Abstract
The recognition of human leukocyte antigen (HLA)-E by the heterodimeric CD94-NKG2 natural killer (NK) receptor family is a central innate mechanism by which NK cells monitor the expression of other HLA molecules, yet the structural basis of this highly specific interaction is unclear. Here, we describe the crystal structure of CD94-NKG2A in complex with HLA-E bound to a peptide derived from the leader sequence of HLA-G. The CD94 subunit dominated the interaction with HLA-E, whereas the NKG2A subunit was more peripheral to the interface. Moreover, the invariant CD94 subunit dominated the peptide-mediated contacts, albeit with poor surface and chemical complementarity. This unusual binding mode was consistent with mutagenesis data at the CD94-NKG2A–HLA-E interface. There were few conformational changes in either CD94-NKG2A or HLA-E upon ligation, and such a “lock and key” interaction is typical of innate receptor–ligand interactions. Nevertheless, the structure also provided insight into how this interaction can be modulated by subtle changes in the peptide ligand or by the pairing of CD94 with other members of the NKG2 family. Differences in the docking strategies used by the NKG2D and CD94-NKG2A receptors provided a basis for understanding the promiscuous nature of ligand recognition by NKG2D compared with the fidelity of the CD94-NKG2 receptors.
MHC-Ia, -Ib, and -I–like molecules play a critical role in regulating the function of NK cells and are recognized by a diverse set of NK receptors (1). In humans, MHC-Ia molecules are recognized by the NK receptors of the Ig superfamily, killer cell Ig-like receptors (KIRs) (2) and Ig-like transcript (3), whereas in mice they are recognized by structurally divergent NK receptors of the C-type lectin superfamily, termed Ly49 (4). In contrast to MHC-Ia molecules, NK cell recognition of MHC-Ib and MHC-I–like molecules is conserved across species and mediated predominantly through the homodimeric NKG2D (5) and heterodimeric CD94-NKG2 receptors (6).
Typically NK cell receptors can recognize multiple MHC-I and MHC-I–like ligands. For example, individual KIRs bind multiple MHC-Ia allotypes (7). Similarly, members of the Ly49 family interact with several distinct MHC-Ia molecules (8), and NKG2D recognizes a diverse array of MHC-I–like ligands, including MICA, MICB, and ULBP1-4 in humans, and H60, RAE-1 proteins, and MULT1 in mice (9). Moreover, there are also examples of interspecies cross-reactivity between mouse NKG2D and the human NKG2D ligands (10) that further highlight the evolutionary conserved nature of these innate interactions. Structural studies on NKG2D, coupled with detailed thermodynamic analyses, have provided insight into how a single receptor can interact with these structurally distinct ligands (11–16).
In contrast to the promiscuity of most NK receptors, the human CD94-NKG2 family solely recognizes the MHC-Ib molecule, HLA-E (17–20). The essentially monomorphic HLA-E has evolved to selectively bind peptides derived from the leader sequences of other class I molecules. Consequently, down-regulation of MHC-I expression leads to a reduction in HLA-E expression at the cell surface. Accordingly, the interaction between CD94-NKG2 and HLA-E represents a central innate mechanism by which NK cells indirectly monitor the expression of other MHC-I molecules within a cell (17, 18).
Although it is unclear how the CD94-NKG2 receptor family interacts with HLA-E, the association of CD94 with NKG2 is critical for the biological activity of this family. Several members of the NKG2 family, including NKG2A, -2B, -2C, and -2E, dimerize with CD94 in vitro (21–23). Members of the CD94-NKG2 family differ in their affinity for HLA-E, with the inhibitory CD94-NKG2A binding more tightly than CD94-NKG2C; yet the structural basis for this observation is unknown (22, 24). To begin to understand the mode of CD94-NKG2A interaction with HLA-E, we have recently reported the structure of the nonligated CD94-NKG2A heterodimer, which resolved the preferential pairing of the CD94-NKG2 subunits (25). Mutagenesis studies have identified residues in HLA-E and CD94-NKG2A that are critical for the interaction and suggested that each subunit made discrete contributions to HLA-E docking (25).
Here, we report the structure of the CD94-NKG2A heterodimer in complex with HLA-E bound to a peptide corresponding to residues 3–11 of the HLA-G leader sequence. Our findings, together with mutational data, provide a basis for understanding the specificity of this interaction as well as the relative contribution of the CD94 and NKG2A subunits in recognizing the monomorphic HLA-E peptide complex.
RESULTS
Overview of the complex
To establish how HLA-E, when bound to the leader sequence peptide of HLA-G (residues 3–11, VMAPRTLFL, HLA-EVMAPRTLFL), was specifically recognized by CD94-NKG2A, we expressed and purified both components, crystallized the complex in space group I4132 with unit cell dimensions a = b = c = 345.2 Å, and determined the structure of the CD94-NKG2A-HLA-EVMAPRTLFL complex to 3.4 Å resolution with an Rfactor and Rfree of 24.8 and 27.9%, respectively (see Materials and methods and Table I). Although the CD94-NKG2A–HLA-EVMAPRTLFL structure was determined to moderate resolution, we had previously determined the structures of the nonligated CD94-NKG2A (25) and HLA-EVMAPRTLFL molecules (26) to 2.5 Å resolution that enabled the structure to be solved readily by molecular replacement, and permitted the ternary complex to be refined well at this resolution. For example, the initial experimental phases clearly showed unbiased electron density for the VMAPRTLFL peptide (Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20072525/DC1), confirming the correctness of the molecular replacement solution. In addition, the electron density at the CD94-NKG2A–HLA-EVMAPRTLFL interface was unambiguous, and side chains at the interface could be modeled into the electron density (Fig. S1, B and C), thus allowing contacts at the interface to be interpreted readily. The crystal contained two virtually indistinguishable ternary complexes within the asymmetric unit (root mean square deviation 0.7 Å). Consequently, structural analyses will be confined to one CD94-NKG2A–HLA-EVMAPRTLFL complex.
Table I.
Data collection and refinement statistics
| Data collection statistics | |
|---|---|
| Temperature (K) | 100 |
| x-ray source | NSLS, Beamline X25 |
| Detector | ADSC Q315 |
| Space group | I4132 |
| Cell dimensions (Å) | 345.2, 345.2, 345.2 |
| Resolution (Å) | 50.0-3.40 |
| Total no. of observations | 680267 |
| No. of unique observations | 48105 (6919) |
| Multiplicity | 14.1 |
| Data completeness (%) | 100.0 (100.0) |
| I/σI | 16.2 (3.0) |
| RPIM (%) | 4.6 (26.5) |
| Refinement statistics | |
| Nonhydrogen atoms | |
| Protein | 10174 |
| Water | 0 |
| Resolution (Å) | 50 – 3.4 |
| Rfactor (%) | 24.8 |
| Rfree (%) | 27.9 |
| r.m.s.d from ideality | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.53 |
| Dihedrals | 26.59 |
| Impropers | 0.963 |
| Ramachandran plot | |
| Most favored | 78.2 |
| Allowed | 20.0 |
| Generous | 1.0 |
| Unfavored | 0.8 |
| B-factors (Å2) | |
| Average per residue | 110 |
The values in parentheses are for the highest resolution bin (approximate interval 0.1 Å). RPIM is Rmerge divided by the root square of the redundancy (reference 53). Rfactor = 100⋅Σhkl | |Fo| - |Fc| | / Σhkl |Fo| for all data except for 5%, which was used for the Rfree calculation.
The CD94-NKG2A docked toward the C-terminal end of the HLA-E antigen-binding cleft, binding at an angle of ∼70° (Fig. 1, A and B, and Figs. S2 and 3, which are available at http://www.jem.org/cgi/content/full/jem.20072525/DC1) in a manner that permitted CD94-NKG2A to sit across both the α1 and α2 helices of HLA-E (Fig. 1). A comparison of the CD94-NKG2A–HLA-EVMAPRTLFL complex with the nonligated CD94-NKG2A (25) and HLA-EVMAPRTLFL structures (26) revealed no significant conformational change in either HLA-E or CD94-NKG2A upon complex formation (root mean square deviation 0.5 and 0.4 Å for CD94-NKG2A and HLA-E, respectively). The one disordered loop of the nonligated CD94-NKG2A heterodimer (residues 199–204 in NKG2A) became ordered in the complex (not depicted), although this observation was attributable to crystal-packing effects, as this loop did not contact HLA-EVMAPRTLFL. Only one residue, Gln 112 of CD94, was re-orientated upon ligation to maximize the complementarity at the interface. Accordingly, this “lock and key” engagement between HLA-EVMAPRTLFL and CD94-NKG2A exemplified the “innate characteristic” of this interaction.
Figure 1.
The CD94-NKG2A–HLA-EVMAPRTLFL complex. NKG2A and CD94 are represented as blue and pale green ribbon structures, respectively. The heavy chain of HLA-E and β2m are shown as violet and cyan ribbons, respectively, with the VMAPRTLFL peptide in orange sticks. (A) Side view of CD94-NKG2A docking onto HLA-EVMAPRTLFL. (B) Top view of CD94-NKG2A docking onto the surface of HLA-EVMAPRTLFL.
CD94-NKG2A binding to HLA-E
The CD94 and NKG2A subunits lay across the antigen-binding cleft, with CD94 and NKG2A almost exclusively interacting with the α1 and α2 helices of HLA-E, respectively. Analysis of the electrostatic surfaces of HLA-E and CD94-NKG2A highlighted a role for charge complementarity at the CD94-NKG2A–HLA-E interface (Fig. 2 A). Namely, a basic region on the α1 helix of HLA-E interacted with an acidic region on CD94 and, conversely, an acidic region on the HLA-E α2 helix docked with a distinct patch of basic charge on NKG2A (Fig. 2 A). The buried surface area (BSA) at the interface was extensive (∼2,100 Å2), with CD94 and NKG2A contributing ∼69 and 31%, respectively (Fig. 2 B). Consistent with the electrostatic complementarity, the CD94-NKG2A–HLA-E interface was characterized by a large number of polar interactions, including 8 salt bridges and 19 H bonds. There was also a small and focused patch of hydrophobic contacts at the interface (Table II). Somewhat surprisingly, the overall shape complementarity (SC) at the interface was relatively poor (SC = 0.63), although the SC for CD94 alone was markedly better (SC = 0.68) compared with NKG2A (SC = 0.31), which further highlighted the greater contribution that CD94 makes to the docking onto HLA-EVMAPRTLFL compared with NKG2A.
Figure 2.
Interaction of CD94-NKG2A subunits to HLA-EVMAPRTLFL. (A) Electrostatics of CD94-NKG2A and HLA-E (represented as surface displaying charged regions) reveal charge complementarity that drives the interaction. (B) CD94-NKG2A interaction footprint on HLA-EVMAPRTLFL. The surface of HLA-E is shown with the peptide as orange space fill. Contacts to HLA-E made by CD94 are represented in pale green, and those made by NKG2A are in blue. Both subunits contact P5-Arg and are shown in teal. (C) Key interactions of CD94 to HLA-E α1 helix. Hydrogen bonds are formed between Gln113 to Asp69 of HLA-E, which also forms a salt bridge with Arg171 of CD94. Asn170 and Glu164 both form hydrogen bonds with Gln72 of HLA-E, whereas Glu89 forms a hydrogen bond with Thr146. (D) Binding of CD94 to HLA-E involves hydrophobic interactions between CD94 Phe114 and Leu162 by Ile73 and Val76 of HLA-E and P8-Phe of the peptide. Leu162 is also involved in VDW interactions further along the α1 helix. In addition, neighboring Asp163 forms a salt bridge with Arg75 of HLA-E, and Ser143 of CD94 forms a hydrogen bond with Arg79. (E) Key NKG2A interactions with HLA-E α2 helix. Arg137 forms a hydrogen bond with Ser151 and a salt bridge with Glu154 of HLA-E. His155 of HLA-E forms a hydrogen bond with Ser172 while making VDW contacts with Pro171. Ala158 of HLA-E makes VDW contacts with Gln212 and Lys217 of NKG2A, the latter of which forms a salt bridge with Asp162 and the α2 helix. Hydrogen bonds are represented as black dashed lines, and salt bridges are in blue.
Table II.
CD94-NKG2A contacts with HLA-E and VMAPRTLFL peptide
| HLA-E contacts | ||
|---|---|---|
| NKG2A | HLA-E | TYPE |
| Arg137NH1 | Ser151Oγ | H bond |
| Arg137NH2 | Glu154Oε1 | Salt bridge |
| Arg137 | Ser151 | VDW |
| Arg137 | Glu154 | VDW |
| Lys164 | Arg65 | VDW |
| Pro171 | His155 | VDW |
| Ser172 | His155 | VDW |
| Lys199Nζ | Asp162Oδ2 | Salt bridge |
| Lys199 | Asp162 | VDW |
| Gln212 | Ala158 | VDW |
| Gln212 | Glu154 | VDW |
| Val213 | His155 | VDW |
| Lys217 | Asp162 | VDW |
| Lys217Nζ | Asp162Oδ2 | H bond |
| Lys217 | Ala158 | VDW |
| Ser223 | Arg131 | VDW |
| Ser224 | Arg131 | VDW |
| CD94 | HLA –E | |
| Ser77Oγ | Arg65NH2 | H bond |
| Ser77 | Arg65 | VDW |
| Gln112 | Ile73 | VDW |
| Gln112Nε2 | Glu152Oε2 | H bond |
| Gln113 | Arg65 | VDW |
| Gln113Oε1 | Asp69Oδ | H bond |
| Gln113 | Asp69 | VDW |
| Phe114 | Gln72 | VDW |
| Phe114 | Val76 | VDW |
| Ser143Oγ | Arg79NH2 | H bond |
| Ser143 | Arg79 | VDW |
| Thr146 | Glu89 | VDW |
| Thr146Oγ1 | Glu89Oε1 | H bond |
| Phe147 | Arg79 | VDW |
| Asn160 | Val76 | VDW |
| Asn160 | Arg79 | VDW |
| Asn160Oδ1 | Arg79NH1 | H bond |
| Asn160° | Arg79NH2 | H bond |
| Ala161 | Val76 | VDW |
| Ala161° | Arg79NH2 | H bond |
| Ala161 | Arg79 | VDW |
| Leu162 | Gln72 | VDW |
| Leu162 | Arg75 | VDW |
| Leu162 | Val76 | VDW |
| Asp163Oδ1 | Arg75NH2 | Salt bridge |
| Asp163 | Arg75 | VDW |
| Glu164 | Glu19 | VDW |
| Glu164Oε1 | Gln72Nε2 | H bond |
| Glu164 | Gln72 | VDW |
| Glu168Oδ2 | Glu19Oε2 | H bond |
| Asn170 | Gln72 | VDW |
| Arg171 | Arg65 | VDW |
| Arg171NH1 | Arg65NH2 | H bond |
| Arg171NH2 | Asp69Oδ2 | Salt bridge |
| PEPTIDE CONTACTS | ||
| NKG2A | PEPTIDE | |
| Pro171 | Arg5 | VDW |
| CD94 | PEPTIDE | |
| Ser110 | Arg5 | VDW |
| Ser110° | Arg5NH2 | H bond |
| Gln112 | Arg5 | VDW |
| Gln112 | Thr6 | VDW |
| Gln112Nε2 | Thr6° | H bond |
| Gln112Oε1 | Thr6N | H bond |
| Gln112 | Phe8 | VDW |
| Phe114 | Phe8 | VDW |
| Asn156 | Phe8 | VDW |
| Asn158 | Phe8 | VDW |
| Asn160 | Phe8 | VDW |
Superscripts refer to atomic nomenclature.
The CD94 footprint on HLA-E was broad, with residues within loops 2, 3, and 5, and β strands 6 and 7 from CD94 interacting with a region spanning residues 65–89 from the α1 helix of HLA-E (Fig. 2 C). Arg65 of HLA-E is situated at the periphery of CD94 subunit and formed van der Waals (VDW) interactions with Arg171. Asp69 of HLA-E appeared to play a central role in the interaction with CD94, as it salt bridged to Arg171 and H bonded to Gln113 (Fig. 2 C). Arg75 formed a salt bridge with Asp163 of CD94, whereas Arg79 of HLA-E H bonded to Ser143 (Fig. 2 C). Glu89 from HLA-E, which is located at the loop C-terminal to the α1 helix, H bonded to Thr146. Gln72 H bonded to Glu164 and Asn170 from CD94, whereas the aliphatic moiety of Gln72 packed against the aromatic ring of Phe114 (Fig. 2 C). Phe114 of CD94 sat in a hydrophobic niche that is formed by Ile73 and Val76 from HLA-E and by P8-Phe from the peptide. Moreover, the neighboring Leu162 of CD94, which formed VDW contacts with Val76 and packed against the aliphatic moiety of Arg75 from HLA-E, extended this hydrophobic patch (Fig. 2 D).
When compared with that of CD94, the footprint of NKG2A on HLA-E was markedly smaller and more focused with loop 3 and β strands 2, 5, and 6 interacting with residues 151–162 of the α2 helix of HLA-E (Fig. 2 E). Nevertheless, analogous to the CD94–HLA-E interactions, the NKG2A–HLA-E contacts were dominated by polar residues. For example, Asp162 from HLA-E formed a salt bridge with Lys217 of NKG2A, whereas Arg137 of NKG2A salt bridged to Glu154 and H bonded to Ser151 of HLA-E. Of the VDW-mediated contacts, the aliphatic moiety of Glu154 interacted with Gln212 of NKG2A. His155 was nestled between Pro171 and Ser172 of NKG2A, and Ala158 interacted with Gln212 and Lys217 of NKG2A (Fig. 2 E).
Accordingly, the large and predominantly polar network of interactions between CD94-NKG2A and HLA-E underscored the specificity of this interaction and highlighted the dominant role of the CD94 subunit with respect to the NKG2A subunit.
CD94-NKG2A peptide-mediated contacts
Overall, the peptide contributed 23% of the BSA at the HLA-E interface, in which the invariant CD94 subunit played a much more marked role (80%) in interacting with the peptide compared with the NKG2A chain (20%). CD94 sat over the P8-Phe position of the peptide while NKG2A was adjacent to P5-Arg, with its contact being limited to a VDW interaction with Pro171 (Fig. 3, A and B). In contrast, CD94 interacted with residues P5-Arg, P6-Thr, and P8-Phe, with Gln112 intercalated between the bulky P5-Arg and P8-Phe and hydrogen bonded to the main chain of P6-Thr. Unexpectedly, three polar residues (Asn156, Asn158, and Asn160) surrounded and contacted the hydrophobic P8-Phe, the latter of which also interacted with Phe114 and Gln112 (Fig. 3 B).
Figure 3.
Peptide-mediated contacts to CD94-NKG2A. (A) The surface of CD94 (pale green) clearly accommodates Arg5 and Phe8 of the peptide (orange sticks) presented on HLA-E (violet ribbon). NKG2A (blue surface) makes minimal contact with the peptide. (B) Pro171 is the only residue of NKG2A that interacts with the peptide, at P5-Arg. CD94 dominates recognition of the peptide making several interactions, including hydrogen bonds of Ser110 to P5-Arg and Gln112 to P6-Thr. A distinct polar pocket created by Asn156, Asn158, Asn160, and Phe114 on CD94 accommodates P8-Phe. (C) KK50.4 TCR interactions to HLA-E–presenting peptide VMAPRTLIL. Interactions are dominated by the Vβ chain of the TCR in a manner analogous to CD94-NKG2A interactions.
The P5-Arg protruded into a cavity between the NKG2A and CD94 subunits (Fig. 3 A), with its aliphatic side chain flanked in an anti-parallel manner with Gln112 of CD94, whereas its guanadinium group H bonded to the main chain of Ser110 of CD94 (Fig. 3 B). However, the P5-Arg did not appear to be well complemented by any charged interactions with the CD94-NKG2A, which may be attributable to it salt bridging to Glu152 of HLA-E.
Accordingly, the interactions with the peptide are dominated by the invariant CD94 subunit, in which the major determinants of the peptide, namely positions P5 and P8, appear poorly matched in terms of the chemical and electrostatic complementarity to CD94-NKG2A.
Verification of the footprint
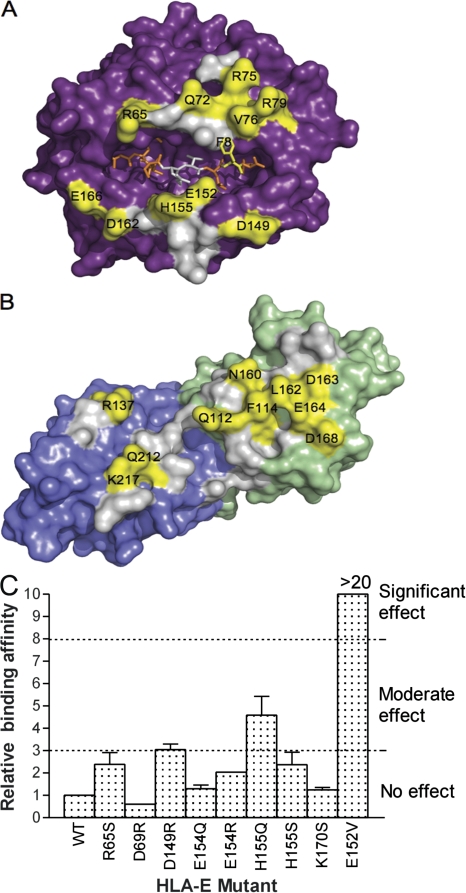
Before determining the structure of the CD94-NKG2A–HLA-E ternary complex, we conducted an alanine-scanning mutagenesis and surface plasmon resonance study at this interface (25). Accordingly, this not only provided us with an opportunity to independently verify the structural footprint from the low-resolution structure reported here, but it also allowed us to address the energetic basis of this specific interaction. First, of the 16 mutations that were made in HLA-E, 8 residues were shown to be important in the interaction. All of these HLA-E residues (Arg65, Arg75, Arg79, Val76, Gln72, Glu152, Asp162, and Glu166), which encompassed residues from the α1 and α2 helices, made contacts with CD94-NKG2A in the ternary complex (Fig. 4 A). Second, of the 21 alanine-scanning mutations made in CD94-NKG2A, 10 residues were shown to be important for the interaction. Again, the structural data indicated that these CD94-NKG2A residues (CD94: Gln112, Phe114, Asn160, Leu162, Asp163, Glu164, Asp168; NKG2A: Gln212, Arg137, Arg215) made contacts with HLA-E, thereby further confirming the interpretation of the CD94-NKG2A–HLA-E footprint (Fig. 4 B). Nevertheless, as expected, there were residues that made contact at the interface that did not contribute significantly to the energetics of the interaction as assessed by surface plasmon resonance (see Discussion).
Figure 4.
Energetically important residues and the corresponding structural footprint. (A) Surface representation of HLA-E with bound peptide (orange). (B) Surface representation of CD94-NKG2A, with CD94 colored green and NKG2A colored blue. Residues that have been shown, via mutagenesis, to influence binding (moderate and significant effects) are shown in yellow. Residues that make contacts but have no demonstrable effect on the affinity of binding are shown in gray. (C) Relative binding of HLA-E mutants to CD94-NKG2A. The binding affinity of each mutant was determined by surface plasmon resonance, and the relative effect on binding was compared with wild-type (WT) HLA-E, which was normalized to a value of 1. There was no effect on binding if there was a less than threefold difference in binding affinity, whereas a greater than eightfold difference represented a significant effect.
To further assess the footprint of CD94-NKG2A on HLA-E, an additional nine mutations were made in HLA-E, and the impact of these mutations was examined using surface plasmon resonance. These mutations (Fig. 4 C) were not alanine-scanning mutations and included charge-reversal mutations (Asp69 to Arg; Glu154 to Arg), which enabled the specificity of the interaction to be further addressed. Only one mutation (Glu152 Val) had a significant effect on the affinity of the interaction, which highlighted that it is the negative charge of Glu152, and not the aliphatic moiety per se, that is a requisite for this interaction. The charge-reversal mutations had no effect on the affinity of the interaction, consistent with the corresponding alanine-scanning mutations at these positions, thereby further confirming that positions 69 and 154 are not critical for determining the specificity of the interaction. Position 155 represents a polymorphic site between HLA-E (His155) and other HLA-Ia molecules (Gln155); surprisingly, substitution of His155 to Gln or Ser had a differential effect, with the former having a moderate effect on the affinity of the interaction, whereas the latter had no significant impact. Moreover, positions 65, 69, and 155, which make contacts with the TCR in all TCR–MHC-I structures determined to date (27, 28), all appear to be dispensable in the interaction with CD94-NKG2A (Fig. 4 C), further highlighting the differing requirements for adaptive and innate receptors interacting with MHC molecules.
Comparison to other footprints
The structure of the CD94-NKG2A–HLA-EVMAPRTLFL complex provided an opportunity to compare it to (a) the αβTCR–HLA-EVMAPRTLIL complex (Fig. 3 C and Figs. S2 and S3) (29), thereby allowing us to address how both innate and adaptive receptors can recognize the same ligand, and (b) the homodimeric NKG2D–MHC-like ligand complexes (11, 12, 14), which enabled us to address the central issue of promiscuous versus specific NK receptor recognition. First, the position of the CD94-NKG2A footprint on HLA-EVMAPRTLFL was similar to the footprint of an HLA-E–restricted TCR (termed KK50.4; Figs. S2 and S3) (29). Similar to CD94-NKG2A, the KK50.4 TCR formed an extensive footprint on HLA-EVMAPRTLIL, focusing on the C-terminal end of the antigen-binding cleft with the TCR docked in a diagonal mode; however, in contrast to CD94-NKG2A recognition of HLA-E, each chain of the αβTCR contributed roughly equally to the interface (Figs. S2 and S3). The KK50.4 TCR interacted predominantly with positions P5-Arg and P8-Ile of the peptide, analogous to that of CD94-NKG2A, suggesting that both innate and adaptive receptors are sensitive to changes to positions P5 and P8 of the peptide. Nevertheless, although the interactions between the peptide and CD94-NKG2A were CD94 centric, both chains of the TCR made substantial contacts with the peptide, with the TCRα chain focused on the peptide positions P4-P6, and the TCRβ chain interacting with P8-Ile (Fig. 3 C). Notably, the interactions between the αβTCR and the peptide exhibited excellent chemical complementarity, which contrasted markedly with the CD94 peptide-mediated interactions (Fig. 3 C).
Second, the location of the CD94-NKG2A footprint on HLA-EVMAPRTLFL was analogous to the positioning of the footprint of human NKG2D on its MHC-like ligands, MICA and, to a lesser extent, ULBP3 (Fig. 5, A and B). Nevertheless, there were notable differences between these footprints, which are attributable to the narrower cleft between the α helices of MICA and ULBP3 compared with HLA-E, and the structural differences between CD94-NKG2A and NKG2D. CD94-NKG2A possessed a flat interacting surface with HLA-E, whereas that of NKG2D was more “saddle-like,” which enabled it to clamp around both α helices of the MHC-like ligands (Fig. 5 A) (11, 12, 14). Moreover, the closer juxtapositioning of the α1 and α2 helices of MICA and ULBP3 enabled each NKG2D subunit within the homodimer to interact with both α helices to some extent, whereas the interactions with the CD94 and NKG2A subunits are virtually polarized to each α helix of HLA-E (Table II). Moreover, although CD94-NKG2A is structurally homologous to NKG2D, different regions within these NK receptors were used to interact with their respective ligands (Fig. 5 C). In contrast to the predominantly electrostatic nature of the CD94-NKG2A–HLA-EVMAPRTLFL interaction, the polar interactions contribute to but do not dominate the interaction between NKG2D and its ligands (Fig. 5, B and C). Specifically, the nonpolar interactions at the NKG2D-MHC–like and CD94-NKG2A–HLA-EVMAPRTLFL interfaces contribute ∼60 and 40% to the BSA at the interface, respectively. This differential usage of polar-mediated contacts may account for the promiscuity in NKG2D recognition compared with a high degree of specificity for ligand displayed by CD94-NKG2A.
Figure 5.
Comparison of polar versus nonpolar interactions by CD94-NKG2A and NKG2D homodimer. Nonpolar interactions are represented in purple, and polar interactions are in blue. In situations where a residue makes both nonpolar interactions as well either a salt bridge or hydrogen bond, it was represented as making a polar interaction. (A) Ribbon representation of CD94-NKG2A in complex with HLA-EVMAPRTLFL and NKG2D homodimers binding to MICA and ULBP3. The subunit of the NKG2D homodimer equivalent to CD94 is represented in pale green, and the subunit equivalent to NKG2A is in blue. The MHC-like molecules, MICA and ULBP3, do not associated with β2m (represented in cyan), and these molecules are represented in violet. (B) “Polarity” footprints onto MHC molecules HLA-E by CD94-NKG2A, and MICA and ULPB3 by NKG2D homodimer. The peptide presented by HLA-E is represented in orange space fill. (C) Residues of CD94-NKG2A and the NKG2D homodimer involved in polar and nonpolar interactions.
Accordingly, the innate NK receptors and the αβTCR focus on a similar region of the HLA-E or MHC-like ligand, regardless of whether the ligand presents peptide or not. However, the characteristics of the footprint deviate between these receptors, thereby providing a basis for the NKG2D promiscuity versus the CD94-NKG2A specificity.
DISCUSSION
We have determined the structure of the CD94-NKG2A–HLA-EVMAPRTLFL complex and thereby provide a structural basis for the specificity of this interaction, which contrasted with the promiscuous recognition of MHC-like ligands by the homodimeric NKG2D receptor. There was little evidence of conformational adjustments of either CD94-NKG2A or HLA-EVMAPRTLFL in the formation of the CD94-NKG2A–HLA-EVMAPRTLFL complex, which is consistent with previous thermodynamic measurements that suggested a “rigid body” mode of recognition (22). This lock and key interaction contrasts with the conformational plasticity that is exemplified by antigen recognition by the adaptive immune system (30). Although CD94 and NKG2A are structurally similar, the distribution of surface electrostatics of CD94-NKG2A differed markedly, dictating the docking orientation of these subunits on HLA-E. A surprising feature of the CD94-NKG2A–HLA-EVMAPRTLFL complex was the degree to which CD94 dominated the interaction with HLA-E and the peptide, as NKG2A was limited to contacting a small region on the α2 helix of HLA-E and a single VDW contact with the peptide.
Our previous alanine-scanning mutagenesis data that assessed the energetic contribution of HLA-E and CD94-NKG2A residues at the interface validated our observed positioning of CD94-NKG2A on HLA-EVMAPRTLFL (25). Virtually all of the HLA-E residues that were critically important to the affinity of the interaction interacted with CD94-NKG2A and vice versa. The role of Glu152 from HLA-E remains unclear because although this residue directly interacted with Gln112 of CD94, the Glu152 Ala and Glu152 Val mutations may also affect the conformation of P5-Arg, which is a critical determinant for CD94-NKG2A recognition (31). Other HLA-E residues that contacted CD94-NKG2A either had a moderate role (Arg65, Arg79, and Glu166) in the energetics interaction or no role at all (Asp69 and Glu154). The alanine-scanning mutagenesis also revealed that CD94 played a notably more dominant role in the energetics of the interaction compared with the NKG2A subunit. Specifically, the CD94 energetic hotspot comprised residues Gln112, Phe114, Asn160, Leu162, Asp163, and Glu164, all of which interacted with HLA-E or the peptide, whereas surprisingly no NKG2A residues were identified as being critical to the interaction. Collectively, these observations also reveal that the energetic footprint of the CD94-NKG2A–HLA-EVMAPRTLFL interaction is smaller than the corresponding structural footprint. Such an observation is consistent with that observed in adaptive immunity (32, 33) and contrasts the more synonymous structural and energetic footprints of the innate NKT TCR–CD1d interaction (34, 35).
Despite the dominant role of CD94, paradoxically there is an approximate sixfold difference in the affinity of CD94-NKG2A (0.7 μM) versus CD94-NKG2C (4.4 μM) for HLA-EVMAPRTLFL (22, 24). The structure of the CD94-NKG2A–HLA-EVMAPRTLFL complex allowed us to begin to address the basis of this observation. Surprisingly, of the sequence differences between NKG2A and NKG2C, none directly contacts HLA-EVMAPRTLFL. However, a region of NKG2A-2C sequence difference spans residues 166–170, which lies at the interface of the CD94-NKG2A heterodimer and has been shown to modulate the affinity for HLA-E (26). This region is in close vicinity to the P5-Arg binding pocket, and accordingly it is conceivable that alterations in the heterodimer interface may impact on the affinity of the interaction. Because none of the residues that was exchanged directly contacts HLA-E, it is also possible that these differences are indirect, namely impacting on the conformation of proximal residues in CD94.
The interactions made by CD94-NKG2A with the HLA-E–bound peptide were suboptimal in chemical, shape, and electrostatic complementarity, in that the P8-Phe and the P5-Arg are accommodated within a polar and a noncharged pocket, respectively. Nevertheless, CD94-NKG2A recognition is sensitive to substitutions at both P5 and P8 positions of the peptide (25, 31). For example, P8-Tyr abrogated recognition, which may be attributed to unfavorable interactions with Phe114 of CD94, and substituting the P8 position to smaller residues, such as P8-Leu, Val, and Ala, also reduced the affinity of the interaction, presumably by creating a cavity at the interface. Similarly, the conservative P5-Arg to P5-Lys mutation abrogated recognition, suggesting either that the P5-Lys-Glu 152 salt bridge is lost, thereby introducing a buried positive charge at the interface, or highlighting the importance of the P5 residue interacting with the main chain of Ser110. Nevertheless, this lack of complementarity is atypical in innate receptor–ligand interactions and is perhaps more characteristic of the “imperfect” interfaces that have been observed in adaptive immunity (27).
Several proteins other than MHC-I molecules have also been shown to encode peptides that can bind to HLA-E. Perhaps the best characterized of these is encoded by UL40 protein of CMV that contains VMAPRTLIL, an exact mimic of a sequence found in most HLA-C alleles, and both binding and functional studies have demonstrated an interaction with CD94-NKG2A (24, 26, 36, 37). Based on the structure of the CD94-NKG2A–HLA-EVMAPRTLFL complex, the presence of an Ile rather than a Phe at P8 would not be predicted to improve the chemical or SC around P8. Consistent with this, binding data indicate that the substitution of Ile for Phe results in a threefold reduction in the affinity of the interaction. In addition, peptides derived from HCV core antigen (YLLPRRGPRL) (38), the ATP-binding cassette transporter multidrug resistance–associated protein 7 (ALALVRMLI) (39), and gliadin (SQQPYLQLQ) (40) have also been shown to bind HLA-E and protect otherwise susceptible target cells via a CD94-NKG2–dependent mechanism. Given the marked differences of these peptides from the canonical MHC-I leader sequence peptide, it will be of interest to establish how these peptides, when bound to HLA-E, interact with CD94-NKG2A.
In addition to being a ligand for the CD94-NKG2 receptors, HLA-E is recognized by CD8 T cells expressing conventional αβTCRs (41–44). The data presented here reveal that the footprint of CD94-NKG2A on HLA-E closely overlaps with that of an HLA-E–restricted TCR (KK50.4) (29). Interestingly CD94-NKG2 receptors are also expressed on these HLA-E–specific T cells (unpublished data) (45). Given the differing affinities between the TCR and CD94-NKG2 receptors for HLA-E, it will be of interest to dissect the biological outcome with these cells. In vitro data suggests that expression of the cognate ligand of the HLA-E–restricted TCR can result in T cell activation despite the expression of the inhibitory CD94-NKG2A receptor (45).
Given the sequence and structural conservation between CD94-NKG2A and NKG2D, as well as their ligands HLA-E, MICA, MICB, and ULBP1-4, it is perhaps not surprising that their footprints were broadly similar. Nevertheless, the molecular basis for ligand recognition differed dramatically with CD94-NKG2A recognition of HLA-E being dominated by charged interactions, whereas hydrophobic interactions played a more significant role in ligand recognition by NKG2D. This dependence on charge in the CD94-NKG2A–HLA-E interaction bears notable similarity with the interaction between KIRs and their class I ligands where disruption of salt bridges abrogated recognition of HLA-C (46, 47).
Although the evolutionary origins of the CD94-NKG2 receptors are not entirely clear, comparison of CD94 and NKG2 sequences in primates and rodents further emphasized the importance of CD94. Although there is significant cross-species variation in the residues that correspond to those in NKG2A that contact HLA-E, 5 of the 13 CD94 residues that contact HLA-E are completely conserved across both primates and rodent species (48–52). Thus, although CD94 by itself is unable to bind HLA-E, it nevertheless dictates the specificity of the interaction, whereas the primary role of the NKG2 subunit is to determine the outcome of ligand recognition.
MATERIALS AND METHODS
Protein expression and purification of CD94-NKG2A and HLA-E.
HLA-E was expressed as inclusion bodies in Escherichia coli and refolded with the VMAPRTLFL peptide according to the previously described method (29). The refolded HLA-EVMARPTLFL complex was further purified using anion exchange and gel filtration chromatography. The extracellular domains of CD94 and NKG2A (CD94 residues 52–179, NKG2A residues 110–223) were expressed as inclusion bodies in E. coli and refolded as described previously (25). The refolded CD94-NKG2A heterodimer was purified by anion exchange and gel filtration chromatography, followed by an additional purification step using anion exchange. The CD94-NKG2A–HLA-EVMARPTLFL complex was generated by mixing the CD94-NKG2A heterodimer with HLA-EVMARPTLFL at a molar ratio of 1.2:1, respectively. The complex was purified via size exclusion chromatography for use in crystallization. The mutagenesis and surface plasmon resonance–based experiments were conducted as described previously (25).
Crystallization, data collection, structure determination, and refinement.
Large crystals (>0.3 mm) of CD94-NKG2A–HLA-EVMARPTLFL complex were grown at 5 mg/ml in 2.0 M NH4SO4, 0.1 M Tris-HCl, pH 7.7–7.9. Drops were made at a ratio of 2:1 (1 μl protein/0.5 μl mother liquor) via the hanging drop vapor diffusion method, and crystals were grown at 18°C. Crystals were flash frozen at 100 K using 20% glycerol as a cyroprotectant. A 3.4 Å dataset was collected at the X25 beamline (Brookhaven) using an ADSC Q315 detector. The data were processed and scaled using the CCP4 suite. The crystals belong to space group I4132, with a = b = c = 345.2 Å, with two complexes per asymmetric unit, with a high solvent content of 80%. Although the crystals diffracted only to 3.4 Å resolution, the I/σI in the highest resolution bin (3.5–3.4 Å) was 3.0, and the overall RPIM (53) value to this resolution was 4.6%. Attempts to dehydrate the crystals or to find an alternative crystallization condition failed to yield better diffracting crystals. The crystal structure of the complex was solved by molecular replacement implemented by Phaser (54) using CD94-NKG2A and HLA-E (minus the bound peptide) as the search models. First, we used CD94-NKG2A (PDB code: 3bdw) as a search model and readily located two molecules within the asymmetric unit. We then fixed this initial solution and searched for the corresponding HLA-E molecules in which the peptide had been deleted (PDB code: 3bze) (unpublished data). Again, two HLA-E molecules were located within the asymmetric unit, with the HLA-E molecules sitting “beneath” the CD94-NKG2A solutions, indicating that we had located two ternary complexes in the asymmetric unit. Searching for a third ternary complex failed to yield a solution. To verify that the molecular replacement solution was correct, a 2 Fo-Fc electron density map was calculated and revealed unbiased electron density for the omitted peptide chain. The progress of refinement was monitored by the Rfree value (5% of the data) with neither a σ nor a low-resolution cutoff being applied to the data, and no water molecules were modeled in the structure. The structure was refined using Phenix (55) interspersed with rounds of model building in “Coot” (56). Given the limited resolution of the data, the refinement proceeded cautiously, with tight noncrystallographic restraints applied to the independent copies within the asymmetric unit. Refinement of B factors led to an ∼4% drop in the Rfactor and Rfree values, thereby justifying the refinement of this parameter. As such, the complex refined very well, presumably aided by the fact that we had previously determined the structures of the nonligated NKG2A and HLA-E components to a 2.5 Å resolution; moreover, there were few conformational changes that took place upon complexation. For the data collection and refinement statistics, see Table I. See Fig. S1 for quality of electron density. All BSA calculations were undertaken using MSD PISA (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). The PDB of the CD94-NKG2A–HLA-E complex has been deposited in the RCSB protein data bank, and the coordinate accession code is 3CDG.
Online supplemental material.
Fig. S1 shows regions of unbiased and final electron density for the complex. Fig. S2 shows a comparison of structural footprints, and Fig. S3 shows the docking mode of CD94-NKG2A and other receptors. Figs. S1–S3 are available at http://www.jem.org/cgi/content/full/jem.20072525/DC1.
Supplemental Material
Acknowledgments
We would like to thank the staff at the Brookhaven Synchrotron for assistance with data collection.
This work was supported by grants from the National Health and Medical Research Council (NHMRC), the Australian Research Council (ARC), an ARC QEII fellowship (to C.S. Clements), Doherty Fellowships from the NHMRC (to L.C. Sullivan and T. Beddoe), an NHMRC Senior Research Fellowship (to M.C.J. Wilce), and an ARC Federation Fellowship (to J. Rossjohn).
The authors have no conflicting financial interests.
Abbreviations used: BSA, buried surface area; KIR, killer cell Ig-like receptor; SC, shape complementarity; VDW, van der Waals.
Emma J. Petrie and Craig S. Clements contributed equally to this work.
Andrew G. Brooks and Jamie Rossjohn contributed equally to this work.
References
- 1.Moretta, L., C. Bottino, D. Pende, R. Castriconi, M.C. Mingari, and A. Moretta. 2006. Surface NK receptors and their ligands on tumor cells. Semin. Immunol. 18:151–158. [DOI] [PubMed] [Google Scholar]
- 2.Boyton, R.J., and D.M. Altmann. 2007. Natural killer cells, killer immunoglobulin-like receptors and human leucocyte antigen class I in disease. Clin. Exp. Immunol. 149:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cella, M., H. Nakajima, F. Facchetti, T. Hoffmann, and M. Colonna. 2000. ILT receptors at the interface between lymphoid and myeloid cells. Curr. Top. Microbiol. Immunol. 251:161–166. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan, K., N. Dimasi, J. Wang, D.H. Margulies, and R.A. Mariuzza. 2002. MHC class I recognition by Ly49 natural killer cell receptors. Mol. Immunol. 38:1023–1027. [DOI] [PubMed] [Google Scholar]
- 5.Gasser, S., and D.H. Raulet. 2006. Activation and self-tolerance of natural killer cells. Immunol. Rev. 214:130–142. [DOI] [PubMed] [Google Scholar]
- 6.Borrego, F., M. Masilamani, J. Kabat, T.B. Sanni, and J.E. Coligan. 2005. The cell biology of the human natural killer cell CD94/NKG2A inhibitory receptor. Mol. Immunol. 42:485–488. [DOI] [PubMed] [Google Scholar]
- 7.Long, E.O., and N. Wagtmann. 1997. Natural killer cell receptors. Curr. Opin. Immunol. 9:344–350. [DOI] [PubMed] [Google Scholar]
- 8.Natarajan, K., N. Dimasi, J. Wang, R.A. Mariuzza, and D.H. Margulies. 2002. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu. Rev. Immunol. 20:853–885. [DOI] [PubMed] [Google Scholar]
- 9.Strong, R.K., and B.J. McFarland. 2004. NKG2D and related immunoreceptors. Adv. Protein Chem. 68:281–312. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, C., N.J. Chalupny, C.L. Sutherland, S. Dosch, P.V. Sivakumar, D.C. Johnson, and D. Cosman. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 197:1427–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, P., D.L. Morris, B.E. Willcox, A. Steinle, T. Spies, and R.K. Strong. 2001. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2:443–451. [DOI] [PubMed] [Google Scholar]
- 12.Li, P., G. McDermott, and R.K. Strong. 2002. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity. 16:77–86. [DOI] [PubMed] [Google Scholar]
- 13.McFarland, B.J., and R.K. Strong. 2003. Thermodynamic analysis of degenerate recognition by the NKG2D immunoreceptor: not induced fit but rigid adaptation. Immunity. 19:803–812. [DOI] [PubMed] [Google Scholar]
- 14.McFarland, B.J., T. Kortemme, S.F. Yu, D. Baker, and R.K. Strong. 2003. Symmetry recognizing asymmetry: analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure. 11:411–422. [DOI] [PubMed] [Google Scholar]
- 15.Radaev, S., B. Rostro, A.G. Brooks, M. Colonna, and P.D. Sun. 2001. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity. 15:1039–1049. [DOI] [PubMed] [Google Scholar]
- 16.Wolan, D.W., L. Teyton, M.G. Rudolph, B. Villmow, S. Bauer, D.H. Busch, and I.A. Wilson. 2001. Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A. Nat. Immunol. 2:248–254. [DOI] [PubMed] [Google Scholar]
- 17.Borrego, F., M. Ulbrecht, E.H. Weiss, J.E. Coligan, and A.G. Brooks. 1998. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell–mediated lysis. J. Exp. Med. 187:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braud, V.M., D.S. Allan, C.A. O'Callaghan, K. Soderstrom, A. D'Andrea, G.S. Ogg, S. Lazetic, N.T. Young, J.I. Bell, J.H. Phillips, et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 391:795–799. [DOI] [PubMed] [Google Scholar]
- 19.Brooks, A.G., F. Borrego, P.E. Posch, A. Patamawenu, C.J. Scorzelli, M. Ulbrecht, E.H. Weiss, and J.E. Coligan. 1999. Specific recognition of HLA-E, but not classical, HLA class I molecules by soluble CD94/NKG2A and NK cells. J. Immunol. 162:305–313. [PubMed] [Google Scholar]
- 20.Lee, N., M. Llano, M. Carretero, A. Ishitani, F. Navarro, M. Lopez-Botet, and D.E. Geraghty. 1998. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA. 95:5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brostjan, C., T. Bellon, Y. Sobanov, M. Lopez-Botet, and E. Hofer. 2002. Differential expression of inhibitory and activating CD94/NKG2 receptors on NK cell clones. J. Immunol. Methods. 264:109–119. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser, B.K., F. Barahmand-Pour, W. Paulsene, S. Medley, D.E. Geraghty, and R.K. Strong. 2005. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J. Immunol. 174:2878–2884. [DOI] [PubMed] [Google Scholar]
- 23.Lazetic, S., C. Chang, J.P. Houchins, L.L. Lanier, and J.H. Phillips. 1996. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 157:4741–4745. [PubMed] [Google Scholar]
- 24.Vales-Gomez, M., H.T. Reyburn, R.A. Erskine, M. Lopez-Botet, and J.L. Strominger. 1999. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 18:4250–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullivan, L.C., C.S. Clements, T. Beddoe, D. Johnson, H.L. Hoare, J. Lin, T. Huyton, E.J. Hopkins, H.H. Reid, M.C.J. Wilce, et al. 2007. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 27:900–911. [DOI] [PubMed] [Google Scholar]
- 26.Hoare, H.L., L.C. Sullivan, C.S. Clements, L.K. Ely, T. Beddoe, K.N. Henderson, J. Lin, H.H. Reid, A.G. Brooks, and J. Rossjohn. 2008. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the cd94-NKG2 natual killer cell receptors. J. Mol. Biol. In press. [DOI] [PubMed]
- 27.Clements, C.S., M.A. Dunstone, W.A. Macdonald, J. McCluskey, and J. Rossjohn. 2006. Specificity on a knife-edge: the alphabeta T cell receptor. Curr. Opin. Struct. Biol. 16:787–795. [DOI] [PubMed] [Google Scholar]
- 28.Tynan, F.E., S.R. Burrows, A.M. Buckle, C.S. Clements, N.A. Borg, J.J. Miles, T. Beddoe, J.C. Whisstock, M.C. Wilce, S.L. Silins, et al. 2005. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 6:1114–1122. [DOI] [PubMed] [Google Scholar]
- 29.Hoare, H.L., L.C. Sullivan, G. Pietra, C.S. Clements, E.J. Lee, L.K. Ely, T. Beddoe, M. Falco, L. Kjer-Nielsen, H.H. Reid, et al. 2006. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol. 7:256–264. [DOI] [PubMed] [Google Scholar]
- 30.Rudolph, M.G., R.L. Stanfield, and I.A. Wilson. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J.D., D.A. Weber, C. Ibegbu, J. Pohl, J.D. Altman, and P.E. Jensen. 2003. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J. Immunol. 171:1369–1375. [DOI] [PubMed] [Google Scholar]
- 32.Borg, N.A., L.K. Ely, T. Beddoe, W.A. Macdonald, H.H. Reid, C.S. Clements, A.W. Purcell, L. Kjer-Nielsen, J.J. Miles, S.R. Burrows, et al. 2005. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat. Immunol. 6:171–180. [DOI] [PubMed] [Google Scholar]
- 33.Manning, T.C., C.J. Schlueter, T.C. Brodnicki, E.A. Parke, J.A. Speir, K.C. Garcia, L. Teyton, I.A. Wilson, and D.M. Kranz. 1998. Alanine scanning mutagenesis of an alphabeta T cell receptor: mapping the energy of antigen recognition. Immunity. 8:413–425. [DOI] [PubMed] [Google Scholar]
- 34.Borg, N.A., K.S. Wun, L. Kjer-Nielsen, M.C. Wilce, D.G. Pellicci, R. Koh, G.S. Besra, M. Bharadwaj, D.I. Godfrey, J. McCluskey, and J. Rossjohn. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49. [DOI] [PubMed] [Google Scholar]
- 35.Scott-Browne, J.P., J.L. Matsuda, T. Mallevaey, J. White, N.A. Borg, J. McCluskey, J. Rossjohn, J. Kappler, P. Marrack, and L. Gapin. 2007. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8:1105–1113. [DOI] [PubMed] [Google Scholar]
- 36.Tomasec, P., V.M. Braud, C. Rickards, M.B. Powell, B.P. McSharry, S. Gadola, V. Cerundolo, L.K. Borysiewicz, A.J. McMichael, and G.W. Wilkinson. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 287:1031. [DOI] [PubMed] [Google Scholar]
- 37.Ulbrecht, M., S. Martinozzi, M. Grzeschik, H. Hengel, J.W. Ellwart, M. Pla, and E.H. Weiss. 2000. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164:5019–5022. [DOI] [PubMed] [Google Scholar]
- 38.Nattermann, J., H.D. Nischalke, V. Hofmeister, G. Ahlenstiel, H. Zimmermann, L. Leifeld, E.H. Weiss, T. Sauerbruch, and U. Spengler. 2005. The HLA-A2 restricted T cell epitope HCV Core35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am. J. Pathol. 166:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wooden, S.L., S.R. Kalb, R.J. Cotter, and M.J. Soloski. 2005. Cutting edge: HLA-E binds a peptide derived from the ATP-binding cassette transporter multidrug resistance-associated protein 7 and inhibits NK cell-mediated lysis. J. Immunol. 175:1383–1387. [DOI] [PubMed] [Google Scholar]
- 40.Terrazzano, G., M. Sica, C. Gianfrani, G. Mazzarella, F. Maurano, B. De Giulio, S. de Saint-Mezard, D. Zanzi, L. Maiuri, M. Londei, et al. 2007. Gliadin regulates the NK-dendritic cell cross-talk by HLA-E surface stabilization. J. Immunol. 179:372–381. [DOI] [PubMed] [Google Scholar]
- 41.Heinzel, A.S., J.E. Grotzke, R.A. Lines, D.A. Lewinsohn, A.L. McNabb, D.N. Streblow, V.M. Braud, H.J. Grieser, J.T. Belisle, and D.M. Lewinsohn. 2002. HLA-E–dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med. 196:1473–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietra, G., C. Romagnani, P. Mazzarino, M. Falco, E. Millo, A. Moretta, L. Moretta, and M.C. Mingari. 2003. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc. Natl. Acad. Sci. USA. 100:10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romagnani, C., G. Pietra, M. Falco, P. Mazzarino, L. Moretta, and M.C. Mingari. 2004. HLA-E-restricted recognition of human cytomegalovirus by a subset of cytolytic T lymphocytes. Hum. Immunol. 65:437–445. [DOI] [PubMed] [Google Scholar]
- 44.Salerno-Goncalves, R., M. Fernandez-Vina, D.M. Lewinsohn, and M.B. Sztein. 2004. Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 173:5852–5862. [DOI] [PubMed] [Google Scholar]
- 45.Romagnani, C., G. Pietra, M. Falco, E. Millo, P. Mazzarino, R. Biassoni, A. Moretta, L. Moretta, and M.C. Mingari. 2002. Identification of HLA-E-specific alloreactive T lymphocytes: a cell subset that undergoes preferential expansion in mixed lymphocyte culture and displays a broad cytolytic activity against allogeneic cells. Proc. Natl. Acad. Sci. USA. 99:11328–11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan, Q.R., E.O. Long, and D.C. Wiley. 2001. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2:452–460. [DOI] [PubMed] [Google Scholar]
- 47.Snyder, G.A., A.G. Brooks, and P.D. Sun. 1999. Crystal structure of the HLA-Cw3 allotype-specific killer cell inhibitory receptor KIR2DL2. Proc. Natl. Acad. Sci. USA. 96:3864–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dissen, E., S.F. Berg, I.H. Westgaard, and S. Fossum. 1997. Molecular characterization of a gene in the rat homologous to human CD94. Eur. J. Immunol. 27:2080–2086. [DOI] [PubMed] [Google Scholar]
- 49.Guethlein, L.A., L.R. Flodin, E.J. Adams, and P. Parham. 2002. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J. Immunol. 169:220–229. [DOI] [PubMed] [Google Scholar]
- 50.LaBonte, M.L., K.L. Hershberger, B. Korber, and N.L. Letvin. 2001. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol. Rev. 183:25–40. [DOI] [PubMed] [Google Scholar]
- 51.Shum, B.P., L.R. Flodin, D.G. Muir, R. Rajalingam, S.I. Khakoo, S. Cleland, L.A. Guethlein, M. Uhrberg, and P. Parham. 2002. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J. Immunol. 168:240–252. [DOI] [PubMed] [Google Scholar]
- 52.Vance, R.E., D.M. Tanamachi, T. Hanke, and D.H. Raulet. 1997. Cloning of a mouse homolog of CD94 extends the family of C-type lectins on murine natural killer cells. Eur. J. Immunol. 27:3236–3241. [DOI] [PubMed] [Google Scholar]
- 53.Diederichs, K., and P.A. Karplus. 1997. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat. Struct. Mol. Biol. 4:269–275. [DOI] [PubMed] [Google Scholar]
- 54.McCoy, A.J. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams, P.D., R.W. Grosse-Kunstleve, L.W. Hung, T.R. Ioerger, A.J. McCoy, N.W. Moriarty, R.J. Read, J.C. Sacchettini, N.K. Sauter, and T.C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 56.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.