691–693. [DOI] [PubMed] [Google Scholar]
- 59.Klinken, S.P., W.S. Alexander, and J.M. Adams. 1988. Hemopoietic lineage switch: v-raf oncogene converts Emu-myc transgenic B cells into macrophages. Cell. 53:857–867. [DOI] [PubMed] [Google Scholar]
- 60.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250–1262. [DOI] [PubMed] [Google Scholar]
- 61.Visvader, J.E., A.G. Elefanty, A. Strasser, and J.M. Adams. 1992. GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J. 11:4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaguchi, Y., L.I. Zon, S.J. Ackerman, M. Yamamoto, and T. Suda. 1998. Forced GATA-1 expression in the murine myeloid cell line M1: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood. 91:450–457. [PubMed] [Google Scholar]
- 63.Seshasayee, D., P. Gaines, and D.M. Wojchowski. 1998. GATA-1 dominantly activates a program of erythroid gene expression in factor-dependent myeloid FDCW2 cells. Mol. Cell. Biol. 18:3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kondo, M., D.C. Scherer, T. Miyamoto, A.G. King, K. Akashi, K. Sugamura, and I.L. Weissman. 2000. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 407:383–386. [DOI] [PubMed] [Google Scholar]
- 65.Iwasaki-Arai, J., H. Iwasaki, T. Miyamoto, S. Watanabe, and K. Akashi. 2003. Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J. Exp. Med. 197:1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heyworth, C., S. Pearson, G. May, and T. Enver. 2002. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 21:3770–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwasaki, H., S. Mizuno, R.A. Wells, A.B. Cantor, S. Watanabe, and K. Akashi. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 19:451–462. [DOI] [PubMed] [Google Scholar]
- 68.Ye, M., H. Iwasaki, C.V. Laiosa, M. Stadtfeld, H. Xie, S. Heck, B. Clausen, K. Akashi, and T. Graf. 2003. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 19:689–699. [DOI] [PubMed] [Google Scholar]
- 69.Iwasaki, H., S. Mizuno, Y. Arinobu, H. Ozawa, Y. Mori, H. Shigematsu, K. Takatsu, D.G. Tenen, and K. Akashi. 2006. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 20:3010–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. [DOI] [PubMed] [Google Scholar]
- 71.Klein, C., H. Bueler, and R.C. Mulligan. 2000. Comparative analysis of genetically modified dendritic cells and tumor cells as therapeutic cancer vaccines. J. Exp. Med. 191:1699–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamaguchi, M., C.S. Lantz, H.C. Oettgen, I.M. Katona, T. Fleming, I. Miyajima, J.P. Kinet, and S.J. Galli. 1997. IgE enhances mouse mast cell FcεRI expression in vitro and in vivo: evidence for a novel amplification mechanism in IgE-dependent reactions. J. Exp. Med. 185:663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiss, M.J., G. Keller, and S.H. Orkin. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 8:1184–1197. [DOI] [PubMed] [Google Scholar]
- 74.Ackerman, K.G., B.J. Herron, S.O. Vargas, H. Huang, S.G. Tevosian, L. Kochilas, C. Rao, B.R. Pober, R.P. Babiuk, J.A. Epstein, et al. 2005. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 1:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.